Short abstract
The discovery of three different enzymes that deubiquitinate histone 2A.
Abstract
Three recent papers have identified distinct enzymes that can remove ubiquitin from mammalian histone 2A (H2A). Functions in transcriptional activation, DNA repair and control of the cell cycle have been proposed for these enzymes.
Ubiquitination is a reversible posttranslational modification of proteins that could rival phosphorylation in its scope and complexity. Removal of ubiquitin is accomplished through the action of deubiquitinating enzymes (DUBs), which fall into five distinct families in the human genome, containing about 84 active members [1,2]. Post-translational modifications of histones include methylation, acetylation, sumoylation and ubiquitination, enabling a combinatorial code to regulate chromatin structure [3] and recruit protein complexes [4]. An example of the interplay between these modifications is provided by the activity of Ubp8, a yeast deubiquitinating enzyme which regulates the methylation of histone 3 (H3) on lysine 4 (K4 methylation) by deubiquitinating histone 2B (H2B) [5]. About 5-15% of histone 2A (H2A) is monoubiquitinated in mammalian cells, making it the most abundant ubiquitinated protein in the nucleus; in fact, it was the first example of a ubiquitinated protein to be described [6]. Monoubiquitination is not linked to protein degradation, but instead plays a role in transcriptional control, DNA repair and other processes. It has also long been known that deubiquitination of histones occurs during metaphase of the cell cycle, coincident with complete condensation of the chromosomes [7,8]. Monoubiquitination of H2A has been linked to Polycomb group complex-dependent gene silencing [9] and X-chromosome inactivation [10]. Furthermore, DNA damage induces monoubiquitination of H2A in the vicinity of DNA lesions after incision of the damaged strand [11].
Identification of H2A-deubiquitinating enzymes
Three recent papers identify distinct DUBs with activity towards H2A and begin to unravel their cellular functions (Figure 1). Zhu et al. [12] identified MYSM1/KIAA1915 (belonging to the JAMM/MPN+ family of metalloprotease DUBs [13]) in a screen for factors regulating androgen-controlled gene expression. The authors argue that MYSM1 is an H2A-specific DUB on the basis of decreased ubiquitination following overexpression of MYSM1 in HEK293T human embryonic kidney cells and the accumulation of ubiquitinated H2A (Ub-H2A) following knockdown of MYSM1.
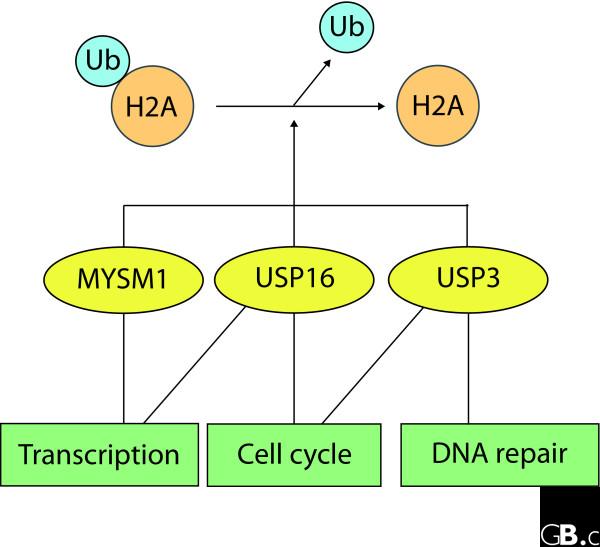
Figure 1.
Common and distinct functional outputs of three enzymes that deubiquitinate H2A. These three enzymes - MYSMI, USP16 and USP3 - have been proposed to remove ubiquitin from Ub-H2A. MYSMI and USP16 share a common role in transcriptional control of potentially distinct gene cohorts, whereas USP16 and USP3 DUB activities have been implicated in cell-cycle progression. USP3 may indirectly influence cycling of cells through effects on DNA damage repair pathways.
The ubiquitin-specific proteases (USPs) comprise the largest family of cysteine protease DUBs. Two members of this family are now proposed to act on H2A. Nicassio et al. [14] have taken chromatin-enriched fractions through affinity purification on ubiquitin-agarose. Analysis of the resultant gel bands by mass spectrometry identified USP3 and USP5/isopeptidase T. USP3 is the human DUB most homologous to Saccharomyces cerevisiae Ubp8, which has previously been shown to promote H2B deubiquitination [15]. Overexpression of USP3 in HeLa cells reduced levels of both Ub-H2A and Ub-H2B, whereas knockdown of USP3 enhanced the ubiquitination of both histones.
In another recent study, Joo et al. [16] have undertaken an epic purification of DUB activity from HeLa nuclear fractions using an in vitro assay of H2A deubiquitination with mononucleosomes as substrate. A silver-stained band identified as USP16/Ubp-M could be correlated with deubiquitinating activity through a six-column purification procedure. Immunodepletion of USP16 from a purified fraction also depleted H2A-DUB activity. Previous work had already shown that overexpression of USP16/Ubp-M in 293 cells leads to H2A deubiquitination [17]. Joo et al. [16] now show that small interfering RNA-mediated knockdown of USP16 in HeLa cells leads to the predicted accumulation of Ub-H2A, but in distinction to USP3, no accumulation of Ub-H2B was found on USP16 knockdown. Interestingly, USP16 showed enhanced in vitro activity against Ub-H2A incorporated into oligonucleosomes and could not act upon Ub-H2A in isolation [16]. Taken together, the data indicate that USP16 is a nucleosomal H2A-specific DUB.
MYSM1 and USP16 activate transcription
Co-immunoprecipitation and peptide mass fingerprinting identified the histone acetylase p300/CBP-associated factor (PCAF) as a MYSM1 binding partner [12]. Subsequent experimental evidence uncovered no cross-talk with histone methylation, but led the authors to propose an interesting model whereby PCAF-dependent acetylation of nucleosomal components precedes MYSM1 deubiquitination of H2A. In turn, this deubiquitination may promote histone 1 (H1) phosphorylation, a trigger for H1 dissociation from nucleosomes that is often linked with gene activation. Zhu et al. [12] provide a clear illustration of this effect, by showing that expression of androgen receptor target genes following dihydrotestosterone (DHT)-induced activation in LNCaP prostate cancer cells is significantly impaired by MYSM1 knockdown. Accordingly, chromatin immunoprecipitation (ChIP) analysis indicated that MYSM1 and PCAF are enriched on the promoter region of the gene for prostate-specific antigen (PSA) gene following DHT treatment. MYSM1 is also enriched at exonic regions of the gene, suggesting a potential role in transcript elongation as well as initiation.
Control of the H2A ubiquitination status has been linked to HOX gene silencing [9,18], leading Joo et al. [16] to examine a role for USP16 in HOX gene expression. They show that HOXD10 transcription is reduced following USP16 knockdown, in a manner that can be rescued by transfection with wild-type enzyme. ChIP analysis revealed binding sites for USP16 within the 5' promoter but not the coding region of HOXD10, which accumulate Ub-H2A following USP16 knockdown. A nice demonstration of the physiological significance of this finding is derived from injection of anti-USP16 into two-cell-stage Xenopus embryos, which leads to reduced Hoxd10 expression and defects in anterior-posterior patterning [16]. Thus both MYSM1 and USP16 can positively regulate the expression of certain genes. This is consistent with their specificity for Ub-H2A, which is thought to be transcriptionally repressive, rather than human Ub-H2B, which activates HOX gene expression [19].
USP3 and USP16 are required for cell-cycle progression
Functional studies suggest roles for both USP3 and USP16 in the control of the cell cycle. Earlier work had shown that USP16 is phosphorylated at the onset of mitosis and dephosphorylated during the metaphase-anaphase transition. Furthermore, USP16 tagged with green fluorescent protein generally presents a cytosolic distribution, but a catalytically inactive form associates with mitotic chromosomes at all stages of cell division and remains nuclear in interphase cells [20]. Joo et al. [16] show that USP16 knockdown reduces the number of cells in M phase. In synchronized control cells, Ub-H2A levels decrease during M phase and recover to normal levels when cells reach G1/S. In USP16 knockdown cells the reduction in Ub-H2A is much less pronounced and entry into mitosis is delayed. Fluctuations in Ub-H2A levels inversely correlate with phospho-H3 levels. Using reconstituted mononuclesomes, Joo et al. [16] could show that the presence of Ub-H2A is inhibitory to Aurora B kinase-mediated phosphorylation of H3 on serine 10 (S10), and that this activity can be restored by treatment with USP16. Thus, H2A deubiquitination may be a prerequisite for H3-S10 phosphorylation; the authors suggest that this is likely to be due to occlusion of the Aurora B binding site by ubiquitin.
Nicassio et al. [14] found that following knockdown of USP3 in an osteosarcoma cell line (U2OS), release from a thymidine block leads to slower progression through S phase and delayed entry into mitosis, as judged by fluorescence-activated cell sorting (FACS) analysis and incorporation of bromodeoxyuridine into DNA. As seen with USP16 knockdown, H3 phosphorylation is delayed. One possible cause of the cell-cycle defects, highlighted by the authors, might be DNA damage leading to the activation of checkpoint controls. They could detect an early signature of this response in USP3-silenced U2OS cells by visualizing the accumulation of phosphorylated H2AX (γ-H2AX) and the checkpoint protein 53BP1 in nuclear foci, together with positive assays for the occurrence of single-strand DNA breaks. Further experiments, monitoring the ability of cells to recover from ionizing-radiation-induced DNA damage, revealed a marked delay in the dissipation of γ-H2AX- and Ub-H2A-positive foci compared with control cells [14].
Other DUBs have also recently been implicated in cell-cycle control. A screen for novel cell-cycle regulators in human cells (using knockdown by short hairpin RNAs) identified USP44 as a critical regulator of the spindle checkpoint, through stabilization of the Mad2-CDC20 complex, which inhibits the anaphase-promoting complex (APC) [21], a ubiquitin ligase that targets certain cell-cycle proteins for destruction. This visual screen containing 63 DUBs (including USP3 and USP16) scored the mitotic index following a 24-hour incubation with taxol. It also identified USP24 as a candidate DUB involved in checkpoint control and CYLD as a pre-mitotic cell-cycle regulator. Subsequent studies of CYLD have suggested that it may act in mitosis by controlling the ubiquitination status of the Polo-like kinase Plk1 [22].
Chromatin binding
MYSM1 is the only DUB in the genome with recognizable chromatin-binding domains, containing both the SWIRM (Swi3p, Rsc8p and Moira) [23] and SANT (SWI-SNF, ADA N-CoR, TFIIIB)/Myb domains [24]. The SANT, but not the SWIRM, domain of MYSM1 can bind directly to DNA [25]. Intriguingly USP3, USP16 and USP44 all possess a zinc-finger domain, ZnF UBP, amino-terminal to their catalytic domain. Several proteins with this motif (for example, USP5/isopeptidase T and HDAC6) have been shown to bind ubiquitin, but in the case of USP5 this requires an unconjugated ubiquitin carboxy-terminal diglycine motif, suggesting that it may primarily process free ubiquitin chains [26] or act as a sensor of free ubiquitin levels. Characterization of the ZnF UBP domain by nuclear magnetic resonance suggests that the same holds true for USP16, despite structural differences from USP5 in the pattern of zinc coordination [27]. An intact ZnF UBP domain is necessary for USP3 association with H2A, as judged by co-immunoprecipitation, but the requirement for ubiquitination is unclear [14]. If the ZnF UBP domain cannot recognize conjugated ubiquitin, then on the basis of the functional studies described above it seems plausible that it might represent a novel recognition motif for as-yet-unidentified chromatin-associated factors.
Three DUBs have now been shown to have profound effects on global Ub-H2A levels (Figure 2). What can this signify? One possibility is that each DUB targets a distinct pool of Ub-H2A. For example, one could propose that USP3 might specifically target the pool of Ub-H2A that accumulates at DNA repair foci, or that MYSM1 and USP16 are recruited to different cohorts of gene promoters. However, the magnitude of changes in the ubiquitinated fraction of H2A seen for each DUB following overexpression or knockdown suggests that the pool accessible to each DUB must be a significant fraction of total H2A. It is currently not known if these effects are additive. Nor at this point can it be definitively concluded that each enzyme directly deubiquitinates Ub-H2A in vivo. Thus, an alternative possibility is that the action of one may be contingent on that of another, through regulation of stability or activity. Other DUBs may yet join this trio, as no systematic screen for DUBs regulating Ub-H2A levels has so far been reported. In fact, while this review was being prepared for publication, Nakagawa et al. [28] reported a role for USP21 in deubiquitinating H2A, resulting in transcriptional activation.
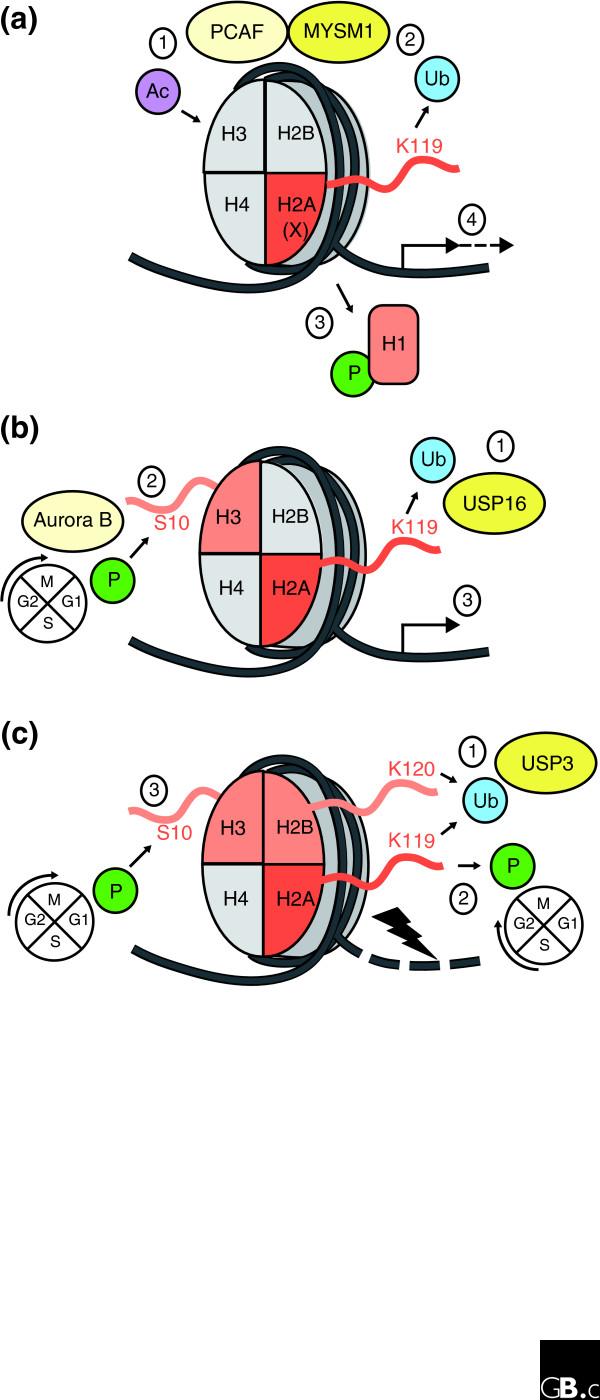
Figure 2.
Proposed models for the role of three H2A-DUBs - MYSM1, USP16 and USP3 - in regulating reversible modifications of histones. (a) Histone acetylation by PCAF (1) is proposed to promote removal of ubiquitin from lysine 119 (K119) in the H2A tail by associated MYSM1 (2). This promotes phosphorylation of H1 and its consequent dissociation from chromatin (3), facilitating transcriptional initiation and elongation of androgen receptor-regulated genes (4). (b) Deubiquitination of Ub-H2A by USP16 (1) leads to phosphorylation of H3 at serine 10 (S10) by the kinase Aurora B and subsequent G2/M cell-cycle progression (2). It also promotes transcriptional initiation of HOX gene transcription (3). (c) USP3 can remove ubiquitin from both H2A K119 and H2B K120 (1), promoting dephosphorylation of the H2AX variant histone and concomitant recovery from the ATM/ATR DNA-damage checkpoint during DNA repair (2). USP3 activity may also promote phosphorylation of H3 at S10, which is associated with entry into M phase (3). (a) adapted from Zhu et al. [12].
Contributor Information
Michael J Clague, Email: clague@liv.ac.uk.
Sylvie Urbé, Email: urbe@liv.ac.uk.
Acknowledgements
SU is a Cancer Research UK senior research fellow.
References
- Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, Bernards R. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- Scheel H. PhD thesis. University of Cologne; 2005. Comparative analysis of the ubiquitin-proteasome system in Homo sapiens and Saccharomyces cerevisiae. [Google Scholar]
- Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu Rev Biochem. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- Ruthenburg AJ, Li H, Patel DJ, Allis CD. Multivalent engagement of chromatin modifications by linked binding modules. Nat Rev Mol Cell Biol. 2007;8:983–994. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla A, Stanojevic N, Duan Z, Sen P, Bhaumik SR. Ubp8p, a histone deubiquitinase whose association with SAGA is mediated by Sgf11p, differentially regulates lysine 4 methylation of histone H3 in vivo. Mol Cell Biol. 2006;26:3339–3352. doi: 10.1128/MCB.26.9.3339-3352.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldknopf IL, Busch H. Isopeptide linkage between nonhistone and histone 2A polypeptides of chromosomal conjugate-protein A24. Proc Natl Acad Sci USA. 1977;74:864–868. doi: 10.1073/pnas.74.3.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller RD, Yasuda H, Hatch CL, Bonner WM, Bradbury EM. Identification of ubiquitinated histones 2A and 2B in Physarum polycephalum. Disappearance of these proteins at metaphase and reappearance at anaphase. J Biol Chem. 1985;260:5147–5153. [PubMed] [Google Scholar]
- Matsui SI, Seon BK, Sandberg AA. Disappearance of a structural chromatin protein A24 in mitosis: implications for molecular basis of chromatin condensation. Proc Natl Acad Sci USA. 1979;76:6386–6390. doi: 10.1073/pnas.76.12.6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- de Napoles M, Mermoud JE, Wakao R, Tang YA, Endoh M, Appanah R, Nesterova TB, Silva J, Otte AP, Vidal M, Koseki H, Brockdorff N. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell. 2004;7:663–676. doi: 10.1016/j.devcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- Bergink S, Salomons FA, Hoogstraten D, Groothuis TA, de Waard H, Wu J, Yuan L, Citterio E, Houtsmuller AB, Neefjes J, Hoeijmakers JH, Vermeulen W, Dantuma NP. DNA damage triggers nucleotide excision repair-dependent monoubiquitylation of histone H2A. Genes Dev. 2006;20:1343–1352. doi: 10.1101/gad.373706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu P, Zhou W, Wang J, Puc J, Ohgi KA, Erdjument-Bromage H, Tempst P, Glass CK, Rosenfeld MG. A histone H2A deubiquitinase complex coordinating histone acetylation and H1 dissociation in transcriptional regulation. Mol Cell. 2007;27:609–621. doi: 10.1016/j.molcel.2007.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maytal-Kivity V, Reis N, Hofmann K, Glickman MH. MPN+, a putative catalytic motif found in a subset of MPN domain proteins from eukaryotes and prokaryotes, is critical for Rpn11 function. BMC Biochem. 2002;3:28. doi: 10.1186/1471-2091-3-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicassio F, Corrado N, Vissers JH, Areces LB, Bergink S, Marteijn JA, Geverts B, Houtsmuller AB, Vermeulen W, Di Fiore PP, Citterio E. Human USP3 is a chromatin modifier required for S phase progression and genome stability. Curr Biol. 2007;17:1972–1977. doi: 10.1016/j.cub.2007.10.034. [DOI] [PubMed] [Google Scholar]
- Henry KW, Wyce A, Lo WS, Duggan LJ, Emre NC, Kao CF, Pillus L, Shilatifard A, Osley MA, Berger SL. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 2003;17:2648–2663. doi: 10.1101/gad.1144003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo HY, Zhai L, Yang C, Nie S, Erdjument-Bromage H, Tempst P, Chang C, Wang H. Regulation of cell cycle progression and gene expression by H2A deubiquitination. Nature. 2007;449:1068–1072. doi: 10.1038/nature06256. [DOI] [PubMed] [Google Scholar]
- Mimnaugh EG, Kayastha G, McGovern NB, Hwang SG, Marcu MG, Trepel J, Cai SY, Marchesi VT, Neckers L. Caspase-dependent deubiquitination of monoubiquitinated nucleosomal histone H2A induced by diverse apoptogenic stimuli. Cell Death Differ. 2001;8:1182–1196. doi: 10.1038/sj.cdd.4400924. [DOI] [PubMed] [Google Scholar]
- Cao R, Tsukada Y, Zhang Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol Cell. 2005;20:845–854. doi: 10.1016/j.molcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Zhu B, Zheng Y, Pham AD, Mandal SS, Erdjument-Bromage H, Tempst P, Reinberg D. Monoubiquitination of human histone H2B: the factors involved and their roles in HOX gene regulation. Mol Cell. 2005;20:601–611. doi: 10.1016/j.molcel.2005.09.025. [DOI] [PubMed] [Google Scholar]
- Cai SY, Babbitt RW, Marchesi VT. A mutant deubiquitinating enzyme (Ubp-M) associates with mitotic chromosomes and blocks cell division. Proc Natl Acad Sci USA. 1999;96:2828–2833. doi: 10.1073/pnas.96.6.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmeier F, Rape M, Draviam VM, Nalepa G, Sowa ME, Ang XL, McDonald ER 3rd, Li MZ, Hannon GJ, Sorger PK, Kirschner MW, Harper JW, Elledge SJ. Anaphase initiation is regulated by antagonistic ubiquitination and deubiquitination activities. Nature. 2007;446:876–881. doi: 10.1038/nature05694. [DOI] [PubMed] [Google Scholar]
- Stegmeier F, Sowa ME, Nalepa G, Gygi SP, Harper JW, Elledge SJ. The tumor suppressor CYLD regulates entry into mitosis. Proc Natl Acad Sci USA. 2007;104:8869–8874. doi: 10.1073/pnas.0703268104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Iyer LM. The SWIRM domain: a conserved module found in chromosomal proteins points to novel chromatin-modifying activities. Genome Biol. 2002;3:research0039.1–0039.7. doi: 10.1186/gb-2002-3-8-research0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aasland R, Stewart AF, Gibson T. The SANT domain: a putative DNA-binding domain in the SWI-SNF and ADA complexes, the transcriptional co-repressor N-CoR and TFIIIB. Trends Biochem Sci. 1996;21:87–88. [PubMed] [Google Scholar]
- Yoneyama M, Tochio N, Umehara T, Koshiba S, Inoue M, Yabuki T, Aoki M, Seki E, Matsuda T, Watanabe S, Tomo Y, Nishimura Y, Harada T, Terada T, Shirouzu M, Hayashizaki Y, Ohara O, Tanaka A, Kigawa T, Yokoyama S. Structural and functional differences of SWIRM domain subtypes. J Mol Biol. 2007;369:222–238. doi: 10.1016/j.jmb.2007.03.027. [DOI] [PubMed] [Google Scholar]
- Reyes-Turcu FE, Horton JR, Mullally JE, Heroux A, Cheng X, Wilkinson KD. The ubiquitin binding domain ZnF UBP recognizes the C-terminal diglycine motif of unanchored ubiquitin. Cell. 2006;124:1197–1208. doi: 10.1016/j.cell.2006.02.038. [DOI] [PubMed] [Google Scholar]
- Pai MT, Tzeng SR, Kovacs JJ, Keaton MA, Li SS, Yao TP, Zhou P. Solution structure of the Ubp-M BUZ domain, a highly specific protein module that recognizes the C-terminal tail of free ubiquitin. J Mol Biol. 2007;370:290–302. doi: 10.1016/j.jmb.2007.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Kajitani T, Togo S, Masuko N, Ohdan H, Hishikawa Y, Koji T, Matsuyama T, Ikura T, Muramatsu M, Ito T. Deubiquitylation of histone H2A activates transcriptional initiation via trans-histone cross-talk with H3K4 di- and trimethylation. Genes Dev. 2008;22:37–49. doi: 10.1101/gad.1609708. [DOI] [PMC free article] [PubMed] [Google Scholar]