Abstract
We report here a second case of coreceptor switch in R5 simian-human immunodeficiency virus SF162P3N (SHIVSF162P3N)-infected macaque CA28, supporting the use of this experimental system to examine factors that drive the change in coreceptor preference in vivo. Virus recovered from CA28 plasma (SHIVCA28NP) used both CCR5 and CXCR4 for entry, but the virus recovered from lymph node (SHIVCA28NL) used CXCR4 almost exclusively. Sequence and functional analyses showed that mutations in the V3 loop that conferred CXCR4 usage in macaque CA28 differed from those described in the previously reported case, demonstrating divergent mutational pathways for change in the coreceptor preference of the R5 SHIVSF162P3N isolate in vivo.
In addition to the requirement for CD4, human immunodeficiency virus type 1 (HIV-1) requires a secondary receptor, CCR5 or CXCR4, for entry into target cells (1). Viruses isolated from newly infected individuals predominantly use CCR5 (R5 viruses), whereas CXCR4-using (X4) viruses emerge in ∼50% of subtype B-infected patients with advanced disease (7). The cause of, mechanistic basis for, and obstacles to coreceptor switch in HIV-1 infection remain incompletely defined. Factors such as immune responses directed at the envelope and target cell availability are likely to influence the probability of X4 virus evolution and selection (8).
We recently reported coreceptor switching in BR24, one of three rhesus macaques (RM) infected with CCR5-tropic simian-human immunodeficiency virus SF162P3N (SHIVSF162P3N) (5), mirroring what happens during the course of HIV-1 infection. Infection of RM with SHIVSF162P3N, therefore, potentially holds promise in providing an experimental model to dissect selective forces that drive the switch from CCR5 to CXCR4 use. To establish the utility of the model, however, it will be necessary to demonstrate that phenotypic switching occurs in other SHIVSF162P3N-infected macaques. Furthermore, genotypic and functional studies of additional switch variants should reveal whether there is only one mutational pathway for evolution toward CXCR4 usage of SHIVSF162P3, or, alternatively, if there are multiple pathways, to identify common envelope properties associated with the switch. To address these issues, six more macaques were inoculated intravenously with the virus. Three of the six macaques were administered the anti-CD8 antibody cM-T807 at the time of virus inoculation to dampen the host immune response and promote virus replication.
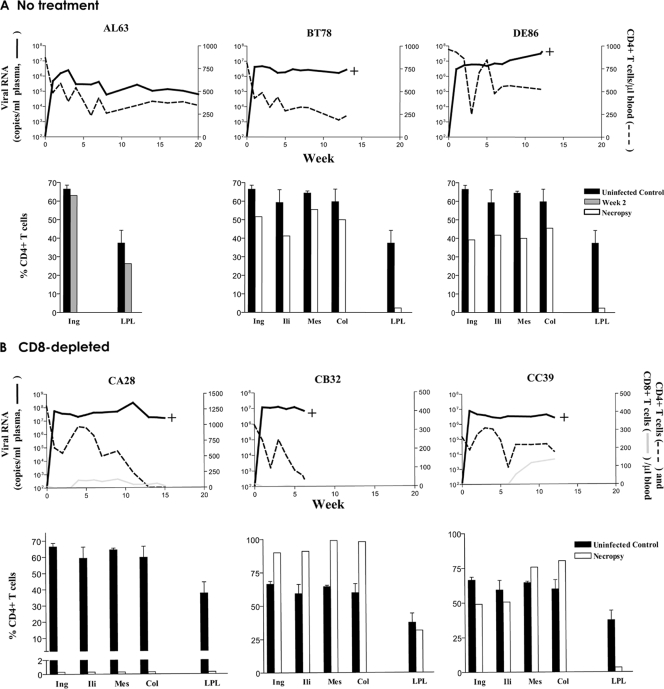
All six inoculated animals were infected, with peak viremia of 106 to 108 RNA copies/ml plasma at 2 weeks postinfection (wpi). Virus replication remained high during the course of infection, with levels that were 1 log higher in the CD8-depleted macaques than in those not receiving antibody treatment (Fig. 1). The exception was AL63 in the no-treatment group. This macaque established a set point that was 1 to 2 logs lower than the set points seen with the others and was the only animal that seroconverted (data not shown). For the three macaques in the no-treatment group (Fig. 1A), peripheral CD4-positive (CD4+) T-cell counts in DE86 recovered to within the normal range after a period of transient acute loss and fluctuated thereafter. The loss was more progressive for macaques AL63 and BT78 but with levels stabilizing at 8 to 10 wpi. BT78 and DE86 developed clinical signs consistent with simian AIDS (SAIDS) and were euthanized at 13 and 12 wpi, respectively, with circulating CD4+ T-cell counts greater than 200 cells/μl of blood. For the three macaques in the CD8-depleted group, partial restoration of peripheral CD8+ T cells was seen only in macaque CC39 (Fig. 1B). All three animals, however, showed substantial CD4+ T-cell rebound after a period of transient loss. But while the CD4+ T-cell count fluctuated for macaque CC39, precipitous decline followed in CA28 and CB32 and was accompanied by a spike in viremia in macaque CA28 but not in CB32 at 11 wpi. The three macaques in the CD8-depleted group all developed clinical signs consistent with SAIDS. Notably, at time of death (15 wpi), no circulating CD4+ T cells could be detected in macaque CA28. Thus, two of the three macaques (BT78 and DE86) infected in the absence and all three macaques (CA28, CB32, and CC39) infected in the presence of the anti-CD8 monoclonal antibody showed a rapid-progressor phenotype that was characterized by sustained high levels of virus replication (106 to 107 RNA copies/ml plasma), weak antiviral antibody response, and early onset of SAIDS.
FIG. 1.
Virologic and immunologic measurements in R5 SHIVSF162P3N-infected RMs. Results for viral load, absolute peripheral CD4+ T counts, and percentages of CD4+ T cells in tissues of macaques in the no-treatment (A) and the CD8-depleted (B) group are shown. Absolute peripheral CD8+ T-cell counts are also shown in Fig. 1B to illustrate the extent and duration of CD8+ T-cell depletion. Tissues analyzed at the time of necropsy included inguinal (Ing), iliac (Ili), mesenteric (Mes), and colonic (Col) lymph nodes (LNs), as well as lamina propria lymphocytes (LPL) from the jejunum. For tissue measurements, baseline values generated from three uninfected macaques are shown for reference, with error bars indicating standard deviations of the means. The greater percentage of CD4 in lymph nodes of CB32 than in those of the controls was due to sustained CD8 depletion in this macaque. +, time of euthanasia.
Analysis of the percentage of CD4+ T cells in tissue compartments of the two macaques (BT78 and DE86) in the no-treatment group at necropsy showed substantial preservation of this target cell population in lymph nodes (≥40%) but not in the lamina propria of the gut (<5%) (Fig. 1A). For the three CD8-depleted rapid progressors, lymphoid CD4+ T cells could still be detected in CB32 and CC39 but not in CA28 at the time of death (Fig. 1B). This pattern of massive CD4+ T-cell loss in lymph nodes of CA28 where CXCR4+ CD4+ naïve and memory T cells were enriched was reminiscent of that seen in macaque BR24 (5) and X4 SHIV-infected macaques (4), prompting us to examine the coreceptor usage of viruses recovered from this animal.
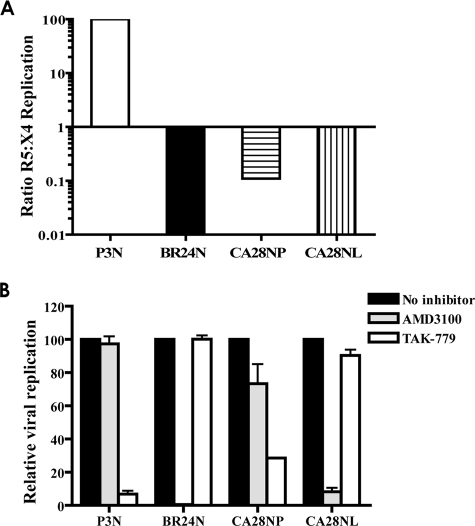
Results showed that viruses recovered from the plasma of CA28 (designated SHIVCA28NP) infected both CCR5- and CXCR4-bearing U87.CD4 target cells but with better growth in the latter, as indicated by an inverse ratio of R5/X4 replication (Fig. 2A). In contrast, viruses recovered from lymphoid tissues (designated SHIVCA28NL), in similarity to the results seen with the X4 variant SHIVBR24N in macaque BR24, showed a clear preference for CXCR4. Entry-blocking experiments with coreceptor inhibitors in TZM-bl cells confirmed the X4 coreceptor preference of the two viruses. Replication of SHIVCA28NP was efficiently blocked by the CCR5 antagonist TAK-779 (>70% inhibition at 1 μM) but could also be inhibited by the CXCR4 inhibitor AMD3100 (∼20% inhibition) (Fig. 2B). In contrast, AMD3100 blocked >90% of replication of the lymph node-derived virus SHIVCA28NL, with little or no inhibition seen with TAK-779. Thus, while the plasma virus from macaque CA28 displayed dual/mixed tropism, the lymph node virus appeared to be specific for CXCR4.
FIG. 2.
Coreceptor usage of SHIVCA28NP and SHIVCA28NL. (A) CXCR4 and CCR5 usage of the inoculating virus SHIVSF162P3N (P3N), the X4 variant SHIVBR24N from macaque BR24 (BR24N), and viruses recovered from plasma (CA28NP) and lymph node (CA28NL) of macaque CA28 at end stage disease was determined by relative replication of the SHIV viruses on U87.CD4.CCR5 and U87.CD4.CXCR4 cells. Data are expressed as the ratio of R5:X4 replication based on p27 antigen production in the two target cell lines after 6 to 7 days of culture. (B) Blocking of SHIV virus entry into TZM-bl cells with 1 μM CXCR4- and CCR5-specific inhibitors AMD3100 and TAK-779. Error bars indicate standard errors of results obtained in experiments using triplicate wells. Results shown are representative of at least two independent experiments.
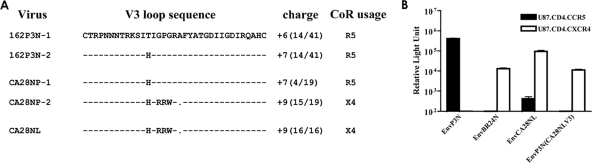
We sequenced the envelope V3 domain of SHIVCA28NP and SHIVCA28NL and compared it to that of the inoculating virus SHIVSF162P3N to identify genetic changes that confer CXCR4 usage. All SHIVCA28NL Env clones sequenced (16 in total) harbored the same five amino acid changes in the V3 loop compared to the sequence of the inoculating strain, which is comprised of two major variants (162P3N-1 and 162P3N-2) (Fig. 3A) and several minor variants (5). These amino acid changes included a threonine-to-histidine change at position 13 and mutations of the GPG sequence at the crown of the loop to RRW, which increased the net positive charge of this domain. Furthermore, the alanine residue immediately downstream of the crown loop sequence was deleted in SHIVCA28NL. In contrast, clonal analysis indicated that virus SHIVCA28NP recovered from plasma comprised a mixture of variants. A total of 15 of 19 clones sequenced showed the RRW substitutions and alanine deletion in the V3 loop, with 4 having sequences similar to that of the inoculating R5 strain.
FIG. 3.
(A) Comparison of V3 loop sequence of SHIVSF162P3N, SHIVCA28NP, and SHIVCA28NL. Dashes denote similarities in sequence, gaps are indicated as dots, and the net positive charges of this region are shown on the right. The numbers in parentheses indicate the number of clones with the indicated sequence referenced to the total number of clones sequenced. (B) Entry into U87.CD4.CCR5 (black bars) and U87.CD4.CXCR4 (white bars) of luciferase reporter viruses expressing the envelope of SHIVSF162P3N (EnvP3N), SHIVBR24N (EnvBR24N), SHIVCA28NL (EnvCA28NL), and V3 loop recombinant EnvP3N(CA28NLV3). All data represent means + standard errors of the means of the results of triplicate experiments.
To investigate whether the changes in the V3 loop of SHIVCA28NL dictate CXCR4 usage, recombinant EnvP3N(CA28NLV3) in which the V3 loop of the parental R5 EnvP3N gp160 was replaced with that of SHIVCA28NL was constructed. In single-round infectivity assays, EnvCA28NL gp160 mediated efficient entry into CXCR4-expressing U87.CD4 cells but with some residual infectivity of CCR5-expressing cells as well (Fig. 3B). In contrast, EnvP3N(CA28NLV3) mediated entry only into CXCR4-expressing indicator cells, albeit with lower efficiency compared to that of EnvCA28NL. These results illustrate that V3 mutations are necessary for coreceptor switching in macaque CA28 but that changes in other regions, such as those in the gp120 V1/V2 and C4 and gp41 domains known to modulate receptor binding and fusion (2, 3, 6, 9-11), likely contribute to optimize SHIVCA28NL entry efficiency. Significantly, the V3 mutations that conferred CXCR4 usage to SHIVCA28NL differed from those previously reported for SHIVBR24N, which included insertions of two basic amino acids (histidine and arginine) immediately upstream of the crown of the V3 loop (5).
We demonstrated X4 virus evolution and emergence in another R5 SHIVSF162P3N-infected RM in this study. Functional data showed that while the SHIVCA28NP virus circulating in macaque CA28 at the time of death was composed of a mixture of CCR5- and CXCR4-using viruses, the SHIVCA28NL virus recovered from lymph nodes used CXCR4 for entry almost exclusively. Mutations in the V3 loop of SHIVCA28NL were largely responsible for the change in coreceptor preference but differed from those previously reported for coreceptor switching in macaque BR24 (5), indicating that there is more than one pathway for CCR5-to-CXCR4 evolution of the R5 SHIVSF162P3N isolate. Both BR24 and CA28 rapidly progressed to disease with high viral load and in the absence of seroconversion. Furthermore, in similarity to the results seen with macaque BR24, tracking experiments showed that X4 virus emerged in CA28 at 11 wpi, following rather than preceding the onset of precipitous CD4+ T-cell decline, and the emerging variant was highly sensitive to soluble CD4 neutralization (data not shown). The similarity in the tempos and conditions of X4 evolution and emergence in the two macaques points to a common mechanistic cause. Our findings obtained with macaque CA28 confirm that the SHIVSF162P3N isolate has an intrinsic tendency to mutate toward CXCR4 usage, providing the tool for an experimental animal model for study of the process and requisites for coreceptor switching.
Nucleotide sequence accession numbers.
The predicted gp160 amino acid sequences of EnvBR24N and EnvCA28NL have been deposited in GenBank (accession numbers EU564103 and EU564104, respectively).
Acknowledgments
We thank Lisa Chakrabarti and Allen Mayer for critical reading of the manuscript and Keith Reimann and Centocor for the anti-human CD8 monoclonal antibody (CM-T807). AMD3100, TAK-779, and TZM-bl cells and the U87.CD4 indicator cell line were obtained through the NIH AIDS Research and Reference Reagent Program.
This work was supported by National Institutes of Health (NIH) grants R01 AI46980 and R37AI41945.
Footnotes
Published ahead of print on 2 April 2008.
REFERENCES
- 1.Berger, E. A. 1997. HIV entry and tropism: the chemokine receptor connection. AIDS 11(Suppl. A)S3-S16. [PubMed] [Google Scholar]
- 2.Chan, D. C., D. Fass, J. M. Berger, and P. S. Kim. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89263-273. [DOI] [PubMed] [Google Scholar]
- 3.Groenink, M., R. A. Fouchier, S. Broersen, C. H. Baker, M. Koot, A. B. van't Wout, H. G. Huisman, F. Miedema, M. Tersmette, and H. Schuitemaker. 1993. Relation of phenotype evolution of HIV-1 to envelope V2 configuration. Science 2601513-1516. [DOI] [PubMed] [Google Scholar]
- 4.Ho, S. H., L. Shek, A. Gettie, J. Blanchard, and C. Cheng-Mayer. 2005. V3 loop-determined coreceptor preference dictates the dynamics of CD4+-T-cell loss in simian-human immunodeficiency virus-infected macaques. J. Virol. 7912296-12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ho, S. H., S. Tasca, L. Shek, A. Li, A. Gettie, J. Blanchard, D. Boden, and C. Cheng-Mayer. 2007. Coreceptor switch in R5-tropic simian/human immunodeficiency virus-infected macaques. J. Virol. 818621-8633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koito, A., G. Harrowe, J. A. Levy, and C. Cheng-Mayer. 1994. Functional role of the V1/V2 region of human immunodeficiency virus type 1 envelope glycoprotein gp120 in infection of primary macrophages and soluble CD4 neutralization. J. Virol. 682253-2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koot, M., I. P. Keet, A. H. Vos, R. E. de Goede, M. T. Roos, R. A. Coutinho, F. Miedema, P. T. Schellekens, and M. Tersmette. 1993. Prognostic value of HIV-1 syncytium-inducing phenotype for rate of CD4+ cell depletion and progression to AIDS. Ann. Intern. Med. 118681-688. [DOI] [PubMed] [Google Scholar]
- 8.Regoes, R. R., and S. Bonhoeffer. 2005. The HIV coreceptor switch: a population dynamical perspective. Trends Microbiol. 13269-277. [DOI] [PubMed] [Google Scholar]
- 9.Rizzuto, C., and J. Sodroski. 2000. Fine definition of a conserved CCR5-binding region on the human immunodeficiency virus type 1 glycoprotein 120. AIDS Res. Hum. Retrovir. 16741-749. [DOI] [PubMed] [Google Scholar]
- 10.Tan, K., J. Liu, J. Wang, S. Shen, and M. Lu. 1997. Atomic structure of a thermostable subdomain of HIV-1 gp41. Proc. Natl. Acad. Sci. USA 9412303-12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wyatt, R., J. Moore, M. Accola, E. Desjardin, J. Robinson, and J. Sodroski. 1995. Involvement of the V1/V2 variable loop structure in the exposure of human immunodeficiency virus type 1 gp120 epitopes induced by receptor binding. J. Virol. 695723-5733. [DOI] [PMC free article] [PubMed] [Google Scholar]