Short abstract
A novel approach for identifying condition-specific regulatory modules in yeast reveals functionally distinct coregulated submodules.
Abstract
We present an approach for identifying condition-specific regulatory modules by using separate units of gene expression profiles along with ChIP-chip and motif data from Saccharomyces cerevisiae. By investigating the unique and common features of the obtained condition-specific modules, we detected several important properties of transcriptional network reorganization. Our approach reveals the functionally distinct coregulated submodules embedded in a coexpressed gene module and provides an effective method for identifying various condition-specific regulatory events at high resolution.
Background
Transcription regulation is a starting point for controlling a variety of biological processes, such as cell cycle progression and adaptive responses to environmental stimuli. Moreover, the regulation is realized by intricate regulatory gene networks that are mainly controlled by transcription factors. In order to appropriately process and respond to environmental changes, cells are likely to use distinct transcriptional regulatory networks by detecting specific features of complex environmental stimuli. Through altering the activities and targets of transcription factors depending on the cellular conditions, rewiring of transcriptional regulatory network occurs to adapt to various stimuli or initiate cellular programs [1]. Therefore, identifying the sophisticated architecture of transcriptional regulatory networks and further deciphering the mechanisms of transcriptional rewiring in response to various conditions would reveal the fundamental aspects of the mechanisms involved in the maintenance of life and adaptation to new environments.
Recently, many studies attempted to address these challenges by examining the transcriptional regulatory networks of Saccharomyces cerevisiae from various complementary perspectives. Luscombe et al. [2] analyzed the dynamics of transcriptional networks by using known transcriptional regulatory information and gene expression profiles of five specific environmental and developmental conditions. They reported that a majority of regulatory interactions among transcription factors and genes are highly condition specific, based on the observation that many of the transcription factors that regulated a large number of target genes in a certain condition did not maintain their regulation in other conditions. They also suggested that the topological properties of the networks differ considerably depending on the types of the conditions, classified as exogenous (for example, environmental stress) and endogenous (for example, cell cycle and sporulation). Harbison et al. [3] attempted to identify the dynamic nature of the transcriptional regulatory networks by conducting genome-wide binding assays for 203 transcription factors under various conditions. They found that, for most of the examined transcription factors, transcription factor binding to a regulatory sequence is highly dependent on the environmental condition of the cells. From these results, it is evident that dynamic alterations in the transcriptional network occur in response to changes in cellular conditions, although the actual mechanisms of rewiring and the detailed descriptions of the condition-specific regulatory networks remain to be explored.
To study all these aspects, we need to identify reliable condition-specific transcriptional regulatory modules. Identification of transcriptional regulatory modules, that is, gene groups sharing common regulatory mechanisms, is a major step toward deciphering the dynamic cellular regulation system more concretely. Many previous studies strived to identify the transcriptional regulatory modules and contributed to the detection of the links between gene expression and gene regulation by suggesting coexpressed gene modules controlled by their own regulators in various manners [4-6]. However, most studies assumed that a transcriptional regulatory network is static and usually defined coexpressed gene groups as the genes displaying similar expression profiles across multiple conditions; this viewpoint prevented the detection of the distinct features of condition-specific regulation. Although other studies employed condition-specific approaches [7-11], they did not clearly show the actual rewiring mechanisms of the condition-specific regulatory networks in response to external or internal signals. Moreover, most of them also presumed that the similarity in expression profiles among several genes implies their coregulation. In fact, stratification based on expression similarity obscures the transcriptional regulation program in many cases because an environmental or biological condition can activate multiple processes in parallel, and similar expression patterns can be elicited under multiple alternative regulatory mechanisms [12].
Here, we present an approach for identifying condition-specific regulatory modules in high resolution by integrating ChIP-chip, mRNA expression and known transcription factor binding motif data. By investigating diverse aspects of the identified modules and their regulators, we tried to dissect the dynamic properties of the condition-dependent regulatory networks and their rewiring mechanism. In this study, we adopted two distinctive strategies to reveal the dynamic transcriptional regulatory modules in detail. First, we identified the modules from each of the selected cellular conditions independently and then compared them in order to reveal the detailed and distinct features of the reorganized transcriptional regulatory network specified in each condition. Our results included various examples of regulatory events occurring in specific conditions that describe the reorganization of the transcriptional regulatory program depending on the change in stimuli conditions. Second, we identified multiple coregulated submodules from each of the coexpressed gene modules in high resolution. In order to obtain coregulated gene groups, we identified small coexpressed gene groups - initial module candidates (IMCs) - that comprised genes sharing common transcription factor binding evidence and employed them to identify the transcriptional regulatory modules. By considering the notion that the same expression can be activated through many independent transcriptional regulatory programs [12], this bottom-up approach allowed the detection of the local regulatory mechanisms that affect only a part of the entire coexpressed genes.
Through specialized strategies, we identified various condition-specific regulatory modules and their designated transcription factors in high resolution by using gene expression data obtained under different experimental conditions: heat shock, nitrogen depletion and mitotic cell cycle [13,14]. Excluding the treatment for cell cycle synchronization, the cell cycle condition can be regarded as a normal condition (YPD medium) with no limitation in cell growth and proliferation. The two stress conditions - heat shock and nitrogen depletion - were selected in order to investigate the distinct effects of environmental stress; the former elicits rapid and massive alterations in gene expression, while the latter is a prolonged nutrient-limiting condition. Although the regulatory modules from the three conditions shared some functional modules, most of them displayed unique functional properties specific to each condition due to the rewiring of the transcriptional regulatory network. In addition, many of the functional gene groups that exhibited distinct expression profiles in other conditions were coexpressed in a certain condition. We also investigated the distinguished condition-specific regulatory roles of the transcription factors by classifying them based on the degree and the manner in which they switch their target genes. Among the results obtained, many clear cases indicated that target switching by a transcription factor depending on the change in conditions entailed alteration of transcription factor combination and nucleosome occupancy on the promoters of the condition-specific target genes; these provided clues to the condition-specific rewiring mechanisms of the dynamic transcriptional regulation programs. We further examined the condition-specific features of the specialized regulatory networks by investigating the structure of the networks among the transcription factors and identifying the feed-forward loops (FFLs). We found that, compared to the cell cycle condition, the stress conditions required a wider propagation of regulatory signals and a substantially larger number of FFLs. Finally, through a case study on an expression pattern module (EPM), we determined a novel regulatory mechanism that can explain how several different transcription factors can induce similar expression profiles of their target genes by suggesting a regulatory hierarchy among the transcription factors.
Results
Identification of regulatory modules
For the condition-specific analysis, we used three different gene expression data sets obtained from experiments performed under the heat shock, nitrogen depletion and cell cycle conditions [13,14]. For each condition, we identified small regulatory units (IMCs) by using the gene expression data and ChIP-chip data [3]. Each IMC comprised genes that are coexpressed under a specific experimental condition and share the same transcription factor binding evidence, as determined by ChIP-chip data (Figure 1a). Since the experimental conditions available in ChIP-chip data are not consistent with those in gene expression data, transcription factor binding evidence in any ChIP-chip data was respected at this step. Due to the augmented evidence by ChIP-chip data, IMCs were more informative than simple gene sets that are grouped by expression similarity alone. Supporting this notion, it has been reported that splitting the coexpressed genes into smaller subsets based on prior knowledge can enhance the identification of new regulatory elements [6]. The similarly expressed IMCs were grouped together and used as the precursors of the expression pattern modules (preEPMs; Figure 1b).
Figure 1.
Overview of the method. (a) Splitting the genome-wide location (ChIP-chip) data into several coexpressed gene sets. Each of the derived target gene sets was called an IMC. Each IMC was named after the transcription factor of the ChIP-chip data followed by a serial number. Gray rectangles indicate the IMCs. Small dots indicate the genes bound to the transcription factor. (b) Generation of preEPMs. The IMCs with similar mean expression patterns were grouped for further analysis. (c) Detecting the regulators in each IMC. Initially, the over-represented motifs in each IMC were detected by the t-test. Next, biologically significant motif evidence and ChIP-chip evidence were selected using a test based on the hypergeometric distribution. Subsequently, in the case of motif evidence, recurrently confirmed motifs in each preEPM were selected. Yellow diamonds and ellipses indicate biologically significant regulators. Gray diamonds and ellipses represent the regulators that were not qualified by the test. Gray curved lines between the regulators indicate synergistic pairs. (d) Identification of an EPM. For each preEPM, the IMCs without a confirmed regulator were eliminated, and the retained IMCs and their corresponding regulators were arranged. Solid lines indicate motif evidence, and dotted lines indicate ChIP-chip evidence. (e) Identification of an RM. Regulators with highly overlapped target genes were united to identify an RM.
In order to detect the plausible regulators of each preEPM, transcription factor binding information from ChIP-chip data [3], known motif data from SCPD [15], TRANSFAC [16] and putative motifs from Harbison et al. [3] were exploited to detect the regulators of each IMC (Figure 1c). First, we examined whether the shared transcription factor of an IMC is a reliable regulator for the IMC. Just the fact that the transcription factor was bound to the genes might not necessarily imply regulation because the gene regulation activity of the transcription factor depends on the condition or cofactors [17,18]. Hence, we performed a hypergeometric test to investigate whether the binding of a transcription factor is associated with gene expression. The hypergeometric test assessed the enrichment of the transcription factor-bound genes among the genes showing expression profiles similar to the mean expression pattern of the IMC in all yeast genes. Throughout the test, we filtered out the transcription factors that were not associated with gene expression. In addition, we employed the transcription factor binding motif data to identify additional regulatory elements. For each IMC, we examined whether a motif was over-represented in the IMC by using the t-test (see Materials and methods). Similar to the relationship between transcription factor binding and gene expression, the presence of a binding site does not guarantee recruitment of transcription factor nor gene regulation. Therefore, we filtered out the motifs that were not significantly associated with expression pattern in the same manner described above. To remove false positives, a motif was considered as the reliable evidence of transcription factor regulation only when it was qualified by the tests for at least two IMCs in a preEPM. As a result, more than half of the initial candidate regulatory evidence was filtered out (Additional data file 1).
Finally, after discarding the IMCs that did not involve any confirmed regulators, EPMs were identified by gathering the retained IMCs in preEPMs. An EPM is defined as a group of genes that share similar expression profiles under a specific condition and their regulators that were confirmed by the statistical examination of the association with the common expression pattern of the EPM. To each regulator identified from the IMCs in the EPM, we allocated the target genes by gathering the genes of the IMCs that had provided confirmatory evidence of the transcription factor (Figure 1d). To further characterize the distinct coregulated gene subgroups in an EPM, we analyzed the combination of regulators in the EPM by examining the overlap level (OL) of their target genes and subsequently defined the regulator-set modules (RMs). A regulator set is a set of transcription factors that share many target genes in an EPM, and the union of their target genes is considered as the member genes of an RM (Figure 1e).
In order to characterize the genes in the EPMs/RMs and the target genes of transcription factors, we conducted a functional category enrichment analysis. Briefly, each gene set was verified for significant enrichment in any of the Gene Ontology (GO) categories [19] (shown in Additional data files 2 and 11). Interestingly, most of our regulatory modules (EPMs and RMs) and the target genes of the transcription factors appeared to have condition-specific functional roles. Moreover, each RM or a combination of multiple RMs appeared to represent a functional part of an EPM. We will discuss the functional enrichment of RMs in detail later in the paper.
Overall results of module analysis
The module analysis described above revealed that several EPMs and RMs differed in the average module size (number of member genes) or in the average number of identified transcription factors depending on the conditions (Table 1). The average number of member genes per EPM was greater in stress conditions, namely, heat shock and nitrogen depletion, whereas that in the cell cycle condition was relatively small. This indicates that a large number of genes are coexpressed in response to stress stimuli, whereas a relatively small number of genes are similarly expressed in response to intrinsic signals for cell cycle progression. A similar tendency was also observed with regard to the number of target genes per transcription factor; on average, 97 genes in the heat shock condition, 78 genes in the nitrogen depletion condition, and 32 genes in the cell cycle condition were found to be regulated by a transcription factor. This tendency is in agreement with the result of a previous report on the properties of condition-specific transcriptional regulatory networks [2], which suggested that a relatively smaller number of target genes are linked to a transcription factor in the cell cycle condition than to regulatory networks in stress conditions.
Table 1.
Number of IMCs, EPMs, RMs and their average number of member genes and regulators
| Average no. of genes/transcription factors | |||||||
|---|---|---|---|---|---|---|---|
| Condition | No. of survived IMCs | No. of EPMs | No. of RMs (average number of RMs per EPM) | IMC | EPM | RM | No. of confirmed transcription factors (average number of targets per transcription factor) |
| Heat shock | 249 | 14 | 88 (6.3) | 9.8/3.2 | 102.7/11.6 | 58.9/3.0 | 67 (96.6) |
| Nitrogen depletion | 340 | 24 | 166 (6.9) | 9.1/3.3 | 77.5/13.1 | 40.3/3.1 | 96 (78.0) |
| Cell cycle | 77 | 9 | 35 (3.9) | 7.5/2.9 | 36.3/7.3 | 26.6/3.0 | 43 (31.5) |
For each condition, we calculated the number of retained IMCs that have at least one confirmed transcription factor. And then, total numbers of EPMs and RMs were counted. We also calculated the average number of genes and transcription factors per IMC, EPM and RM.
Interestingly, the average number of transcription factors per RM was quite similar across all the three conditions. We have previously noted that an RM is a coregulated functional unit for the coexpressed genes. The number of regulators in each functional unit was approximately three in all the conditions, implying that, on average, three transcription factors participate in the gene regulation of a specific functional unit, regardless of the condition. However, the average number of RMs per EPM displayed a clear difference; the EPMs in the stress conditions tended to have more RMs than those in the cell cycle condition. On average, seven RMs in the nitrogen depletion condition, six RMs in the heat shock condition, and four RMs in the cell cycle condition were included in an EPM. This implies that EPMs in the stress conditions include more diverse functional units than those in the cell cycle condition. Accordingly, the average number of transcription factors per EPM in the two stress conditions was significantly larger than that in the cell cycle condition. This might be the result of a more intensive need for cooperation among various functional gene groups in order to respond to stress stimuli. We will describe the detailed examples of this cooperation later in the paper.
Condition-specific organization of regulatory modules
Our results showed that the transcriptional regulatory modules were largely reorganized depending on the cellular conditions. As expected, the difference between the normal condition (for example, cell cycle) and the environmental stress conditions (for example, heat shock and nitrogen depletion) was conspicuous. In the cell cycle condition, periodic changes in the gene expression levels along cell cycle progression were reflected in the organization of relatively small EPMs. On the other hand, in the environmental stress conditions, an evident symmetry of expression profiles appeared between stress-induced EPMs and stress-repressed EPMs. Moreover, clear differences in the reorganizing patterns between the EPMs under the heat shock condition and those under the nitrogen depletion condition were observed, although they shared some common features of general response to stress. Regarding the average expression profiles of the EPMs, the heat stress-induced or the heat stress-repressed EPMs displayed transient but significant changes in their transcription levels, whereas the genes in the nitrogen depletion-induced EPMs showed induction or repression over an extended period. Besides, there were many unique features of the organized condition-specific modules depending on the type of the stimulus.
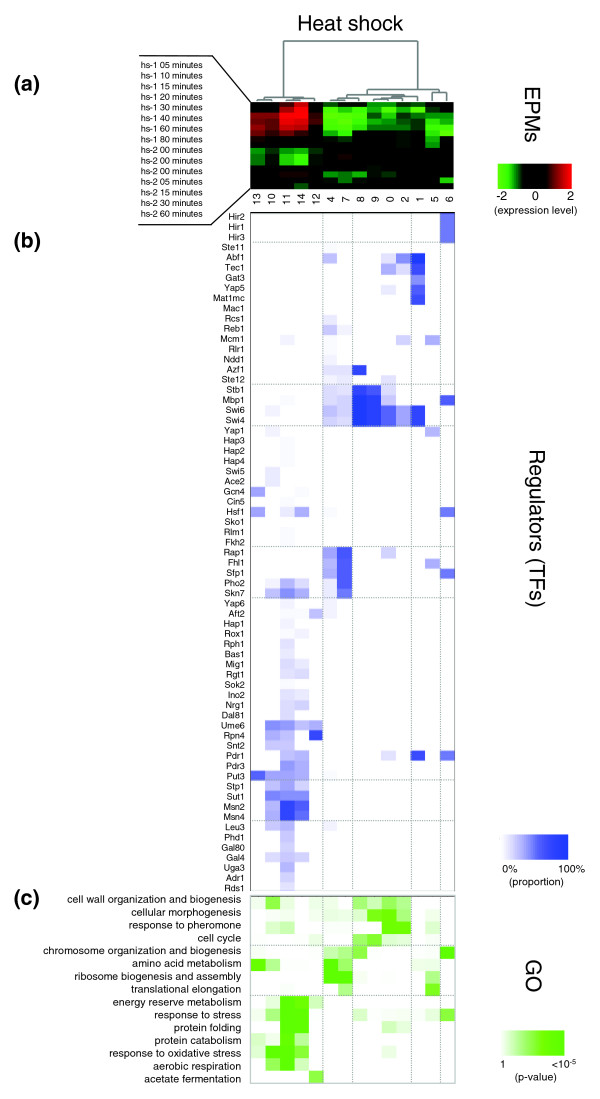
In the heat shock condition, two large clusters of EPMs exhibited reciprocal expression profiles: one comprised upregulated EPMs and the other comprised downregulated EPMs. Further, the EPMs in each of the clusters could be distinguished based on their distinct peak points (Figure 2a and Additional data file 3). In the upregulated EPMs (heat shock EPMs 10-14), various stress-response genes (for example, protein folding and degradation, oxidative stress response, and energy reserve metabolism-related genes) were included together with the genes for energy derivation (for example, aerobic respiration and fermentation genes) (Figure 2c). These results are consistent with several known facts: first, the concurrent induction of protein folding/degradation genes and aerobic respiration genes supports the notion that chaperones and proteolytic proteins require large amounts of ATP [20] that can be supplied by aerobic respiration and fermentation; second, it has also been reported that the levels of major energy reserves (for example, glycogen and trehalose) increase in response to the heat shock condition [21]; and third, heat stress produces oxidative stress that involves mitochondrial respiratory electron carriers [22]. The downregulated EPMs were largely organized into two groups: one comprised the genes related to cell cycle, mating and cell wall (heat shock EPMs 0, 2, 8 and 9), and the other comprised the genes involved in ribosome biogenesis and protein biosynthesis (heat shock EPMs 4 and 7). Their expression profiles exhibited the process of adaptation to the heat shock condition, that is, initially they are highly repressed, but after significant time has elapsed, their expression levels start increasing [23] (more detailed descriptions are provided in Additional data file 3).
Figure 2.
EPMs identified in the heat shock condition. (a) The result by hierarchical clustering of the average expression patterns of EPMs in the heat shock condition. The numbers indicate the EPM indices. (b) Regulator matrix whose entries represent the percentage of genes controlled by each transcription factor in the EPM. The names of transcription factors are shown on the left side. (c) Gene annotation enrichment matrix whose entries represent the enrichment levels of each EPM in the GO 'biological process' categories shown on the left side. For efficient explanation and visualization, only selected GO categories are shown. EPMs identified in the nitrogen depletion and the cell cycle conditions are shown in Additional data file 2.
In the nitrogen depletion condition, a wide range of functional gene groups displayed various expression profiles, and a number of EPMs were organized; these demonstrated interesting condition-specific features. There were four EPMs related to amino acid metabolism, and they could be divided into two groups - amino acid biosynthetic EPMs (nitrogen depletion EPMs 0, 1 and 2) and amino acid catabolic EPMs (nitrogen depletion EPM 25) (see Additional data file 2). In the microarray experiments for nitrogen depletion, a medium containing a small amount of a nitrogen source but neither amino acids nor nucleotides was used [14]. Until the depletion of the nitrogen source, the cells behaved as if they were under amino acid starvation. Genes in the amino acid biosynthetic EPMs (EPM 0, 1 and 2) were induced as long as the nitrogen source was available but displayed an abrupt decline after the depletion of the nitrogen source. On the other hand, EPM 25, which included amino acid catabolic genes and the genes responsible for the nitrogen starvation response, displayed a reverse pattern; they were quiescent while the nitrogen source was available but started to be induced after the depletion of the nitrogen source. It appears that amino acid catabolic EPMs contribute to increasing the turnover rate of amino acids in response to nitrogen starvation. Moreover, the expression profiles of ribosome biogenesis EPMs (nitrogen depletion EPMs 11, 12 and 19) fluctuated depending on the availability of amino acids; their expression levels were upregulated when amino acids were available (Additional data file 3).
In the cell cycle condition, several phase-specific cell cycle EPMs (cell cycle EPMs 1, 5 and 6) were identified, and their regulators were largely in agreement with those mentioned in the previous reports (Additional data file 4). In addition, we detected ribosome biogenesis EPMs (cell cycle EPMs 0 and 4), an energy generation-related EPM (cell cycle EPM 7) and an amino acid metabolism-related EPM (cell cycle EPM 8) (Additional data file 2). The expression levels of all these EPMs commonly peaked at the G1 phase and the G2/M transition, although their overall expression profiles were distinguishable (Additional data file 3). This result indicates that the roles of these EPMs are particularly important during the G1 phase and the G2/M transition; this finding is supported by the previous studies wherein genes controlling ribosome biogenesis and protein translation have been identified as the critical regulators of cell growth and cell cycle in yeast [24-26] and by the studies demonstrating that the critical cell size requirement is fulfilled in the G1/S and G2/M transitions [27,28]. Unexpectedly, a stress response-related EPM was also detected (cell cycle EPM 3). The presence of this EPM appears to reflect the experimental condition adopted by Cho et al. [13]; they employed the heat shock treatment for cell cycle synchronization before their measurements. The average expression of this EPM displayed a peak at the beginning of the experiments but abruptly decreased later, implying that the influence of the heat shock treatment vanishes with time. The phase-specific cell cycle EPMs are discussed in more detail in Additional data file 4.
Comparison of modules across conditions
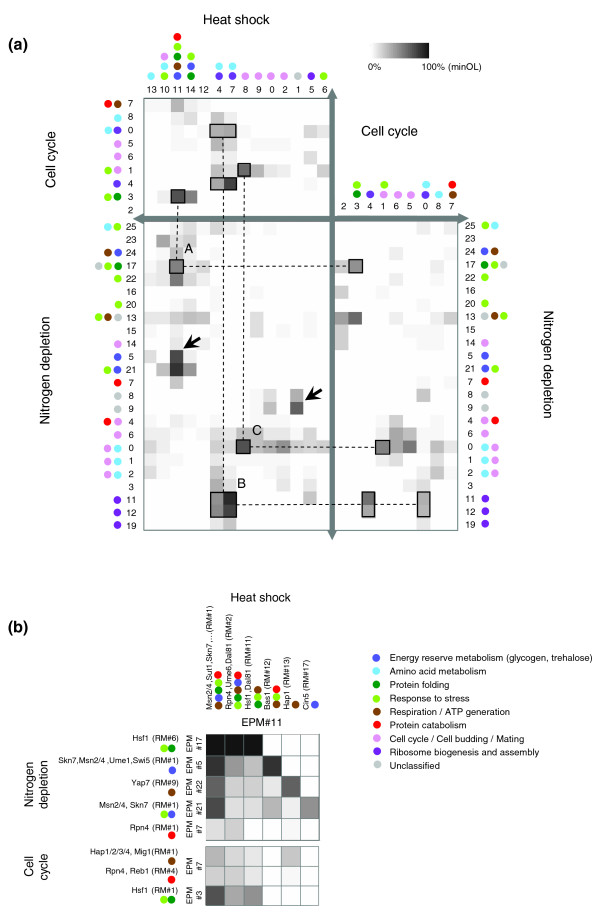
To further investigate the differences and similarities among EPMs from the three tested conditions, the member genes in the EPMs were compared across conditions. Although the shapes of the reorganized EPMs differed among the three conditions, the following three highly overlapped EPM clusters were detected in all the conditions (Figure 3a): EPMs of stress response (heat shock EPM 11, nitrogen depletion EPM 17 and cell cycle EPM 3), EPMs of ribosome biogenesis (heat shock EPMs 4 and 7, nitrogen depletion EPMs 11 and 12 and cell cycle EPMs 0 and 4) and EPMs of the cell cycle (heat shock EPM 8, nitrogen depletion EPM 0 and cell cycle EPM 1). These modules shared some common transcription factors, and we conjecture that the regulation of these modules would be conserved in various physiological conditions.
Figure 3.
Overlap matrices of regulatory modules. (a) Overlap matrices between EPMs in all the three conditions. The OLs were calculated as the proportion of the intersection genes in the smaller EPM (minOL). The enriched GO categories of each EPM are also shown as several colored dots. Black-lined boxes represent the EPMs that are significantly overlapped across all the three conditions. 'A' indicates the overlapped stress-related EPMs represented by the three boxes linked by dashed lines. They have the common regulators Msn2/4 and Hsf1. Identically, the EPMs indicated as 'B' have the common regulators Rap1, Sfp1 and Fhl1. The EPMs indicated as 'C' have Mbp1, Swi4, Swi6 and Stb1 as their common regulators. Black arrows indicate EPMs that are highly overlapped between the heat shock and nitrogen depletion conditions. (b) Overlap matrices between RMs (minOL). Several RMs, which were included in the distinct EPMs in the nitrogen depletion and cell cycle conditions, are significantly overlapped with the RMs in heat shock EPM 11.
Some functional EPMs were detected in only the two environmental stress conditions. For instance, genes for energy reservation (for example, generating glycogen and trehalose) were included only in the EPMs in the heat shock (EPMs 11 and 14) and nitrogen depletion (EPMs 5 and 21) conditions. All these EPMs were commonly regulated by Msn2/4 and Skn7 (Figure 2b), which are well-known stress-response regulators [29-31]. Furthermore, both heat shock EPM 1 and nitrogen depletion EPM 9 were enriched with 'biological process unknown' genes and contained several common regulators (Yap5, Gat3, Swi4/6, Tec1, Mat1-Mc and Abf1) and were found to overlap significantly; however, these EPMs did not overlap with any cell cycle EPMs. These EPMs may be related to some unknown functions that are commonly involved in heat shock and nitrogen depletion response.
By analyzing the overlap of several RMs, we found that various gene groups involved in several distinct EPMs in other conditions converged to form a single EPM in a specific condition. For example, several stress-response gene groups and energy generation-related gene groups, which showed diverse expression patterns and were organized into several independent EPMs in the nitrogen depletion or cell cycle condition, were coexpressed under the heat shock condition and formed an integrated EPM (Figure 3b). Among the nitrogen depletion EPMs, the crucial parts of the EPMs for energy reserve metabolism (nitrogen depletion EPMs 5 and 21), protein folding and degradation (nitrogen depletion EPMs 17 and 7, respectively) and respiration (nitrogen depletion EPM 22) converged into a single heat-shock EPM (heat shock EPM 11). Similarly, many genes for protein folding, protein degradation and respiration in the EPMs in the cell cycle condition (cell cycle EPMs 3 and 7) were found to be included together in the heat shock EPM 11. Nitrogen depletion EPM 0 also exhibited coexpression of multiple functional gene groups that were included in several different EPMs in other conditions (Additional data file 5).
It is also notable that the list of target genes of Rpn4, a transcription factor for heat shock EPM 11 and known as a transcriptional activator of genes encoding proteasomal subunits [32], was expanded to include the protein folding-related genes, while Rpn4 retained its regulatory role on the genes related to protein degradation in the heat shock condition. Similarly, in addition to the previously characterized stress response-related target genes, energy generation-related genes were included in the target genes of Msn2/4 and Skn7, which are the major regulators of heat shock EPM 11. From these examples, we conjecture that some coordinated regulation might operate for a more efficient response to the heat stress. In the heat shock condition, protein folding and protein degradation might be coherently regulated because the failure of the protein folding process often entails degradation of the misfolded proteins. In addition, the coupling of energy generation and protein folding (and degradation) would enhance the response to heat stress because the latter process requires considerable energy, as mentioned before. Several previous studies support our inferences. It has been reported that molecular chaperones assist in not only protein refolding but also protein degradation by interacting with protein degradation systems; when chaperones fail in their functions of protein folding, assembly or translocation, they facilitate degradation of the mishandled proteins [33,34]. Our results and experimental evidence suggest that cells can respond to a stimulus more rapidly and efficiently by co-inducing the energy-consuming stress response genes and the energy-providing genes.
Specified regulatory roles of transcription factors depending on conditions
A total of 109 transcription factors were confirmed as regulators of all the EPMs and RMs identified from the three conditions; 67, 96 and 43 transcription factors were confirmed in the identified modules from the heat shock, the nitrogen depletion and the cell cycle conditions, respectively. There were 33 transcription factors common in all the three conditions (Additional data file 6). In order to investigate the overall regulatory roles of the transcription factors in each condition, we identified all the target genes of each transcription factor and analyzed their enriched functional GO categories (Additional data file 7). Of the 33 common transcription factors, 20 appeared to retain at least one of their regulatory roles in all the conditions. Among the 109 total transcription factors, 69 exhibited their known regulatory roles in at least one condition. Considering that we conducted the analysis for only three conditions and that many transcription factors exhibit their roles only under specific conditions, we believe that the number of transcription factors that agree with their experimentally proven roles would increase if more diverse conditions were analyzed.
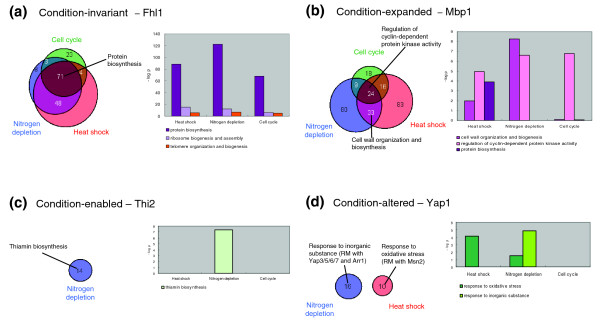
Similar to the classification of the transcription factor binding patterns into four types based on the change in conditions by Harbison et al. [3], we attempted to classify the transcription factors based on the alterations in target genes as follows: 'condition-invariant', in which the transcription factor target genes are highly conserved across the conditions; 'condition-expanded', in which the list of target genes in one condition is further expanded to include more target genes in other condition; 'condition-enabled', in which the transcription factor regulates some target genes in one specific condition but not in other; and 'condition-altered', in which different sets of target genes are regulated by the same transcription factor in different conditions. We found that most transcription factors could be classified into one or more of these groups, and the overall OL between the target genes of transcription factors in different conditions indirectly reflected their types (Figure 4 and Additional data file 7).
Figure 4.
Condition-specific types of transcription factor. The transcription factors were classified into four types based on the alteration in the target genes: (a) condition-invariant, (b) condition-expanded, (c) condition-enabled and (d) condition-altered. The venn diagrams show the overlapped target genes of the representative transcription factors among the three conditions. In the bar graph, the y-axis represents the significance of the p value for the enriched functional categories of the target genes in each condition.
The transcription factors Rap1, Fhl1, and Sfp1, which are the well-established ribosome biogenesis-related regulators [35,36], were classified into the 'condition-invariant' group; they retained most of their regulatory roles (protein biosynthesis, ribosome biogenesis and assembly, and telomere maintenance) in all the three conditions (Figure 4a). Mbp1, a renowned cell cycle regulator, could be categorized as a 'condition-expanded' transcription factor; it expanded its targets to include the cell wall biosynthesis-related genes under the two environmental stress conditions (Figure 4b). Many other cell cycle-related transcription factors, including Swi4/6 and Stb1, showed a similar expansion of targets to regulate the cell wall biosynthesis-related genes under the two stress conditions. Rpn4 was another good example of 'condition-expanded' transcription factors. As mentioned earlier, the target gene list of Rpn4 was expanded to include the protein folding-related genes in response to heat shock, while Rpn4 retained its own regulatory role of protein degradation. Many transcription factors could be categorized as 'condition-enabled' transcription factors; Thi2, a transcriptional activator of thiamin biosynthetic genes [37], appeared to exhibit its known role only under the nitrogen depletion condition (Figure 4c). Zap1, a zinc-responsive transcription factor that activates the zinc transporter genes [38], was confirmed as a regulator of zinc transportation-related genes only under the cell cycle condition. Snt2, a previously uncharacterized DNA-binding protein, was predicted to control the genes related to ATP synthesis and energy reserve metabolism only under the heat shock condition. There were some 'condition-altered' transcription factors whose regulatory roles changed completely depending on the condition. Interestingly, the change in regulatory roles often necessitated the alteration of partner transcription factors. For example, Yap1, a regulator of genes related to the response to oxidative stress and inorganic substances [39,40], was predicted to regulate the genes related to oxidative stress response along with Msn2 under the heat shock condition (RM 9 of heat shock EPM 10), whereas it appeared to control the genes involved in response to inorganic substances along with Yap3/5/6/7 and Arr1 (Yap8) under the nitrogen depletion condition (RM 18 of nitrogen depletion EPM 13); all these roles detected in this study were largely in agreement with their known functions (Figure 4d) [41,42]. Similarly, Uga3 appeared to regulate the genes related to protein biosynthesis and ribosome biogenesis along with Sfp1, Fhl1, Skn7 and Sut1 under the cell cycle condition (RM 3 of cell cycle EPM 0), while it was predicted to regulate the branched chain amino acid biosynthetic genes along with Leu3 under the nitrogen depletion condition (RM 3 of nitrogen depletion EPM 2). Moreover, Uga3 along with Msn2/4 and many other transcription factors exerted its regulatory role on the genes related to ATP generation and energy reserve metabolism under the heat shock condition (RMs 2 and 3 of heat shock EPM 11).
In order to validate the prediction of 'condition-altered' regulators, we examined whether the accessibility of the predicted target promoters undergoes changes depending on the conditions by using an independent data set that represents the genome-wide occupancy of nucleosomes and transcriptional machinery under the heat shock condition [18]. Compared to the normal condition, the promoter regions of Uga3's and Yap1's target genes predicted by our analysis of heat shock response exhibited lower occupancy of histones and higher occupancy of most transcriptional machinery components (for example, RNA polymerase II) under the heat shock condition; on the contrary, the occupancy on the promoters of Uga3's target genes predicted by our analysis of cell cycle condition exhibited the opposite pattern (Additional data file 8). This result indicates that the predicted target genes of Uga3 and Yap1 become accessible to the transcriptional machinery to different extents depending on the conditions, supporting the predictions by our analysis.
Condition-specific regulatory networks among transcription factors
To further investigate the specific properties of the transcriptional regulatory networks in each condition, we analyzed the relationship among the transcription factors by identifying the transcription factors that regulate the expression levels of other transcription factors. We could detect some condition-dependent topological and functional features in the networks. The complexity of the regulatory networks among the transcription factors (for example, the number of target transcription factors of a transcription factor) in the stress conditions was higher than that in the cell cycle condition; on average, 4.5, 3.7 and 1 transcription factor-encoding genes were found to be regulated by a transcription factor under the heat shock, the nitrogen depletion and the cell cycle conditions, respectively (see Figure 5a for the cell cycle condition and Additional data file 9 for the two stress conditions). It appears that the number of target transcription factors and the overall topology of networks are related to the different responses to different types of stimuli. Under the heat shock condition, fast and global signal propagation for adapting to the new environment would be required; therefore, a highly dispersed transcription factor hierarchy would be efficient for such a response. On the other hand, such widespread regulatory signal propagation would not be necessary under the cell cycle condition because cell cycle progression requires only periodic control of some specific functional genes.
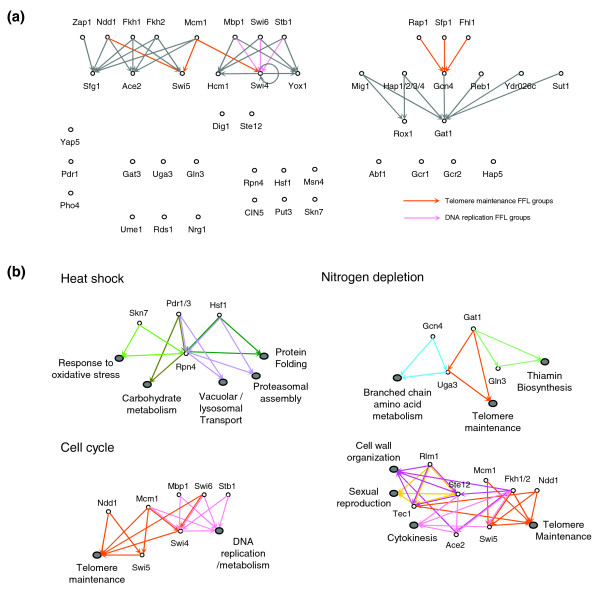
Figure 5.
Transcriptional regulation among transcription factors. (a) Regulatory network among the transcription factors in the cell cycle condition. Each arrow represents transcriptional regulation. Two transcription factors linked by each colored arrow form an FFL group with the genes that are enriched in some specific functional category. The colors of the arrows imply the enriched functional categories. The networks in the two stress conditions are shown in Additional data file 9. (b) Condition-specific FFL groups. Each gray ellipse represents the overlapped target genes of the two transcription factors in an FFL group. Only selected FFL groups are shown (see Additional data file 10 for the complete list of the enriched functional categories of the FFL groups).
We found many FFLs in the condition-specific transcriptional regulatory networks. An FFL is composed of two input transcription factors, one of which regulates the other and both jointly regulate a specific target gene. It responds only to a persistent stimulus and is often detected under the conditions wherein an external signal causes many systems to respond rapidly [43]. Moreover, the combination of two input transcription factors may contribute to the gain of specificity for certain functional target genes. We defined an 'FFL group' when two transcription factors shared some functionally coherent target genes enriched in at least one of the GO categories. The numbers of the identified FFL groups were consistent with the properties of the FFLs, and considerably more FFL groups were identified in the two stress conditions than the cell cycle condition. In total, we identified 91 FFL groups in the heat shock condition, 132 in the nitrogen depletion condition and 9 in the cell cycle condition, reflecting the characteristics of the cellular conditions; nitrogen depletion generates prolonged nitrogen starvation signals, while the signals in cell cycle progression are relatively transient. Moreover, the gene expression data of the heat shock condition indicated that a number of genes undergo an abrupt change in expression level, implying that extremely rapid and widespread regulatory signal propagation occurs during this condition.
The target genes of the FFL groups reflected specifically activated (or repressed) functions that were related to the conditions in which the FFL groups were identified. Recently, it has been reported that Hsf1 and Pdr1/3 participate in the regulation of Rpn4 that in turn activates the expression of proteasomal genes, particularly under the heat shock condition [44]. Our results from the heat shock condition were in good agreement with this experimental evidence; Hsf1 and Rpn4 were predicted to constitute an FFL group whose target genes are involved in proteosomal assembly and protein folding, and Pdr1 and Pdr3 appeared to form FFL groups along with Rpn4 to regulate the genes related to vacuolar and lysosomal transport, respectively (Figure 5b, heat shock). In the nitrogen depletion condition, we observed many FFL groups that specified their targets by altering the partner transcription factors; Gcn4 and Uga3 were predicted to form an FFL group that regulates the branched chain amino acid metabolic genes, while Gat1 and Uga3 constituted another FFL group controlling the genes involved in telomere maintenance. Gat1 also appeared to participate in an FFL group along with Gln3 to regulate the thiamin biosynthetic genes (Figure 5b, nitrogen depletion). Besides, we detected many cases in which the FFLs reflected the known roles of the participating transcription factors. For example, Ste12, Tec1 and Rlm1, all of which are reported to participate in the cell wall integrity signaling pathway [45-47], appeared to constitute the FFL groups regulating the cell wall biosynthetic genes. In the cell cycle condition, all the identified FFL groups were involved in cell cycle control or regulation of telomere maintenance; Mbp1, Swi6, Stb1 and Mcm1 appeared to regulate the genes involved in the cell cycle along with Swi4 as a second transcription factor, while Ndd1 and Mcm1 were found to regulate the expression of Swi5, leading to a coordinated regulation of the genes related to telomere maintenance (Figure 5b, cell cycle).
High-resolution regulatory modules - a case study with nitrogen depletion EPM 2
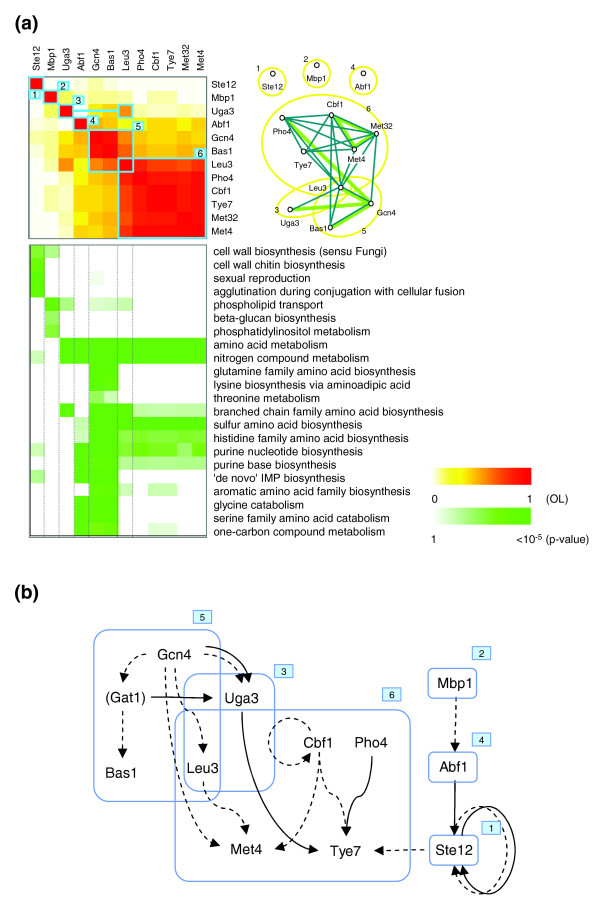
One of our interesting results is that each coexpressed gene module (EPM) appears to include several separate submodules (RMs) that are regulated by different sets of transcription factors but yet share the same expression pattern. Figure 6a shows how well RMs are defined as functional parts in nitrogen depletion EPM 2. In this EPM, we detected six RMs that were largely involved in cell wall biosynthesis (RMs 1 and 2) and amino acid metabolism (RMs 3-6). Each of the RMs further included distinguishable detailed functional parts. Notably, each or various combinations of the RMs 3-6 were specifically enriched with the functional categories related to amino acid metabolism (for example, branched chain family amino acid biosynthesis, sulfur amino acid biosynthesis). In addition, most of the predicted regulators of the RMs in this EPM have relevant experimental evidence (more detailed explanations and literature evidence are described in Additional data file 4).
Figure 6.
Case study: RMs in nitrogen depletion EPM 2. (a) EPM 2 of the nitrogen depletion condition is represented by two matrices. The upper matrix represents the overlap between the target genes of the regulators in the EPM, and blue boxes represent the RMs. The lower matrix represents the enrichment of the target genes per regulator in the GO 'biological process' categories. For simplicity, only selected categories are shown. A more detailed explanation is presented in Additional data file 4, and the complete matrices with all significantly enriched categories (p value < 0.01) are presented in Additional data file 11. In the graph, yellow ellipses indicate the RMs. Blue lines represent a significant overlap (OL ≥ 0.5) between the target genes of the two transcription factors at nodes, and green lines represent synergistic links identified by the hypergeometric test for transcription factor pairs (see Materials and methods). The detailed information about other EPMs and RMs is available in Additional data file 11. (b) Potential regulatory scheme among the regulators in the nitrogen depletion EPM 2. Solid arrows indicate the regulator-target gene relationship from the prediction under the nitrogen depletion condition, and dotted arrows indicate the transcription factor binding information from ChIP-chip data. Gat1 is included for completion of the hierarchy, although it is not among the predicted regulators of the nitrogen depletion EPM 2.
An interesting issue raised by our results is how several different transcription factors in an EPM can drive a similar expression pattern of their target genes. It is presumable that there are 'master regulators' that, by hierarchically controlling distinct sets of transcription factors, allow coordinated transcriptional responses of a large set of genes that are not directly coregulated by the same transcription factors. By combining our transcription factor-target gene prediction and ChIP-chip data [3], we could detect a potential hierarchical structure of the regulators within nitrogen depletion EPM 2 (Figure 6b). Except Met32, 11 out of the 12 transcription factors participated in this potential regulatory scheme, and Gcn4, Cbf1, Pho4 and Mbp1 were located at the top of the hierarchy, suggesting that these four transcription factors are the putative 'master regulators' for this EPM. Across all the EPMs, on average 56% of the regulators in an EPM (60%, 59% and 41% under the heat shock, the nitrogen depletion and the cell cycle conditions, respectively) appeared to participate in the potential regulatory network among the regulators in the EPM (Additional data file 12).
Comparison with other methods
To evaluate our method, we compared our modules with several other predicted gene regulatory modules, including GRAM [5] and COGRIM [48], which were predicted using similar types of data but different approaches. The GRAM algorithm identified 106 modules by combining the ChIP-chip and gene expression data, and COGRIM predicted 2,298 gene-transcription factor interactions by integrating ChIP-chip, transcription factor binding motif and gene expression data. First, for comparison of the coverage of the results from different approaches, we compared the numbersof distinct genes andtranscription factors that are included in the predicted modules. As shown in Table 2, the EPMs and RMs included much more genes (2,099) and transcription factors (109) than GRAM and COGRIM, indicating that our method provides more information about transcriptional regulatory events than other methods, despite that we used gene expression data from only 42 experiments while other methods employed approximately 500 expression experiments.
Table 2.
Overall comparison with other methods
| GO functional enrichment level | |||||||
|---|---|---|---|---|---|---|---|
| Overall modules | |||||||
| Method | No. of modules | No. of distinct genes | No. of distinct TFs | BP | MF | CC | Target genes of common 38 TFs |
| EPM | 47 | 2,099 | 109 | 14.36 (8.63) | 12.94 (8.53) | 15.32 (7.80) | 17.86 (21.49) |
| RM | 289 | 2,099 | 109 | 7.56 (3.43) | 6.51 (3.24) | 7.23 (4.95) | |
| GRAM | 106 | 655 | 69 | 8.54 (1.05) | 7.77 (4.64) | 8.73 (2.18) | 12.28 (19.47) |
| COGRIM (B-/C+) | 39 | 841 | 39 | 8.67 (2.97) | 6.81 (4.24) | 7.43 (2.72) | 8.81 (13.43) |
| COGRIM (B+/C+) | 39 | 936 | 39 | 5.3 (0.68) | 4.55 (0.18) | 4.86 (0.68) | 5.19 (4.81) |
The number of modules obtained from each method and the number of distinct genes and transcription factors (TFs) that constitute modules identified by each method are shown. In addition, GO functional enrichment levels are shown. The GO (BP, biological process; MF, molecular function; CC, cellular process) enrichment p values were transformed to negative log values and averaged over all modules. We also performed a functional enrichment test for the target genes of 38 transcriptional factors that were commonly predicted in all the three methods. The standard deviations are shown in parentheses.
We then compared the average GO enrichment level of the predicted regulatory modules by calculating a negative-log transformed p value. As shown in Table 2, the average enrichment levels of EPMs were higher than those of the other predicted modules. This implies that EPMs comprise more functionally coherent genes according to the GO annotation. For a more objective comparison, we also analyzed the enrichment levels of the target genes of 38 transcription factors that are commonly included in all the predictions. The result showed that our method has the highest enrichment level (Table 2).
In order to compare how well the modules can explain the detailed parts of a function, we further analyzed the enrichment levels of the modules related to ribosome biogenesis and assembly. We found that our EPMs and RMs showed higher enrichment levels than other methods in 11 out of 14 GO categories related to the function (Additional data file 13). Furthermore, we compared nitrogen depletion EPM 12 with several modules from GRAM, all of which are significantly related to ribosome biogenesis. In GRAM, 17 transcription factors were linked to 21 ribosome biogenesis-related gene modules. However, many of their modules were found to highly overlap with each other; therefore, they could not be distinguished in terms of function. In contrast, nitrogen depletion EPM 12 contained ten transcription factors (eight transcription factors overlapped with GRAM) and was divided into five RMs that clearly described the detailed cellular process of ribosomal biogenesis, from rRNA synthesis to ribosome assembly (Additional data file 13).
In general, a high coverage can be obtained by simply adopting lenient criteria for identifying modules. However, the more lenient criteria are applied, the more false positives are incorporated in the analysis result, leading to lower specificity that would yield poor functional enrichment. Remarkably, comparison with other methods suggests that our results are not only functionally more relevant but also cover a wider range of the whole yeast genome. We believe that higher enrichment level and coverage of our results could be accomplished by the bottom-up approach that initially identified small coexpressed and coregulated gene groups by efficiently combining the ChIP-chip and expression data. In contrast to other methods that used combined expression data over a number of experimental conditions, we used separate experimental units of expression data for identifying condition-specific regulatory modules; this enabled us to obtain the target genes of transcription factors that are specific to each condition and functionally more coherent. Through this bottom-up approach, it was possible to identify many distinct submodules included in a coexpressed gene group and describe the detailed multiple alternative regulatory mechanisms involved in the coexpression.
Discussion
In this study, we elucidated the coexpressed gene modules (EPMs) and their intrinsic functional submodules (RMs) along with their regulators through a sensitive and robust method using ChIP-chip, known motif and microarray data. We presented several condition-specific EPMs and described the differences between the EPMs reorganized depending on the change in conditions. Substantial parts of the results are consistent with the previously reported condition-specific regulatory events and well-characterized regulatory mechanisms, suggesting that our strategy and results are reliable.
Our results revealed several important features about the dynamic nature of the condition-specific reorganization of the transcriptional regulatory network. Certain functions appeared predominantly in the organized gene regulatory modules in each condition. For example, various stress response- and energy-related genes constituted a large proportion of the modules detected in the heat shock condition, while genes for amino acid and nucleotide metabolism were organized into many modules in the nitrogen depletion condition. In the cell cycle condition, many gene regulatory modules responsible for the internal periodic phase-specific cell cycle signals were identified. Besides, in the two stress conditions, the change in condition triggered the coexpression of many distinct functional groups related to the specific stimuli, and this coexpression might contribute to the efficient response to the stress. Our results also suggest that the roles of transcription factors can be altered depending on the condition. Most transcription factors expanded or altered their targets in order to regulate the genes that are required for the specific condition, and this alteration of targets often entailed the switching of partner transcription factors and a change in nucleosome occupancy, which provide clues for understanding the rewiring mechanism of transcriptional network. Several possible factors might contribute to the condition-specific transcriptional regulation of diverse genes. For regulation of a specific group of genes, the promoters of the genes should become accessible and transcriptional machinery should be recruited to them. As we demonstrated, condition-specific binding of transcription factors is related to the change in nucleosome occupancy, which can be either a cause or a result of changing promoter specificity of the transcription factors. The change in nucleosome occupancy induced by chromatin remodeling in response to the specific cellular condition might cause the condition-dependent transcription factor binding [49]. On the other hand, the target specificity might be obtained by variable association with other transcription factors that can affect the nucleosome occupancy. Supporting this notion, some transcription factors, such as Rap1 and Msn2, are known to have a role in influencing the accessibility of promoters [50,51].
Interestingly, our results indicated distinct and interesting hypotheses regarding the condition-specific regulatory mechanisms, which are somewhat different from those of the previous studies. As mentioned before, Luscombe et al. [2] employed an invaluable approach for revealing the dynamic properties of transcriptional regulatory networks. Their findings are partially consistent with ours; enormous alteration of the targets of transcription factors occurred depending on the conditions, and greater numbers of target genes per transcription factor were observed in exogenous conditions (for example, stress response condition) than in endogenous conditions (for example, cell cycle condition). However, there was some discrepancy between our results and those of Luscombe et al. [2]. In our results, constancy in the number of transcription factors for regulating a certain functional gene group across all conditions was observed. In contrast, Luscombe et al. [2] concluded that endogenous conditions require more complex transcription factor combinations than exogenous conditions based on the greater 'in-degree' of endogenous conditions, implying that a larger number of transcription factors regulate a target gene. They also found significantly more FFLs in the endogenous transcriptional regulatory networks, while we found a much larger number of functional FFL groups in the exogenous conditions. This discrepancy might be due to the different definition of the 'stress response condition'. While we separately used only the small subsets of the microarray experiments (heat shock and nitrogen depletion) for analyzing the detailed features of the selected stress conditions, Luscombe et al. [2] considered all the various stress conditions included in the experiments of Gasch et al. [14] as the stress response condition; therefore, their results might reflect the regulatory events corresponding to 'general stress responses', which occur commonly in all stress conditions [14].
Although our approach deciphered the condition-specific regulatory mechanisms efficiently, it has some limitations. Our methodology depends largely on the quality and abundance of the known transcription factor binding motif information and ChIP-chip data; hence, the results would reflect only a part of the events actually happening in a cell. Moreover, the number of our tested conditions is too small to depict a wide variety of alteration in the regulatory mechanisms corresponding to various conditions. In the future, we would like to apply this method to other important conditions and perform putative motif analysis to identify additional novel regulatory factors. The increasing knowledge of regulatory mechanisms in various conditions together with in-depth studies focused on the comparison of the 'condition-specific' transcriptional modules revealed by more diverse condition-specific results will shed light on the highly sophisticated and elaborate regulatory mechanisms of transcription in yeast cells. Furthermore, with increasing ChIP-chip, mRNA expression data and reliable genome information about higher eukaryotes, the approach used in this work can be readily extended to study condition-, tissue- and developmental stage-specific transcriptional regulatory networks in diverse organisms.
Conclusion
In this study, we aimed at deciphering the transcriptional regulatory mechanisms in yeast with two major perspectives; we focused on unveiling the dynamic nature of transcriptional rewiring entailed by the change in conditions and investigating multiple distinct coregulated functional parts existing in a coexpressed gene group. Consequently, we could detect several important features of the condition-specific transcriptional regulatory networks. First, the organization of the coexpressed gene modules is altered depending on the cellular conditions; many functional modules unique to each condition were identified, although some modules related to fundamental cellular processes were sustained over multiple conditions. Second, there are some specific functions emphasized in each condition, and the genes related to these functions tend to be coexpressed and, therefore, constitute relatively larger EPMs. Third, the reorganization of the transcriptional regulatory modules entail the alteration of targets and partner regulators of transcription factors, suggesting that rewiring of the transcriptional regulatory networks occurs due to the dynamic properties of the transcription factors. In fact, many FFL groups, which include a common transcription factor, exhibited distinct functional roles depending on various secondary transcription factors, leading to a conclusion that alteration of partner transcription factors can determine target specificity. Furthermore, our results indicate that several different transcription factors in an EPM can drive a similar expression pattern of their target genes, most probably by the involvement of 'master regulators' that hierarchically control distinct sets of transcription factors. Besides, many EPMs and RMs suggest novel regulatory mechanisms of various transcription factors, including their partnerships and target genes, in a condition-specific manner. These results provide reliable hypotheses for unveiling the condition-specific transcriptional regulatory networks and for studying the regulation of biological processes induced under specific conditions.
Materials and methods
Identification of initial module candidates
The first step in our method was integration of the genome-wide location data (ChIP-chip) and the gene expression data to identify IMCs. In this study, we used the ChIP-chip data [3] that contain the binding information of 204 transcription factors (although Harbison et al. [3] describe only 203 transcription factors), and the gene expression data under three experimental conditions - mitotic cell cycle (17 experiments) from Cho et al. [13] and heat shock (15 experiments) and nitrogen depletion (10 experiments) conditions from Gasch et al. [14]. Each of the identified IMCs satisfied three requirements: first, all genes should have common transcription factor binding evidence with a p value < 10-3; second, the pair-wise Pearson correlation coefficient (PCC) between two expression profiles should be >0.7; and third, there should be at least five element genes (Figure 1a). Eventually, the IMCs with similar mean expression profiles (PCC > 0.7) were grouped together and called preEPMs (Figure 1b). Determination of the appropriate cut-off values are described in Additional data file 13.
Detecting over-represented motifs
To identify the over-represented motifs in each IMC, we used 152 known or putative motifs (corresponding to 110 transcription factors) obtained from SCPD (23 matrices) [15], TRANSFAC (27 matrices) [16] and Harbison et al. (102 matrices) [3]. A motif was considered over-represented in an IMC if the p value was found to be <0.01 in the t-test examining the difference between the distribution of maximum log-odds scores calculated for the promoters of the genes in the IMC and those calculated for the promoters of the genes that did not have sufficient binding evidence regarding the transcription factor of the IMC (p value > 0.95). In this study, the promoter region was defined as the 750 bp upstream sequences from the transcription start site. As usual, the log-odds score of a motif for a promoter sequence was calculated using the position-weight matrix constructed for each motif.
Confirming motif evidence and ChIP-chip evidence
From the over-represented motifs, we further identified the motifs associated with the expression coherence in each IMC (Figure 1c). Specifically, we performed the hypergeometric test with the number of genes in the yeast genome (G), the number of yeast genes that contained a motif in their promoters (B), the number of yeast genes whose expression profiles were similar to that of the IMC (g), and the number of yeast genes whose expression profiles were similar to that of the IMC and contained the motif in their promoters simultaneously (b) using the equation:
This hypergeometric test measured the level of enrichment in a gene group whose expression profiles were similar to that of an IMC by genes that contained the motif in their promoters against the average number of genes that contained the motif in their promoters for a randomly selected gene group. Motifs with a p value < 0.01 were considered as biologically significant in this study.
For the ChIP-chip transcription factor that was used for identifying the IMC, we also examined the association between transcription factor binding and expression coherence using the hypergeometric test. In this case, B and b were calculated for the transcription factor binding target genes with a p value < 0.001 in the ChIP-chip data instead of the genes containing the motifs (Figure 1c).
Identification of EPMs and RMs
All the IMCs without biologically significant regulators were eliminated, and the selected genes of a preEPM were combined to form an EPM (Figure 1d). The RMs were identified by grouping the regulators in each EPM according to the degree of overlap between their target genes (Figure 1e). We listed the non-redundant regulators in the IMCs and calculated the OL between the target gene sets for all the possible pairs of regulators in each EPM using the equation:
where Si and Sj denote the target gene sets of regulators i and j, respectively.
An RM was identified by gathering the regulators and their target genes in an EPM, where all the OLs between any pair of target gene sets were >0.5. The cut-off value (OL of 0.5) guaranteed a sufficient diversity in functional enrichment for explaining the functional parts in detail and the low redundancy level of the RMs (Additional data file 14).
Analysis of synergistic pairs
We defined a combination of transcription factors as a synergistic pair when the genes having evidence of both transcription factors showed a significantly more coherent gene expression pattern than the genes with evidence for only one transcription factor. There is a clear difference between sharing target genes (RMs) and synergism of a transcription factor pair; synergism might involve physical or genetic interaction between the transcription factors ('AND' logic), whereas overlap of target genes implies that the transcription factors might have similar regulatory roles regardless of significant dependency between them ('OR' logic). To identify synergistic pairs, all the pairs of over-represented motifs in each IMC were examined using the hypergeometric test. For a motif pair 'a and b', P(b|a, X) was considered the p value in the hypergeometric test examining the enrichment of genes with motif 'b' in the group of genes that have motif 'a' and show a similar expression pattern with an IMC X against the genes that have motif 'a' alone. We defined the motif pair 'a and b' as a synergistic pair when the value of P(b|a, X) or P(a|b, X) was <0.01. For each of the two motifs, we used the mean maximal log-odds score of the IMC X as the threshold score for deciding whether the motif exists in the promoter of a gene. The motif synergistic pairs detected in more than one IMC of each preEPM were considered reliable in this study. We also searched for transcription factor synergistic pairs by applying the same method to a pool of IMCs with a similar expression pattern and their related transcription factors.
Functional enrichment analysis
We examined whether the genes of each EPM and the assigned target genes of each regulator in the EPM were significantly enriched by specific functional categories using the GO categories. Categories with a p value of <0.01, as revealed by the hypergeometric test, were considered statistically significant in this study, and the redundant categories were trimmed (for example, when 'response to pheromone', 'conjugation with cellular fusion' and 'response to pheromone during conjugation with cellular fusion' were simultaneously enriched, we adopted only one proper category). For Figure 2, we used only the GO 'biological process' categories, and a subset of the attributes was selected for their diversity.
Abbreviations
ChIP-chip, chromatin immunoprecipitation microarray; EPM, expression pattern module; FFL, feed-forward loop; GO, Gene Ontology; IMC, initial module candidate; OL, overlap level; PCC, Pearson correlation coefficient; preEPM, precursor of expression pattern module; RM, regulator-set module.
Authors' contributions
HGL and HSL conceived the study and developed the algorithm. WKH, SHJ and YSL managed and coordinated the project. WKH and THC contributed to drafting the manuscript by ongoing discussions. HGL, HSL and WKH participated in writing and revising the final manuscript. All authors read and approved the final manuscript.
Additional data files
The following additional data are available with the online version of this paper. Additional data file 1 is a diagram describing the filtering steps for obtaining reliable candidate regulators. Additional data file 2 contains the matrices representing overall information about EPMs of the nitrogen depletion and the cell cycle conditions. Additional data file 3 contains detailed descriptions of condition-specific features of EPMs with their average gene expression profiles. Additional data file 4 contains the case studies that describe the RMs and their regulators in several cell cycle EPMs, heat shock EPM 0 and nitrogen depletion EPM 2. Additional data file 5 contains the matrices describing nitrogen depletion EPM 0, which includes various functional gene groups that formed several EPMs in other conditions. Additional data file 6 is a Venn diagram showing the numbers of overlapped regulators among three conditions. Additional data file 7 is a table listing enriched functional categories for the target genes of all confirmed transcription factors. Additional data file 8 includes bar graphs representing the relative nucleosome occupancy levels on the promoters of condition-specific target genes of Uga3 and Yap1. Additional data file 9 is a diagram showing the regulatory networks among transcription factors under the heat shock and the nitrogen depletion conditions. Additional data file 10 is a table listing enriched functional categories for the targets of FFL groups. Additional data file 11 contains the matrices describing all EPMs and RMs, including lists of synergistic pairs of regulators. Additional data file 12 is a table listing possible regulatory relationships among transcription factors in the same EPM. Additional data file 13 contains detailed descriptions of RMs in nitrogen depletion EPM 12 and a comparison with the modules determined using other algorithms. Additional data file 14 contains detailed descriptions of the criteria for choosing appropriate cut-off values.
Supplementary Material
Filtering steps for obtaining reliable candidate regulators.
Matrices representing overall information about EPMs of the nitrogen depletion and the cell cycle conditions.
Detailed descriptions of condition-specific features of EPMs with their average gene expression profiles
Case studies that describe the RMs and their regulators in several cell cycle EPMs, heat shock EPM 0 and nitrogen depletion EPM 2.
Matrices describing nitrogen depletion EPM 0, which includes various functional gene groups that formed several EPMs in other conditions.
Numbers of overlapped regulators among three conditions.
Enriched functional categories for the target genes of all confirmed transcription factors.
Relative nucleosome occupancy levels on the promoters of condition-specific target genes of Uga3 and Yap1
Regulatory networks among transcription factors under the heat shock and the nitrogen depletion conditions.
Enriched functional categories for the targets of FFL groups.
Matrices describing all EPMs and RMs, including lists of synergistic pairs of regulators.
Possible regulatory relationships among transcription factors in the same EPM.
Detailed descriptions of RMs in nitrogen depletion EPM 12 and a comparison with the modules determined using other algorithms.
Criteria for choosing appropriate cut-off values.
Contributor Information
Hun-Goo Lee, Email: btwall@gmail.com.
Hyo-Soo Lee, Email: lee.hyosoo@gmail.com.
Sang-Hoon Jeon, Email: jeon@donga.co.kr.
Tae-Hoon Chung, Email: thchung@tgen.org.
Young-Sung Lim, Email: lysoo@donga.co.kr.
Won-Ki Huh, Email: wkh@snu.ac.kr.
Acknowledgements
We thank all those who helped in the preparation of this manuscript, and, in particular, Ernest Fraenkel, who provided substantial and helpful comments on the earlier draft of the manuscript. We also thank Jung-Hye Roe, Sung Hee Baek, Jongsik Chun, Jun-Jae Chung, Sejin Kim and Tae-Hyung Kim for their critical reading of the manuscript. We are also grateful to Young-Chul Kim whose management facilitated the success of this study. This work was supported by Dong-a Seetech Co. Ltd, the 21C Frontier Microbial Genomics and Application Center Program, Ministry of Science and Technology (MG05-0203-2-0), the SRC/ERC Program of MOST/KOSEF (R11-2005-009-02002-0), and the second stage of the Brain Korea 21 Project in 2007, Republic of Korea.
References
- Balazsi G, Oltvai ZN. Sensing your surroundings: how transcription-regulatory networks of the cell discern environmental signals. Sci STKE. 2005;2005:pe20. doi: 10.1126/stke.2822005pe20. [DOI] [PubMed] [Google Scholar]
- Luscombe NM, Babu MM, Yu H, Snyder M, Teichmann SA, Gerstein M. Genomic analysis of regulatory network dynamics reveals large topological changes. Nature. 2004;431:308–312. doi: 10.1038/nature02782. [DOI] [PubMed] [Google Scholar]
- Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, Yoo J. et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beer MA, Tavazoie S. Predicting gene expression from sequence. Cell. 2004;117:185–198. doi: 10.1016/S0092-8674(04)00304-6. [DOI] [PubMed] [Google Scholar]
- Bar-Joseph Z, Gerber GK, Lee TI, Rinaldi NJ, Yoo JY, Robert F, Gordon DB, Fraenkel E, Jaakkola TS, Young RA. et al. Computational discovery of gene modules and regulatory networks. Nature biotechnology. 2003;21:1337–1342. doi: 10.1038/nbt890. [DOI] [PubMed] [Google Scholar]
- Vandepoele K, Casneuf T, Van de Peer Y. Identification of novel regulatory modules in dicotyledonous plants using expression data and comparative genomics. Genome biology. 2006;7:R103. doi: 10.1186/gb-2006-7-11-r103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hvidsten TR, Wilczynski B, Kryshtafovych A, Tiuryn J, Komorowski J, Fidelis K. Discovering regulatory binding-site modules using rule-based learning. Genome research. 2005;15:856–866. doi: 10.1101/gr.3760605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihmels J, Friedlander G, Bergmann S, Sarig O, Ziv Y, Barkai N. Revealing modular organization in the yeast transcriptional network. Nature genetics. 2002;31:370–377. doi: 10.1038/ng941. [DOI] [PubMed] [Google Scholar]
- Segal E, Shapira M, Regev A, Pe'er D, Botstein D, Koller D, Friedman N. Module networks: identifying regulatory modules and their condition-specific regulators from gene expression data. Nature genetics. 2003;34:166–176. doi: 10.1038/ng1165. [DOI] [PubMed] [Google Scholar]
- Kim H, Hu W, Kluger Y. Unraveling condition specific gene transcriptional regulatory networks in Saccharomyces cerevisiae. BMC bioinformatics. 2006;7:165. doi: 10.1186/1471-2105-7-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garten Y, Kaplan S, Pilpel Y. Extraction of transcription regulatory signals from genome-wide DNA-protein interaction data. Nucleic acids research. 2005;33:605–615. doi: 10.1093/nar/gki166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shatkay H, Edwards S, Wilbur WJ, Boguski M. Genes, themes and microarrays: using information retrieval for large-scale gene analysis. Proceedings/International Conference on Intelligent Systems for Molecular Biology; ISMB. 2000;8:317–328. [PubMed] [Google Scholar]
- Cho RJ, Campbell MJ, Winzeler EA, Steinmetz L, Conway A, Wodicka L, Wolfsberg TG, Gabrielian AE, Landsman D, Lockhart DJ. et al. A genome-wide transcriptional analysis of the mitotic cell cycle. Molecular cell. 1998;2:65–73. doi: 10.1016/S1097-2765(00)80114-8. [DOI] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. Genomic expression programs in the response of yeast cells to environmental changes. Molecular biology of the cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Zhang MQ. SCPD: a promoter database of the yeast Saccharomyces cerevisiae. Bioinformatics. 1999;15:607–611. doi: 10.1093/bioinformatics/15.7.607. [DOI] [PubMed] [Google Scholar]
- Matys V, Fricke E, Geffers R, Gossling E, Haubrock M, Hehl R, Hornischer K, Karas D, Kel AE, Kel-Margoulis OV. et al. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer A, Workman C, Hollunder J, Radke D, Moller U, Wilhelm T, Ideker T. Integrated assessment and prediction of transcription factor binding. PLoS computational biology. 2006;2:e70. doi: 10.1371/journal.pcbi.0020070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanton SJ, Pugh BF. Full and partial genome-wide assembly and disassembly of the yeast transcription machinery in response to heat shock. Genes & development. 2006;20:2250–2265. doi: 10.1101/gad.1437506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT. et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature genetics. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallouk Y, Vayssier-Taussat M, Bonventre JV, Polla BS. Heat shock protein 70 and ATP as partners in cell homeostasis (Review). International journal of molecular medicine. 1999;4:463–474. doi: 10.3892/ijmm.4.5.463. [DOI] [PubMed] [Google Scholar]
- Parrou JL, Teste MA, Francois J. Effects of various types of stress on the metabolism of reserve carbohydrates in Saccharomyces cerevisiae: genetic evidence for a stress-induced recycling of glycogen and trehalose. Microbiology (Reading, England) 1997;143:1891–1900. doi: 10.1099/00221287-143-6-1891. [DOI] [PubMed] [Google Scholar]
- Davidson JF, Schiestl RH. Mitochondrial respiratory electron carriers are involved in oxidative stress during heat stress in Saccharomyces cerevisiae. Mol Cell Biol. 2001;21:8483–8489. doi: 10.1128/MCB.21.24.8483-8489.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch AP, Werner-Washburne M. The genomics of yeast responses to environmental stress and starvation. Functional & integrative genomics. 2002;2:181–192. doi: 10.1007/s10142-002-0058-2. [DOI] [PubMed] [Google Scholar]
- Jorgensen P, Nishikawa JL, Breitkreutz BJ, Tyers M. Systematic identification of pathways that couple cell growth and division in yeast. Science. 2002;297:395–400. doi: 10.1126/science.1070850. [DOI] [PubMed] [Google Scholar]
- Zhang J, Schneider C, Ottmers L, Rodriguez R, Day A, Markwardt J, Schneider BL. Genomic scale mutant hunt identifies cell size homeostasis genes in S. cerevisiae. Curr Biol. 2002;12:1992–2001. doi: 10.1016/S0960-9822(02)01305-2. [DOI] [PubMed] [Google Scholar]
- Grewal SS, Edgar BA. Controlling cell division in yeast and animals: does size matter? Journal of biology. 2003;2:5. doi: 10.1186/1475-4924-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupes I. Checking cell size in yeast. Trends Genet. 2002;18:479–485. doi: 10.1016/S0168-9525(02)02745-2. [DOI] [PubMed] [Google Scholar]
- Harvey SL, Kellogg DR. Conservation of mechanisms controlling entry into mitosis: budding yeast wee1 delays entry into mitosis and is required for cell size control. Curr Biol. 2003;13:264–275. doi: 10.1016/S0960-9822(03)00049-6. [DOI] [PubMed] [Google Scholar]
- Martinez-Pastor MT, Marchler G, Schuller C, Marchler-Bauer A, Ruis H, Estruch F. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE). Embo J. 1996;15:2227–2235. [PMC free article] [PubMed] [Google Scholar]
- Boy-Marcotte E, Perrot M, Bussereau F, Boucherie H, Jacquet M. Msn2p and Msn4p control a large number of genes induced at the diauxic transition which are repressed by cyclic AMP in Saccharomyces cerevisiae. J Bacteriol. 1998;180:1044–1052. doi: 10.1128/jb.180.5.1044-1052.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Godon C, Lagniel G, Spector D, Garin J, Labarre J, Toledano MB. Yap1 and Skn7 control two specialized oxidative stress response regulons in yeast. J Biol Chem. 1999;274:16040–16046. doi: 10.1074/jbc.274.23.16040. [DOI] [PubMed] [Google Scholar]
- Mannhaupt G, Schnall R, Karpov V, Vetter I, Feldmann H. Rpn4p acts as a transcription factor by binding to PACE, a nonamer box found upstream of 26S proteasomal and other genes in yeast. FEBS letters. 1999;450:27–34. doi: 10.1016/S0014-5793(99)00467-6. [DOI] [PubMed] [Google Scholar]
- Hayes SA, Dice JF. Roles of molecular chaperones in protein degradation. The Journal of cell biology. 1996;132:255–258. doi: 10.1083/jcb.132.3.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macario AJ, Conway de Macario E. Molecular chaperones: multiple functions, pathologies, and potential applications. Front Biosci. 2007;12:2588–2600. doi: 10.2741/2257. [DOI] [PubMed] [Google Scholar]
- Rudra D, Warner JR. What better measure than ribosome synthesis? Genes & development. 2004;18:2431–2436. doi: 10.1101/gad.1256704. [DOI] [PubMed] [Google Scholar]
- Zhao Y, McIntosh KB, Rudra D, Schawalder S, Shore D, Warner JR. Fine-structure analysis of ribosomal protein gene transcription. Mol Cell Biol. 2006;26:4853–4862. doi: 10.1128/MCB.02367-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura H, Kawasaki Y, Kaneko Y, Nosaka K, Iwashima A. Cloning and characteristics of a positive regulatory gene, THI2 (PHO6), of thiamin biosynthesis in Saccharomyces cerevisiae. FEBS letters. 1992;297:155–158. doi: 10.1016/0014-5793(92)80349-L. [DOI] [PubMed] [Google Scholar]
- Zhao H, Eide DJ. Zap1p, a metalloregulatory protein involved in zinc-responsive transcriptional regulation in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:5044–5052. doi: 10.1128/mcb.17.9.5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuge S, Jones N, Nomoto A. Regulation of yAP-1 nuclear localization in response to oxidative stress. The EMBO journal. 1997;16:1710–1720. doi: 10.1093/emboj/16.7.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A, Wemmie JA, Edgington NP, Goebl M, Guevara JL, Moye-Rowley WS. Yeast bZip proteins mediate pleiotropic drug and metal resistance. The Journal of biological chemistry. 1993;268:18850–18858. [PubMed] [Google Scholar]
- Hasan R, Leroy C, Isnard AD, Labarre J, Boy-Marcotte E, Toledano MB. The control of the yeast H2O2 response by the Msn2/4 transcription factors. Molecular microbiology. 2002;45:233–241. doi: 10.1046/j.1365-2958.2002.03011.x. [DOI] [PubMed] [Google Scholar]
- Rodrigues-Pousada C, Nevitt T, Menezes R. The yeast stress response. Role of the Yap family of b-ZIP transcription factors. The PABMB Lecture delivered on 30 June 2004 at the 29th FEBS Congress in Warsaw. The FEBS journal. 2005;272:2639–2647. doi: 10.1111/j.1742-4658.2005.04695.x. [DOI] [PubMed] [Google Scholar]
- Shen-Orr SS, Milo R, Mangan S, Alon U. Network motifs in the transcriptional regulation network of Escherichia coli. Nature genetics. 2002;31:64–68. doi: 10.1038/ng881. [DOI] [PubMed] [Google Scholar]
- Hahn JS, Neef DW, Thiele DJ. A stress regulatory network for co-ordinated activation of proteasome expression mediated by yeast heat shock transcription factor. Molecular microbiology. 2006;60:240–251. doi: 10.1111/j.1365-2958.2006.05097.x. [DOI] [PubMed] [Google Scholar]
- Jung US, Levin DE. Genome-wide analysis of gene expression regulated by the yeast cell wall integrity signalling pathway. Molecular microbiology. 1999;34:1049–1057. doi: 10.1046/j.1365-2958.1999.01667.x. [DOI] [PubMed] [Google Scholar]
- Garcia R, Bermejo C, Grau C, Perez R, Rodriguez-Pena JM, Francois J, Nombela C, Arroyo J. The global transcriptional response to transient cell wall damage in Saccharomyces cerevisiae and its regulation by the cell integrity signaling pathway. The Journal of biological chemistry. 2004;279:15183–15195. doi: 10.1074/jbc.M312954200. [DOI] [PubMed] [Google Scholar]
- Madhani HD, Galitski T, Lander ES, Fink GR. Effectors of a developmental mitogen-activated protein kinase cascade revealed by expression signatures of signaling mutants. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:12530–12535. doi: 10.1073/pnas.96.22.12530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Jensen ST, Stoeckert CJ Jr. Clustering of genes into regulons using integrated modeling-COGRIM. Genome biology. 2007;8:R4. doi: 10.1186/gb-2007-8-1-r4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck MJ, Lieb JD. A chromatin-mediated mechanism for specification of conditional transcription factor targets. Nature genetics. 2006;38:1446–1451. doi: 10.1038/ng1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin C, Tice-Baldwin K, Shore D, Arndt KT. RAP1 is required for BAS1/BAS2- and GCN4-dependent transcription of the yeast HIS4 gene. Mol Cell Biol. 1991;11:3642–3651. doi: 10.1128/mcb.11.7.3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deckert J, Struhl K. Histone acetylation at promoters is differentially affected by specific activators and repressors. Mol Cell Biol. 2001;21:2726–2735. doi: 10.1128/MCB.21.8.2726-2735.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaldis P, Sutton A, Solomon MJ. The Cdk-activating kinase (CAK) from budding yeast. Cell. 1996;86:553–564. doi: 10.1016/S0092-8674(00)80129-4. [DOI] [PubMed] [Google Scholar]
- Koch C, Nasmyth K. Cell cycle regulated transcription in yeast. Curr Opin Cell Biol. 1994;6:451–459. doi: 10.1016/0955-0674(94)90039-6. [DOI] [PubMed] [Google Scholar]
- Ho Y, Costanzo M, Moore L, Kobayashi R, Andrews BJ. Regulation of transcription at the Saccharomyces cerevisiae start transition by Stb1, a Swi6-binding protein. Mol Cell Biol. 1999;19:5267–5278. doi: 10.1128/mcb.19.8.5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer VR, Horak CE, Scafe CS, Botstein D, Snyder M, Brown PO. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature. 2001;409:533–538. doi: 10.1038/35054095. [DOI] [PubMed] [Google Scholar]
- Nasmyth K, Dirick L. The role of SWI4 and SWI6 in the activity of G1 cyclins in yeast. Cell. 1991;66:995–1013. doi: 10.1016/0092-8674(91)90444-4. [DOI] [PubMed] [Google Scholar]
- Koch C, Moll T, Neuberg M, Ahorn H, Nasmyth K. A role for the transcription factors Mbp1 and Swi4 in progression from G1 to S phase. Science. 1993;261:1551–1557. doi: 10.1126/science.8372350. [DOI] [PubMed] [Google Scholar]
- Chang VK, Fitch MJ, Donato JJ, Christensen TW, Merchant AM, Tye BK. Mcm1 binds replication origins. J Biol Chem. 2003;278:6093–6100. doi: 10.1074/jbc.M209827200. [DOI] [PubMed] [Google Scholar]
- Amon A, Tyers M, Futcher B, Nasmyth K. Mechanisms that help the yeast cell cycle clock tick: G2 cyclins transcriptionally activate G2 cyclins and repress G1 cyclins. Cell. 1993;74:993–1007. doi: 10.1016/0092-8674(93)90722-3. [DOI] [PubMed] [Google Scholar]
- Surana U, Robitsch H, Price C, Schuster T, Fitch I, Futcher AB, Nasmyth K. The role of CDC28 and cyclins during mitosis in the budding yeast S. cerevisiae. Cell. 1991;65:145–161. doi: 10.1016/0092-8674(91)90416-V. [DOI] [PubMed] [Google Scholar]
- Althoefer H, Schleiffer A, Wassmann K, Nordheim A, Ammerer G. Mcm1 is required to coordinate G2-specific transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:5917–5928. doi: 10.1128/mcb.15.11.5917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson KA, Nelson C, Tai G, Hung W, Yong C, Astell C, Sadowski I. Two regulators of Ste12p inhibit pheromone-responsive transcription by separate mechanisms. Mol Cell Biol. 2000;20:4199–4209. doi: 10.1128/MCB.20.12.4199-4209.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koranda M, Schleiffer A, Endler L, Ammerer G. Forkhead-like transcription factors recruit Ndd1 to the chromatin of G2/M-specific promoters. Nature. 2000;406:94–98. doi: 10.1038/35017589. [DOI] [PubMed] [Google Scholar]
- Zhu G, Spellman PT, Volpe T, Brown PO, Botstein D, Davis TN, Futcher B. Two yeast forkhead genes regulate the cell cycle and pseudohyphal growth. Nature. 2000;406:90–94. doi: 10.1038/35017581. [DOI] [PubMed] [Google Scholar]
- Hollenhorst PC, Pietz G, Fox CA. Mechanisms controlling differential promoter-occupancy by the yeast forkhead proteins Fkh1p and Fkh2p: implications for regulating the cell cycle and differentiation. Genes Dev. 2001;15:2445–2456. doi: 10.1101/gad.906201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, Reynolds DM, Shevchenko A, Shevchenko A, Goldstone SD, Dalton S. Forkhead transcription factors, Fkh1p and Fkh2p, collaborate with Mcm1p to control transcription required for M-phase. Curr Biol. 2000;10:896–906. doi: 10.1016/S0960-9822(00)00618-7. [DOI] [PubMed] [Google Scholar]
- Jorgensen P, Tyers M. The fork'ed path to mitosis. Genome biology. 2000;1:REVIEWS1022. doi: 10.1186/gb-2000-1-3-reviews1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pic A, Lim FL, Ross SJ, Veal EA, Johnson AL, Sultan MR, West AG, Johnston LH, Sharrocks AD, Morgan BA. The forkhead protein Fkh2 is a component of the yeast cell cycle transcription factor SFF. The EMBO journal. 2000;19:3750–3761. doi: 10.1093/emboj/19.14.3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon I, Barnett J, Hannett N, Harbison CT, Rinaldi NJ, Volkert TL, Wyrick JJ, Zeitlinger J, Gifford DK, Jaakkola TS. et al. Serial regulation of transcriptional regulators in the yeast cell cycle. Cell. 2001;106:697–708. doi: 10.1016/S0092-8674(01)00494-9. [DOI] [PubMed] [Google Scholar]
- Mai B, Miles S, Breeden LL. Characterization of the ECB binding complex responsible for the M/G(1)-specific transcription of CLN3 and SWI4. Mol Cell Biol. 2002;22:430–441. doi: 10.1128/MCB.22.2.430-441.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInerny CJ, Partridge JF, Mikesell GE, Creemer DP, Breeden LL. A novel Mcm1-dependent element in the SWI4, CLN3, CDC6, and CDC47 promoters activates M/G1-specific transcription. Genes & development. 1997;11:1277–1288. doi: 10.1101/gad.11.10.1277. [DOI] [PubMed] [Google Scholar]
- Lyons TJ, Gasch AP, Gaither LA, Botstein D, Brown PO, Eide DJ. Genome-wide characterization of the Zap1p zinc-responsive regulon in yeast. Proc Natl Acad Sci USA. 2000;97:7957–7962. doi: 10.1073/pnas.97.14.7957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falchuk KH, Fawcett DW, Vallee BL. Role of zinc in cell division of Euglena gracilis. J Cell Sci. 1975;17:57–78. doi: 10.1242/jcs.17.1.57. [DOI] [PubMed] [Google Scholar]
- Prasad AS, Beck FW, Endre L, Handschu W, Kukuruga M, Kumar G. Zinc deficiency affects cell cycle and deoxythymidine kinase gene expression in HUT-78 cells. J Lab Clin Med. 1996;128:51–60. doi: 10.1016/S0022-2143(96)90113-4. [DOI] [PubMed] [Google Scholar]
- Cabib E, Mol PC, Shaw JA, Choi WJ. Biosynthesis of cell wall and septum during yeast growth. Arch Med Res. 1993;24:301–303. [PubMed] [Google Scholar]
- Peters JM. The anaphase-promoting complex: proteolysis in mitosis and beyond. Mol Cell. 2002;9:931–943. doi: 10.1016/S1097-2765(02)00540-3. [DOI] [PubMed] [Google Scholar]
- Chernomordik L, Kozlov MM, Zimmerberg J. Lipids in biological membrane fusion. J Membr Biol. 1995;146:1–14. doi: 10.1007/BF00232676. [DOI] [PubMed] [Google Scholar]
- Emoto K, Umeda M. An essential role for a membrane lipid in cytokinesis. Regulation of contractile ring disassembly by redistribution of phosphatidylethanolamine. J Cell Biol. 2000;149:1215–1224. doi: 10.1083/jcb.149.6.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emoto K, Umeda M. Membrane lipid control of cytokinesis. Cell Struct Funct. 2001;26:659–665. doi: 10.1247/csf.26.659. [DOI] [PubMed] [Google Scholar]
- Lin LH, Lee HC, Li WH, Chen BS. Dynamic modeling of cis-regulatory circuits and gene expression prediction via cross-gene identification. BMC Bioinformatics. 2005;6:258. doi: 10.1186/1471-2105-6-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buehrer BM, Errede B. Coordination of the mating and cell integrity mitogen-activated protein kinase pathways in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:6517–6525. doi: 10.1128/mcb.17.11.6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oehlen L, Cross FR. The mating factor response pathway regulates transcription of TEC1, a gene involved in pseudohyphal differentiation of Saccharomyces cerevisiae. FEBS letters. 1998;429:83–88. doi: 10.1016/S0014-5793(98)00568-7. [DOI] [PubMed] [Google Scholar]
- Baetz K, Moffat J, Haynes J, Chang M, Andrews B. Transcriptional coregulation by the cell integrity mitogen-activated protein kinase Slt2 and the cell cycle regulator Swi4. Mol Cell Biol. 2001;21:6515–6528. doi: 10.1128/MCB.21.19.6515-6528.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin DE. Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2005;69:262–291. doi: 10.1128/MMBR.69.2.262-291.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cid VJ, Duran A, del Rey F, Snyder MP, Nombela C, Sanchez M. Molecular basis of cell integrity and morphogenesis in Saccharomyces cerevisiae. Microbiological reviews. 1995;59:345–386. doi: 10.1128/mr.59.3.345-386.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valtz N, Herskowitz I. Pea2 protein of yeast is localized to sites of polarized growth and is required for efficient mating and bipolar budding. The Journal of cell biology. 1996;135:725–739. doi: 10.1083/jcb.135.3.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igual JC, Johnson AL, Johnston LH. Coordinated regulation of gene expression by the cell cycle transcription factor Swi4 and the protein kinase C MAP kinase pathway for yeast cell integrity. The EMBO journal. 1996;15:5001–5013. [PMC free article] [PubMed] [Google Scholar]
- Bouquin N, Johnson AL, Morgan BA, Johnston LH. Association of the cell cycle transcription factor Mbp1 with the Skn7 response regulator in budding yeast. Molecular biology of the cell. 1999;10:3389–3400. doi: 10.1091/mbc.10.10.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidlingmaier S, Weiss EL, Seidel C, Drubin DG, Snyder M. The Cbk1p pathway is important for polarized cell growth and cell separation in Saccharomyces cerevisiae. Mol Cell Biol. 2001;21:2449–2462. doi: 10.1128/MCB.21.7.2449-2462.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore P, Sharrocks AD. The MADS-box family of transcription factors. European journal of biochemistry/FEBS. 1995;229:1–13. doi: 10.1111/j.1432-1033.1995.tb20430.x. [DOI] [PubMed] [Google Scholar]
- Roberts CJ, Nelson B, Marton MJ, Stoughton R, Meyer MR, Bennett HA, He YD, Dai H, Walker WL, Hughes TR. et al. Signaling and circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles. Science. 2000;287:873–880. doi: 10.1126/science.287.5454.873. [DOI] [PubMed] [Google Scholar]
- Lengeler KB, Davidson RC, D'Souza C, Harashima T, Shen WC, Wang P, Pan X, Waugh M, Heitman J. Signal transduction cascades regulating fungal development and virulence. Microbiol Mol Biol Rev. 2000;64:746–785. doi: 10.1128/MMBR.64.4.746-785.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, Zeitlinger J, Schreiber J, Hannett N, Kanin E. et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- Kollar R, Reinhold BB, Petrakova E, Yeh HJ, Ashwell G, Drgonova J, Kapteyn JC, Klis FM, Cabib E. Architecture of the yeast cell wall. Beta(1-->6)-glucan interconnects mannoprotein, beta(1-->)3-glucan, and chitin. J Biol Chem. 1997;272:17762–17775. doi: 10.1074/jbc.272.28.17762. [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG. Evidence for translational regulation of the activator of general amino acid control in yeast. Proceedings of the National Academy of Sciences of the United States of America. 1984;81:6442–6446. doi: 10.1073/pnas.81.20.6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosch HU, Scheier B, Lahti R, Mantsala P, Braus GH. Transcriptional activation of yeast nucleotide biosynthetic gene ADE4 by GCN4. The Journal of biological chemistry. 1991;266:20453–20456. [PubMed] [Google Scholar]
- Rolfes RJ, Hinnebusch AG. Translation of the yeast transcriptional activator GCN4 is stimulated by purine limitation: implications for activation of the protein kinase GCN2. Mol Cell Biol. 1993;13:5099–5111. doi: 10.1128/mcb.13.8.5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan K, Meyer MR, Jackson BM, Slade D, Roberts C, Hinnebusch AG, Marton MJ. Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol Cell Biol. 2001;21:4347–4368. doi: 10.1128/MCB.21.13.4347-4368.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt KT, Styles C, Fink GR. Multiple global regulators control HIS4 transcription in yeast. Science. 1987;237:874–880. doi: 10.1126/science.3303332. [DOI] [PubMed] [Google Scholar]
- Tice-Baldwin K, Fink GR, Arndt KT. BAS1 has a Myb motif and activates HIS4 transcription only in combination with BAS2. Science. 1989;246:931–935. doi: 10.1126/science.2683089. [DOI] [PubMed] [Google Scholar]
- Rolfes RJ, Zhang F, Hinnebusch AG. The transcriptional activators BAS1, BAS2, and ABF1 bind positive regulatory sites as the critical elements for adenine regulation of ADE5,7. The Journal of biological chemistry. 1997;272:13343–13354. doi: 10.1074/jbc.272.20.13343. [DOI] [PubMed] [Google Scholar]
- Friden P, Schimmel P. LEU3 of Saccharomyces cerevisiae activates multiple genes for branched-chain amino acid biosynthesis by binding to a common decanucleotide core sequence. Mol Cell Biol. 1988;8:2690–2697. doi: 10.1128/mcb.8.7.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Cooper TG, Kohlhaw GB. The Saccharomyces cerevisiae Leu3 protein activates expression of GDH1, a key gene in nitrogen assimilation. Mol Cell Biol. 1995;15:52–57. doi: 10.1128/mcb.15.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talibi D, Grenson M, Andre B. Cis- and trans-acting elements determining induction of the genes of the gamma-aminobutyrate (GABA) utilization pathway in Saccharomyces cerevisiae. Nucleic acids research. 1995;23:550–557. doi: 10.1093/nar/23.4.550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebora K, Laloo B, Daignan-Fornier B. Revisiting purine-histidine cross-pathway regulation in Saccharomyces cerevisiae: a central role for a small molecule. Genetics. 2005;170:61–70. doi: 10.1534/genetics.104.039396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes WB, Appling DR. Cloning and characterization of methenyltetrahydrofolate synthetase from Saccharomyces cerevisiae. The Journal of biological chemistry. 2002;277:20205–20213. doi: 10.1074/jbc.M201242200. [DOI] [PubMed] [Google Scholar]
- Kastanos EK, Woldman YY, Appling DR. Role of mitochondrial and cytoplasmic serine hydroxymethyltransferase isozymes in de novo purine synthesis in Saccharomyces cerevisiae. Biochemistry. 1997;36:14956–14964. doi: 10.1021/bi971610n. [DOI] [PubMed] [Google Scholar]
- Blaiseau PL, Thomas D. Multiple transcriptional activation complexes tether the yeast activator Met4 to DNA. The EMBO journal. 1998;17:6327–6336. doi: 10.1093/emboj/17.21.6327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell KF, Baker RE. Possible cross-regulation of phosphate and sulfate metabolism in Saccharomyces cerevisiae. Genetics. 1992;132:63–73. doi: 10.1093/genetics/132.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham TH, Clemente JC, Satou K, Ho TB. Computational discovery of transcriptional regulatory rules. Bioinformatics. 2005;21(Suppl 2):ii101–ii107. doi: 10.1093/bioinformatics/bti1117. [DOI] [PubMed] [Google Scholar]
- Hollunder J, Friedel M, Beyer A, Workman CT, Wilhelm T. DASS: efficient discovery and p value calculation of substructures in unordered data. Bioinformatics. 2006. [DOI] [PubMed]
- Saccharomyces Genome Database. http://www.yeastgenome.org http://www.yeastgenome.org
- Della Seta F, Ciafre SA, Marck C, Santoro B, Presutti C, Sentenac A, Bozzoni I. The ABF1 factor is the transcriptional activator of the L2 ribosomal protein genes in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:2437–2441. doi: 10.1128/mcb.10.5.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevitt T, Pereira J, Rodrigues-Pousada C. YAP4 gene expression is induced in response to several forms of stress in Saccharomyces cerevisiae. Yeast. 2004;21:1365–1374. doi: 10.1002/yea.1188. [DOI] [PubMed] [Google Scholar]
- Pic-Taylor A, Darieva Z, Morgan BA, Sharrocks AD. Regulation of cell cycle-specific gene expression through cyclin-dependent kinase-mediated phosphorylation of the forkhead transcription factor Fkh2p. Mol Cell Biol. 2004;24:10036–10046. doi: 10.1128/MCB.24.22.10036-10046.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn JS, Hu Z, Thiele DJ, Iyer VR. Genome-wide analysis of the biology of stress responses through heat shock transcription factor. Mol Cell Biol. 2004;24:5249–5256. doi: 10.1128/MCB.24.12.5249-5256.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuller HJ. Transcriptional control of nonfermentative metabolism in the yeast Saccharomyces cerevisiae. Curr Genet. 2003;43:139–160. doi: 10.1007/s00294-003-0381-8. [DOI] [PubMed] [Google Scholar]
- Vyas VK, Berkey CD, Miyao T, Carlson M. Repressors Nrg1 and Nrg2 regulate a set of stress-responsive genes in Saccharomyces cerevisiae. Eukaryot Cell. 2005;4:1882–1891. doi: 10.1128/EC.4.11.1882-1891.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nourani A, Wesolowski-Louvel M, Delaveau T, Jacq C, Delahodde A. Multiple-drug-resistance phenomenon in the yeast Saccharomyces cerevisiae: involvement of two hexose transporters. Mol Cell Biol. 1997;17:5453–5460. doi: 10.1128/mcb.17.9.5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fingerman I, Nagaraj V, Norris D, Vershon AK. Sfp1 plays a key role in yeast ribosome biogenesis. Eukaryotic cell. 2003;2:1061–1068. doi: 10.1128/EC.2.5.1061-1068.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ness F, Bourot S, Regnacq M, Spagnoli R, Berges T, Karst F. SUT1 is a putative Zn[II]2Cys6-transcription factor whose upregulation enhances both sterol uptake and synthesis in aerobically growing Saccharomyces cerevisiae cells. Eur J Biochem. 2001;268:1585–1595. doi: 10.1046/j.1432-1327.2001.02029.x. [DOI] [PubMed] [Google Scholar]
- Mitchell AP, Magasanik B. Regulation of glutamine-repressible gene products by the GLN3 function in Saccharomyces cerevisiae. Mol Cell Biol. 1984;4:2758–2766. doi: 10.1128/mcb.4.12.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slattery MG, Liko D, Heideman W. The function and properties of the Azf1 transcriptional regulator change with growth conditions in Saccharomyces cerevisiae. Eukaryot Cell. 2006;5:313–320. doi: 10.1128/EC.5.2.313-320.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeast Deletion Project and Proteomics of Mitochondria Database. http://www-deletion.stanford.edu/YDPM/ http://www-deletion.stanford.edu/YDPM/
- Zhu Y, Xiao W. Pdr3 is required for DNA damage induction of MAG1 and DDI1 via a bi-directional promoter element. Nucleic Acids Res. 2004;32:5066–5075. doi: 10.1093/nar/gkh838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung US, Sobering AK, Romeo MJ, Levin DE. Regulation of the yeast Rlm1 transcription factor by the Mpk1 cell wall integrity MAP kinase. Mol Microbiol. 2002;46:781–789. doi: 10.1046/j.1365-2958.2002.03198.x. [DOI] [PubMed] [Google Scholar]
- GeneDB. http://www.genedb.org http://www.genedb.org
- Mendizabal I, Rios G, Mulet JM, Serrano R, de Larrinoa IF. Yeast putative transcription factors involved in salt tolerance. FEBS Lett. 1998;425:323–328. doi: 10.1016/S0014-5793(98)00249-X. [DOI] [PubMed] [Google Scholar]
- Crespo JL, Powers T, Fowler B, Hall MN. The TOR-controlled transcription activators GLN3, RTG1, and RTG3 are regulated in response to intracellular levels of glutamine. Proc Natl Acad Sci USA. 2002;99:6784–6789. doi: 10.1073/pnas.102687599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cade RM, Errede B. MOT2 encodes a negative regulator of gene expression that affects basal expression of pheromone-responsive genes in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:3139–3149. doi: 10.1128/mcb.14.5.3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai B, Breeden L. Xbp1, a stress-induced transcriptional repressor of the Saccharomyces cerevisiae Swi4/Mbp1 family. Mol Cell Biol. 1997;17:6491–6501. doi: 10.1128/mcb.17.11.6491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook WJ, Chase D, Audino DC, Denis CL. Dissection of the ADR1 protein reveals multiple, functionally redundant activation domains interspersed with inhibitory regions: evidence for a repressor binding to the ADR1c region. Mol Cell Biol. 1994;14:629–640. doi: 10.1128/mcb.14.1.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boorsma A, de Nobel H, ter Riet B, Bargmann B, Brul S, Hellingwerf KJ, Klis FM. Characterization of the transcriptional response to cell wall stress in Saccharomyces cerevisiae. Yeast. 2004;21:413–427. doi: 10.1002/yea.1109. [DOI] [PubMed] [Google Scholar]
- Reeder RH. rRNA synthesis in the nucleolus. Trends Genet. 1990;6:390–395. doi: 10.1016/0168-9525(90)90298-K. [DOI] [PubMed] [Google Scholar]
- Sun C, Woolford JL Jr. The yeast nucleolar protein Nop4p contains four RNA recognition motifs necessary for ribosome biogenesis. The Journal of biological chemistry. 1997;272:25345–25352. doi: 10.1074/jbc.272.40.25345. [DOI] [PubMed] [Google Scholar]
- Miyoshi K, Miyakawa T, Mizuta K. Repression of rRNA synthesis due to a secretory defect requires the C-terminal silencing domain of Rap1p in Saccharomyces cerevisiae. Nucleic acids research. 2001;29:3297–3303. doi: 10.1093/nar/29.16.3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann-Le Denmat S, Werner M, Sentenac A, Thuriaux P. Suppression of yeast RNA polymerase III mutations by FHL1, a gene coding for a fork head protein involved in rRNA processing. Mol Cell Biol. 1994;14:2905–2913. doi: 10.1128/mcb.14.5.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee N, Zhang MQ. Identifying cooperativity among transcription factors controlling the cell cycle in yeast. Nucleic Acids Res. 2003;31:7024–7031. doi: 10.1093/nar/gkg894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagamine N, Kawada Y, Sakakibara Y. Identifying cooperative transcriptional regulations using protein-protein interactions. Nucleic Acids Res. 2005;33:4828–4837. doi: 10.1093/nar/gki793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmona-Saez P, Chagoyen M, Rodriguez A, Trelles O, Carazo JM, Pascual-Montano A. Integrated analysis of gene expression by Association Rules Discovery. BMC bioinformatics. 2006;7:54. doi: 10.1186/1471-2105-7-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Filtering steps for obtaining reliable candidate regulators.
Matrices representing overall information about EPMs of the nitrogen depletion and the cell cycle conditions.
Detailed descriptions of condition-specific features of EPMs with their average gene expression profiles
Case studies that describe the RMs and their regulators in several cell cycle EPMs, heat shock EPM 0 and nitrogen depletion EPM 2.
Matrices describing nitrogen depletion EPM 0, which includes various functional gene groups that formed several EPMs in other conditions.
Numbers of overlapped regulators among three conditions.
Enriched functional categories for the target genes of all confirmed transcription factors.
Relative nucleosome occupancy levels on the promoters of condition-specific target genes of Uga3 and Yap1
Regulatory networks among transcription factors under the heat shock and the nitrogen depletion conditions.
Enriched functional categories for the targets of FFL groups.
Matrices describing all EPMs and RMs, including lists of synergistic pairs of regulators.
Possible regulatory relationships among transcription factors in the same EPM.
Detailed descriptions of RMs in nitrogen depletion EPM 12 and a comparison with the modules determined using other algorithms.
Criteria for choosing appropriate cut-off values.