Abstract
Noroviruses are positive-sense, single-stranded RNA viruses that cause acute gastroenteritis. They recognize human histo-blood group antigens as receptors in a strain-specific manner. The structures presented here were analyzed in order to elucidate the structural basis for differences in ligand recognition of noroviruses from different genogroups, the prototypic Norwalk virus (NV; GI-1) and VA387 (GII-4), which recognize the same A antigen but differ in that NV is unable to bind to the B antigen. Two forms of the receptor-binding domain of the norovirus coat protein, the P domain and the P polypeptide, that were previously shown to differ in receptor binding and P-particle formation properties were studied. Comparison of the structures of the NV P domain with and without A trisaccharide and the NV P polypeptide revealed no major ligand-induced changes. The 2.3-Å cocrystal structure reveals that the A trisaccharide binds to the NV P domain through interactions with the residues Ser377, Asp327, His329, and Ser380 in a mode distinct from that previously reported for the VA387 P-domain-A-trisaccharide complex. Mutational analyses confirm the importance of these residues in NV P-particle binding to native A antigen. The α-GalNAc residue unique to the A trisaccharide is buried deeply in the NV binding pocket, unlike in the structures of A and B trisaccharides bound to VA387 P domain, where the α-fucose residue forms the most protein contacts. The A-trisaccharide binding mode seen in the NV P domain complex cannot be sterically accommodated in the VA387 P domain.
Noroviruses are a major cause of epidemics of acute gastroenteritis and thus pose a widespread health risk (6, 10). These viruses have an ∼7.7-kb single-stranded, positive-sense RNA genome comprised of three open reading frames (ORFs); ORF1 encodes a polyprotein precursor that is posttranslationally processed into several nonstructural proteins, while the other two ORFs encode the major (VP1) and minor (VP2) capsid proteins. When expressed in vitro, VP1 can assemble into virus-like particles (VLPs) morphologically similar to the native capsid (11). The atomic structure of the recombinant Norwalk virus (NV) VLP has been reported (15) to be a T = 3 icosahedral structure made up of 180 VP1 monomers. Each VP1 has two major domains, a shell (S) domain, which forms the core of the icosahedral shell (1), and a protruding (P) domain, which forms arches extending from the shell. These two domains are linked through a 10-residue hinge. Since the P domain is located at the most exterior surface of the viral particle and contains the most variable sequence, it is presumed that the P domain is responsible for host interactions and immune responses. In the assembled viral particle, the P domain also participates in dimeric interactions that are believed to stabilize the capsid (1). Furthermore, previous studies suggested that VP1 may be processed proteolytically into a soluble ∼30-kDa protein (referred to as the P polypeptide), which could play a role in the replication, host immune response, and pathogenesis of noroviruses (19). The P domain can be further divided into the subdomains P1 and P2, which correspond to the leg and head, respectively, of the arch-like P dimer (15). A P particle comprised of 12 P dimers was also observed when the P domain was expressed in vitro (18). Curiously, the P polypeptide, which differs from the P domain only in the absence of a 4- to 7-residue arginine-rich sequence at the C terminus, is unable to form P particles but can form dimers (19).
Human histo-blood group antigens (HBGAs) have been shown to be host receptors for noroviruses, and eight distinct receptor-binding patterns have been described (reviewed in reference 17). HBGAs are complex carbohydrates linked to proteins or lipids present on epithelial cells in the gastrointestinal and respiratory tracts, on the surfaces of red blood cells, and as free antigens in body fluids such as saliva, blood, and intestinal contents. Noroviruses are genetically highly diverse: at least 27 genetic clusters or genotypes in five genogroups have been described. The prototypic NV belonging to genogroup I (GI) binds A and H antigens, while VA387 of GII binds A, B, and H antigens. The crystal structures of the VA387 P domain bound to A and B trisaccharides have been reported (3). In this study, we present the crystal structures of the P domain of NV, both free and bound to an A trisaccharide, as well as the structures of the P polypeptides of NV and VA387. Comparison of these structures provides direct evidence of different receptor-binding modes responsible for different binding patterns of noroviruses. This information is of fundamental importance to future studies of norovirus biology and in the development of strategies to control and prevent norovirus-associated disease.
MATERIALS AND METHODS
Protein expression, purification, and crystallization.
The DNA fragments encoding the NV P domain with the hinge (amino acids 218 to 530), the NV P polypeptide (amino acids 228 to 523), and VA387 P polypeptide (amino acids 224 to 535) were PCR amplified and cloned into the vector pGEX-4T-1 (Amersham Bioscience, Piscataway, NJ) at the BamHI and NotI sites (16, 17, 19). The constructs were expressed in Escherichia coli strain BL21, and protein expression was induced at room temperature overnight with 0.5 mM isopropyl-β-d-thiogalactopyranoside. The recombinant proteins were purified using glutathione-Sepharose 4 Fast Flow (Amersham Bioscience), thrombin-cleaved directly on the resin, and then further purified by gel filtration with a Superdex 200 size exclusion column (19). The purified proteins were concentrated to about 10 mg/ml and stored in 20 mM Tris-HCl (pH 8.0) and 150 mM NaCl at −80°C. The proteins were crystallized by the hanging-drop vapor diffusion method at room temperature, with drops consisting of 1 μl of the protein solutions and 1 μl of the reservoir solutions. The reservoir solution for VA387-P-polypeptide contained 0.1 M Tris (pH 8.5), 22% polyethylene glycol (PEG) 4000, and 3% 1,5-diaminopentane and yielded needle-shaped crystals. NV P polypeptide and NV P domain crystallized from a reservoir solution containing 13% (wt/vol) PEG 4000, 0.2 M (NH4)2SO4, and 0. 1 M sodium acetate (pH 4.5). For the A-trisaccharide-NV P-domain complex crystallization, 10 mg/ml NV P domain was mixed with a 60-fold molar excess of the A trisaccharide and incubated at 4°C for 5 h. Crystallizations were then set up at room temperature using a reservoir solution containing 13% (wt/vol) PEG 4000, 0.2 M ammonium sulfate, and 0.1 M sodium acetate pH 4.5.
Data collection, processing, structure determination, and refinement.
X-ray diffraction data were collected from flash-cooled crystals on a RAXIS IV++ image plate system after soaking the crystal in cryoprotectant solution. The cryoprotectant solution for the NV P-domain, P-polypeptide, and P-domain-A-trisaccharide complex crystals was 35% (wt/vol) PEG 4000, 0.2 M ammonium sulfate, and 0.1 M sodium acetate (pH 4.5). The cryoprotectant for the VA387 P polypeptide crystals was 35% (wt/vol) PEG 4000, 0.1 M Tris (pH 8.5), and 3% 1,5-diaminopentane. Data were processed, scaled, and merged using the HKL program package (14). The crystal structures were all solved by molecular replacement using CNS (2). The search models used were generated from the VA387 P-domain structure (2OBR.PDB) (3) for the solution of the VA 387 P-polypeptide structure and from residues Phe230 to Arg520 from the NV capsid structure (1IHM.PDB) (15) for the solution of the structures of NV P domain and P polypeptide. Multiple cycles of rebuilding guided by electron density maps were conducted using the program O (12), and refinement was conducted with CNS (2). At the final stages of refinement, composite omit maps were calculated to check for model bias, and water molecules with good hydrogen-bonding potentials were located in peaks (>2.5τ) of the Fo-Fc map. Data processing and structural refinement statistics are summarized in Tables 1 and 2. The refined structures were evaluated with the program PROCHECK (13).
TABLE 1.
Data collection statistics
| Statistic | VA387 P polypeptide | NV P polypeptide | NV P domain | NV P domain-A trisaccharide |
|---|---|---|---|---|
| Space group | C2221 | I212121 | I212121 | P3121 |
| Cell dimensions (Å) | ||||
| a | 54.36 | 82.80 | 83.75 | 83.01 |
| b | 96.80 | 83.77 | 82.95 | 83.01 |
| c | 118.73 | 102.23 | 102.74 | 162.98 |
| Resolution (Å)a | 100-2.3 (2.38-2.3) | 100-2.6 (2.69-2.6) | 100-2.7 (2.8-2.7) | 100-2.3 (2.39-2.3) |
| Measured reflections | 782,145 | 196,457 | 218,506 | 429,920 |
| Unique reflections | 8,941 | 8,335 | 10,119 | 26,460 |
| Redundancya | 3.5 (3.1) | 1.9 (2.0) | 3.3 (3.2) | 3.8 (3.9) |
| I/σ(I)a | 17.50 (3.67) | 13.65 (5.50) | 12.22 (4.57) | 26.19 (14.43) |
| Completeness (%)a | 97.3 (87.9) | 89.8 (84.2) | 99.3 (99.2) | 90 (88.6) |
| Rsym (%)a,b | 7.1 (23.4) | 5.5 (15.5) | 8.4 (25.7) | 3.9 (12.1) |
Overall (last shell).
Rsym = Σ Ii − <I>/Σ<I>, where <I> is the mean intensity of the reflections with intensity Ii and common indices h, k, and l.
TABLE 2.
Structure refinement
| Statistic | VA387 P polypeptide | NV P polypeptide | NV P domain | NV P domain-A trisaccharide |
|---|---|---|---|---|
| No. of reflections in working set | 8,737 | 8,083 | 9,678 | 26,272 |
| No. of reflections in test set | 901 | 861 | 997 | 2,594 |
| R factor (%)a | 22.70 | 21.24 | 20.45 | 22.32 |
| Rfree (%)b | 26.71 | 27.08 | 25.32 | 26.22 |
| RMSD | ||||
| Bond lengths (Å) | 0.011 | 0.006 | 0.006 | 0.006 |
| Bond angles (°) | 1.6 | 1.2 | 1.3 | 1.5 |
| Average B factors (Å2) | 47.1 | 42.6 | 40.1 | 40.6 |
| Residues in Ramachandran plot (%) | ||||
| Most favored regions | 84.5 | 84.9 | 84.0 | 87.3 |
| Additionally allowed regions | 15.1 | 14.2 | 15.1 | 12.3 |
| Generously allowed regions | 0.4 | 0.9 | 0.9 | 0.5 |
| Disallowed regions | 0 | 0 | 0 | 0 |
R factor = Σhkl| |Fobs| − k |Fcal|/ΣhklFobs, where Fobs and Fcal are observed and calculated structure factors, respectively.
For Rfree, the sum is extended over a subset of reflections (10%) excluded from all stages of refinement.
Construction of mutant P particles.
The wild-type NV P particle (CNGRC-H/P) was expressed in bacteria as described previously (18, 19). Mutant P particles with the single-residue mutations D327A, H329A, S338A, and S377A at the A-trisaccharide binding site were constructed by site-directed mutagenesis using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) and the corresponding primer pairs (D327A, CCTCGGTGGTTGTGCTTGGCATATCAATATG and CATATTGATATGCCAAGCACAACCACCGAGG; H329A, GGTGGTTGTGATTGGGCTATCAATATGACACAG and CTGTGTCATATTGATAGCCCAATCACAACCACC; S338A, ACACAGTTTGGCCATGCTAGCCAGACCCAGTATG and ATACTGGGTCTGGCTAGCATGGCCAAACTGTGTC; S377A, AGTTCTTAGCTGGATTGCCCCCCCATCACAC and GTGTGATGGGGGGGCAATCCAGCTAAGAAC). The formation of P particles or P dimers was determined by gel filtration using a Superdex 200 size exclusion column (GE Health Care Bio-Sciences, Piscataway, NJ) powered by an AKTA-FPLC system (model 920; GE Health Care Bio-Sciences) followed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, in which the P particles form a peak at ∼830 kDa and the P dimer forms a peak at ∼69 kDa (18, 19).
HBGA-binding assays.
Saliva-based binding assays were performed basically as described elsewhere (9, 18). The affinity column-purified P particles and P dimers were first diluted to 1 mg/ml as the starting solutions; then they were diluted further in a threefold series to 10 μg/ml (100×), 3.3 μg/ml (300×), 1.1 μg/ml (900×), 0.37 μg/ml (2,700×), 0.12 μg/ml (8,100×), 0.04 μg/ml (2,430×), and 0.01 μg/ml (72,900×), directly on enzyme-linked immunosorbent assay plates. Three well-characterized saliva samples that contained high levels of A, B, and H antigens were used for measuring the receptor-binding capability of the P particles and P dimers (9, 18).
Protein Data Bank accession numbers.
The coordinates and structure factors for the NV P domain (3BYI.PDB), NV P polypeptide (3BY2.PDB), NV P domain with A trisaccharide (3BY3.PDB), and VA387 P polypeptide (3BQJ.PDB) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ.
RESULTS
Structures of the P domain and P polypeptide.
Crystal structures of the P domain and the P polypeptide of NV and of the P polypeptide of VA387 were determined using molecular replacement. The final refined models for the P domain and the P polypeptide of the NV included residues 230 to 516, with residues 398 to 399 and 452 to 454 not modeled due to a lack of electron density. In both structures, the P protein can be divided into two subdomains, P1 and P2 (Fig. 1a and b). Subdomain P1, residues 230 to 285 and 406 to 516 (NV P protein), has a mixed α/β structure made up of two twisted antiparallel β-sheets and a single α-helix. Subdomain P2 is an insertion in P1 consisting of residues 286 to 405 and folds into a six-stranded antiparallel β-barrel. The two subdomains are connected by a tight interface stabilized by both hydrophobic interactions and hydrogen bonds. Sequestered in this interface is a water molecule that is hydrogen-bonded to the side chain and backbone nitrogen of Asp322 in the P2 subdomain and to the carbonyl backbone O of residue 245 and the side chain of Asn 267 in P1. The exposed surfaces of P1 and P2 are largely loops and include the three short stretches of amino acids for which electron density was not visible. These surface loops are also the regions of the lowest sequence conservation within the P domain. The VA387 P polypeptide refined model contained residues 225 to 529, with residues 296 to 297, 372 to 374, and 392 to 393 missing due to poor electron density.
FIG. 1.
(a) Structure-based superimposition of the amino acid sequences of NV P domain and VA387 P domain. Identical residues are highlighted with a red background, while similar residues are in red type. Secondary structures present in the P polypeptide structures of NV and VA387 are indicated above and below the sequences, respectively; arrows represent β-strands, and coils represent helices (either 310 [η] or alpha [α]). Blue circles indicate residues involved in A-trisaccharide interaction with the NV P domain, and pink circles indicate residues involved in A-trisaccharide interaction with the VA387 P domain. Sequence alignment was done with ESPript (8), and secondary structure assignment was done with STRIDE (7). (b) Ribbon diagram showing one subunit of the NV P domain (generated with RIBBONS [4]). Strands (β) are shown in pink, α-helices (α) in cyan, 310 helices (η) in green, coils and turns in gray. Secondary structural elements are labeled as in panel a. The N terminus of the structure (Pro230) is indicated in blue, and a dotted arrow indicates the location of the S domain in the intact VP1 protein. The C terminus of the crystallographic model (Val516) is indicated, along with the residues not visible in the experimental density map (red sequence). (c) Superimposition of the NV (blue) and VA387 (pink) P domain structures. Green boxes highlight regions where the secondary structure is distinctly different.
The atomic structures of the P domain and the P polypeptide of both NV and VA387 are very similar (Cα root mean square deviation [RMSD] of 0.544 Å for the NV structures and Cα RMSD of 0.626 Å for the VA387 structures). Notably, the C-terminal residues that distinguish the P domain from the P polypeptide for either NV or VA387 cannot be seen in the electron density maps. For this reason, we present a comparison of the structures of only the P polypeptides of NV and VA387 here (Fig. 1c). The structure of the NV P polypeptide is very similar to the corresponding region in the previously determined 3.4-Å crystal structure of the NV VLP (15), with an RMSD of 0.78 Å for 281 superimposed Cα atoms. While the folds of the P polypeptides of NV and VA387 are similar, superimposition of the structures yields an overall Cα RMSD for the two P polypeptide structures of 4.2 Å, largely attributable to the P2 subdomain (the Cα RMSD for 123 atoms is 5 Å, while RMSD for 156 Cα atoms of the P1 subdomain is 2 Å). The most prominent differences are seen in the loops on the surfaces not involved in dimer interactions (Fig. 1c). The 310 helices η2, η3, and η5 in the NV P polypeptide are not present in the VA387 P polypeptide (Fig. 1a and c).
Dimerization and oligosaccharide binding by the P domain.
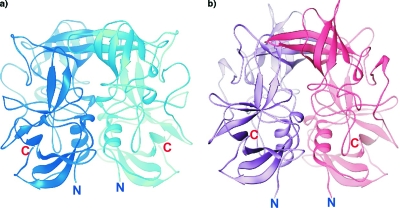
The P domains and the P polypeptides of NV and VA387 are dimers related by crystallographic twofold axes in the structures described here and in a previous study (3) (Fig. 2a and b). The dimer interface is extensive in all structures; for example, it occludes 3,360 Å2 of total surface area in the NV P domain dimer. There is a large cavity with a volume of 61 Å3 in the NV P dimer between the P1 and P2 subdomains. The P1 dimer interface is dominated by hydrophobic interactions and hydrogen bonds formed largely by residues in the α1 helix of each monomer, while the P2 dimer interface is predominantly composed of hydrogen bonds between side chains in strands β4 and β5 of each monomer.
FIG. 2.
Dimers of the NV P domain (a) and the VA387 P domain (2OBR.PDB) (3) (b). In each case the two subunits are shown in different colors. The N and C termini of the structures are indicated.
Cocrystallization of the NV P dimer with the A trisaccharide yielded a new trigonal crystal form with a dimer in the asymmetric unit. The structure was solved by molecular replacement using the crystallographic dimer of the NV P domain dimer as a model. Refinement of the protein model followed by calculation of an Fo-Fc map yielded clear density that could be modeled and refined as the A trisaccharide (Fig. 3a). The binding mode of the NV P domain with the A trisaccharide differs from that previously reported for the VA387 P dimer with the same A trisaccharide or with the B trisaccharide (3). In the NV P-domain-sugar complex, the sugar is snugly placed in a shallow binding pocket lined almost entirely by the P2 subdomain (Fig. 3b). There is a small degree of asymmetry in sugar placement in the two monomers of the NV P dimer (Fig. 4a and b). All three sugar rings of the A trisaccharide are visible in the electron density map. The α-GalNAc ring is inserted deeply into the binding pocket and makes the most protein contacts (Fig. 3c and 4a and b). It is held firmly in place by hydrogen bonds between the sugar O3a and O4a hydroxyls and the side chain hydroxyl of Ser377, and the O4a hydroxyl and the backbone carbonyl oxygen of Pro378. The side chain of Ser377 is in turn held rigidly by a hydrogen bond with the imidazole ring of His329, which in one subunit also hydrogen-bonds with the O3a of the α-GalNAc. In one monomer, O3a is further hydrogen-bonded to the side chain of Asp327, which in turn is held in place by a hydrogen bond with the hydroxyl side chain and backbone N of Ser380. Ser380 itself hydrogen-bonds with the O3a hydroxyl of the α-fucose ring in both subunits. In one subunit, water-mediated hydrogen bonds are observed between the acetyl amide of the α-GalNAc and the side chain oxygen of Asp344 and between the side chain hydroxyl of Ser338 from the noncognate subunit. The central β-galactose unit of the trisaccharide does not participate in any interactions in either subunit. The asymmetry seen in interactions with the two subunits (Fig. 4a and b) is due almost entirely to a slightly altered placement of the trisaccharide in the binding pocket.
FIG. 3.
(a) Stereo view of the electron density for the A trisaccharide in the experimental Fo-Fc map calculated after the initial molecular replacement solution for the NV P-domain-A-trisaccharide complex structure was refined with only protein coordinates. The coordinates of the final refined A trisaccharide are also shown. The map is contoured at 1.5σ. (b) Ribbon diagram showing superimposition of the A-trisaccharide-bound NV P domain (blue) and the A-trisaccharide-bound VA387 P domain (pink; 2OBS.PDB) superimposed. The A trisaccharide in the NV complex is shown as a ball-and-stick model in blue, and the A trisaccharide in the VA387 complex is in red, demonstrating the different binding sites occupied by the receptor oligosaccharides. (c) Stereo view of the A-trisaccharide binding site in the NV P domain. Dashed lines indicate hydrogen bonds, “W” indicates solvent molecules, and the apostrophe in Ser338′ indicates an interaction with the noncognate subunit. The image was generated with PyMOL (PyMOL Graphics System; DeLano Scientific, San Carlos, CA).
FIG. 4.
Schematic representations of contacts between the A trisaccharide and the NV P domain seen in the two noncrystallographically related subunits of the NV P domain dimer (a and b) and the contacts between the A trisaccharide and the VA387 P domain identified in the previously published crystal structure (2OBS.PDB) (3) (c). Dashed lines indicate hydrogen bonds, and “W” indicates solvent molecules. The apostrophe in Ser338′ indicates an interaction with the noncognate subunit. Obb and Nbb refer to contacts with the protein backbone. H-bond distances are indicated.
Confirmation of the oligosaccharide-binding sites by mutagenesis.
To investigate the functional involvement of the four critical A-trisaccharide-interacting NV P-domain residues identified in the structural studies, Asp327, His329, Ser338, and Ser377, they were each replaced by an alanine. The resulting mutant P particles were tested for their ability to bind to native HBGAs by saliva-binding assays using the NV P particle as model (9, 18, 19). The D327A and S377A mutants lost the binding ability completely, while the H329A mutant bound with lower affinity to the A- and H-positive saliva (Fig. 5), confirming that these residues are important for binding to native HBGAs. The simultaneous loss of binding to both A and H antigens suggests that the binding sites for these two antigens are close to each other or overlap. Interestingly, replacement of Ser338 with an alanine did not cause a significant change in saliva binding. Ser338 makes solvent-mediated contact with the N-acetyl moiety of the A trisaccharide in only one of the two dimer-related protein-sugar interfaces seen in the dimeric complex. The mutagenesis would suggest that this interaction is dispensable for the interaction.
FIG. 5.
HBGA-binding assays of wild-type and mutant P particles with single amino acid mutations at the A-trisaccharide-binding site. The x axes show the protein concentrations of the P particles, while the y axes indicate measurements of optical density at 450 (OD450) nm. Data are averages from triplicate experiments. O, A, and N, type O (containing H antigen), A, and nonsecretor saliva, respectively.
DISCUSSION
The structural analyses presented here were undertaken in order to address an outstanding question regarding norovirus-host interaction: what is the stereochemical basis for the differences and similarities in host receptor specificity of different norovirus strains? We determined the structures of the major surface proteins of the P domain alone and in the presence of HBGA carbohydrates from two strains representing two genotypes in two genogroups, the prototypic NV, belonging to GI, genotype 1(GI-1), and VA387, belonging to GII, genogroup 4 (GII-4) (3). Our data showed that although both strains bind the same A and H antigens and the binding interfaces are located on the same region of the P-2 domain, the binding mode and the amino acids involved in the interaction are completely different. These data support the genetic classification indicating that NV and VA387 are not from the same evolutionary lineage in terms of selection by the human host HBGAs (17).
The structure of the NV P domain bound to the A trisaccharide (representative of the type A antigens of the ABO family of human HBGAs) reveals a binding pocket in the P2 subdomain of both subunits of the P dimer that would be on the most exposed surface of the viral capsid (Fig. 3b). Interestingly, there are distinct differences between this structure and the structure of the A trisaccharide bound to the P domain of VA387 (Fig. 3b and 4). The two most striking features are that the trisaccharide is inserted into the binding pocket in different orientations in the two structures and that the trisaccharide is bound to different parts of the same surface pocket in the two complexes. In the previously reported VA387 complex, each trisaccharide in the dimer is contacted by both subunits of the protein (3). In the NV P domain complex there is only a single water-mediated contact with the noncognate subunit (Fig. 4b). Unlike in the case of VA387 P domain complex with the A trisaccharide, the central β-galactose makes no contact with the P domain of NV and the α-GalNAc moiety is involved in an intricate pattern of hydrogen-bonding with the P2 subdomain, while the α-fucose ring participates in only a single hydrogen bond with the protein.
All side chains involved in A trisaccharide binding by the NV P domain are buttressed by intraprotein interactions (Fig. 4a and b), which are also present in the structure of the apo-NV P domain. Thus, there is very little structural accommodation involved in trisaccharide binding by the NV P domain. Not surprisingly, the N-acetylgalactosamine residue, which is the distinguishing feature between the A and B antigens, participates in potentially discriminating NV P protein interactions to a much greater extent than the invariant α-fucose residue. NV is unable to bind to the B trisaccharide, which differs only by the absence of the acetyl amide group present in the α-GalNAc residue. The N-acetyl amide group is not involved in direct contact with the P domain but makes water-mediated hydrogen bonds with the side chains of Asp344 and Ser338. Interestingly, the contact between the A trisaccharide and Ser338 represents the only interaction made by the sugar across the dyad axis of the NV P domain dimer. However, replacement of Ser338 of the NV P domain with an alanine does not affect binding to the A antigen (Fig. 5). His329 and Ser377 of the NV P domain, which are involved in critical A-trisaccharide interactions, were identified previously in an evolutionary-trace analysis as being in a GI class-specific patch (CS-4) likely to be involved in carbohydrate interactions (5).
The less stringent P domain, that of VA387, has the α-fucose residue inserted deepest into the binding pocket and mediating the bulk of the capsid-sugar interactions. The α-fucose is identical in A, B, and H antigens, and interactions seen in the crystal structures of VA387 P domain with both A and B trisaccharides are similar (6). Modeling an NV-type binding mode (i.e., inserting the α-GalNAc unit most deeply into the binding pocket) for either A or B trisaccharide in the VA387 P domain reveals significant steric hindrance (Fig. 6); close contacts are seen between the O6a hydroxyl of the terminal sugar moiety (α-GalNAc in A trisaccharide and α-galactose in B trisaccharide) and the Ile389-Gly392 region of VA387, which is the start of a large surface loop region between β7 and β8. Thus, for steric reasons, the receptor binding site of the NV P domain is different from that of the VA387 P domain. Furthermore, none of the sugar-contacting amino acid residues in the NV P domain are conserved in the VA387 protein. Both NV and VA387 bind the H antigen, which is characterized by the presence of α-fucose on C-2 of the terminal galactose (as in the A and B antigens), and the absence of additional sugars linked to the galactose. Structures of the H disaccharide bound to either NV or VA387 P domains are not yet available, but modeling suggests that binding of this oligosaccharide is sterically compatible with either binding mode.
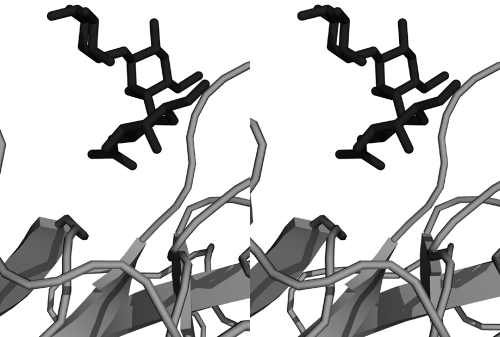
FIG. 6.
The structures of the NV P domain-A trisaccharide complex and the VA387 P domain-A trisaccharide complex (2OBS.PDB) were superimposed. Shown here, as a stereo view, is the A trisaccharide from the NV P domain complex and that from the VA387 protein, demonstrating steric clashes that prevent an NV-type carbohydrate binding mode for the VA387 P domain.
In previous studies we showed that P polypeptide is unable to form P particles and bind to HBGA receptors in conventional saliva-based binding assays (19). The structures described here of the P domain containing the Arg cluster are nearly identical to that of the P polypeptide lacking the Arg cluster. The C-terminal Arg-cluster-containing residues would extend away from the dimer interface and distal to the sugar-binding pocket of the P dimer (Fig. 1b and 2). Hence, it is unlikely for the Arg cluster to be involved in either dimerization or sugar binding. In fact, neither the P domain nor the P polypeptide revealed electron density in the crystal structures for residues beyond Val516 for NV and Gly530 for VA387, suggesting that the Arg cluster may be disordered. Another possibility is suggested by the previous crystallographic analysis of the VA387 P domain (3), where mass spectrometry of the VA387 P-domain-oligosaccharide complex crystals provided evidence of proteolytic degradation, leaving only the P polypeptide. Taken together, these observations again raise the question of whether the P polypeptide can bind to HBGAs. The previous conclusion that P polypeptide does not bind to HBGAs was based on conventional saliva-binding assays using a P polypeptide at a low concentration ranging from 0.01 to 2.5 μg/ml. However, the cocrystallization of both the VA387 and NV P domains with oligosaccharides was conducted at much higher concentrations of protein and in the presence of a >60-fold molar excess of trisaccharide. Saliva-binding assays have not been conducted under these conditions. It is also possible that the Arg cluster participates in interdimer interactions which stabilize the P particle and that increased avidity of sugar binding by the P particle is the stoichiometric result of the presence of multiple sugar binding sites. The structure of the P particle elucidated by cryo-EM (6a) supports this possibility by suggesting that the region of the Arg cluster may be involved in inter-P-dimer interactions.
Understanding how viruses recognize their receptors and enter cells is essential to an understanding of tissue and host specificity, mechanisms of virulence, and consequently the outcome of a viral infection. Structural studies have thus far provided important new insights regarding the mechanisms by which NV and VA387 recognize their host receptors. These could provide the basis for the design of therapeutic and prophylactic agents. In addition, human noroviruses are diverse, and eight receptor binding patterns associated with the human ABO, secretor, and Lewis types have been described. The two strains characterized in this study represent only two of the eight binding patterns; there is much to be learned, and our data provide a good starting point and a framework with which to design more detailed experiments probing norovirus-host interactions.
Acknowledgments
This work was supported by Public Health Service grant RO1 AI055649 from the National Institute of Allergy and Infectious Diseases.
Footnotes
Published ahead of print on 2 April 2008.
REFERENCES
- 1.Bertolotti-Ciarlet, A., L. J. White, R. Chen, B. V. Prasad, and M. K. Estes. 2002. Structural requirements for the assembly of Norwalk virus-like particles. J. Virol. 764044-4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brunger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54(Pt. 5)905-921. [DOI] [PubMed] [Google Scholar]
- 3.Cao, S., Z. Lou, M. Tan, Y. Chen, Y. Liu, Z. Zhang, X. C. Zhang, X. Jiang, X. Li, and Z. Rao. 2007. Structural basis for the recognition of blood group trisaccharides by norovirus. J. Virol. 815949-5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carson, M. 1987. Ribbon models of macromolecules. J. Mol. Graphics 5103-106. [Google Scholar]
- 5.Chakravarty, S., A. M. Hutson, M. K. Estes, and B. V. Prasad. 2005. Evolutionary trace residues in noroviruses: importance in receptor binding, antigenicity, virion assembly, and strain diversity. J. Virol. 79554-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Estes, M. K., B. V. Prasad, and R. L. Atmar. 2006. Noroviruses everywhere: has something changed? Curr. Opin. Infect. Dis. 19467-474. [DOI] [PubMed] [Google Scholar]
- 6a.Fang, P. -A., T. Chachiyo, M. Tan, M. Xia, X. Jiang, and W. Jiang. 2007. Octahedral and tetrahedral structures of the protruding domain of the norovirus capsid proteins, abstr. W9-7. 26th Annu. Meet. Am. Soc. Virol. 2007. Oregon State University, Corvallis, OR.
- 7.Frishman, D., and P. Argos. 1995. Knowledge-based protein secondary structure assignment. Proteins 23566-579. [DOI] [PubMed] [Google Scholar]
- 8.Gouet, P., E. Courcelle, D. I. Stuart, and F. Metoz. 1999. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics 15305-308. [DOI] [PubMed] [Google Scholar]
- 9.Huang, P., T. Farkas, S. Marionneau, W. Zhong, N. Ruvoen-Clouet, A. L. Morrow, M. Altaye, L. K. Pickering, D. S. Newburg, J. LePendu, and X. Jiang. 2003. Noroviruses bind to human ABO, Lewis, and secretor histo-blood group antigens: identification of 4 distinct strain-specific patterns. J. Infect. Dis. 18819-31. [DOI] [PubMed] [Google Scholar]
- 10.Hutson, A. M., R. L. Atmar, and M. K. Estes. 2004. Norovirus disease: changing epidemiology and host susceptibility factors. Trends Microbiol. 12279-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang, X., M. Wang, D. Y. Graham, and M. K. Estes. 1992. Expression, self-assembly, and antigenicity of the Norwalk virus capsid protein. J. Virol. 666527-6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones, T. A., J.-Y. Zou, S. W. Cowan, and M. Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47110-119. [DOI] [PubMed] [Google Scholar]
- 13.Laskowski, R. A., M. W. MacArthur, D. S. Moss, and J. M. Thornton. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26283-291. [Google Scholar]
- 14.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276307-326. [DOI] [PubMed] [Google Scholar]
- 15.Prasad, B. V., M. E. Hardy, T. Dokland, J. Bella, M. G. Rossmann, and M. K. Estes. 1999. X-ray crystallographic structure of the Norwalk virus capsid. Science 286287-290. [DOI] [PubMed] [Google Scholar]
- 16.Tan, M., R. S. Hegde, and X. Jiang. 2004. The P domain of norovirus capsid protein forms dimers and binds to histo-blood group antigen receptors. J. Virol. 786233-6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tan, M., and X. Jiang. 2005. Norovirus and its histo-blood group antigen receptors: an answer to a historical puzzle. Trends Microbiol. 13285-293. [DOI] [PubMed] [Google Scholar]
- 18.Tan, M., and X. Jiang. 2005. The P domain of norovirus capsid protein forms a subviral particle that binds to histo-blood group antigen receptors. J. Virol. 7914017-14030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tan, M., J. Meller, and X. Jiang. 2006. C-terminal arginine cluster is essential for receptor binding of norovirus capsid protein. J. Virol. 807322-7331. [DOI] [PMC free article] [PubMed] [Google Scholar]