Abstract
Krüppel-like transcription factors (Klfs) modulate fundamental cell processes. Cardiac myocytes are terminally-differentiated, but hypertrophy in response to stimuli such as endothelin-1. H2O2 or cytokines promote myocyte apoptosis. Microarray studies of neonatal rat myocytes identified several Klfs as endothelin-1-responsive genes. We used quantitative PCR for further analysis of Klf expression in neonatal rat myocytes. In response to endothelin-1, Klf2 mRNA expression was rapidly increased (∼ 9-fold; 15–30 min) with later increases in expression of Klf4 and Klf6 (∼ 5-fold; 30–60 min). All were regulated as immediate early genes (cycloheximide did not inhibit the increases in expression). Klf5 expression was increased at 1–2 h (∼ 13-fold) as a second phase response (cycloheximide inhibited the increase). These increases were transient and attenuated by U0126. H2O2 increased expression of Klf2, Klf4 and Klf6, but interleukin-1β or tumor necrosis factor α downregulated Klf2 expression with no effect on Klf4 or Klf6. Of the Klfs which repress transcription, endothelin-1 rapidly downregulated expression of Klf3, Klf11 and Klf15. The dynamic regulation of expression of multiple Klf family members in cardiac myocytes suggests that, as a family, they are actively involved in regulating phenotypic responses (hypertrophy and apoptosis) to extracellular stimuli.
Keywords: Cardiac myocytes, Endothelin-1, Immediate early genes, Krüppel-like factors, Gene expression, Cytokines
1. Introduction
C2H2 zinc finger transcription factors possess two cysteine and two histidine residues which co-ordinate Zn2+ within each finger to form a conserved DNA-binding structure. Sp1-like proteins and Krüppel-like factors (Klfs) each have three C2H2 zinc fingers at their C-termini, and Klfs are distinguished by a highly conserved interfinger space sequence [1]. The seventeen mammalian Klfs which have been identified are now designated Klf1-17 [2,3], though many have alternative names. Several were named according to the tissues in which they were originally shown to be enriched (e.g. Klf1 is erythroid Eklf, Klf2 is lung Lklf, Klf4 is gut Gklf, Klf5 is intestinal Iklf and Klf15 is kidney Kklf [2]). Other Klfs are widely expressed including Klf3 (basic Bklf), Klf6 and Klf7 (ubiquitous Uklf). Klf9 was identified as a basal transcription element binding (BTEB) protein, with Klf5 and Klf13 as homologues (BTEB2 and BTEB3, respectively). Klf10 and Klf11 were identified as early genes induced by transforming growth factor β and were named TIEG and TIEG2, respectively.
Klfs bind to consensus GC-rich or CACCC sequences through the three C-terminal zinc fingers [1]. The frequency of such sequences in gene promoters, coupled with the high sequence identity between Klf proteins in their DNA-binding domains, raises questions relating to the specificity of any Klf for a particular promoter and potential functional redundancy. Nevertheless, Klfs can be clustered according to whether they act primarily as transcriptional repressors and/or activators, and the domain structure of the N-terminal regulatory regions. Klf3, Klf8 and Klf12 recruit C-terminal binding proteins (CtBPs) to repress transcription, whereas Klf9, Klf10, Klf11, Klf13 and Klf16 repress transcription through interaction with mSIN3a [1,4]. Klf15 also represses transcription, though the mechanism is not clear. Klf2, Klf4, Klf5, Klf6, and Klf7 are largely transcriptional activators [1], but may suppress gene expression in specific circumstances (e.g. Klf5 negatively regulates expression of Klf4 [5]). In addition to interactions with other proteins, post-translational modifications (e.g. phosphorylation or acetylation) regulate the transactivating activities of different Klfs [1,6]. Globally, Klfs regulate fundamental cellular responses such as growth, apoptosis, angiogenesis and proliferation. For example, Klf5 overexpression is associated with cell proliferation, whereas Klf2, Klf4 and Klf6 are more consistently implicated in cell cycle arrest [7–9]. Klf4 and Klf5 may both promote apoptosis induced by oxidative stress though the mechanism is unclear [9].
Mammalian cardiac myocytes (the contractile cells of the heart) become terminally-differentiated shortly after birth. Subsequent growth of the heart results from an increase in size of individual cardiac myocytes and, in the adult, myocytes may hypertrophy in order to maintain or increase cardiac output. This is associated with physiological and morphological changes (increases in cell size and myofibrillogenesis) and changes in gene expression including increased expression of immediate early genes (IEGs), and re-expression of genes normally expressed in early development [10]. Stimuli such as endothelin-1 (ET-1), which activate Gq protein-coupled receptors are particularly implicated in cardiac myocyte hypertrophy, and promote both the physiological/morphological changes and the changes in gene expression associated with the response [11]. In contrast, oxidative stresses or pro-inflammatory cytokines may induce cardiac myocyte apoptosis [12]. Many studies have examined the intracellular signalling pathways which are activated by various stimuli in cardiac myocytes, and these are presumed to lead to changes in gene and protein expression to promote hypertrophy or to facilitate myocyte death [13].
Many Klfs are expressed in adult hearts to a degree although the cell types in which they are expressed cannot be ascertained. For example, Klf2 is associated with endothelial cells [14,15], and cardiac endothelial cells could account for expression of Klf2 in whole heart extracts [16]. As reviewed by Haldar et al. [17], Klf13 and Klf15 are the only Klfs which have so far been shown to play a role in cardiac myocytes. Klf13 is highly expressed in adult hearts [18] and is required for normal cardiac development in Xenopus [19]. Klf15 is highly expressed in adult hearts and in cardiac myocytes [20,21]. It appears to be anti-hypertrophic since it is downregulated during hypertrophy, and overexpression of Klf15 in cardiac myocytes suppresses the morphological changes and changes in gene expression induced by hypertrophic stimuli [21]. Our microarray studies highlight the dynamic changes in expression of many genes (including Klfs) induced by H2O2 (an example of oxidative stress) or ET-1 in cardiac myocytes over 4 h [22–25]. Here, we explore further the regulation of Klf family members in cardiac myocytes in response to ET-1, H2O2 and pro-inflammatory cytokines [interleukin 1β (IL-1β) and tumour necrosis factor α (TNFα)]. The dynamic regulation of expression of multiple Klf family members suggests that, as a family, they are actively involved in regulating phenotypic responses of cells to extracellular stimuli.
2. Materials and methods
2.1. Primary culture of neonatal rat cardiac myocytes and preparation of polysomes
Myocytes were dissociated from the ventricles of 2- to 4-day-old Sprague‑Dawley rat hearts by an adaptation of the method of Iwaki et al. [26] as previously described [27]. Cells were plated at 4 × 106 cells/60 mm Primaria culture dish (BD Biosciences) for 18 h in 15% (v/v) foetal calf serum, then serum was withdrawn for 24 h before experimentation. Myocytes were exposed to ET-1 (100 nM), H2O2 (0.2 mM), IL-1β (100 ng/ml) or TNFα (10 ng/ml) with or without pre-treatment (10 min) with inhibitors (10 μM U0126; 50 μM LY294002; 5 μM SB203580). Cardiac myocyte polysomes were prepared by sucrose density centrifugation (0.8–1.6 M sucrose gradients; 5 ml) as previously described [25]. Fractions were collected by upward displacement (fraction 1 from the top of the gradient, 0.8 M sucrose; fraction 12 from the bottom of the gradient, 1.6 M sucrose) whilst monitoring absorbance at 254 nm.
2.2. RNA preparation and quantitative PCR (qPCR)
Total RNA and polysome RNA (fractions 6–11 of the sucrose gradients) were extracted and cDNA synthesized using reverse transcription as previously described [22,25]. qPCR was performed using a Real‑Time PCR System (Model 7500, Applied Biosystems) as described [25] using primers designed to amplify mRNA sequences across an intron (Table 1). qPCR analysis of glyceraldehyde 3-phosphate dehydrogenase (Gapdh) was performed as a control and the relative quantification protocol was used. PCR conditions were 50 °C for 2 min, 95 °C for 10 min (Jump-Start Taq polymerase activation step), followed by 40 cycles of 95 °C for 15 s and 59 °C for 60 s. Following qPCR, dissociation curve analysis was routinely performed to check for aberrant amplification products (e.g. primer‑dimers).
Table 1.
Primers used for QPCR validation of microarray data
| Gene | Accession no. | Size (bp) | Forward primer | Reverse primer |
|---|---|---|---|---|
| Gapdh | NM_017008 | 83 | GCTGGCATTGCTCTCAATGACA (1738–1759) | TCCACCACCCTGTTGCTGTA (1801–1820) |
| Il1rl1 | NM_013037 | 84 | GTCTCAAGAGATCGTCTGAAG (418–438) | CGATTCAGGGCTTCTGATAAC (481–501) |
| Irs2 | XM_573948 | 107 | CACCTACGCAAGCATCGACT (3921–3940) | GATTCAGAGTCTTCGACGAG (4008–4027) |
| Klf2 | NM_001007684 | 173 | ACTTGCAGCTACACCAACTG (805–824) | CTGTGACCCGTGTGCTTG (960–977) |
| Klf3 | NM_001105742 | 165 | TCATGTACACCAGCCACCTG (691–710) | TAGTCAGTCCTCTGTGGTTC (837–856) |
| Klf4 | NM_053713 | 175 | TCAAGAGCTCATGCCACCGG (1180–1199) | CTCGCCTGTGTGAGTTCGCA (1335–1354) |
| Klf5 | NM_053394 | 212 | AGCTCACCTGAGGACTCATA (1437–1456) | GTGCGCAGTGCTCAGTTCT (1631–1649) |
| Klf6 | NM_031642 | 126 | CCTTACAGATGCTCTTGGGA (854–873) | GGAGAAACACCTGTCACAGT (960–979) |
| Klf9 | NM_057211 | 124 | TGGCTGTGGGAAAGTCTATGG (954–974) | CTCGTCCGAGCGCGAGAACT (1058–1077) |
| Klf10 | NM_031135 | 201 | TACTGATGTCTTCACCTACAG (1079–1099) | GTACCACAAACATGACCGTG (1260–1279) |
| Klf11 | NM_001037354 | 165 | AACAGAATCACCTTAGCAGAG (1499–1519) | AACAGCCCAGAGACCATGG (1645–1663) |
| Klf15 | NM_053536 | 164 | GATGAGTTGTCACGGCACC (1371–1389) | CACTGCGCTCAGTTGATGG (1516–1534) |
Nucleotide positions in transcripts are shown in parentheses for each primer. mRNA sequences (accession numbers provided) for established genes were obtained from the Rat Genome Database (http://rgb.mcw.edu, viewed at http://www.ncbi.nlm.nih.gov/entrez).
2.3. Western blotting
Cardiac myocyte nuclear extracts were prepared and Western blotting performed essentially as described [28]. Extracts from 1.8 × 106 cells were analysed with separation of proteins on 10% (v/v) polyacrylamide gels. Nitrocellulose blots were probed with Klf6 rabbit polyclonal antibodies (Santa Cruz Biotechnology Inc.; Klf6(R-173), sc-7158, 1/500 dilution). Bands were detected and analysed by scanning densitometry as described [29].
3. Results
3.1. ET-1 regulates expression of multiple klf family members in cardiac myocytes
Our microarray studies (with Affymetrix rat genome 230 2.0 arrays) of the effects of ET-1 on cardiac myocyte gene expression over 4 h [25] identified Klfs as a family of transcription factors which appeared to be dynamically regulated at the mRNA level. Mining the data specifically for Klfs, we identified rapid and transient increases in expression of Klf2, Klf4, Klf5, Klf6, Klf9 and Klf10, with concomitant downregulation of Klf3, Klf11 and Klf15 (Table 2). There was no statistically significant change in expression of Klf13 or Klf16, although Klf13 exhibited a small (non-significant; one-way ANOVA with Tukey post-test) increase in expression over 2– 4 h. Klf1 (probeset 1382033_at), Klf7 (probesets 1380363_at, 1377618_at, 1384497_at) and Klf12 (probeset 1385545_at) were not consistently called “present” with low fluorescence values and are thus expressed at minimal levels. We could not identify Klf8, Klf14 (Sp6) or Klf17 on the arrays.
Table 2.
Regulation of Klf isoform expression by ET-1 in cardiac myocytes (microarray analysis)
| Gene symbol | Probeset ID | Time (h) |
|||
|---|---|---|---|---|---|
| 0.5 | 1 | 2 | 4 | ||
| Significantly changed | |||||
| Klf2(p) | 1376569_at, 1386041_a_at, 1394068_x_at | 3.50 | 3.22 | 0.98 | 1.20 |
| Klf3 | 1378332_at, 1389479_at | 0.88 | 0.65 | 0.60 | 1.08 |
| Klf4 | 1387260_at | 1.98 | 3.60 | 2.69 | 1.79 |
| Klf5 | 1368363_at, 1394039_at | 1.02 | 2.68 | 4.61 | 1.74 |
| Klf6 | 1387060_at, 1388986_at, 1395557_at | 2.36 | 4.25 | 2.05 | 1.71 |
| Klf9 | 1370209_at, 1371864_at | 1.03 | 1.22 | 1.61 | 1.27 |
| Klf10 | 1368650_at | 0.77 | 0.85 | 2.14 | 1.50 |
| Klf11 | 1379914_at | 0.89 | 0.63 | 0.56 | 0.48 |
| Klf15 | 1368249_at, 1381396_s_at | 0.86 | 0.53 | 0.27 | 0.45 |
| Not significantly changed | |||||
| Klf13 | 1375177_at, 1383013_at | 1.07 | 1.10 | 1.58 | 1.67 |
| Klf16 | 1374231_at | 1.06 | 1.12 | 1.47 | 1.11 |
Cardiac myocytes were exposed to 100 nM endothelin-1 for the times indicated. RNA was extracted and gene expression profiling was performed using Affymetrix Rat Genome 230 2.0 arrays. Results are the mean expression relative to controls for n = 3 (0.5 or 4 h), 6 (2 h) or 8 (1 h) separate sets of samples. For multiple probesets, the average of the values is given. (p), predicted gene. Klf1, Klf7 and Klf12 were not expressed at any significant level in cardiac myocytes. Klf8 and Klf17 were not identified on the rat microarrays. Klf14 is now classified as Sp6 [1], but we could not identify this on the rat microarrays.
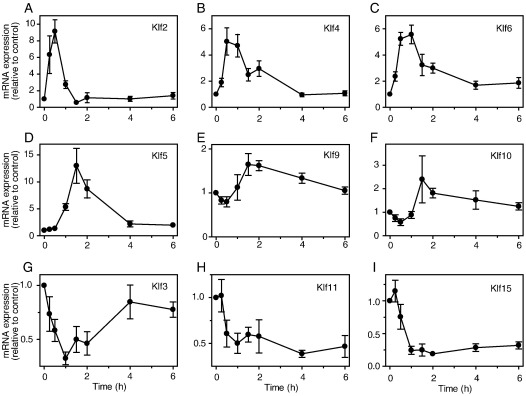
Using qPCR, we examined more carefully the effects of 100 nM ET-1 on Klf mRNA expression. Klf2 mRNA expression was significantly increased within 15 min, with maximal expression (∼ 9-fold relative to controls) at 30 min (Fig. 1A). Klf4 and Klf6 mRNAs were also upregulated (Fig. 1, B and C), though the relative stimulation was less (∼ 5-fold relative to controls) and the response was delayed relative to Klf2 (maximal expression at 0.5–1 h). Klf5 mRNA was upregulated from 1 h, with maximal expression at 1.5 h (∼ 13-fold relative to controls) (Fig. 1D). For these Klfs, the increase in expression was transient, with expression of Klf2 returning to baseline within 90 min and expression of Klf4, Klf5 and Klf6 declining to basal levels within 4 h. The increases in expression of Klf9 and Klf10 were relatively small (maximal expression < 2-fold at 1.5–2 h) although levels remained elevated over 4–6 h (Fig. 1, E and F). Consistent with our microarray data, Klf3, Klf11 and Klf15 were downregulated in response to ET-1. Whereas the decrease in expression of Klf3 was transient with minimal expression at ∼ 30 min (Fig. 1G), the decrease in expression of Klf11 and Klf15 was sustained over at least 6 h (Fig. 1, H and I). It is notable that the Klfs which were downregulated and those with only a small increase in expression are largely associated with transcriptional repression, whereas the Klfs with the greatest increases in expression (Klf2, Klf4, Klf5 and Klf6) are mostly associated with transcriptional activation [1,4,6]. Further studies focused on this latter group.
Fig. 1.
Regulation of Klf expression by ET-1. Cardiac myocytes were exposed to 100 nM ET-1 for the times indicated. RNA was extracted and expression of mRNAs for different Klf family members (A, Klf2; B, Klf4; C, Klf6; D, Klf5; E, Klf9; F, Klf10; G, Klf3; H, Klf11; I, Klf15) analysed by qPCR. Results are expressed relative to unstimulated controls and are means ± S.E.M. for at least 4 independent preparations of myocytes.
3.2. Regulation of Klf2, Klf4, Klf5 and Klf6 by ET-1
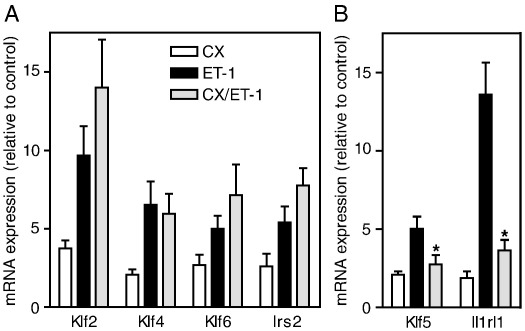
To determine whether Klf2, Klf4, Klf5 and Klf6 mRNAs were upregulated as IEGs (i.e. the increase in expression is regulated by pre-existing transcription factors and does not require synthesis of new proteins), we examined the effects of 20 μM cycloheximide (to inhibit protein synthesis). Cycloheximide alone increased the expression of Klf2, Klf4 and Klf6 mRNAs (Fig. 2A; probably due to activation of stress-activated signalling pathways [30,31]), but did not inhibit the increase in expression induced by ET-1. However, the increase in expression of Klf5 by ET-1 was inhibited by cycloheximide (Fig. 2B), indicating that de novo protein synthesis is required for upregulation of Klf5 mRNA. Thus, Klf2, Klf4 and Klf6 are regulated as IEGs in response to ET-1, whereas Klf5 is a second phase gene.
Fig. 2.
Klf2, Klf4 and Klf6, but not Klf5, are upregulated as immediate early genes by ET-1. Cardiac myocytes were unstimulated or exposed to 20 μM cycloheximide (CX, open bars), 100 nM ET-1 (black bars), or ET-1 in the presence of cycloheximide (grey bars) for 0.5 h (Klf2, Klf4, Klf6) or 1 h (Irs2, Klf5, Il1rl1). RNA was extracted and expression of mRNAs for Klf2, Klf4, Klf6 and Irs2 (A, immediate early genes) or Klf5 and Il1rl1 (B, second phase genes) analysed by qPCR. Results are expressed relative to unstimulated controls and are means ± S.E.M. for 3 or 4 independent preparations of myocytes. ⁎p < 0.01 relative to ET-1 alone (one-way ANOVA repeated measures with TUKEY post-test).
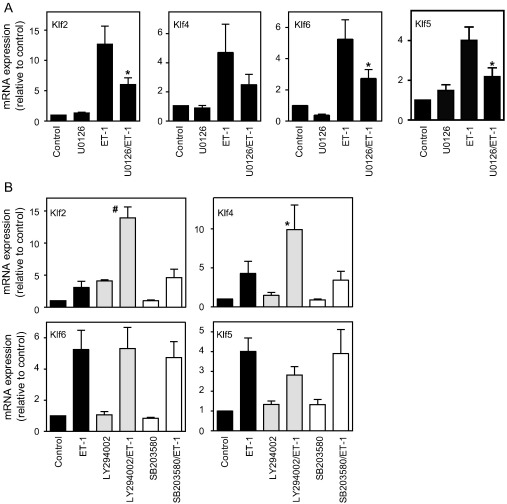
ET-1 potently activates the extracellular signal-regulated kinases 1/2 (ERK1/2) in cardiac myocytes [32]. We used 10 μM U0126 (a selective inhibitor of the ERK1/2/5 cascades [33–35], though ERK5 is not significantly activated by ET-1 [24]) to determine whether the increase in expression of Klf2, Klf4, Klf5 and Klf6 requires ERK1/2 signalling. U0126 minimally affected basal expression of each Klf, but partially inhibited the ET-1-induced increase in expression of Klf2 (56 ± 4% inhibition at 30 min; mean ± SEM, n = 5), Klf4 (43 ± 9% inhibition at 1 h; n = 4), Klf5 (72 ± 8% inhibition at 1 h; n = 4) and Klf6 (45 ± 8% inhibition at 1 h; n = 4) (Fig. 3A). These data suggest that ERK1/2 signalling is required, at least in part, for the upregulation of Klf2, Klf4, Klf5 and Klf6 mRNAs by ET-1.
Fig. 3.
Upregulation of Klf2, Klf4, Klf6 and Klf5 by ET-1 is mediated in part through the ERK1/2 cascade. Cardiac myocytes were unstimulated (Control), or exposed to inhibitors alone (10 μM U0126, 50 μM LY294002, 5 μM SB203580), to 100 nM ET-1 alone or to ET-1 in the presence of each inhibitor for 0.5 or 1 h. RNA was extracted and expression of mRNAs for Klf2, Klf4, Klf6 or Klf5 analysed by qPCR. A, Effects of U0126 on expression of Klf mRNAs at 0.5 (Klf2) or 1 h (Klf4, Klf5, Klf6). B, Effects of LY294002 or SB203580 on expression of Klf mRNAs at 1 h. Results are expressed relative to unstimulated controls and are means ± S.E.M. for 4 or 5 independent preparations of myocytes. ⁎p < 0.05, #p < 0.001 relative to ET-1 alone (one-way ANOVA repeated measures with TUKEY post-test).
Signalling through phosphoinositide 3′ kinase (PI3K) may increase expression of Klf2 [36], and p38-MAPK is activated by ET-1 in cardiac myocytes [37], so we examined the effects of 50 μM LY294002 or 5 μM SB203580 (selective inhibitors of PI3K and p38-MAPK, respectively [34]). Surprisingly, LY294002 increased basal expression of Klf2 and Klf4 in cardiac myocytes and promoted further the increase in expression induced by ET-1 (Fig. 3B), indicating that PI3K signalling is not required for the upregulation induced by ET-1 and suggesting that basal PI3K signalling negatively regulates Klf2/Klf4 expression. LY294002 did not affect Klf6 expression, but attenuated the increase in expression of Klf5 by ET-1. Since basal PI3K signalling is required for protein synthesis in cardiac myocytes [38], the effect of LY294002 on the increase in Klf5 induced by ET-1 (which requires de novo protein synthesis; Fig. 2B) may be a reflection of this. SB203580 had a minimal effect on the basal expression of Klf2, Klf4, Klf5 or Klf6, or on the increase in expression induced by ET-1 (Fig. 3B). It is unlikely that p38-MAPK signalling plays a significant role in the response.
3.3. Translation-state analysis of Klf2, Klf4, Klf5 and Klf6 mRNA expression
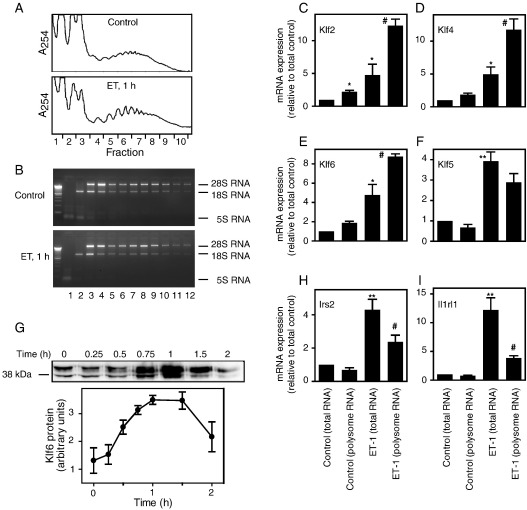
Given the increasing evidence for translational regulation of specific mRNAs [39,40], we performed translation-state analysis of Klf2, Klf4, Klf5 or Klf6 mRNAs. We used qPCR to compare the relative levels of expression in the total RNA pool and in RNAs associated with cardiac myocyte polysomes (prepared by sucrose density centrifugation; Fig. 4, A and B). Klf2, Klf4 and Klf6 mRNAs were marginally enriched in cardiac myocyte polysomes in unstimulated cells and, following stimulation with ET-1 (1 h), the relative increase in the polysomes was at least as great as in the total RNA pool (Fig. 4, C–E). Klf2 was increased 5.6-fold in the polysomes (4.8-fold in total RNA), Klf4 was increased 6.4-fold (5.0-fold in total RNA) and Klf6 was increased 4.6-fold (4.8-fold in total RNA). Klf5 mRNA was marginally decreased in the polysomes relative to the total pool, but the increase in expression induced by ET-1 in the polysomes (4.3-fold) was slightly higher than in the total pool (3.9-fold) (Fig. 4F). We therefore expect all these Klfs to be efficiently translated into protein. Consistent with this, Klf6 protein (detected as bands of ∼ 38–40 kDa on Western blots) was increased in cardiac myocytes exposed to ET-1, with maximal expression at ∼ 1 h (Fig. 4G). It could be argued that early genes should be efficiently translated and this may be a property of all such genes. However, Irs2 was upregulated as an IEG by ET-1 (no inhibition by cycloheximide, Fig. 2A), but the mRNA was not enriched in the polysomes and the increase in the polysomes was significantly less than that in the total RNA pool (Fig. 4H). Similarly, for interleukin 1 receptor-like 1 (Il1rl1), a second phase gene (upregulation was inhibited by cycloheximide; Fig. 2B), the increase in expression in the polysomes was significantly less than in the total RNA pool (Fig. 4I). It is therefore notable that Klf2, Klf4, Klf5 and Klf6 mRNAs are efficiently recruited to the polysomes for translation into protein.
Fig. 4.
Translation-state analysis of Klf mRNAs. Cardiac myocytes were unstimulated (Control) or exposed to 100 nM ET-1 (1 h). RNA was extracted for the total RNA pool. Polysomes were prepared from the same myocyte preparations using sucrose density centrifugation. A, A254 profiles for sucrose density gradients. B, Agarose gel electrophoresis of RNA from each fraction with ethidium bromide staining to highlight 28 S, 18S and 5S ribosomal RNAs. Fractions 6–11 were pooled for polysomal RNA. Expression of mRNAs for Klf2 (C), Klf4 (D), Klf6 (E), Klf5 (F), Irs2 (H) or Il1rl1 (I) in total RNA and polysome RNA pools was analysed by qPCR. Results are expressed relative to levels in total RNA from unstimulated cells and are means ± S.E.M. for 3 or 4 independent preparations of myocytes. ⁎p < 0.05, ⁎⁎p < 0.001 relative to Control (total RNA); #p < 0.01 relative to ET-1 (total RNA) (one-way ANOVA with TUKEY post-test). G, Western blotting of Klf6 protein in cardiac myocytes exposed to ET-1 for the times indicated. A representative image is shown in the upper panel, with densitometric analysis in the lower panel (results are means ± S.E.M. for 3 independent myocyte preparations).
3.4. Regulation of Klf2, Klf4 and Klf6 by H2O2 and pro-inflammatory cytokines
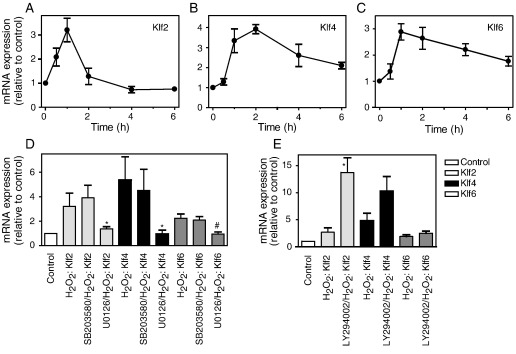
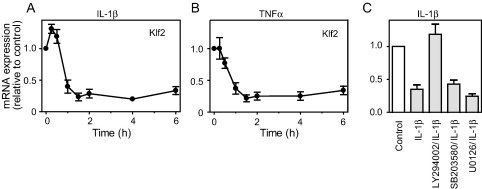
We considered whether other stimuli also regulate expression of Klf2, Klf4 or Klf6 mRNAs. H2O2 (0.2 mM) promotes cardiac myocyte apoptosis [41], but also stimulates ERK1/2 (and other pathways) [42], and stimulated a transient increase in expression of Klf2 (Fig. 5A). However, the response was less than that of ET-1 (∼ 3.6-fold) and was delayed (maximal increase at 1 h) (Fig. 1A). Consistent with our previous report [23], 0.2 mM H2O2 upregulated Klf4 and Klf6 mRNAs with maximal expression (5.0-fold) from 1 h and sustained expression over at least 6 h (Fig. 5, B and C). We have observed a similar delay in induction of other IEGs by H2O2 compared with ET-1 (e.g. Fos family members, unpublished data), which may reflect the delayed time course for activation of ERK1/2 signalling (maximal activation of ERK1/2 by ET-1 is at ∼ 5 min [43]; maximal activation by H2O2 is at 15–30 min [42]). Consistent with a role for ERK1/2 signalling, U0126 inhibited the increase in expression of Klf2, Klf4 and Klf6 by H2O2 (Fig. 5D). As with ET-1, SB203580 did not affect the response to H2O2 (Fig. 5D), whereas LY294002 enhanced the response (Fig. 5E). In endothelial cells, Klf2 is downregulated by TNFα [15]. IL-1β or TNFα profoundly and rapidly (from ∼ 1 h) downregulated Klf2 mRNA expression, a response which was sustained over at least 6 h (Fig. 6, A and B). Neither cytokine significantly affected the expression of Klf4 or Klf6 (data not shown). The decrease in expression of Klf2 in response to IL-1β was not affected by either SB203580 or U0126, although LY294002 (which itself increased expression of Klf2 mRNA, Fig. 3B) restored the baseline levels of expression (Fig. 6C).
Fig. 5.
H2O2 increases expression of Klf2, Klf4 and Klf6 mRNA in cardiac myocytes. A–C, Cardiac myocytes were exposed to 0.2 mM H2O2 for the times indicated. RNA was extracted and expression of mRNAs for Klf2 (A), Klf4 (B), or Klf6 (C) analysed by qPCR. Results are expressed relative to unstimulated controls and are means ± S.E.M. for at least 4 independent preparations of myocytes. D and E, Cardiac myocytes were unstimulated (Control) or exposed to 0.2 mM H2O2 (1 h) in the absence or presence of 5 μM SB203580 or 10 μM U0126 (D), or 50 μM LY294002 (E). Expression of mRNAs for Klf2 (pale grey bars), Klf4 (black bars), or Klf6 (dark grey bars) was analysed by qPCR. Results are expressed relative to levels in unstimulated myocytes and are means ± S.E.M. for 5 (D) or 7 (E) independent preparations of myocytes. ⁎p < 0.001, #p < 0.01 relative to H2O2 alone (one-way ANOVA with TUKEY post-test).
Fig. 6.
Il-1β or TNFα promote downregulation of Klf2. Cardiac myocytes were exposed to 100 ng/ml Il-1β (A) or 10 ng/ml TNFα (B) for the times indicated. RNA was extracted and expression of Klf2 mRNA analysed by qPCR. Results are expressed relative to unstimulated controls and are means ± S.E.M. for at least 4 independent preparations of myocytes. C, Cardiac myocytes were unstimulated (Control) or exposed to 100 ng/ml Il-1β (1 h) in the absence or presence of 50 μM LY294002, 5 μM SB203580 or 10 μM U0126 and expression of Klf2 mRNA analysed by qPCR. Results are expressed relative to levels in unstimulated myocytes and are means ± S.E.M. for 3 independent preparations of myocytes. ⁎p < 0.01 relative to Il-1β alone (one-way ANOVA with TUKEY post-test).
4. Discussion
4.1. Regulation of gene expression by Klf family members
Many studies of individual Klfs relate to specific cells/tissues in which they are highly expressed or for which the phenotype of a transgenic mouse suggests they may play some role. Thus, most studies of Klf2 focus on its regulation in lung tissue, endothelial cells or T cells, whereas many studies of Klf4 are in gut, endothelial cells or smooth muscle cells, and Klf5 is studied largely in gut or smooth muscle cells. Although there are indications that Klfs are transcriptionally regulated in these various cells (e.g. Klf2 expression is increased by shear stress in endothelial cells [14]), and there is an indication of interplay between Klf family members (e.g. Klf4 and Klf5 appear to operate in an antagonistic manner to regulate cell cycle progression [8]), few studies consider the regulation and function of the Klfs as a family. Our data demonstrate substantial and dynamic regulation of nine Klfs by ET-1 in a single cell type (Fig. 1) with at least three regulated as IEGs (Fig. 2A), suggesting that Klfs potentially play a much more significant role in the phenotypic responses of more cell types than has so far been considered.
Klfs are defined by the high degree of homology in their C-terminal DNA-binding domains [1,6]. It is probably not surprising, therefore, that all bind to similar consensus DNA sequences or, with 17 family members, that such elements are relatively common in gene promoters. The Klfs form two principal groups of transcriptional repressors (acting through CtBPs or mSin3a) plus a group which are primarily transcriptional activators, and it could be argued that the different Klf groups all do bind to similar sequences but the effect depends on whether the Klf is an activator or repressor and, if the latter, the mode of action. An additional factor could be that different cells express different Klfs and interactions with other cell-specific transcription factors may result in differential gene expression. In cardiac myocytes, we detected expression of at least 11 Klfs (Table 2), with regulation of several family members from each of the activator/repressor groups by ET-1 (Fig. 1), suggesting that (at least in this system) neither tissue specificity nor classification according to global function is responsible for functional differences between individual Klfs. Since there is evidence for functional specificity in other cells [44,45], minor variations in primary and secondary structure may be sufficient for individual Klfs to have differing affinities for precise sequences in various gene promoters. Expression of any individual gene may therefore reflect the balance of Klfs expressed at any particular time in relation to the precise DNA-binding sites which can be accessed by them.
4.2. Regulation of expression of Klf2, Klf4, Klf5 and Klf6
Of the Klfs which were upregulated by ET-1, Klf2 mRNA expression increased very rapidly and transiently (Fig. 1A), and it was regulated as an IEG (Fig. 2A). Partial inhibition of the response by U0126 (Fig. 3A) suggests that ERK1/2 signalling is required to some degree. ERK1/2 signalling was also required for upregulation of Klf2 by H2O2 (Fig. 5D). In contrast to endothelial cells subjected to shear stress [36], the increase in Klf2 expression by LY294002 and enhanced response to ET-1 in the presence of LY294002 (Fig. 3B) indicated that PI3K signalling negatively regulates Klf2 expression in cardiac myocytes. ERK5 phosphorylation of the transcription factor MEF2 promotes Klf2 expression in other cells [46], but ERK5 is not significantly activated in cardiac myocytes by ET-1 [24], ERK1/2 do not efficiently activate MEF2 [47] and inhibition of p38-MAPKs (which also phosphorylate MEF2 [47]) by SB203580 had no effect on Klf2 expression induced by ET-1 (Fig. 3B), suggesting that MEF2 is not the principal factor involved in this context. Of the transcription factors associated with the Klf2 promoter identified by Ahmed and Lingrel [48], the most probable candidate for regulating Klf2 expression by ET-1 in our study is CREB. CREB is rapidly (within 5–10 min) phosphorylated in cardiac myocytes exposed to ET-1, a response which is inhibited by U0126 [49].
Like Klf2, the increases in expression of Klf4 and Klf6 (Fig. 1, B and C) were inhibited by U0126, (Figs. 3A and 5D), and both were regulated as IEGs (Fig. 2A). Our data are consistent with other systems in which Klf4 is regulated as an IEG by 15-deoxy-Δ12,14 prostaglandin J2 [50] or platelet-derived growth factor [51], and Klf6 is regulated as an IEG by phorbol esters or serum [52,53]. However, for either Klf4 or Klf6, the potential transcription factors or mRNA stabilisation factors which promote the increase in expression are not known. The increase in expression of Klf5 induced by ET-1 was inhibited by cycloheximide (Fig. 2B), so the principal factors promoting its expression must be newly-synthesized proteins, and the response was inhibited by U0126 (Fig. 3A), implicating ERK1/2 signalling in the response. These data are consistent with other studies in which phorbol esters or fibroblast growth factor increase Klf5 expression in smooth muscle cells in an ERK1/2-dependent manner [54] and, in fibroblasts, the increase in expression induced by phorbol esters is inhibited by cycloheximide [55].
Studies of mRNA expression raise the question of whether or not changes in mRNA expression equate to changes in protein expression. In cardiac myocytes treated with ET-1, translation-state analysis indicated that mRNAs for Klf2, Klf4, Klf5 and Klf6 were increased to a similar or greater extent in the polysomes than in the total RNA pool (Fig. 4). Since ET-1 also increases the global rate of protein synthesis in cardiac myocytes [56] (illustrated by the increase in A254 profile of polysome fractions 6–11 with concomitant decrease of monosome fractions 2–4; Fig. 4A), overall, we would predict that the rate of synthesis of each of the Klf proteins should be increased at least in proportion to the mRNA. Consistent with this, we detected a significant, transient increase in expression of Klf6 protein following stimulation with ET-1 (Fig. 4G). Although cellular stresses such as oxidative stress can generally inhibit translation, 0.2 mM H2O2 does not have a significant effect on global protein synthesis over 4 h, and our microarray data indicate that Klf2, Klf4, and Klf6 are increased to a similar extent in the polysomes as in total RNA (data not shown).
4.3. Regulating Klf-dependent gene expression in cardiac myocytes
Downregulation of Klf15 is associated with the hypertrophic response of cardiac myocytes and may be required for hypertrophy to develop [21]. However, our data suggest that the situation regarding Klf-mediated regulation of gene expression in cardiac myocytes is more complex. In cardiac myocytes exposed to ET-1, we detected a rapid decrease in expression of mRNAs for three established repressors of gene expression (Klf3, Klf11 and Klf15; Fig. 1, G, H and I), with simultaneous rapid increases in expression of four Klfs more commonly associated with transcriptional activation (Klf2, Klf4, Klf5 and Klf6; Fig. 1, A–D). In response to ET-1, therefore, there appears to be an immediate overall switch towards increasing Klf-directed gene expression. However, the increases in expression of Klf2, Klf4, Klf5 and Klf6, and decrease in expression of Klf3 were transient, with a delayed increase in expression of Klf9 and Klf10, suggesting that the balance of Klf-regulated gene expression continues to change. A similar effect was detected with H2O2, with an early increase in expression of Klf2, Klf4, Klf5 and Klf6 and simultaneous decrease in expression of Klf3, Klf11 and Klf15 (Fig. 5 and [23]). In contrast, Il-1β or TNFα promoted downregulation of Klf2 expression (Fig. 6) with little effect on Klf4 or Klf6. Clearly, the genes which each of the Klfs regulates in cardiac myocytes remain to be established.
Acknowledgements
We thank Dr. Thomais Markou for her assistance in preparing the polysomes. This work was supported by grants from the British Heart Foundation.
References
- 1.Kaczynski J., Cook T., Urrutia R. Sp1- and Krüppel-like transcription factors. Genome Biol. 2003;4:206. doi: 10.1186/gb-2003-4-2-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Suske G., Bruford E., Philipsen S. Mammalian SP/KLF transcription factors: bring in the family. Genomics. 2005;85:551–556. doi: 10.1016/j.ygeno.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 3.van Vliet J., Crofts L.A., Quinlan K.G.R., Czolij R., Perkins A.C., Crossley M. Human KLF17 is a new member of the Sp/KLF family of transcription factors. Genomics. 2006;87:474–482. doi: 10.1016/j.ygeno.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 4.Lomberk G., Urrutia R. The family feud: turning off Sp1 by Sp1-like Klf proteins. Biochem. J. 2005;392:1–11. doi: 10.1042/BJ20051234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dang D.T., Zhao W., Mahatan C.S., Geiman D.E., Yang V.W. Opposing effects of Krüppel-like factor 4 (gut-enriched Krüppel-like factor) and Krüppel‑like factor 5 (intestinal-enriched Krüppel-like factor) on the promoter of the Krüppel-like factor 4 gene. Nucleic Acids Res. 2002;30:2736–2741. doi: 10.1093/nar/gkf400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bieker J.J. Krüppel-like factors: three fingers in many pies. J. Biol. Chem. 2001;276:34355–34358. doi: 10.1074/jbc.R100043200. [DOI] [PubMed] [Google Scholar]
- 7.Suzuki T., Aizawa K., Matsumura T., Nagai R. Vascular implications of the Krüppel-like family of transcription factors. Arterioscler. Thromb. Vasc. Biol. 2005;25:1135–1141. doi: 10.1161/01.ATV.0000165656.65359.23. [DOI] [PubMed] [Google Scholar]
- 8.Ghaleb A.M., Nandan M.O., Chanchevalap S., Dalton W.B., Hisamuddin I.M., Yang V.W. Krüppel-like factors 4 and 5: the yin and yang regulators of cellular proliferation. Cell Res. 2005;15:92–96. doi: 10.1038/sj.cr.7290271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang Y., Goldstein B.G., Chao H.H., Katz J.P. KLF4 and KLF5 regulate proliferation, apoptosis and invasion in esophageal cancer cells. Cancer Biol. Ther. 2005;4:1216–1221. doi: 10.4161/cbt.4.11.2090. [DOI] [PubMed] [Google Scholar]
- 10.Dorn G.W., Robbins J., Sugden P.H. Phenotyping hypertrophy; eschew obfuscation. Circ. Res. 2003;92:1171–1175. doi: 10.1161/01.RES.0000077012.11088.BC. [DOI] [PubMed] [Google Scholar]
- 11.Sugden P.H. An overview of endothelin signaling in the cardiac myocyte. J. Mol. Cell. Cardiol. 2003;35:871–886. doi: 10.1016/s0022-2828(03)00153-6. [DOI] [PubMed] [Google Scholar]
- 12.Clerk A., Cole S.M., Cullingford T.E., Harrison J.G., Jormakka M., Valks D.M. Regulation of cardiac myocyte cell death. Pharmacol. Ther. 2003;97:223–261. doi: 10.1016/s0163-7258(02)00339-x. [DOI] [PubMed] [Google Scholar]
- 13.Clerk A., Cullingford T.E., Fuller S.J., Giraldo A., Markou T., Pikkarainen S., Sugden P.H. Signaling pathways mediating cardiac myocyte gene expression in physiological and stress responses. J. Cell. Physiol. 2007;212:311–322. doi: 10.1002/jcp.21094. [DOI] [PubMed] [Google Scholar]
- 14.Feinberg M.W., Lin Z., Fisch S., Jain M.K. An emerging role for Krüppel-like factors in vascular biology. Trends Cardiovasc. Med. 2004;14:241–246.. doi: 10.1016/j.tcm.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 15.Atkins G.B., Jain M.K. Role of Krüppel-like transcription factors in endothelial biology. Circ. Res. 2007;100:1686–1695. doi: 10.1161/01.RES.0000267856.00713.0a. [DOI] [PubMed] [Google Scholar]
- 16.Wani M.A., Conkright M.D., Jeffries S., Hughes M.J., Lingrel J.B. cDNA isolation, genomic structure, regulation, and chromosomal localization of human lung Kruppel-like factor. Genomics. 1999;60:78–86. doi: 10.1006/geno.1999.5888. [DOI] [PubMed] [Google Scholar]
- 17.Haldar S.M., Ibrahim O.A., Jain M.K. Kruppel-like factors (KLFs) in muscle biology. J. Mol. Cell. Cardiol. 2007;43:1–10. doi: 10.1016/j.yjmcc.2007.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scohy S., Gabant P., Van Reeth T., Hertveldt V., Drèze P.-L., Van Vooren P., Rivière M., Szpirer J., Szpirer C. Identification of KLF13 and KLF14 (SP6), novel members of the SP/XKLF transcription factor family. Genomics. 2000;70:93–101. doi: 10.1006/geno.2000.6362. [DOI] [PubMed] [Google Scholar]
- 19.Lavallée G., Andelfinger G., Nadeau M., Lefebvre C., Nemer G., Horb M.E., Nemer M. The Krüppel-like transcription factor KLF13 is a novel regulator of heart development. EMBO J. 2006;25:5201–5213. doi: 10.1038/sj.emboj.7601379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uchida S., Tanaka Y., Ito H., Saitoh‑Ohara F., Inazawa J., Yokoyama K.K., Sasaki S., Marumo F. Transcriptional regulation of the CLC-K1 promoter by myc-associated zinc finger protein and kidney-enriched Krüppel-like factor, a novel zinc finger repressor. Mol. Cell. Biol. 2000;20:7319–7331. doi: 10.1128/mcb.20.19.7319-7331.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fisch S., Gray S., Heymans S., Haldar S.M., Wang B., Pfister O., Cui L., Kumar A., Lin Z., Sen-Banerjee S., Das C.A., Petersen H., Mende U., Burleigh B.A., Zhu Y., Pinto Y.M., Liao R., Jain M.K. Kruppel-like factor 15 is a regulator of cardiomyocyte hypertrophy. Proc. Natl. Acad. Sci. U. S. A. 2007;104:7074–7079. doi: 10.1073/pnas.0701981104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kemp T.J., Causton H.C., Clerk A. Changes in gene expression induced by H2O2 in cardiac myocytes. Biochem. Biophys. Res. Commun. 2003;307:416–421. doi: 10.1016/s0006-291x(03)01215-4. [DOI] [PubMed] [Google Scholar]
- 23.Clerk A., Kemp T.J., Zoumpoulidou G., Sugden P.H. Cardiac myocyte gene expression profiling during H2O2-induced apoptosis. Physiol. Genomics. 2007;29:118–127. doi: 10.1152/physiolgenomics.00168.2006. [DOI] [PubMed] [Google Scholar]
- 24.Kennedy R.A., Kemp T.J., Sugden P.H., Clerk A. Using U0126 to dissect the role of the extracellular signal-regulated kinase 1/2 (ERK1/2) cascade in the regulation of gene expression by endothelin-1 in cardiac myocytes. J. Mol. Cell. Cardiol. 2006;41:236–247. doi: 10.1016/j.yjmcc.2006.04.011. [DOI] [PubMed] [Google Scholar]
- 25.Cullingford T.E., Markou T., Fuller S.J., Giraldo A., Pikkarainen S., Zoumpoulidou G., Alsafi A., Ekere C., Kemp T.J., Dennis J.L., Game L., Sugden P.H., Clerk A. Temporal regulation of expression of immediate early and second phase transcripts by endothelin-1 in cardiomyocytes. Genome Biol. 2008;9:R32.. doi: 10.1186/gb-2008-9-2-r32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iwaki K., Sukhatme V.P., Shubeita H.E., Chien K.R. α- and β-Adrenergic stimulation induces distinct patterns of immediate early gene expression in neonatal rat myocardial cells. fos/jun expression is associated with sarcomere assembly; Egr-1 induction is primarily an α1-mediated response. J. Biol. Chem. 1990;265:13809–13817. [PubMed] [Google Scholar]
- 27.Bogoyevitch M.A., Clerk A., Sugden P.H. Activation of the mitogen-activated protein kinase cascade by pertussis toxin-sensitive and -insensitive pathways in cultured ventricular cardiomyocytes. Biochem. J. 1995;309:437–443. doi: 10.1042/bj3090437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clerk A., Harrison J.G., Long C.S., Sugden P.H. Pro-inflammatory cytokines stimulate mitogen‑activated protein kinases, increase phosphorylation of c-Jun and ATF2 and upregulated c-Jun protein in neonatal rat ventricular myocytes. J. Mol. Cell. Cardiol. 1999;31:2087–2099. doi: 10.1006/jmcc.1999.1040. [DOI] [PubMed] [Google Scholar]
- 29.Markou T., Cullingford T.E., Giraldo A., Weiss S.C., Alsafi A., Fuller S.J., Clerk A., Sugden P.H. Glycogen synthase kinases 3α and 3β in cardiac myocytes: regulation and consequences of their inhibition. Cell. Signal. 2008;20:206–218. doi: 10.1016/j.cellsig.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 30.Kyriakis J.M., Avruch J. pp54 Microtubule-associated protein 2 kinase. A novel serine/threonine kinase regulated by phosphorylation and stimulated by poly-l-lysine. J. Biol. Chem. 1990;265:17355–17363. [PubMed] [Google Scholar]
- 31.Moriguchi T., Kuroyanagi N., Yamaguchi K., Gotoh Y., Irie K., Kano T., Shirakabe K., Muro Y., Shibuya H., Matsumoto K., Nishida E., Hagiwara M. A novel kinase cascade mediated by mitogen‑activated protein kinase kinase 6 and MKK3. J. Biol. Chem. 1996;271:13675–13679. doi: 10.1074/jbc.271.23.13675. [DOI] [PubMed] [Google Scholar]
- 32.Clerk A., Aggeli I.-K.S., Stathopoulou K., Sugden P.H. Peptide growth factors signal differentially through protein kinase C to extracellular signal-regulated kinase in neonatal cardiomyocytes. Cell. Signal. 2006;18:225–235. doi: 10.1016/j.cellsig.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 33.Favata M.F., Horiuchi K.Y., Manos E.J., Daulerio A.J., Stradley D.A., Feeser W.S., Van Dyk D.E., Pitts W.J., Earl R.A., Hobbs F., Copeland R.A., Magolda R.L., Scherle P.A., Trzaskos J.M. Identification of a novel inhibitor of mitogen-activated protein kinase kinase. J. Biol. Chem. 1998;273:18623–18632. doi: 10.1074/jbc.273.29.18623. [DOI] [PubMed] [Google Scholar]
- 34.Davies S.P., Reddy H., Caivano M., Cohen P. Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem. J. 2000;351:95–105. doi: 10.1042/0264-6021:3510095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mody N., Leitch J., Armstrong C., Dixon J., Cohen P. Effects of MAP kinase cascade inhibitors on the MKK5/ERK5 pathway. FEBS Lett. 2001;502:21–24. doi: 10.1016/s0014-5793(01)02651-5. [DOI] [PubMed] [Google Scholar]
- 36.van Thienen J.V., Fiedderus J.O., Dekker R.J., Rohlena J., van Ijzendoorn G.A., Kootstra N.A., Pannekoek H., Horrevoets A.J. Shear stress sustains atheroprotective endothelial KLF2 expression more potently than statins through mRNA stabilization. Cardiovasc. Res. 2006;72:231–240. doi: 10.1016/j.cardiores.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 37.Clerk A., Michael A., Sugden P.H. Stimulation of the p38 mitogen-activated protein kinase pathway in neonatal rat ventricular myocytes by the G protein-coupled receptor agonists, endothelin‑1 and phenylephrine: a role in cardiac myocyte hypertrophy? J. Cell Biol. 1998;142:523–535. doi: 10.1083/jcb.142.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pham F.H., Sugden P.H., Clerk A. Regulation of protein kinase B and 4E-BP1 by oxidative stress in cardiac myocytes. Circ. Res. 2000;86:1252–1258. doi: 10.1161/01.res.86.12.1252. [DOI] [PubMed] [Google Scholar]
- 39.Bilanges B., Stokoe D. Mechanisms of translational deregulation in human tumors and therapeutic intervention strategies. Oncogene. 2007;26:5973–5990. doi: 10.1038/sj.onc.1210431. [DOI] [PubMed] [Google Scholar]
- 40.Leung A.K., Sharp P.A. Function and localization of microRNAs in mammalian cells. Cold Spring Harbor Symp. Quant. Biol. 2006;71:29–38. doi: 10.1101/sqb.2006.71.049. [DOI] [PubMed] [Google Scholar]
- 41.Cook S.A., Sugden P.H., Clerk A. Regulation of Bcl-2 family proteins during development and in response to oxidative stress in cardiac myocytes: association with changes in mitochondrial membrane potential. Circ. Res. 1999;85:940–949. doi: 10.1161/01.res.85.10.940. [DOI] [PubMed] [Google Scholar]
- 42.Clerk A., Michael A., Sugden P.H. Stimulation of multiple mitogen-activated protein kinase sub-families by oxidative stress and phosphorylation of the small heat shock protein, HSP25/27, in neonatal ventricular myocytes. Biochem. J. 1998;333:581–589. doi: 10.1042/bj3330581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clerk A., Bogoyevitch M.A., Andersson M.B., Sugden P.H. Differential activation of protein kinase C isoforms by endothelin-1 and phenylephrine and subsequent stimulation of p42 and p44 mitogen-activated protein kinases in ventricular myocytes cultured from neonatal rat hearts. J. Biol. Chem. 1994;269:32848–32857. [PubMed] [Google Scholar]
- 44.Boon R.A., Fledderus J.O., Volger O.L., van Wanrooij E.J.A., Pardali E., Weesie F., Kuiper J., Pannekoek H., ten Dijke P., Horrevoets A.J.G. KLF2 suppresses TGF-β signaling in endothelium through induction of Smad7 and inhibition of AP-1. Arterioscler. Thromb. Vasc. Biol. 2007;27:532–539. doi: 10.1161/01.ATV.0000256466.65450.ce. [DOI] [PubMed] [Google Scholar]
- 45.Hamik A., Lin Z., Kumar A., Balcells M., Sinha S., Katz J., Feinberg M.W., Gerszten R.E., Edelman E.R., Jain M.K. Kruppel-like factor 4 regulates endothelial inflammation. J. Biol. Chem. 2007;282:13769–13779. doi: 10.1074/jbc.M700078200. [DOI] [PubMed] [Google Scholar]
- 46.Sohn S.J., Li D., Lee L.K., Winoto A. Transcriptional regulation of tissue-specific genes by ERK5 mitogen-activated protein kinase. Mol. Cell. Biol. 2005;25:8553–8566. doi: 10.1128/MCB.25.19.8553-8566.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ornatsky O.I., Cox D.M., Tangirala P., Andreucci J.J., Quinn Z.A., Wrana J.L., Prywes R., Yu Y.T., McDermott J.C. Post-translational control of the MEF2A transcriptional regulatory protein. Nucleic Acids Res. 1999;27:2646–2654. doi: 10.1093/nar/27.13.2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ahmad N., Lingrel J.B. Kruppel-like factor 2 transcriptional regulation involves heterogeneous nuclear ribonucleoproteins and acetyltransferases. Biochemistry. 2005;44:6276–6285. doi: 10.1021/bi050018s. [DOI] [PubMed] [Google Scholar]
- 49.Harrison J.G., Sugden P.H., Clerk A. Endothelin-1 promotes phosphorylation of CREB transcription factor in primary cultures of neonatal rat cardiac myocytes: implications for the regulation of c-jun expression. Biochim. Biophys. Acta. 2004;1644:17–25. doi: 10.1016/j.bbamcr.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 50.Chen Z.Y., Tseng C.-C. 15-Deoxy-Δ12,14 prostaglandin J2 up-regulates Krüppel-like factor 4 expression independently of peroxisome proliferators-activated receptor γ by activating the mitogen-activated protein kinase kinase/extracellular signal-regulated kinase signal transduction pathway in HT-29 colon cancer cells. Mol. Pharmacol. 2005;68:1203–1213. doi: 10.1124/mol.105.014944. [DOI] [PubMed] [Google Scholar]
- 51.Liu Y., Sinha S., McDonald O.G., Shang Y., Hoofnagle M.H., Owens G.K. Kruppel-like factor 4 abrogates myocardin-induced activation of smooth muscle gene expression. J. Biol. Chem. 2005;280:9719–9727. doi: 10.1074/jbc.M412862200. [DOI] [PubMed] [Google Scholar]
- 52.Inuzuka H., Wakao H., Masuho Y., Tojo H., Nanbu-Wakao R. cDNA cloning and expression analysis of mouse zf9, a Krüppel-like transcription factor gene that is induced by adipogenic hormonal stimulation in 3T3-L1 cells. Biochim. Biophys. Acta. 1999;1447:199–207. doi: 10.1016/s0167-4781(99)00161-x. [DOI] [PubMed] [Google Scholar]
- 53.Kojima S., Hayashi S., Shimokado K., Suzuki Y., Shimada J., Crippa M.P., Friedman S.L. Transcriptional activation of urokinase by the Krüppel-like factor Zf9/COPEB activates latent TGF-β1 in vascular endothelial cells. Blood. 2000;95:1309–1316. [PubMed] [Google Scholar]
- 54.Kawai-Kowase K., Kurabayashi M., Hoshino Y., Ohyama Y., Nagai R. Transcriptional activation of the zinc finger transcription factor BTEB2 gene by Egr-1 through mitogen-activated protein kinase pathways in vascular smooth muscle cells. Circ. Res. 1999;85:787–795.. doi: 10.1161/01.res.85.9.787. [DOI] [PubMed] [Google Scholar]
- 55.Sun R., Chen X., Yang V.W. Intestinal-enriched Krüppel-like factor (Krüppel-like factor 5) is a positive regulator of cellular proliferation. J. Biol. Chem. 2001;276:6897–6900. doi: 10.1074/jbc.C000870200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pham F.H., Cole S.M., Clerk A. Regulation of cardiac myocyte protein synthesis through phosphatidylinositol 3′ kinase and protein kinase B. Adv. Enzyme Regul. 2001;41:73–86. doi: 10.1016/s0065-2571(00)00007-8. [DOI] [PubMed] [Google Scholar]