Abstract
The human endogenous retrovirus K (HERV-K) family of endogenous retroviruses consists of ≈50 proviral copies per haploid human genome. Herein, the HERV-Ks are shown to encode a sequence-specific nuclear RNA export factor, termed K-Rev, that is functionally analogous to the HIV-1 Rev protein. Like HIV-1 Rev, K-Rev binds to both the Crm1 nuclear export factor and to a cis-acting viral RNA target to activate nuclear export of unspliced RNAs. Surprisingly, this HERV-K RNA sequence, which is encoded within the HERV-K long terminal repeat, is also recognized by HIV-1 Rev. These data provide surprising evidence for an evolutionary link between HIV-1 and a group of endogenous retroviruses that first entered the human genome ≈30 million years ago.
During the retroviral replication cycle, the initial genome length viral transcript must be expressed in both fully spliced and incompletely spliced forms. However, cells have developed mechanisms to prevent the nuclear export of RNAs that retain splice sites, i.e., pre-mRNAs, via the endogenous mRNA export pathway. Retroviruses have therefore evolved in ways that enable them to target their incompletely spliced transcripts to alternative nuclear export pathways (reviewed in refs. 1 and 2). Thus, HIV-1 encodes a protein termed Rev that recruits a cellular nuclear export factor, termed Crm1, to the Rev response element (RRE), a structured RNA target sequence encoded within the viral genome (3–6). Nuclear export of incompletely spliced viral RNAs requires the recruitment of multiple Rev and Crm1 molecules to the RRE and depends on the GTP-bound form of the cellular Ran protein, which mediates the Rev–Crm1 interaction (4, 6). Although most other complex retroviruses also encode Rev-like proteins, simple retroviruses lack a Rev equivalent and instead rely on the direct recruitment of a host nuclear RNA export factor to a cis-acting RNA target termed a constitutive transport element (CTE; ref. 7). In the case of the Mason–Pfizer monkey virus (MPMV) CTE, this factor has been identified as the host protein Tap (8, 9). Importantly, CTE/Tap-mediated nuclear RNA export is independent of Crm1 function (9–11).
Endogenous retroviruses are encoded within the genomes of all higher eukaryotes and arose from the infection of germ cells by exogenous retroviruses (12, 13). These infections generally occurred in the distant past and most endogenous retroviruses have suffered inactivating mutations or deletions over time. The human endogenous retrovirus K (HERV-K) family is represented in the human genome by ≈50 more-or-less full-length proviruses and over 1,000 solitary long terminal repeats (LTRs) (12). The HERV-Ks first entered the primate genome shortly after the split of new world and old world monkeys (i.e., ≥30 million years ago) with some evolutionarily more recent HERV-K insertions postdating the human/chimpanzee division (i.e., ≈5 million years ago; ref. 14). The HERV-Ks have no known close exogenous relative, although they do show some homology to mouse mammary tumor virus and to the recently described human retrovirus 5 (12, 15). Importantly, individual HERV-K proviruses encode functional forms of several viral proteins, and the HERV-Ks are expressed in certain human tumor cell lines where they can produce viral particles (12, 13).
Analysis of the RNA expression pattern of full-length HERV-K has identified, not only the genome length and singly spliced Env mRNA seen in all retroviruses, but also a doubly spliced RNA that has the ability to encode a 105-aa protein of unknown function, termed central ORF (c-orf; refs. 16 and 17). Although c-orf and HIV-1 Rev (H-Rev) lack any sequence homology, three lines of evidence led to the suggestion by Löwer et al. (17) that c-orf could be the HERV-K homolog of the H-Rev nuclear RNA export factor. First, the c-orf is formed by splicing the initiation codon and the most N-terminal residues of Env to a short coding exon that overlaps with Env but in a different reading frame (17). This process is how Rev is encoded in several lentiviruses, including Visna maedi virus (VMV), although not HIV-1. Second, c-orf contains a stretch of arginine residues reminiscent of the arginine-rich RNA binding motif seen in Rev (1, 18). Finally, c-orf expression results in the partial localization of this protein to the nucleolus, as is also seen with H-Rev (18). It is important to note, however, that the presence of an arginine-rich RNA binding motif and a predominantly nucleolar localization are equally characteristic of the HIV-1 Tat protein (19) and that a possible role for c-orf in mediating nuclear RNA export has not been addressed previously. In this article, we report that the c-orf protein is indeed able to function as a Crm1-dependent, sequence-specific nuclear RNA export factor that is functionally equivalent to the Rev protein seen in HIV-1. This report therefore describes the identification of a regulatory protein encoded by a human endogenous retrovirus and has significant implications for the study of the evolution of present day exogenous retroviruses, including HIV-1.
Materials and Methods
Construction of Molecular Clones.
The pBC12/CMV expression plasmid and derivatives pBC12/CMV/ΔCAN, pBC12/CMV/β-galactosidase (β-GAL), pcRev (expressing H-Rev), and pcL [expressing VMV Rev (V-Rev)] have been described (6, 18, 20). A synthetic DNA encoding the reported 105-aa c-orf/HERV-K Rev (K-Rev) sequence (17) was prepared containing a 5′ BspHI and a 3′ XhoI overhang. This DNA was then cloned into pBC12/CMV and into a pBC12/CMV-based VP16 fusion protein expression plasmid (6) that then expressed K-Rev either unfused or fused, respectively, to the VP16 transcription activation domain. K-Rev was also cloned into pSG424 (21) for expression as a GAL4 DNA binding domain fusion. The pDM128/PL indicator plasmid and variants containing the HIV-1 RRE (H-RRE), the VMV RRE (V-RRE), or the MPMV CTE also have been described (6, 22, 23). Candidate HERV-K RRE sequences were PCR amplified from total HeLa cell DNA by using primers that inserted 5′ BglII and 3′ XbaI sites and then were introduced into these same sites in the polylinker present in pDM128/PL. The pDM128/HERV-K RRE (K-RRE) plasmid described contains a 434-bp HERV-K sequence (GenBank accession no. AF179225), extending from residue 8,719 to 9,152 according to the sequence of Ono et al. (24), and differs from our published sequence at 10 of 434 residues. The same 434-bp K-RRE BglII–XbaI fragment also was cloned into the BamHI and XbaI sites present in the in vitro RNA transcription plasmid pGEM-3ZF(+), and the K-RRE then was transcribed by using T7 RNA polymerase after linearization at the polylinker SalI site. Similar in vitro expression plasmids containing the H-RRE or the V-RRE have been described (20, 25).
Mammalian Two-Hybrid Assays.
Two-hybrid assays were performed in human 293T cells essentially as described (6); 293T cells (35-mm cultures) were transfected with the pG6(−31)HIVLTRΔTAR indicator plasmid (26) and pSG424-based (21) expression plasmids encoding either the GAL4 DNA binding domain alone or that domain fused to the full-length Crm1 or K-Rev (6, 27). Cells were also cotransfected with a pBC12/CMV-based plasmid expressing a fusion of K-Rev to the VP16 transcription activation domain (6) or pBC12/CMV as a negative control. pBC12/CMV/β-GAL served as an internal control. At ≈48 h after transfection, cells were lysed, and induced chloramphenicol acetyl transferase (CAT) and β-GAL activities were determined (6).
In Vitro Protein Binding Experiments.
Recombinant glutathione S-transferase (GST)-CRM1 protein was coupled to Affi-gel 10 active ester agarose beads (Bio-Rad) as described (28). 35S-Met-labeled K-Rev protein was translated in vitro (T7 TNT kit, Promega) and partially purified over Q-Sepharose ion exchange and single-stranded DNA-cellulose columns (Amersham Pharmacia). RanQ69L, a Ran mutant resistant to GTP hydrolysis (29), was expressed, purified, and cleaved from GST as described (28). Microaffinity columns with 10-μl bed volumes that contained GST, GST-CRM1 only, GST-CRM1 preincubated with RanQ69L plus 5 mM GTP, or GST-CRM1 preincubated with RanQ69L plus 5 mM GDP were prepared as described (28). Equal aliquots of K-Rev were loaded over each column in 100 μl of AC buffer [100 mM NaCl/10 mM Hepes, pH 7.4/10% (vol/vol) glycerol/1 mM DTT]. The columns were then washed with 200 μl of AC buffer and eluted with 50 μl of 500 mM MgCl2/0.05% SDS. The eluate from each column was loaded on a 10–20% polyacrylamide/SDS gradient gel (ReadyGel, Bio-Rad), and bound K-Rev was visualized by fluorography.
Results
K-Rev Specifically Interacts with the Crm1 Nuclear Export Factor.
To test whether c-orf, which we will henceforth term K-Rev, is indeed an H-Rev homolog, we first asked whether K-Rev could bind specifically to the Crm1 nuclear export factor in the human cell nucleus. For this purpose, we used a previously described (6) mammalian two-hybrid assay based on an indicator construct that contains a minimal HIV-1 LTR promoter element, flanked 5′ by GAL4 DNA binding sites and linked to the cat indicator gene. Cotransfection of this indicator plasmid with a vector expressing either a fusion protein consisting of the VP16 transcription activation domain linked to K-Rev (K-Rev/VP16) or of the GAL4 DNA binding domain linked to Crm1 (GAL4/Crm1) failed to induce CAT expression (Table 1). In contrast, coexpression in 293T cells of both K-Rev/VP16 and GAL4/Crm1 resulted in a specific, ≈8-fold induction in CAT activity, consistent with a Crm1–K-Rev interaction. Because multimerization is critical for H-Rev function (25, 30), we also asked whether K-Rev would multimerize in vivo. As shown in Table 1, K-Rev indeed proved able to form, at minimum, homodimers in this human-cell-based protein–protein interaction assay.
Table 1.
K-Rev binds to CRM1 and to itself in vivo
| Transfected effector plasmids | Relative activation |
|---|---|
| GAL4 + K-Rev/VP16 | 1.0 |
| GAL4/Crm1 only | 0.7 ± 0.4 |
| GAL4/Crm1 + K-Rev/VP16 | 8.2 ± 2.0 |
| GAL4/K-Rev only | 0.7 ± 0.1 |
| GAL4/K-Rev + K-Rev/VP16 | 27.4 ± 2.4 |
Mammalian two-hybrid assays were performed in 293T cells as described (6). These data (±SEM) represent the average of four experiments and are expressed relative to the culture transfected with plasmids expressing GAL4 and K-Rev/VP16, which was set arbitrarily at 1.0.
As noted above, the interaction of H-Rev with Crm1 occurs efficiently only in the presence of the GTP-bound form of Ran, i.e., the form that is concentrated in the nucleus (4). To test whether the interaction of K-Rev with Crm1 is also Ran-GTP dependent, we prepared 35S-labeled K-Rev by in vitro translation and then measured binding to a recombinant GST-Crm1 fusion protein in the absence or presence of Ran-GTP. As shown in Fig. 1A, specific binding of GST-Crm1 by K-Rev in vitro indeed proved to be Ran-GTP dependent.
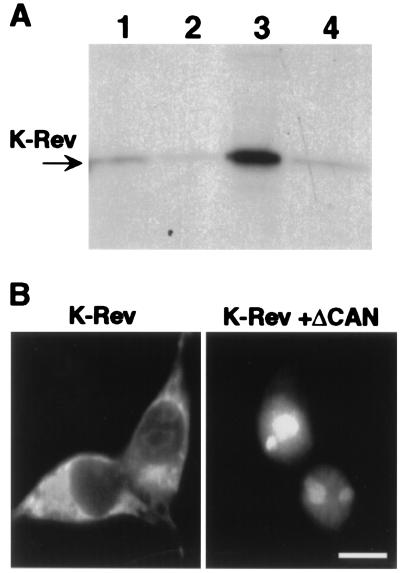
Figure 1.
Functional interaction between Crm1 and K-Rev. (A) Microaffinity columns containing GST (lane 1), GST-Crm1 only (lane 2), GST-Crm1 preincubated with Ran-GTP (lane 3), or GST-Crm1 preincubated with Ran-GDP (lane 4) were prepared as described (6, 28). Equal aliquots of 35S-labeled K-Rev were then loaded onto each column, and bound proteins were eluted by using 0.05% SDS. The eluates were resolved by gel electrophoresis and visualized by fluorography. (B) Human 293T cells (35-mm cultures) were transfected with 500 ng of pBC12/CMV/K-Rev and 1 μg of pBC12/CMV/ΔCAN or the parental pBC12/CMV plasmid by using the calcium phosphate procedure. At ≈48 h after transfection, the cells were fixed and stained as described (6) by using a 1:500 dilution of a rabbit polyclonal anti-K-Rev antiserum and a 1:2,000 dilution of a FITC-conjugated donkey anti-rabbit antiserum (The Jackson Laboratory). No staining was observed in mock-transfected cultures. (Bar ≈20 μm.)
To examine the subcellular localization of K-Rev, we raised a rabbit polyclonal antiserum against recombinant K-Rev and then performed immunofluorescence analysis on 293T cells transfected with a K-Rev expression plasmid. As shown in Fig. 1B, we observed a predominantly cytoplasmic localization for K-Rev with only modest nuclear/nucleolar fluorescence, in contrast to a previous report suggesting a predominantly nuclear/nucleolar localization (17). If K-Rev is indeed a nucleocytoplasmic shuttle protein, then the primarily cytoplasmic localization observed for K-Rev would indicate that the nuclear import of K-Rev is less efficient than the Crm1-dependent nuclear export. The inhibition of Crm1 function should then result in the nuclear accumulation of K-Rev. Previously, we and others have described a truncated form of the nucleoporin Nup214/CAN, termed ΔCAN, that selectively blocks Crm1-dependent, but not Crm1-independent, nuclear export by binding to Crm1 (6, 27, 31). As shown in Fig. 1B, coexpression of ΔCAN indeed induced a nuclear and intensely nucleolar localization for K-Rev, consistent with the hypothesis that the normally cytoplasmic steady-state localization of K-Rev in transfected 293T cells requires ongoing Crm1-mediated nuclear export, i.e., that K-Rev is a nucleocytoplasmic shuttle protein.
K-Rev Is a Sequence-Specific Nuclear RNA Export Factor.
If K-Rev is the HERV-K homolog of H-Rev, then HERV-K should encode an RNA target for K-Rev (K-RRE) equivalent to the H-RRE. Based on the precedent of known retroviral RNA export targets (1, 2), we predicted that the K-RRE would be located in the env gene or in the 3′ LTR of HERV-K. To identify the K-RRE, we inserted HERV-K sequences obtained by PCR of human genomic DNA into the polylinker present in pDM128/PL, which contains a cat gene flanked by 5′ and 3′ splice sites (22, 23). CAT expression, therefore, depends on the nuclear export of an unspliced cat RNA, and this export occurs efficiently only when a CTE or RRE is inserted into the pDM128/PL plasmid, in the latter case only in the presence of the cognate viral RNA export factor.
Insertion of a sequence containing the predicted HERV-K LTR U3/R region into pDM128/PL in fact conferred K-Rev-dependent CAT expression. This result is shown in Fig. 2A. The pDM128/PL derivative (pDM128/K-RRE) contains a 434-bp HERV-K LTR sequence extending from residue 8,719 to 9,152, according to the proviral sequence of Ono et al. (24). Other pDM128/PL variants containing the H-RRE or the V-RRE were also analyzed in the presence or absence of plasmids expressing K-Rev, H-Rev, or V-Rev (6). As may be observed, the pDM128/H-RRE indicator is activated by H-Rev but not by either K-Rev or V-Rev. Similarly, the pDM128/V-RRE plasmid gives elevated CAT activity on coexpression of V-Rev but is not affected by either H-Rev or K-Rev. Finally, pDM128/K-RRE is activated, not only by the cognate K-Rev protein, but also by the H-Rev protein. However, pDM128/K-RRE is not affected by V-Rev expression. To confirm that the induction of CAT expression from the pDM128/K-RRE indicator plasmid, on expression of K-Rev, was indeed at the level of unspliced nuclear RNA export, we performed an RNase protection assay by using nuclear and cytoplasmic RNA fractions (9). As shown in Fig. 2B, coexpression of K-Rev indeed induced the cytoplasmic expression of an unspliced CAT mRNA while exerting little effect on cytoplasmic spliced RNA expression or on the nuclear expression of either spliced or unspliced pDM128/K-RRE-derived RNAs. This result, therefore, shows that K-Rev is indeed acting by selectively inducing the nuclear export of an unspliced CAT mRNA that contains the K-RRE sequence.
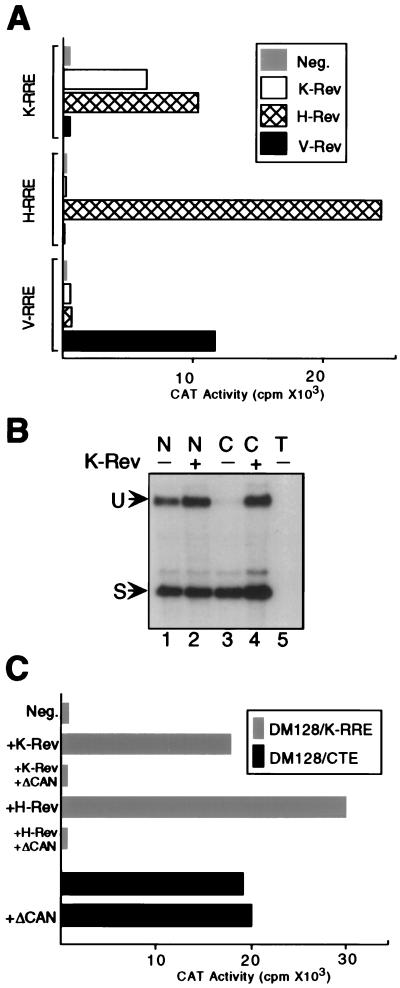
Figure 2.
K-Rev activates the cytoplasmic expression of an unspliced mRNA bearing the K-RRE. (A) Cells (293T) were cotransfected with a pDM128-based indicator construct bearing the K-RRE, H-RRE, or V-RRE, together with a pBC12/CMV-based expression vector encoding K-Rev, H-Rev, or V-Rev or with pBC12/CMV as a negative (Neg) control. pBC12/CMV/β-GAL was cotransfected as an internal control. At ≈48 h after transfection, cells were harvested, and induced CAT and β-GAL activities were determined. The presented data were corrected for minor variations in β-GAL activity and are representative of four independent experiments. (B) Cells (293T) were transfected with pDM128/K-RRE together with either pBC12/CMV/K-Rev (+) or the parental pBC12/CMV plasmid (−). At ≈48 h after transfection, nuclear (N) and cytoplasmic (C) RNA fractions were isolated and analyzed by RNase protection assay by using a previously described (9) 32P-labeled RNA probe that traverses the 3′ splice site. Lane 5 contains total (T) RNA from mock-transfected 293T cells. U, unspliced RNA; S, spliced RNA. (C) Cells (293T) were transfected with pDM128/K-RRE and pBC12/CMV (Neg), pBC12/CMV/K-Rev, or pBC12/CMV/H-Rev, as described for A, except that an additional 500 ng of either pBC12/CMV/ΔCAN or pBC12/CMV was cotransfected. The pDM128/CTE indicator plasmid contains the MPMV CTE. Induced CAT and β-GAL activities were determined as described for A. ΔCAN does not significantly affect expression of the β-GAL internal control (6). These data are representative of three independent experiments.
Because K-Rev, like H-Rev, binds to Crm1 (Table 1; Fig. 1A), we would predict that K-Rev-induced CAT mRNA expression should be blocked by expression of ΔCAN, a selective inhibitor of Crm1 function that does not affect Crm1-independent RNA export, such as the export of cellular mRNAs (6, 27, 31). As shown in Fig. 2C, coexpression of ΔCAN indeed entirely inhibited both K-Rev- and H-Rev-induced expression of the unspliced CAT mRNA encoded by pDM128/K-RRE. In contrast, ΔCAN had no effect on the expression of a pDM128-derived unspliced RNA containing the MPMV CTE, which is known to be exported via a Crm1-independent pathway (8–11). Therefore, we conclude that K-Rev, like H-Rev, indeed uses the Crm1 export factor to mediate nuclear RNA export.
Sequence-Specific RNA Binding by K-Rev.
H-Rev-dependent nuclear RNA export requires the direct interaction of multiple H-Rev molecules with the H-RRE (25, 30). To test whether K-Rev can bind the K-RRE specifically, we performed a gel retardation assay by using a recombinant GST-K-Rev fusion protein and 32P-labeled RNA probes encoding the K-RRE or the V-RRE (20). As shown in Fig. 3A, GST-K-Rev indeed proved able to bind the K-RRE, but not the V-RRE, specifically in vitro. The observation of at least two retarded bands may imply the binding of more than one GST-K-Rev molecule by the K-RRE. To confirm the specificity of the K-RRE/K-Rev interaction further, we also performed an in vitro competition experiment by using unlabeled K-RRE or H-RRE RNA (25). As shown in Fig. 3B, only the K-RRE competitor was able to inhibit the binding of K-Rev to the labeled K-RRE probe. Therefore, the ability of K-Rev to bind to a particular viral RRE in vitro (Fig. 3) correlates with the ability of K-Rev to activate the expression of an unspliced RNA containing that same RNA element in vivo (Fig. 2A). The ability of K-Rev to bind specifically to the K-RRE, but not to irrelevant RNA elements, has also been confirmed in vivo by using the yeast three-hybrid assay (data not shown).
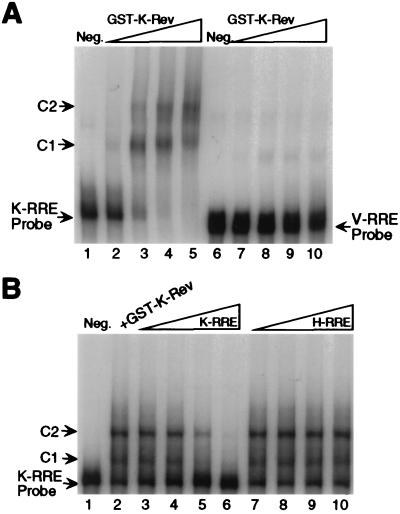
Figure 3.
Direct interaction between recombinant K-Rev and the K-RRE in vitro. (A) The RNA gel-shift procedure used has been described (25). All reactions contained 1 μg of yeast tRNA and 4 μg of rRNA as nonspecific competitors. A 32P-labeled 434-nt K-RRE probe or a 293-nt V-RRE probe was prepared by in vitro transcription, and ≈1 ng (4 × 104 cpm) of probe was then added to a premade in vitro reaction containing no (lanes 1 and 6, Neg.), ≈3 ng (lanes 2 and 7), ≈9 ng (lanes 3 and 8), ≈27 ng (lanes 4 and 9), or ≈81 ng (lanes 5 and 10) of GST-K-Rev. After incubation for 10 min at 4°C, reaction products were resolved on a 5% native polyacrylamide gel and visualized by autoradiography. C1 and C2 indicate distinct protein complexes. (B) The procedure was performed as described for A, except that the reaction mix, containing 9 ng of GST-K-Rev, was preincubated for 10 min at 4°C with an unlabeled K-RRE or H-RRE competitor RNA. (lane 1, Neg.) No added GST-K-Rev; (lane 2) no additional RNA competitor; (lanes 3 and 7) ≈3 ng of RNA competitor; (lanes 4 and 8) ≈12 ng of RNA competitor; (lanes 5 and 9) ≈50 ng of RNA competitor; (lanes 6 and 10) ≈200 ng of RNA competitor.
Discussion
In this manuscript, we show that at least some members of the HERV-K family of endogenous human retroviruses encode a nuclear RNA export factor that is mechanistically similar to the Rev protein found in HIV-1 and other lentiviruses. Specifically, K-Rev, like H-Rev, can bind to both the Crm1 nuclear export factor (Table 1; Fig. 1A) and to a cis-acting viral RNA response element (Fig. 3) and can thereby target unspliced RNAs containing this sequence for nuclear export (Fig. 2). In addition, K-Rev and H-Rev also share the ability to form homomultimers (Table 1). The functional similarity between H-Rev and K-Rev is underlined further by the observation that both proteins can induce the cytoplasmic expression of an mRNA bearing the K-RRE (Fig. 2 A and C). Nevertheless, we do not believe that K-Rev and H-Rev have the same RNA sequence specificity. Thus, K-Rev is not able to bind the H-RRE (Figs. 2A and 3B) and a preliminary analysis of the K-RRE has allowed the functional interaction with K-Rev or H-Rev to be mutationally segregated.
Although the ability of H-Rev to interact functionally with the K-RRE is therefore probably fortuitous, it does emphasize how similar K-Rev is to known lentiviral Rev proteins. This similarity—not only in terms of function, but also in terms of location within the retroviral genome—raises the possibility that lentiviral Rev proteins may have been captured originally by recombination with an ancient HERV-K like retrovirus. Alternatively, both viral families may have obtained their Rev genes independently by recombination with an as yet unidentified cellular Rev homolog. A third possibility (i.e., that lentiviruses evolved from a HERV-K like retrovirus) seems unlikely, given the very limited sequence homology that is observed (12, 15).
The ability of H-Rev to interact functionally with the K-RRE raises the possibility that HIV-1 infection might promote HERV-K structural protein expression in humans. Expression of HERV-K transcripts is detected readily in certain tumors, including teratocarcinomas and testicular cancers, and low levels are also observed in normal placenta and testicular tissue (12, 32). Although the expression of HERV-K RNAs in cells that are targets for HIV-1 infection, such as lymphocytes and macrophages, has not yet been examined, it is intriguing that up to 70% of HIV-1-positive patients were reported to express antibodies able to recognize HERV-K structural proteins, whereas little or no reactivity was observed among HIV-1-negative blood donors (12). Although this finding could reflect a low level of cross-reactivity between the structural proteins of HIV-1 and HERV-K, this result does raise the possibility that HIV-1 infection, and thus perhaps H-Rev expression, is indeed able to induce the expression of HERV-K structural proteins. How this might affect the pathogenic potential of HIV-1 is, of course, at present unclear.
We report the identification of a fully functional Rev-like nuclear RNA export factor encoded by the HERV-K family of HERVs. Our report describes a functional regulatory protein encoded by an endogenous retrovirus and the third example of a retrovirally encoded RNA export factor (the others being the lentiviral Rev proteins and the Rex proteins encoded by T-cell leukemia viruses; refs. 1 and 2). The observation of a Rev equivalent in endogenous retroviruses that date back ≥30 million years may imply that the acquisition of this activity occurred early in retroviral evolution.
Abbreviations
- β-GAL
β-galactosidase
- CAT
chloramphenicol acetyl transferase
- c-orf
central ORF
- CTE
constitutive transport element
- VMV
Visna Maedi virus
- GST
glutathione S-transferase
- HERV
human endogenous retrovirus
- H-Rev
HIV-1 Rev
- V-Rev
VMV Rev
- K-Rev
HERV-K Rev
- RRE
Rev response element
- H-RRE
HIV-1 RRE
- K-RRE
HERV-K RRE
- V-RRE
VMV RRE
- LTR
long terminal repeat
- MPMV
Mason–Pfizer monkey virus
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF179225).
References
- 1.Pollard V W, Malim M H. Annu Rev Microbiol. 1998;52:491–532. doi: 10.1146/annurev.micro.52.1.491. [DOI] [PubMed] [Google Scholar]
- 2.Cullen B R. Virology. 1998;249:203–210. doi: 10.1006/viro.1998.9331. [DOI] [PubMed] [Google Scholar]
- 3.Stade K, Ford C S, Guthrie C, Weis K. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- 4.Fornerod M, Ohno M, Yoshida M, Mattaj I W. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 5.Neville M, Stutz F, Lee L, Davis L I, Rosbash M. Curr Biol. 1997;7:767–775. doi: 10.1016/s0960-9822(06)00335-6. [DOI] [PubMed] [Google Scholar]
- 6.Bogerd H P, Echarri A, Ross T M, Cullen B R. J Virol. 1998;72:8627–8635. doi: 10.1128/jvi.72.11.8627-8635.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bray M, Prasad S, Dubay J W, Hunter E, Jeang K-T, Rekosh D, Hammarskjöld M-L. Proc Natl Acad Sci USA. 1994;91:1256–1260. doi: 10.1073/pnas.91.4.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grüter P, Tabernero C, von Kobbe C, Schmitt C, Saavedra C, Bachi A, Wilm M, Felber B K, Izaurralde E. Mol Cell. 1998;1:649–659. doi: 10.1016/s1097-2765(00)80065-9. [DOI] [PubMed] [Google Scholar]
- 9.Kang Y, Cullen B R. Genes Dev. 1999;13:1126–1139. doi: 10.1101/gad.13.9.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pasquinelli A E, Ernst R K, Lund E, Grimm C, Zapp M L, Rekosh D, Hammarskjöld M-L, Dahlberg J E. EMBO J. 1997;16:7500–7510. doi: 10.1093/emboj/16.24.7500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saavedra C, Felber B, Izaurralde E. Curr Biol. 1997;7:619–628. doi: 10.1016/s0960-9822(06)00288-0. [DOI] [PubMed] [Google Scholar]
- 12.Löwer R, Löwer J, Kurth R. Proc Natl Acad Sci USA. 1996;93:5177–5184. doi: 10.1073/pnas.93.11.5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patience C, Wilkinson D A, Weiss R A. Trends Genet. 1997;13:116–120. doi: 10.1016/s0168-9525(97)01057-3. [DOI] [PubMed] [Google Scholar]
- 14.Medstrand P, Mager D L. J Virol. 1998;72:9782–9787. doi: 10.1128/jvi.72.12.9782-9787.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Griffiths D J, Venables P J W, Weiss R A, Boyd M T. J Virol. 1997;71:2866–2872. doi: 10.1128/jvi.71.4.2866-2872.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Löwer R, Boller K, Hasenmaier B, Korbmacher C, Müller-Lantzsch N, Löwer J, Kurth R. Proc Natl Acad Sci USA. 1993;90:4480–4484. doi: 10.1073/pnas.90.10.4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Löwer R, Tönjes R R, Korbmacher C, Kurth R, Löwer J. J Virol. 1995;69:141–149. doi: 10.1128/jvi.69.1.141-149.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malim M H, Böhnlein S, Hauber J, Cullen B R. Cell. 1989;58:205–214. doi: 10.1016/0092-8674(89)90416-9. [DOI] [PubMed] [Google Scholar]
- 19.Hauber J, Malim M H, Cullen B R. J Virol. 1989;63:1181–1187. doi: 10.1128/jvi.63.3.1181-1187.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tiley L S, Cullen B R. J Virol. 1992;66:3609–3615. doi: 10.1128/jvi.66.6.3609-3615.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sadowski I, Ptashne M. Nucleic Acids Res. 1989;17:7539. doi: 10.1093/nar/17.18.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fridell R A, Partin K M, Carpenter S, Cullen B R. J Virol. 1993;67:7317–7323. doi: 10.1128/jvi.67.12.7317-7323.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hope T J, Huang X, McDonald D, Parslow T G. Proc Natl Acad Sci USA. 1990;87:7787–7791. doi: 10.1073/pnas.87.19.7787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ono M, Yasunaga Y, Miyata T, Ushikubo H. J Virol. 1986;60:589–598. doi: 10.1128/jvi.60.2.589-598.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malim M H, Cullen B R. Cell. 1991;65:241–248. doi: 10.1016/0092-8674(91)90158-u. [DOI] [PubMed] [Google Scholar]
- 26.Southgate C D, Green M R. Genes Dev. 1991;5:2496–2507. doi: 10.1101/gad.5.12b.2496. [DOI] [PubMed] [Google Scholar]
- 27.Fornerod M, van Deursen J, van Baal S, Reynolds A, Davis D, Murti K G, Fransen J, Grosveld G. EMBO J. 1997;16:807–816. doi: 10.1093/emboj/16.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Truant R, Cullen B R. Mol Cell Biol. 1999;19:1210–1217. doi: 10.1128/mcb.19.2.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bischoff F R, Klebe C, Kretschmer J, Wittinghofer A, Ponstingl H. Proc Natl Acad Sci USA. 1994;91:2587–2591. doi: 10.1073/pnas.91.7.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zapp M L, Hope T J, Parslow T G, Green M R. Proc Natl Acad Sci USA. 1991;88:7734–7738. doi: 10.1073/pnas.88.17.7734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zolotukhin A S, Felber B K. J Virol. 1999;73:120–127. doi: 10.1128/jvi.73.1.120-127.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sauter M, Schommer S, Kremmer E, Remberger K, Dölken G, Lemm I, Buck M, Best B, Neumann-Haefelin D, Mueller-Lantzsch N. J Virol. 1995;69:414–421. doi: 10.1128/jvi.69.1.414-421.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]