Abstract
We argue that the large-dimensional dynamical systems which frequently occur in biological models can sometimes be effectively reduced to much smaller ones. We illustrate this by applying projection operator techniques to a mean-field model of an infectious disease spreading through a population of households. In this way, we are able to accurately approximate the dynamics of the system in terms of a few key quantities greatly reducing the number of equations required. We investigate linear stability in this framework and find a new way of calculating the familiar threshold criterion for household systems.
Keywords: model reduction, projection operators, household epidemics
1. Introduction
In mathematical models of biological dynamics, the state space can easily become large, causing a proliferation in the number of equations needed. This can make models difficult to deal with and insight unlikely. Usually, one is interested only in the behaviour of a few special quantities describing the system. It would be useful if one could find an approximate description of such systems only in terms of these quantities of interest.
Projection operator techniques (e.g. Grabert 1982; Rau and Müller 1996) were developed in the context of statistical mechanics for extracting the dynamics of coarse-grained thermodynamic variables, given the underlying laws on a much larger state space. From a dynamical system governing the vectors X and Y,
one can write down a dynamical law for the vector X which refers only to X itself, at the price of acquiring history dependence (e.g. Givon et al. 2004),
| (1.1) |
The rewrite equation (1.1) is no more tractable than the original problem, but in situations where this approach works, the function is relatively simple and is thought of as dominating the dynamics. The function N, which depends only on time and the initial conditions, is called the noise term. This is typically rapidly decaying and is often neglected altogether. The integral part of equation (1.1) is known as the memory term and is often well approximated by a kernel function integrated against some function of X. Typically, the ‘memory’ decays rapidly, and the first port of call is a Markovian approximation, with the kernel approximated as proportional to a δ function.
We apply this approach to a susceptible–infected–recovered (SIR) model which describes the spread of an infectious disease through a population of smaller subgroups. Any individual can infect any other, but those in the same group have an enhanced hazard of doing so. Such multilevel mixing models have been much studied recently with household structure in mind (e.g. Ball et al. 1997; Becker et al. 2003; Ball & Becker 2006). Results have centred around final epidemic sizes, threshold behaviour and static vaccination policies. We show that the dynamics can be rewritten in the form
| (1.2) |
where Sx and Ix are the total number of susceptibles and infecteds, respectively, at time x; N is the noise function as above; and k is the kernel which we will go on to approximate. The advantage is that while the number of equations needed to describe this system increases like n2 (where n is the subgroup size), this formulation only ever needs two equations. Indeed, if the population consists of subgroups of different sizes, then the number of equations increases like the maximum group size cubed. These two equations are those of the usual freely mixing SIR model, but with corrections which account for the underlying inhomogeneity of mixing.
We go on to analyse linear stability about the disease-free equilibrium and obtain a stability discriminant which can be identified with the household reproduction number, R*. This R* is loosely defined as the expected number of houses infected by one singly infected household and determines whether a disease can spread or not, according to whether it is greater or less than unity.
2. Basic model
Let yij be the number of households which contain i susceptible individuals, j infecteds and, by implication, n−i−j recovered individuals. Our dynamical law is specified by
| (2.1) |
where is the total number of infecteds in the population; β is the global infection strength; ν is the recovery rate; and δ is the difference between the local (within-group) infection strength and β. Where i and j imply a negative number of susceptible, infected or recovered, the corresponding y is taken to be zero. This can be thought of as the mean-field version of the master equation, which is exact in the infinite population limit, or as a deterministic starting point in its own right (though a deterministic model could be valid only for large total populations).
Taking a linear combination of these equations, we obtain the rate laws for the total number of infecteds and the total number of susceptibles, ,
| (2.2) |
where v is the vector whose elements are given by vij=ij. When the difference between local and global infection strengths, δ, vanishes, the group structure dissolves and we are left with the usual mass action SIR equations. We will choose to eliminate yn,0 and y0,n in favour of S and I, using their definitions. Henceforth, the vector y is understood with these two elements deleted. If C is the vector with all elements zero except Cn−1,1=1, the dynamics of y take the form
which can be formally solved with a matrix integrating factor U(t, s), such that U(t, t)=1 and dU/dt=−ΛU, to give
| (2.3) |
This brings equations (2.2) into the advertised form of equation (1.2), with the kernel given by k=vTUC. We will restrict to initial conditions only with singly infected households, for which N(t)∝k(t, 0). The equations (2.2) are exactly equivalent to the full set of equations in equation (2.1), but are still difficult to compute because the time dependence of Λ leads U to depend on repeated time-integrals of I. We introduce an approximation (A1), equivalent to assuming that the part of Λ proportional to ν commutes with the rest. This simplifies things significantly because vT turns out to be a right eigenvector of both the ν-part of Λ and the part proportional to I. The upshot is that one may approximate the kernel of equation (1.2) by
where is the cumulative number of infections (equal to R/ν in the case without vaccination). More details can be found in the electronic supplementary material. The functions, fn, are determined in a standard way by solving a linear set of constant coefficient differential equations. These differential equations are equivalent to the master equation of the SI model, and a general solution is given in Bailey (1975). For n=1–5, we have
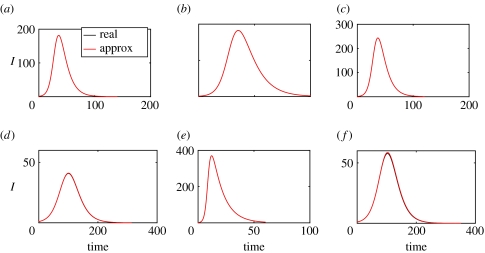
Representative performance of approximation (A1) is depicted in figure 1. More details on the error behaviour can be found in the supplementary information. In Bailey (1975), an asymptotic solution for large n is also given, which could be adapted to provide a convenient large-n asymptotic approximation to fn.
Figure 1.
Comparison of approximate (red) and real (black) epidemic curves for populations of m houses of size n. Cases have (n, δ/β, Rg=βS0/ν, m) given by: (a) (2,2,3,300); (b) (3,2,3,200); (c) (4,2,3,200); (d) (2,8,1.5,300); (e) (3,8,1.5,200) and (f) (4,8,1.5,200). m is chosen so as to accentuate errors.
3. Linear stability and thresholds
To study the linear stability of the disease-free equilibrium, one considers the linearized version of the differential equation for I given by equations (2.2) and (2.3). Not all elements of y are dynamical at linear order in I. Let L denote Λ with I=0, and the rows and columns referring to the non-dynamical elements of y excised. The matrix L is invertible and a linear function of ν and δ. To determine stability, one Laplace transforms the linearized dynamics and asks whether the solution's transform has any poles with positive real part. In the electronic supplementary material, we show that such poles are absent (implying stability), if
(where Rg=βS0/ν is the reproductive ratio for only global infections and S0 is the initial number of susceptibles) and that if this inequality is reversed, the system is unstable. The quantity can be identified with the household reproduction ratio R* and provides a single compact formula which is easy to implement. Given this, one can compare the threshold behaviour of the full system and the system under approximation (A1) for different ν and δ, and see that their thresholds are very similar. The case n=2 is degenerate, with the full and approximate systems sharing the same threshold. For concreteness, the n=2 and n=3 cases yield (for more details, see the electronic supplementary material)
4. Distributions of household sizes
More realistically, one can consider a population of households of different sizes. Let yi,j;n be the number of houses of size n with i susceptibles and j infecteds. If there is no death, there is no mechanism to change the total number of individuals in a household, and the governing equations form parallel sets with the same structure as equation (2.1), but with global infections at each n still forced by the total number of infecteds, I. In addition to approximation (A1), we will make the approximation (A2) that Sn, the number of susceptibles residing in houses of size n, is a constant fraction of the total number of susceptibles, i.e.
| (3.1) |
While not particularly accurate in its own right, this approximation does allow us to obtain a good approximation to the dynamics, once again in the form equation (1.2), where now the noise and kernel are given by
with in being the number of houses of size n singly infected at t=0. We note that one could easily extend this procedure to the case where different household sizes have a different within-house infection strength, δn. Such variability has been shown to give a better fit to the real data for influenza (Cauchemez 2004).
5. Control measures
Many globally applied control measures can be represented as a change in transmission parameters β and δ, brought about, for example, by social distancing measures or mass prophylaxis. Most simply, β and/or δ become functions of time. More realistically, they could be modelled as being functions of I or J, if a response is initiated by a threshold level of infections. None of these possibilities spoils the structure which has allowed us develop our approximate dynamics. Instead of βJ, one would have ; and the functions fn would be computed from differential equations with time-varying coefficients.
One might also be interested in interventions which are household-based responses to infection: interventions, such as vaccination, applied to a household on finding an infection there. Let us assume that an intervention transfers the remaining susceptible individuals of a house directly to the removed class and is applied after detection of the index case (or not at all). In terms of the y variables, one can represent this scenario by the addition of −κyn−1,1 to the yn−1,1 rate equation, and κyn−1,1 to the y0,n rate equation. The new parameter κ contains information about the coverage of the measure, and the rate with which it is applied. The cumulative number of cases J and R/ν now need to be distinguished. One can proceed very much as before, but with two kernels—one proportional to δ and another proportional to κ—which appear asymmetrically in the and equations (both in the S equation; but only the δ kernel in the I equation; more details are provided in the supplementary information). One can again approximate the dynamics, so it depends only on S, I and J, though the accuracy is worse than for the simple case.
6. Discussion
In this paper, we have often used the word ‘household’. This should not be taken too literally—our approach is particularly convenient for large subgroup sizes n, such as might be appropriate to the description of animal populations. Equations of the form (1.2) can then be efficiently solved by the analogues of Runge–Kutta techniques (Lubich 1982), and one avoids dealing with the n2 or n3 equations of the conventional approach.
Projection operator techniques should be applicable to a wide range of biological problems beyond the example of this paper. In their full generality, they can produce equations of motion of the form (1.1) for mean variables of stochastic systems. Their use requires an insightful choice of relevant variables, and some effort in setting up the formalism for the problem of interest; but they can provide a completely fresh starting point for approximations and a substantial gain in simplicity.
Acknowledgments
This work was supported by the UK BBSRC. The authors would like to thank Christophe Fraser and Lorenzo Pellis for their useful conversations.
Supplementary Material
This provides more details of the calculations, and further discussion of accuracy and control measures
References
- Bailey N.T.J. Charles Griffin and Co; London, UK: 1975. The mathematical theory of infectious diseases and its applications. [Google Scholar]
- Ball F.G, Becker N.G. Control of transmission with two types of infection. Math. Biosci. 2006;200:170–187. doi: 10.1016/j.mbs.2005.12.024. [DOI] [PubMed] [Google Scholar]
- Ball F.G, Mollison D, Scalia-Tomba G. Epidemics with two levels of mixing. Ann. Appl. Probab. 1997;7:46–89. doi: 10.1214/aoap/1034625252. [DOI] [Google Scholar]
- Becker N.G, Britton T, O'Neill P.D. Estimating vaccine effects on transmission of infection from household outbreak data. Biometrics. 2003;59:467–475. doi: 10.1111/1541-0420.00056. [DOI] [PubMed] [Google Scholar]
- Cauchemez S, et al. A Bayesian MCMC approach to study transmission of influenza: application to household longitudinal data. Stat. Med. 2004;23:3469–3487. doi: 10.1002/sim.1912. [DOI] [PubMed] [Google Scholar]
- Givon D, Kupferman R, Stuart A.M. Extracting macroscopic dynamics: model problems and algorithms. Nonlinearity. 2004;17:R55–R127. doi: 10.1088/0951-7715/17/6/R01. [DOI] [Google Scholar]
- Grabert H. Springer; Berlin, Germany: 1982. Projection operator techniques in nonequilibrium statistical mechanics. [Google Scholar]
- Lubich Ch. Runge–Kutta theory of volterra integrodifferential equations. Numerische Mathematik. 1982;40:119–135. doi: 10.1007/BF01459081. [DOI] [Google Scholar]
- Rau J, Müller B. From reversible quantum microdynamics to irreversible quantum transport. Phys. Rept. 1996;272:1–59. doi: 10.1016/0370-1573(95)00077-1. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This provides more details of the calculations, and further discussion of accuracy and control measures