Abstract
In prokaryotic nucleotide excision repair, UvrA recognizes DNA perturbations and recruits UvrB for the recognition and processing steps in the reaction. One of the most remarkable aspects of UvrA is that it can recognize a wide range of DNA lesions that differ in chemistry and structure. However, how UvrA interacts with DNA is unknown. To examine the role that the UvrA C-terminal zinc finger domain plays in DNA binding, an eleven amino acid deletion was constructed (ZnG UvrA). Biochemical characterization of the ZnG UvrA protein was carried out using UvrABC DNA incision, DNA binding and ATPase assays. Although ZnG UvrA was able to bind dsDNA slightly better than wild-type UvrA, the ZnG UvrA mutant only supported 50–75% of wild type incision. Surprisingly, the ZnG UvrA mutant, while retaining its ability to bind dsDNA, did not support damage-specific binding. Furthermore, this mutant protein only provided 10% of wild-type Bca UvrA complementation for UV survival of an uvrA deletion strain. In addition, ZnG UvrA failed to stimulate the UvrB DNA damage-associated ATPase activity. Electrophoretic mobility shift analysis was used to monitor UvrB loading onto damaged DNA with wild-type UvrA or ZnG UvrA. The ZnG UvrA protein showed a 30–60% reduction in UvrB loading as compared with the amount of UvrB loaded by wild-type UvrA. These data demonstrate that the C-terminal zinc finger of UvrA is required for regulation of damage-specific DNA binding.
Nucleotide excision repair (NER)2 is the primary mechanism cells use to repair a diverse set of DNA lesions. In prokaryotes, nucleotide excision repair requires the collaborative action of UvrA, UvrB, and UvrC. The fundamental process of DNA damage recognition is central to the function of UvrA and the overall repair reaction. It is believed that UvrA initially recognizes the damage-induced distortion in the DNA, and then hands off the damaged DNA to UvrB, so that UvrB can make a more detailed assessment of the nature of the helical perturbation. Once on DNA, UvrB is responsible for preparing the DNA for incision by the two nuclease centers of UvrC (for recent reviews see Refs. 1–3). In a general way, NER proteins fulfill all the criteria of a kinetic proofreading mechanism for damage processing: 1) damage specificity is not absolute; 2) ATP is consumed to generate irreversible intermediates and to delay initial binding and processing from incision, and 3) dissociation of UvrA or UvrA/UvrB from DNA or lack of stimulating the UvrC nuclease domains help safeguard against inappropriate incision (4). However, the precise mechanism by which UvrA and UvrB effectively recognize DNA damage in a sea of non-damaged DNA is unknown and of critical importance.
UvrA is a large protein, ~105 kDa, possessing two ATP-binding cassette-type ATPases (ABC ATPase) and two zinc finger domains (5, 6) and was recently reviewed in Ref. 3. The conserved motifs of the ABC ATPase are not contiguous, but are interrupted after the Walker A and before the signature sequence, LSGG, with an insertion sequence containing the conserved zinc fingers. The two ABC ATPases are connected through a flexible protease-sensitive linker region. Therefore, both the N- and C-terminal ABC ATPase domains are separable, and each has been shown to possess some DNA binding capacity (7). Because of the presence of the ATPase domains, DNA binding by UvrA is regulated by ATP binding and hydrolysis. Whereas nucleotide binding by UvrA is not absolutely required for DNA binding, ATP binding promotes the dimerization of UvrA (8), which in turn facilitates DNA binding (9). In contrast, ATP hydrolysis is believed to drive UvrA dimer dissociation and consequently reduces DNA binding (10, 11). Despite this general picture, the precise mechanistic details of how nucleotide binding and hydrolysis regulate the UvrA DNA binding are not known.
Zinc fingers are a common structural element utilized by sequence-specific DNA-binding proteins to interact with DNA (12). However, zinc fingers also mediate protein-protein, protein-RNA, and protein-ligand interactions (13). Currently, there are 64 families of zinc fingers listed in the PROSITE data base, yet neither of UvrA zinc fingers shares strong homology to any of these family members. Previously, investigators mutagenized the zinc-coordinating cysteines of Escherichia coli UvrA C-terminal zinc finger producing bacteria that were UV-sensitive and NER-defective (14–16).
In this study, we re-investigated the role of the UvrA C-terminal zinc finger in the NER reaction. A mutant of Bacillus caldotenax UvrA (Bca UvrA) was generated by substituting a glycine for eleven of the highly conserved amino acids within the C-terminal zinc finger. The mutant protein, ZnG UvrA, was purified and characterized. Our results demonstrate that the mutant ZnG UvrA can bind dsDNA, but has lost its damage-specific dsDNA binding, and cannot complement for in vivo UV resistance.
EXPERIMENTAL PROCEDURES
Proteins
The C-terminal ABC ATPase domain (CABC fragment) of uvrABca was amplified using the following primers (5′-GCG ACC GGA TCC ATG CTG GCC GCG GAC TAT TTG and 5′-GAG AGA GCG GCC GCT TAC GCC TTC ACC GCT TCA TAT TG) and the Pfu turbo DNA polymerase (Stratagene). The CABC fragment includes nucleotides 1643 to the end of the uvrABca gene; thus this fragment includes the linker region and the C-terminal ABC motifs, amino acids 549–952. The PCR fragment was cloned into a Topo cloning vector, pCRBII-TOPO, then transferred into the pGEX4T1 vector by digestion with BamH1 and NotI to create pGEX4T1-Wt CABC.
Construction of the ZnG deletion mutant of pGEX-CABC was performed with the QuikChange site-directed mutagenesis kit from Stratagene using pGEX4T1-Wt CABC as template, primers (5′-CAT GGC GAT GGC ATC ATC GGT GTC CCG TGC GAA GTG TGC CAC and 5′-GTG GCA CAC TTC GCA CGG GAC ACC GAT GAT GCC ATC GCC ATG) and Pfu turbo DNA polymerase. An alignment of the zinc finger region showing which amino acids are deleted is depicted in Fig. 1.
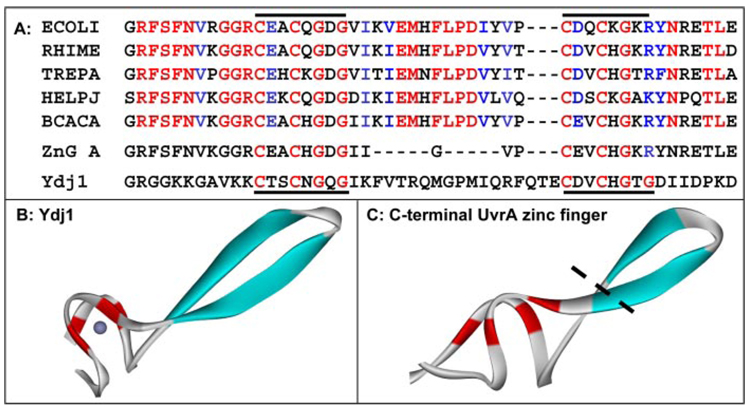
FIGURE 1. Sequence alignment and homology modeling of the C-terminal zinc finger of UvrA.
A, sequence alignment of the C-terminal zinc finger found in five UvrA homologues. Color coding of the alignment is based on the homology among 22 UvrA proteins in the seed alignment for UvrA within the HAMAP project (record MF_00205, Ref. 45). Red indicates at least 95% conservation; blue denotes that the amino acid is functionally conserved (K/R, D/E, F/Y, M/V/I/L) in 95% of 22 homologues. The red letters within ZnG UvrA and Ydj1 and the black bars denote the position of the conserved motif. The Entrez protein accession numbers are as follows: E. coli UvrA, (ECOLI) P0A698; Rhizobium meliloti UvrA, (RHIME) P56899; Treponema pallidum UvrA, (TREPA) O83527; Helicobacter pylori J99 UvrA, (HELPJ) Q9ZLD6, and B. caldotenax UvrA,(BCACA) AAK29748. B, crystal structure of the ZnII domain of Ydj1, the yeast dnaJ homologue is shown (PDB accession code 1NLT). C, model of the C-terminal zinc finger of Bca UvrA containing 35 residues. The black dotted line represents where the zinc finger would be truncated in the ZnG UvrA mutant. The protein images were generated with DS ViewerPro (Accelrys).
To place the ZnG mutation into the full-length uvrA gene, the BseR1 and NcoI fragment from pGEX4T1-ZnG CABC was removed, gel-purified, and ligated into the BseR1/NcoI (New England Biolabs) double-digested, artic phosphatase-treated pTYB1-Wt uvrABca vector, thus completing the construction of pTYB1-ZnG uvrABca.
The plasmid coding for GST-ZnF, pGEX6p1-ZnF was created by PCR amplification of nucleotides 2002–2463 by Pfu turbo DNA polymerase primers (5′-ATA GGG ATC CGG CGA GCA CCG CGA CAT TC and 5′-ACA CGC GGC CGC GCC GAG CTT CAT ATA ACC). The PCR fragment was cloned into a Topo cloning vector, pCRBII-TOPO, then transferred into the pGEX6p1 vector by digestion with BamH1 and NotI to create pGEX6p-ZnF.
The inserts of all vectors were sequenced. Upon re-sequencing the pTYB1-Wt uvrABca expression vector, we discovered several sequence variations from the original Bca UvrA sequence deposited in Entrez, accession number AAK29748. There were a total of fourteen amino acid changes; however none reside within any conserved sequence elements, (sequence conservation defined as 70% or more identity among 24 UvrA homologues). The majority of the variations, 13 of the 14, were in the poorly conserved linker region. Of all the variations, only linker region substitution Phe600 to Thr600 is unique to the new Bca UvrA sequence. The remaining amino acid changes exist in at least one or more UvrA homologues. The Wt UvrA, ZnG UvrA, Wt GST-CABC, and ZnG GST-CABC proteins used in this study share these fourteen amino acid substitutions.
Expression and Purification of the Proteins-Bca Wt UvrA, Bca ZnG UvrA, Bca Wt UvrB, Bca Δβ-hairpin UvrB, and Thermatoga maritima Wt UvrC (Tma UvrC)
All proteins were expressed in BL21 (DE3) RIL cells (Stratagene) and purified using the T7 IMPACTtm system (New England Biolabs) by standard procedures. The GST-containing proteins were expressed in BL21 (DE3) RIL cells (Stratagene) and purified using glutathione-Sepharose 4B resin (GE Healthcare). The GST-containing proteins were eluted from the column with reduced glutathione (10 mm) in 20 mm Tris-HCl, pH 8, 0.5 m NaCl, 0.1 mm EDTA, 5 mm DTT, and 0.25% Triton X-100, then dialyzed into storage buffer. UvrA, UvrB, and the GST fusion proteins were maintained at −20 °C in storage buffer (50 mm Tris-HCl, pH 7.5, 500 mm KCl, 0.1 mm EDTA, and 50% glycerol) until use.
DNA Substrates
DNA substrates were synthesized by Sigma-Genosys. The DNA sequence of the 50-mer double-stranded substrate containing a single internal fluorescein (FldT) adduct was: F2650 5′-GAC TAC GTA CTG TTA CGG CTC CAT C[FldT]C TAC CGC AAT CAG GCC AGA TCT GC-3′ while the non-damaged complementary bottom strand was NDB, 5′-GCA GAT CTG GCC TGA TTG CGG TAG CGA TGG AGC CGT AAC AGT ACG TAG TC-3′. The non-damaged strand, NDT, has the same sequence as F2650 except it contains a dT at the position of FldT. The DNA was 5′ end-labeled using T4 polynucleotide kinase and [γ-32P]ATP (3000 Ci/mmol, Amersham Biosciences) according to standard procedures. The reaction was terminated by the addition of EDTA (20 mm), and the enzyme was heat-denatured by incubation for 10 min at 65 °C. Unincorporated radioactive nucleotides were removed by gel filtration chromatography (Biospin-6, Bio-Rad). The labeled oligonucleotide was annealed with equimolar amounts of the complementary oligonucleotide.
When referring to the oligonucleotide duplexes, the strand listed first is the one that is 5′ end-labeled. The double-stranded character of the oligonucleotide duplex was analyzed on a native 10% polyacrylamide gel.
UV Survival Assay
WP2 uvrA− trp− cells (Mol Tox, Inc., Boone, NC) were incubated with 50 ng of pT7pol26 plasmid (Gentaur, Belgium) and 50 ng of pTYB1 (New England Biolabs), pTYB1-Wt UvrABca or pTYB1-ZnG UvrABca for 10 min. Wt WP2 cells (MolTox, Inc.) were incubated with 50 ng of pT7pol26 and 50 ng of pTYB1 or pTYB1-ZnG for 10 min. Transformations were carried out in cuvettes with a 0.2-cm electrode gap using a Gene Pulser II (Bio-Rad) electroporator with peak discharge at 2.4 kV, resistance set at 100 ohms and capacitance set to 25 microfarads. Immediately following transformation, cells were transferred into 250 µl of SOC broth. After 1 h of shaking at 37 °C, the entire culture was spread onto LB plates containing 100 µg/ml ampicillin and 50 µg/ml kanamycin. Individual colonies were selected and grown to an A600 of ~1.0. Then the cell culture was diluted 2-fold, and the proteins were induced by addition of IPTG (0.1 mm) for 1 h at 37 °C. This concentration of IPTG gave low, but detectable levels of UvrA protein. In duplicate, three serial dilutions of each sample of culture (100 µl) were spread onto LB plates containing 50 µg/ml kanamycin and 100 µg/ml ampicillin and UV-irradiated. The appropriate UV dose was calculated by measuring the fluency from an 8-watt 254-nm germicidal lamp using a 254-nm UVX Radiometer (UVP Inc.). Serial dilutions of unirradiated cultures were also plated and used to determine the plating efficiency of each transformant. The number of colonies obtained after 20 h of incubation at 37 °C was recorded, and the percent survival calculated from the plating efficiency of the non-irradiated controls. Two or three independent experiments were performed for each sample. The mean survival of two-three independent experiments is plotted as a function of UV fluence.
Incision Assay
Prior to initiation of the incision assay, the UvrABC proteins were heated to 65 °C for 10 min. The 5′-end-labeled duplex DNA (2 nm, F2650/NDB) was treated with UvrABC (20 nm Wt or ZnG Bca UvrA, 100 nm Bca UvrB, and 50 nm Tma UvrC) in 20 µl of UvrABC buffer (50 mm Tris-HCl, pH 7.5, 50 mm KCl, 10 mm MgCl2, 1 mm ATP, and 5 mm DTT) at 55 °C for the indicated time. For those reactions containing supercoiled undamaged plasmid DNA, varying concentrations of pUC19 DNA (New England Biolabs) were included as labeled in the figure legend. The reactions were terminated by addition of EDTA (20 mm). Ten percent of the reaction was removed, denatured with formamide and heated to 85 °C for 5 min. Incision products were resolved on a 10% denaturing polyacrylamide gel, and electrophoresis was performed at 325 V in Tris borate-EDTA buffer (89 mm Tris, 89 mm boric acid, and 2 mm EDTA) for 40 min. Gels were dried and exposed to a PhosphorImager screen (Molecular Dynamics) overnight. The percent of the DNA incised was calculated using the Molecular Dynamics software, ImageQuant to determine the band intensities within each lane. The percentage of DNA incised is reported as the mean ± S.D. (n = 3).
Electrophoretic Mobility Shift Assay (EMSA)
For UvrA·DNA EMSAs, the binding reactions were performed with duplexed DNA substrate (2 nm) and protein (concentrations as indicated in the figure legend) in 20-µl reaction buffer (50 mm Tris-HCl, pH 7.5, 100 mm KCl, 10 mm MgCl2, 1 mm ATP, 5 mm DTT, and 1 µm bovine serum albumin) for 15 min at 37 °C or 55 °C (as indicated in the figure legend). The reactions were loaded onto a 3.5% native polyacrylamide gel (29:1; acrylamide:bis). For EMSAs containing ATP and magnesium, the gels and running buffer contained 44.5 mm Tris pH 8.3, 44.5 mm boric acid, and 1 mm EDTA, 1 mm ATP and 10 mm MgCl2. For EMSAs without ATP and magnesium, the gels and running buffer contained 44.5 mm Tris, 44.5 mm boric acid, and 1 mm EDTA. Electrophoresis was carried out for 1 h at 100 V with the gel rigs at 4 °C. The gels were dried and exposed to a phosphorimager screen. The percent of DNA bound in the various protein-DNA complexes was calculated based on the total radioactivity in the lane. The percentage is reported as the mean ± S.D. (n = 3). Data are reported as the fraction of DNA-bound, including higher order multimers, versus protein concentration and fitted by nonlinear regression analysis (17).
For the UvrA·UvrB·DNA EMSAs, the enzymes were preheated to 65 °C for 10 min prior to initiation of the reactions and UvrA·UvrB reaction mixtures. Binding reactions were performed with 2 nm F2650/NDB and Wt UvrA (20 nm) or ZnG UvrA (20 nm) and Wt UvrB (100 nm) in 20 µl of reaction buffer (50 mm Tris-HCl, pH 7.5, 100 mm KCl, 10 mm MgCl2, 1 mm ATP, 5 mm DTT, and 1 µm bovine serum albumin) for 30 min at 55 °C. For those lanes that contain additional plasmid DNA, a 100-fold molar excess of pUC DNA (molar excess relative to the concentration of oligonucleotide in base pairs) was added prior to adding the oligonucleotide to the reactions. The reactions were loaded onto a 4% native polyacrylamide gel (29:1, acrylamide:bis) and subjected to electrophoresis at 100 V for 1 h, 4 °C. The gels and buffers contained 44.5 mm Tris, 44.5 mm boric acid and 1 mm EDTA, 1 mm ATP, and 10 mm MgCl2.
ATP Hydrolysis Assay
The conversion of ATP to ADP by the UvrAB system was monitored using a coupled enzyme assay system consisting of pyruvate kinase and lactic dehydrogenase to couple the hydrolysis of ATP to the oxidation of NADH (ε340 nm = 6220 m−1 cm−1). ATP (Roche Applied Science) was added to a final concentration of 1 mm in a 100-µl reaction mixture containing 50 mm Tris-HCl, pH 7.5, 55 mm KCl, 4 mm MgCl2, 1 mm DTT, 12.6 units/ml l-lactic dehydrogenase (Sigma), 10 units/ml pyruvate kinase (Sigma), 2 mm phosphoenol pyruvate (Roche Applied Science), 0.15 mm NADH (Roche Applied Science), 50 nm Bca UvrA (Wt or mutants), and 100 nm Bca UvrB. The Bca proteins were preheated to 65 °C for 10 min prior to initiation of the reactions. Each protein was assayed in the absence of DNA as well as in the presence of supercoiled pUC19 (SC DNA, 10 ng/µl) or UV-irradiated pUC18 DNA (UV DNA, 10 ng/µl) substrate. The UV-damaged DNA was prepared by exposure of 1 µg/µl pUC18 plasmid DNA to 200 J/m2 for 1 min. The rate of hydrolysis was calculated from the linear change in absorbance at 340 nm at either 37 or 55 °C over a 20–30-min period, using a Beckman DU-640 spectrophotometer. In addition, the rates were blank corrected for the oxidation of NADH (+ATP) in the absence of additional proteins. For ATPase reactions at 37 °C, the proteins were not preheated prior to the assay. For those reactions at 55 °C, the data are reported as the mean rate (M/min) ± S.D. (n = 3 or 4).
Creation of the Zinc Finger Structural Model
The C-terminal zinc finger domain was modeled based on an alignment with the coordinates from the solved structure of Ydj1. Ydj1, the Saccharomyces cerevisiae dnaJ homologue, is a protein chaperone involved in the regulation of Hsp90 and Hsp70 (18–20). The structure of Ydj1 was solved in complex with its peptide substrate (PDB code 1NLT) (21). The Ydj1 zinc finger structure was selected as the template to model the UvrA zinc finger as they share a conserved CXXCXGXG sequence, which is common to this zinc finger protein family, referred to as DNAJ_CXXCXGXG (Pfam record PF00684, Ref. 22). In the DnaJ family of proteins, CXXCXGXG is repeated four times. In the C-terminal domain of UvrA, the sequence CXXCXGX(R/K) is repeated twice. The lysine-containing consensus is also found among the DnaJ proteins. From the 20 DnaJ protein sequences that form the Pfam seed alignment for Pfam record PF00684, the CXXCXGX(R/K) is found 30% of the time within the fourth DnaJ repeat. The sequence alignment of Ydj1 and several UvrA proteins used to develop the structural model for the UvrA C-terminal zinc finger is shown in Fig. 1.
Development of a Structural Model for the ABC ATPase within the C-terminal Domain of UvrA, Residues 603–713 and 801–935
A comparative model was developed using structural (DALI/FSSP, VAST) and sequence (T-Coffee, ClustalW) alignments of solved ABC transporter proteins including the MalK ATPase subunit of maltose ABC transporter (PDB code 1G29, Ref. 23), the MJ0796 ATP-binding cassette protein (PDB code 1L2T, Ref. 24), and the ATP binding subunit of histidine permease (PDB code 1B0U, Ref. 25). These structures were selected because they were solved at high resolution (1.9, 1.7, and 1.5 Å, respectively) and shared the highest sequence similarity with UvrA. Coordinates from all three solved structures contributed to the model; however, the majority was from 1B0U, which shares 32% sequence identity with UvrA in the ABC transporter domain regions. The RMS deviation between the solved ABC transporter structures used to develop the UvrA model was 3.7–3.9Å (backbone Cα). The ATP was placed in the model UvrA structure based on the position of ATP within the histidine permease solved structure (PDB code 1B0U). The distances between the ATP atoms and atoms of the ABC ATPase-conserved motifs (Walker A, Q-loop, ABC signature, Walker B, and His-loop) were optimized. The model was manually edited using the Accelrys InsightII Homology Module Software. The final model was subjected to minimization and short (200 ps) molecular dynamics runs to resolve discontinuities with CHARMm. The RMS deviation between the final UvrA ABC transporter monomer model and the 1B0U PDB structure is 3.2 Å.
A model for the UvrA C-terminal dimer was constructed using the solved Rad50 dimer structure (PDB codes: 1F2U, 1F2T) as a model template for the interactions between the two monomeric C-terminal ABC transporter domains of UvrA. Each UvrA monomer was superimposed onto each of the Rad50 monomers so that the UvrA dimer structural interactions would simulate that of the Rad50 dimer organization. ATP atoms and the conserved atoms belonging to the ABC transporter motifs were used for the superposition of each of the ABC monomeric units with the Rad50 monomer to create UvrA C-terminal dimer model coordinates. The RMS deviation between the Rad50 dimer and UvrA C-terminal dimer is 4.8 Å.
RESULTS
Design and Construction of ZnG UvrA
Prior research on the UvrA C-terminal zinc finger focused on disrupting the zinc-coordinating amino acids. Specifically, substitution of E. coli UvrA C-terminal zinc finger cysteine, C763F, produced a protein that was unable to bind dsDNA; therefore, this led to the hypothesis that the C-terminal zinc finger region of UvrA was responsible for DNA binding (16). Visse et al. (14) also mutated one of the C-terminal cysteines and noted that the structural stability of the mutant protein was impaired. The zinc ion plays an important role in the structural architecture of zinc fingers and mutagenesis of the zinc-anchoring cysteines probably disrupts the global fold of the C-terminal ABC ATPase domain.
Unlike many other zinc finger DNA-binding proteins (13), UvrA is not a sequence-specific DNA-binding protein. Furthermore, close inspection of the amino acids within and around the C-terminal zinc finger reveals that the conserved amino acids tend to be hydrophobic, whereas only a few are positively charged residues, which may be available to interact with the DNA phosphate backbone. For these reasons, we re-investigated the role of the C-terminal zinc finger domain of UvrA.
A multiple sequence alignment of the UvrA C-terminal zinc finger region is displayed in Fig. 1A. Inspection of the sequences reveals a common motif: C(D/E)XCXGXGX3(I/V)EMXFLPDX4 C(D/E)XCXG. Note that there are conserved glycines after each set of cysteines. The dnaJ family of proteins possess a similar zinc finger signature; however dnaJ proteins have 4 repeats of CX2CXGX(G/K). The structure of the yeast DnaJ homolog, Ydj1, has been solved (PDB 1NLT, Ref. 21), and its model is shown in Fig. 1B. A model of the UvrA C-terminal zinc finger was created based on the structure of the ZnII domain of Ydj1 because it contains 22 amino acids between the conserved cysteines, similar to the UvrA spacing of 19 amino acids, (Fig. 1C). Based on the structural model and sequence alignments, a deletion mutant of UvrA was created. This deletion mutant replaced eleven of the amino acids within the C-terminal finger with a single glycine residue (Fig. 1A).
In Vivo Complementation and Survival
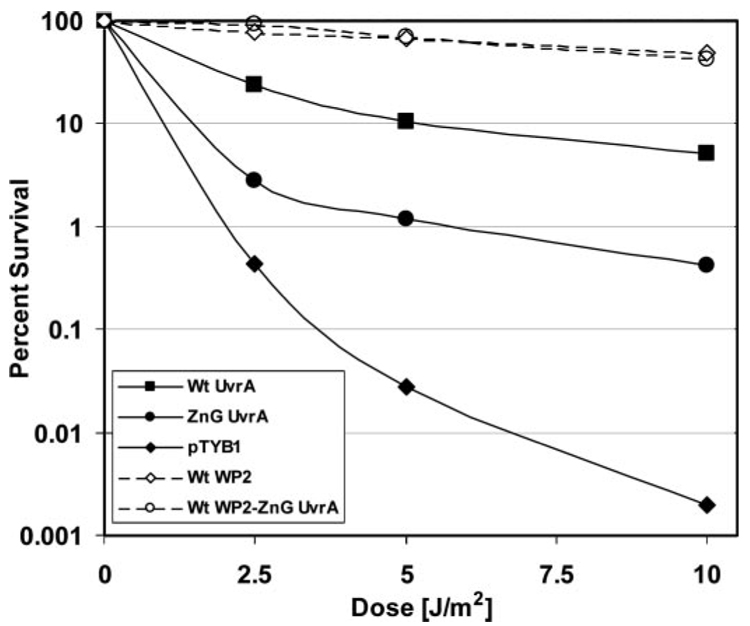
In vivo complementation and UV survival studies were conducted in the WP2 uvrA− strain. Transformation of WP2 uvrA− cells with plasmids encoding T7 polymerase and Bca Wt UvrA increases the cells UV survival by more than 382-fold, pTYB1 versus pTYB1-Wt UvrA, when exposed to 5 J/m2 of UV. These results indicate that at 37 °C, Bca Wt UvrA can complement the E. coli system. In contrast, transformation of pTYB1-ZnG UvrA versus pTYB1 only confers a 43-fold increase in UV survival following 5 J/m2 of UV. The UV survival differences between Wt UvrA and ZnG UvrA transformed samples varied between 8.6–12.4-fold. Clearly, the ZnG UvrA protein is defective at some step in the NER reaction because it provides a lower level of UV protection than Wt Bca UvrA.
Wild-type WP2 cells were also included in the UV survival analysis. Even though Wt Bca UvrA provided significant protection from UV, it did not provide the same level of defense as endogenous E. coli UvrA, compare Wt WP2 with Wt UvrA in Fig. 2. Bca UvrA was 3–10-fold less effective after UV than endogenous E. coli UvrA. The ZnG UvrA containing vector was also transformed into the Wt WP2 cells to determine if ZnG UvrA would be a dominant negative mutation. The Wt WP2 cells expressing ZnG UvrA do not display a greater sensitivity to UV than cells transformed with empty vector (Fig. 2). Therefore, ZnG UvrA does not exert a dominant negative affect on endogenous E. coli UvrA.
FIGURE 2. UV survival of E. coli WP2 strains.
WP2 (trp−, uvrA−) cells were transformed with pT7pol26, a plasmid encoding an IPTG-inducible T7 polymerase, and pTYB1 or pTYB1-Wt UvrA or pTYB1-ZnG UvrA. In addition, WP2 (trp−) cells with endogenous UvrA were transformed with pT7pol26 and pTYB1 or pTYB1-ZnG UvrA. After individual colonies were selected and grown to an A600 of ~1.0, the cell culture was diluted 2-fold, and the proteins were induced by addition of IPTG (0.1 mm) for 1 h at 37 °C. Serial dilutions of each sample of culture (100 µl) was then spread onto a nutrient rich-media and UV-irradiated with a 254-nm germicidal light source. The numbers of colonies visible after 20 h of growth at 37 °C was recorded, and the fraction of cells surviving after each dose of UV was calculated based on the plating efficiency of the unirradiated controls. The mean of two or three independent experiments is reported. Solid lines indicate the WP2 (trp−, uvrA−) strain and dashed lines indicate the wild-type WP2 (trp−) strain. Transformants contained pT7pol26 and pTYB1 (diamonds), or pTYB1-Wt UvrA (squares), or pTYB1-ZnG UvrA (circles).
ZnG UvrA Supports Less Incision
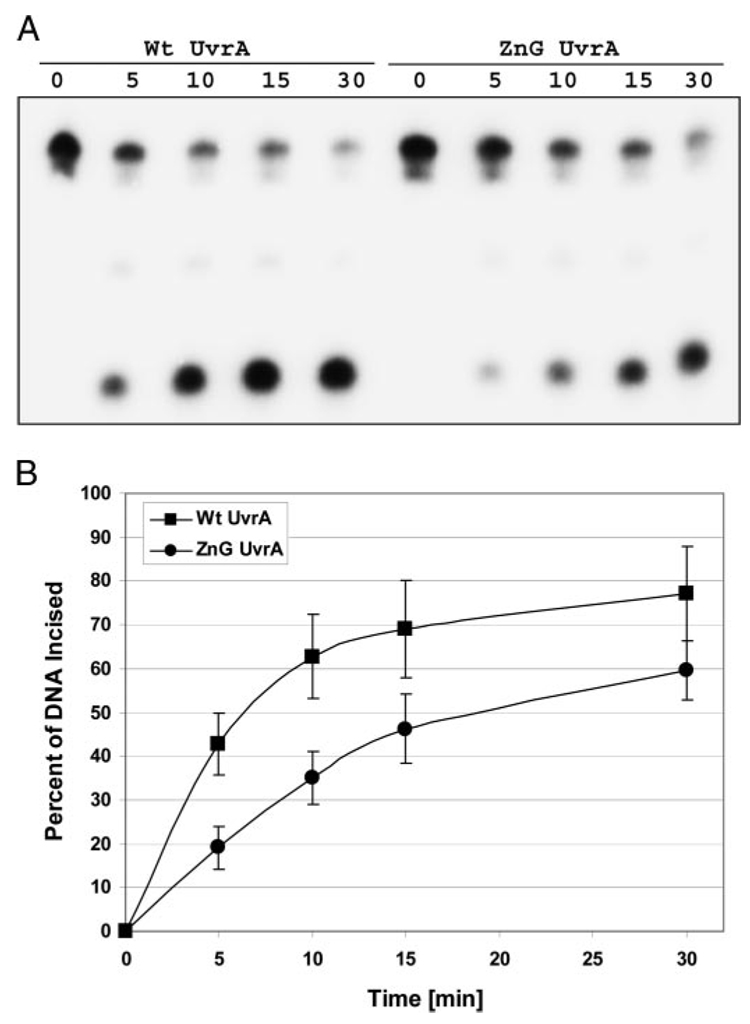
To test whether ZnG UvrA is capable of promoting nucleotide excision, a 50-bp double-stranded DNA (dsDNA) duplex containing a centrally located fluorescein adducted thymine was used to monitor the rate of incision in our oligonucleotide incision assay. The ZnG UvrA mutant displayed normal 5′ and 3′ incision patterns; however, the rate of the reaction was slower (Fig. 3). The greatest difference between the ZnG and Wt UvrA proteins was observed at the 5-min time point with ZnG UvrA displaying ~45% of Wt UvrA activity (Fig. 3B). Given the UV survival results, the relatively high level of incision observed from the ZnG UvrA samples was unexpected.
FIGURE 3. ZnG UvrA supports reduced incision activity.
A, incision of the 5′-end-labeled substrate (F2650/NDB) was monitored over time. The fluorescein adducted 50-bp duplex (F2650/NDB) was incubated with UvrB (100 nm), UvrC (50 nm) and 20 nm of the indicated UvrA protein for varying times at 55 °C in reaction buffer. The reactions were terminated with stop buffer, and the incision products were analyzed on a 10% denaturing polyacrylamide gel. B, graphic representation of the incision activity at various times using the indicated UvrA proteins. Data are reported as the mean ± S.D. (n = 4).
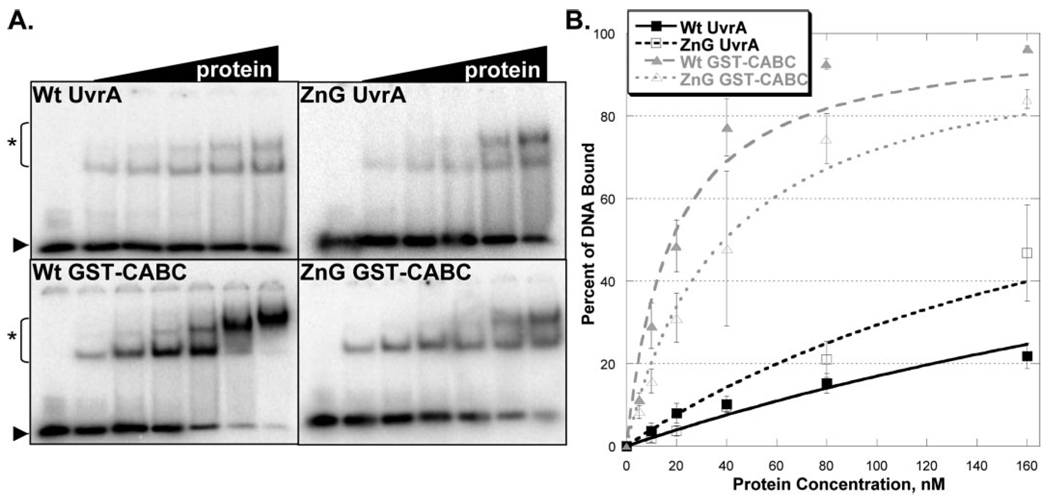
dsDNA Binding Is Not Defective in ZnG UvrA
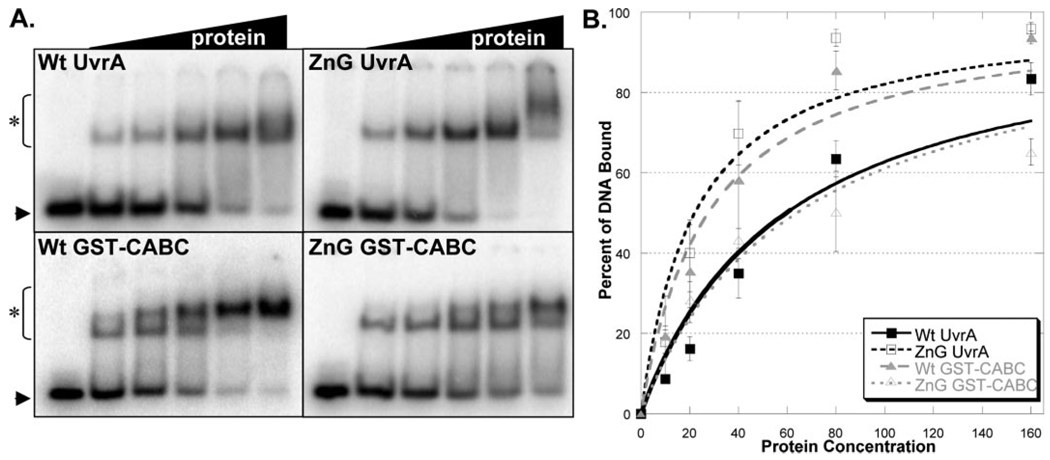
EMSAs are routinely used to determine the relative DNA binding affinities of proteins. The damaged dsDNA binding capacity of Wt and ZnG UvrA proteins were tested at various protein concentrations (10–160 nm) in the presence of ATP (1 mm) and MgCl2 (10 mm), Fig. 4A. The EMSA gels and binding isotherms, (Fig. 4B), indicate that ZnG UvrA, surprisingly, binds ~2-fold more tightly to dsDNA than Wt UvrA. The apparent equilibrium dissociation constant, Kd, for binding to damaged DNA by Wt UvrA and ZnG UvrA are 59 and 21.6 nm, respectively. These results are in stark contrast to the results obtained with the E. coli C763F UvrA, which had completely lost its ability to interact with DNA (16).
FIGURE 4. Damaged DNA binding profiles in the presence of ATP and magnesium.
A, EMSA was used to monitor the DNA binding properties of the proteins. Increasing amounts of protein (10–160 nm) were incubated with 2 nm NDB/F2650 duplex DNA in reaction buffer containing ATP (1 mm) and MgCl2 (10 mm) for 15 min at 37 °C. The reaction mixtures were separated on 3.5% polyacrylamide native gels in the presence of 1 mm ATP and 10 mm MgCl2. Asterisk (*) denotes protein-DNA complexes, and arrow denotes migration of free DNA. The data are reported as the mean ± S.D. (n = 3). B, quantitation of EMSAs in A. Binding isotherms were fitted by nonlinear regression analysis using Kaleidegraph® and the method of Schofield (17).
As noted previously, UvrA is believed to possess two DNA binding domains, one at the N-terminal and the other is at the C-terminal end of the protein (7). Therefore, the observed dsDNA binding of ZnG UvrA may have resulted from enhanced binding mediated by the UvrA N-terminal domain. To evaluate the C-terminal dsDNA binding independent of the UvrA N-terminal domain, glutathione S-transferase (GST) was fused to the N terminus of the C-terminal ABC domain of UvrA, the CABC domain. Wt GST-CABC and the corresponding ZnG GST-CABC protein, containing the eleven amino acid deletion, were created. Because dimerization is critical for UvrA and likewise for CABC (data not shown), the GST domain supplies the essential dimerization capacity for these chimeric proteins.
Wt and ZnG GST-CABC proteins were evaluated for their dsDNA binding abilities (5–160 nm). Both proteins readily formed protein-DNA complexes (Fig. 4A). At low protein concentrations (10–20 nm) and in the presence of ATP, the Wt GST-CABC and ZnG GST-CABC proteins showed little difference with regard to dsDNA binding (Fig. 4B). However, as the protein concentration increased the amount of DNA retained by ZnG GST-CABC was reduced relative to that for Wt GST-CABC, which is reflected in the apparent equilibrium dissociation constants, Kd values of 27 and 63 nm for Wt GST-CABC and ZnG GST-CABC, respectively.
dsDNA Binding Is Greatest for the GST Fusion Proteins in the Absence of ATP
Because ATP can modulate UvrA DNA binding, it was important to check the relative DNA binding in the absence of ATP. As can be seen, the relative amount of protein-DNA complexes generated by Wt and ZnG UvrA is severely impaired in the absence of ATP, compare Fig. 5A to Fig. 4A. The dramatic differences between the apparent Kd for Wt UvrA (488 nm) and ZnG UvrA, (240 nm), versus Wt GST-CABC (17 nm) and ZnG GST-CABC (38 nm) are consistent with the notion that, in the full-length proteins, the binding of ATP favors dimer formation and thus dsDNA binding (9). In sharp contrast, the two GST-CABC proteins, both Wt and ZnG GST-CABC, show significantly elevated dsDNA binding, relative to the full-length proteins, in the absence of ATP (Fig. 5). This is most likely because of the fact that a stable dimeric protein was created with the addition of GST to the CABC domain and therefore these proteins are less influenced by the presence or absence of nucleotide co-factor. Both GST-tagged proteins bind dsDNA efficiently; however, the ZnG GST-CABC possesses slightly less robust DNA binding capacity than Wt GST-CABC.
FIGURE 5. Damaged DNA binding profiles in the absence of ATP and magnesium.
A, EMSAs contained an increasing amount of protein (5–160 nm) and 2 nm NDB/F2650 duplex DNA in reaction buffer without ATP and MgCl2. Proteins were allowed to incubate with DNA for 15 min at 37 °C then were loaded onto a 3.5% native polyacrylamide gel. Asterisk (*) denotes protein-DNA complexes and arrow denotes migration of free DNA. The data are reported as the mean ± S.D. (n = 3). B, quantitation of EMSAs in A. Binding isotherms were fitted by nonlinear regression analysis using Kaleidegraph® and the method of Schofield (17).
Specific DNA Binding Is Compromised in ZnG UvrA, GST-CABC, and ZnG GST-CABC
Damage-specific binding was evaluated for each protein by comparing the observed binding to the fluorescein-containing duplex DNA with an undamaged DNA duplex of the same sequence, Table 1. Wt UvrA had a 2.2-fold greater affinity for damaged DNA than non-damaged DNA. In contrast, none of the other proteins displayed a significant difference in their relative DNA binding affinities.
TABLE 1. Apparent dissociation constants (Kd) for nonspecific and specific DNA binding.
Samples containing different amounts of the indicated protein were incubated with damaged (NDB/F2650) or non-damaged (NDB/NDT) duplex (2 nm) in the presence of ATP(1 mm) and MgCl2 (10 mm) and fractioned by electrophoresis as in Fig. 4. The apparent dissociation constant reported represents the mean of three or more independent determinations with associated error. The errors for the relative affinities were derived from the equation error = 1/M1 − 1/(M1 + S.E.), where M1 is the Ka, and the S.E. was derived from the curve fitting by Kaleidegraph® using the method of Schofield (17).
|
Kd(app) ± error |
||
|---|---|---|
| Non-damaged DNA | Damaged DNA | |
| nm | nm | |
| Wt UvrA | 129 ± 23 | 59 ± 8 |
| ZnG UvrA | 28 ± 4 | 22 ± 4 |
| Wt GST-CABC | 18 ± 3 | 27 ± 4 |
| ZnG GST-CABC | 77 ± 13 | 63 ± 7 |
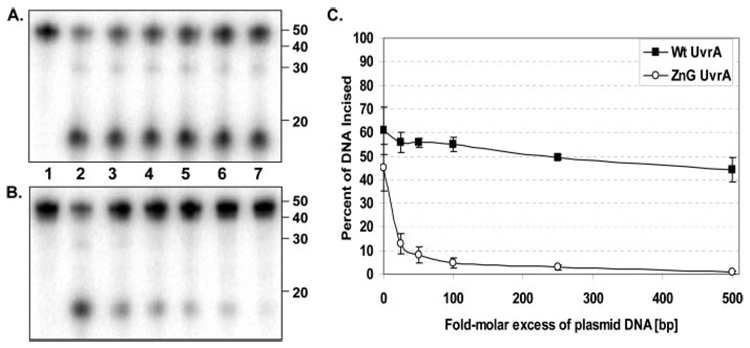
Oligonucleotide Incision in the Presence of Plasmid DNA
The loss of specific dsDNA binding for ZnG UvrA may explain why the protein cannot complement the wp2 uvrA− cells and why the rate of incision for the ZnG UvrA-containing sample is reduced relative to the Wt UvrA-containing sample. To test the hypothesis that the ZnG UvrA mutant nonspecific dsDNA binding was the cause for the decrease in incision, supercoiled undamaged plasmid DNA was titrated into the incision reaction. If the ZnG UvrA protein is unable to discriminate undamaged from damaged DNA, then addition of excess plasmid DNA should reduce the rate and extent of the oligonucleotide incision. As seen in Fig. 6, the addition of plasmid (25–500-fold molar excess of base pairs) to the incision reaction had little adverse effect on the rate of the reaction initiated by Wt UvrA but dramatically inhibited incision by ZnG UvrA. These results support our hypothesis that nonspecific DNA binding by ZnG UvrA renders the protein dysfunctional.
FIGURE 6. Excess plasmid DNA inhibits incision assays initiated by ZnG UvrA.
A, Wt UvrA (20 nm) or B, ZnG UvrA (20 nm) was incubated with increasing concentrations of pUC19 DNA (25–500 molar excess in bp DNA, relative to oligonucleotide concentration) for 10 min at room temperature prior to addition of 2 nm 5′ end-labeled "damaged" oligonucleotide duplex (F2650/NDB), UvrB (100 nm), and UvrC (50 nm) in reaction buffer. Proteins were incubated for 30 min at 55 °C then the reactions were terminated with stop buffer, and the incision products were analyzed on a 10% denaturing polyacrylamide gel. C, graphic representation of the incision activity with various concentrations of excess plasmid DNA. Data are reported as the mean ± S.D. (n = 3).
EMSA Reveals ZnG UvrA Loads UvrB onto DNA Less Efficiently
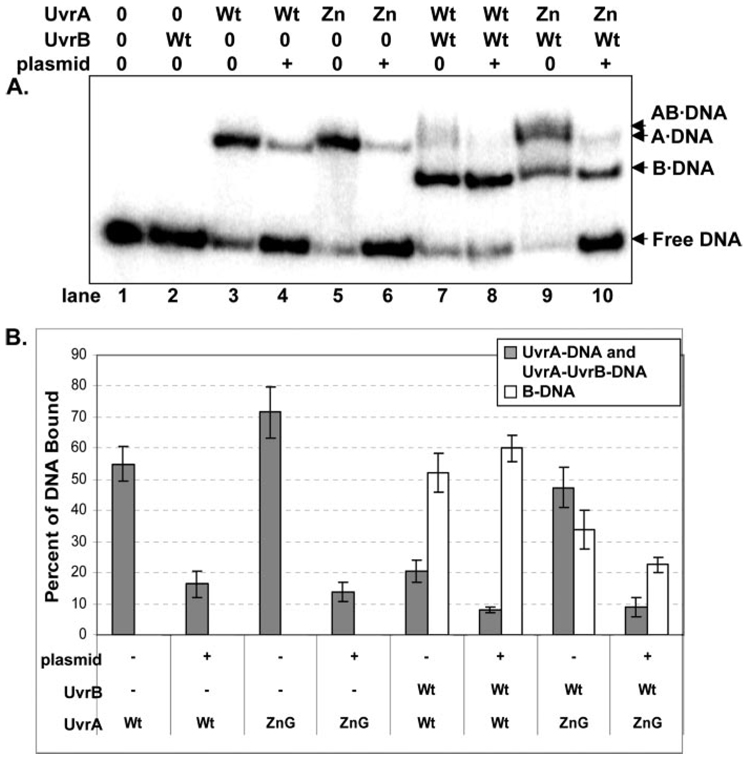
In the nucleotide excision repair reaction scheme, the Uvr proteins employ a kinetic proof reading mechanism for damage processing (4). UvrA first makes contact with DNA, detects the DNA damage, and then passes the DNA off to UvrB so that UvrB can verify whether a bona fide lesion is present (2, 26). If a lesion is not present, UvrB will not load onto non-damaged DNA. Thus, UvrB is believed to signal to UvrA to dissociate, and both proteins dissociate to continue the search for damage. If on the other hand a lesion is present, UvrB will take possession of the DNA, activate its cryptic ATPase site, and signal UvrA to dissociate (1, 2, 26).
If ZnG UvrA binds DNA nonspecifically, then it will be defective in loading UvrB onto DNA. As can be seen, ZnG UvrA is impaired in its ability to load UvrB onto a damaged oligonucleotide (Fig. 7A, compare lanes 7 and 9; Fig. 7B). ZnG UvrA retains a greater proportion of the DNA than Wt protein, therefore 37% less DNA is transferred to Wt UvrB. The presence of an excess amount of undamaged DNA should impede the rate at which ZnG UvrA is capable of searching for the damaged oligonucleotide and further diminish the amount of DNA transferred to UvrB. Addition of excess plasmid DNA (100-fold molar excess of base pairs of DNA) to the EMSA reactions reduced the amount of DNA bound by each protein individually (Fig. 7A, compare lanes 4 and 6). The DNA transfer reaction between Wt UvrA and Wt UvrB was essentially unaffected by the addition of plasmid, between 50 and 60% of the DNA is transferred to UvrB (compare lane 7 and 8). Whereas in the ZnG UvrA-containing lane, lane 9, the DNA transfer reaction to UvrB is inhibited by 35% relative to the wild-type proteins, lane 7, because the DNA is captured in non-productive ZnG UvrA complexes. Upon adding exogenous DNA, the total amount of oligonucleotide engaged in a protein-DNA complex is reduced by 53%, relative to the total protein-DNA complexes in lane 8. When plasmid DNA is present, ZnG UvrA cannot proceed through its catalytic cycle as efficiently as Wt UvrA, and therefore these results suggest that the excess plasmid DNA retained ZnG UvrA and consequently reduced the overall interaction with the oligonucleotide.
FIGURE 7. Reduced loading of UvrB onto sites of DNA damage by ZnG UvrA.
A, Wt UvrA (Wt, 20 nm) or ZnG UvrA (Zn, 20 nm) were incubated alone or with Wt UvrB protein (100 nm) for 30 min at 55 °C in the presence of 2 nm F2650/NDB duplex DNA. The protein-DNA complexes were separated on 4% native polyacrylamide gels containing ATP (1 mm) and MgCl2 (10 mm). B, quantitation of EMSAs in A, reporting the percent of DNA bound to the various protein-DNA complexes. The data are reported as the mean ± S.D. (n = 3). Gray bars indicate the percentage of DNA bound to UvrA either as the A2·DNA or AB·DNA complex, while white bars represent the B·DNA complexes.
Our data suggest that ZnG UvrA binds to the DNA nonspecifically and attempts to present undamaged DNA to UvrB. It is believed that UvrB will not load onto undamaged DNA. To confirm that Bca UvrB will not load onto undamaged DNA, Wt and ZnG UvrA were incubated with undamaged oligonucleotide in the presence of Wt UvrB. Neither protein was able to load more than 2% of Wt UvrB onto the undamaged oligonucleotide (data not shown).
One reason for the poor loading of UvrB by the ZnG UvrA mutant could be due to an impaired protein-protein interaction between these two proteins. Domain 2 of Bca UvrB is the main interaction domain for Bca UvrA (27).3 Previously, we have shown that a domain 2 mutant of UvrB, R183E UvrB, was not loaded onto damaged DNA by UvrA and failed to activate its ATPase because of an UvrA-UvrB protein-protein interaction defect (27). Therefore, to rule out a protein-protein interaction defect, the Δβ-hairpin UvrB mutant was used in the EMSA experiments to demonstrate that the ZnG UvrA is fully competent in its ability to recruit UvrB to the UvrA·DNA complex. The Δβ-hairpin UvrB mutant has no means to discriminate damaged from undamaged DNA, is not capable of taking possession of the DNA from UvrA (26), but possesses the cryptic UvrB ATPase activity (28). Utilization of the Δβ-hairpin UvrB mutant provides us the opportunity to trap the UvrA proteins in a ternary complex (UvrA·Δβ-hairpin UvrB·DNA) because the complex dissociates slowly. The presence of the slower migrating band in Fig. 8 is indicative of the ternary complex. Fig. 8 also shows that the ZnG UvrA protein is capable of recruiting UvrB to the oligonucleotide as well as Wt UvrA. In addition, we have conducted pull-down experiments between a GST-UvrB domain 2 construct and Wt UvrA or ZnG UvrA. This GST-UvrB domain 2 construct was capable of pulling down either the Wt or ZnG UvrA proteins (data not shown). Together these two experiments indicate that ZnG UvrA can interact with UvrB, but falls to load UvrB at the site of damage.
FIGURE 8. ZnG UvrA forms normal protein-protein interactions with Δβ-Hairpin UvrB.
Wt UvrA (20 nm) or ZnG UvrA (20 nm) were incubated alone or with Δβ-Hairpin UvrB (ΔβH, 100 nm) for 15 min at 55 °C in the presence of 2 nm NDB/F2650 duplex DNA in reaction buffer. The protein-DNA complexes were separated on 4% native polyacrylamide gels containing ATP (1 mm) and MgCl2 (10 mm). Representative gel (n = 3).
ZnG UvrA Fails to Activate the UvrB Cryptic ATPase
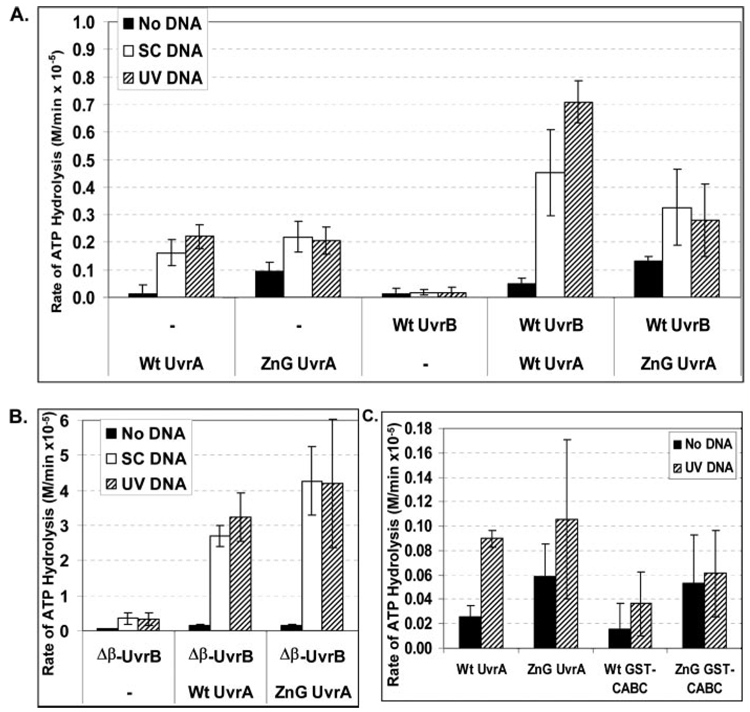
The binding and hydrolysis of nucleotides by UvrA alters its DNA binding and protein-protein interaction capabilities. When ATPase assays are performed with E. coli UvrA, UvrB, and unmodified DNA, a modest amount of ATPase activity is observed and, generally, it is less than 2-fold more than that observed with UvrA alone (29–31). Therefore, if ZnG UvrA is binding to non-damaged DNA sites it should not activate the UvrB-damaged DNA-dependent ATPase. Using an enzyme-coupled assay (27), the relative ATP hydrolysis rates for all of the proteins were measured in the absence of DNA and in the presence of supercoiled plasmid DNA and UV-irradiated plasmid DNA.
When the reactions were conducted at 55 °C and in the absence of DNA, ZnG UvrA possessed ~8-fold higher basal ATPase activity than Wt UvrA, see Fig. 9. In the presence of DNA, either supercoiled (SC DNA) or UV-irradiated DNA (UV DNA), the individual UvrA proteins had a similar level of ATPase activity. However, ZnG UvrA failed to activate UvrB ATPase in the presence of supercoiled or UV-irradiated DNA. Fig. 9A. The inability of ZnG UvrA to engage the UvrB ATPase is not because of a protein-protein interaction defect, as shown in Fig. 8 because ZnG UvrA is capable of activating the ATPase of the Δβ-hairpin UvrB mutant (Fig. 9B).
FIGURE 9. ZnG UvrA fails to unlock UvrB's crytpic ATPase.
A, conversion of ATP to ADP by Wt UvrA (50 nm), ZnG UvrA (50 nm), and Wt UvrB (100 nm) at 55 °C was monitored using a coupled enzyme assay system consisting of pyruvate kinase and lactic dehydrogenase, which links the hydrolysis of ATP to the oxidation of NADH (see "Experimental Procedures"). The data are reported as the mean ± S.D. (n = 3 or 4). B, ATPase activity of Wt UvrA or ZnG UvrA (50 nm) in the presence of Δβ-Hairpin UvrB (100 nm) was assayed at 55 °C. The data are reported as the mean ± S.D. (n = 3) C, ATPase activity of Wt UvrA, ZnG UvrA, Wt GST-CABC, or ZnG GST-CABC (50 nm) was examined at 37 °C. Data are reported as the mean and range, n = 2. Black bar indicates ATPase assayed in the absence of DNA (No DNA), white bar indicates addition of supercoiled plasmid DNA (SC DNA, 10 ng/µl) and hatched bar indicated addition of 10 ng/µl UV-irradiated plasmid DNA.
To investigate the ATPase activity of our GST fusion proteins, we conducted a series of ATPase reactions at 37 °C because GST is a thermolabile protein (32). At this temperature and in the presence of UV DNA, Wt UvrA retains 40% of its ATPase activity relative to the ATPase activity observed at 55 °C. In addition, in the presence of Wt UvrA and Wt UvrB, the UV DNA-stimulated ATPase activity (3.3-fold) was still observed. At 37 °C and in the presence of UV irradiated DNA, ZnG UvrA possesses 51% of its ATPase activity relative to the ATPase activity observed at 55 °C and in combination with UvrB did not exhibit the UV DNA-stimulated ATPase, as was the case at 55 °C (Fig. 9A).
At 37 °C, the overall ATPase activity for Wt GST-CABC and ZnG GST-CABC was reduced relative to the ATPase activity of the full-length proteins (Fig. 9C). Compared with full-length UvrA at 37 °C, Wt GST-CABC retained 59% (no DNA) and 41% (UV DNA) activity, whereas ZnG GST-CABC displayed 90% (no DNA) and 58% (UV DNA) of the ZnG UvrA ATPase activity. In the absence of DNA, the ZnG GST-CABC mutant displayed a 2.3-fold higher level of ATPase activity than Wt GST-CABC. In the presence of UV irradiated DNA, Wt GST-CABC and ZnG GST-CABC had a similar amount of activity. If ATP hydrolysis is required for dissociation, then the lower ATPase activity of the GST-CABC proteins could contribute to their observed tighter DNA binding. In addition, these results support the conclusion that UvrA folds into two functional domains and that the C-terminal domain remains a functional ATPase when the N-terminal domain is replaced with an artificial dimerization domain, GST.
The C-terminal Zinc Finger Domain Is Insufficient for DNA Binding
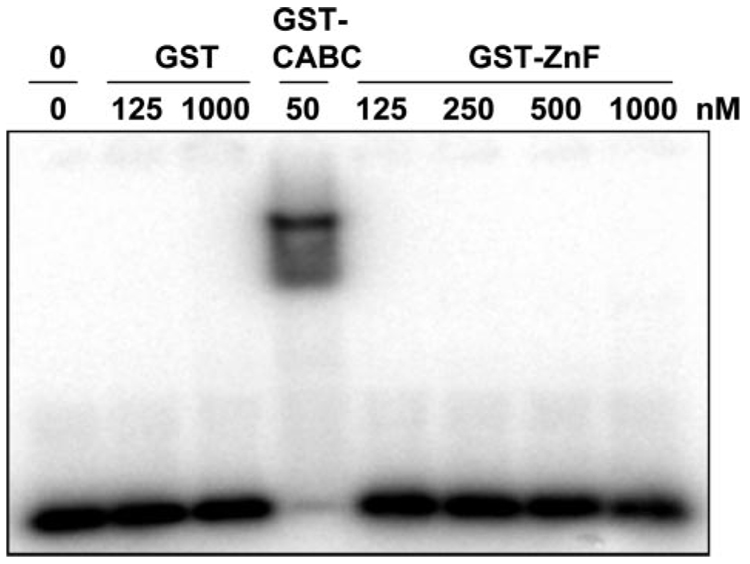
Our data show that deletion of the eleven amino acids within the C-terminal zinc finger reduces the observed DNA binding mediated by ZnG GST-CABC (compare Wt GST-CABC and ZnG GST-CABC DNA binding in Fig. 5 and Table 1). Because isolated zinc finger domains have been shown to bind to DNA separately, a construct containing residues 667–820 was designed to determine if the UvrA C-terminal zinc finger domain possessed a DNA binding ability, independent of the remainder of the protein. This domain was chosen for analysis because we reasoned it might be an autonomously folded unit, which lies between the conserved Walker A and signature sequences of the ABC ATPase motifs. Overexpression of this fragment of UvrA fused to GST produced a highly soluble protein; however, our attempts to make smaller fragments from within this region yielded mostly insoluble proteins. The protein produced, GST-ZnF, was purified and analyzed by EMSA. As can be seen in Fig. 10, we tested a range of protein concentrations (125 nm to 1 µm). However, the GST-ZnF protein showed no detectable DNA binding under our reaction conditions. In addition, we conducted protein-DNA cross-linking experiments in an attempt to detect weak or transient GST-ZnF·DNA interactions. We have successfully used this strategy on UvrA and UvrB (26), however we were unable to detect a GST-ZnF·DNA protein cross-link using the same technique (data not shown).
FIGURE 10. No apparent DNA binding by GST-ZnF.
GST (125 nm or 1 µm), GST-CABC (50 nm) or increasing concentrations of GST-ZnF (125 nm to 1 µm) were incubated with F2650/NDB in reaction buffer for 15 min at 37 °C. The samples were then loaded onto a 4% native polyacrylamide gel, and electrophoresis was carried out for 1 h at 100 V.
DISCUSSION
Nucleotide excision repair is quite remarkable among DNA repair mechanisms because it possesses the ability to recognize and repair a wide set of structurally unrelated DNA lesions (1, 33). UvrA was the first protein identified as being important for the removal of UV-induced DNA lesions, yet little is known about its structure and mechanism of damage recognition. In an attempt to elucidate how UvrA mediates damage recognition, we have created and evaluated an eleven amino acid deletion within the UvrA C-terminal zinc finger domain. We have shown that ZnG UvrA, a protein with an eleven amino acid deletion within the C-terminal zinc finger, binds dsDNA better than Wt UvrA and that the C-terminal zinc finger region does not display detectable DNA binding. These results suggest that, contrary to previously published work (7, 16), the C-terminal zinc finger does not interact with DNA directly but rather, may regulate UvrA DNA binding by some indirect mechanism.
Specific DNA binding by UvrA is a dynamic process that involves the ABC ATPase motifs. It is thought that specific binding is achieved by limiting the amount of time UvrA resides on undamaged DNA, i.e. the dissociation rate from undamaged DNA (34, 35). It has been shown that damage recognition is regulated by the ATPase motifs because mutagenesis of the C-terminal Walker A motif generates UvrA proteins with reduced specific DNA binding properties (11, 34, 35). Given the fact that the ABC ATPase motifs regulate the DNA binding properties of UvrA, our data are consistent with the conclusion that deletion of the zinc finger amino acids alters the intrinsic ATPase properties of the ZnG UvrA protein and indirectly its DNA binding properties.
Other laboratories have investigated the role of the UvrA C-terminal zinc finger. A previous study, in which one of the zinc-coordinating cysteine residues was substituted with phenylalanine, demonstrated that the mutant protein lost its DNA binding ability (16). Therefore the authors concluded that the C-terminal zinc finger region is responsible for the UvrA DNA binding capacity (16). In another study, in vivo survival after UV irradiation was monitored for two C-terminal cysteine mutants, C763S and C763G. Both failed to complement the uvrA deletion strain (14). In this study, both proteins could not be purified because of poor solubility and therefore could not be characterized biochemically. Our data clearly show that the C-terminal domain of UvrA can bind DNA in the absence of the amino acids within the zinc finger. Furthermore, experiments using the isolated zinc finger region, and a sensitive protein-DNA cross-linking technique, do not support the hypothesis that the UvrA protein makes direct contact with DNA via its C-terminal zinc finger.
The XPA protein is believed to be important for damage recognition in the mammalian NER reaction (36). The UvrA zinc fingers have been compared with XPA zinc finger since they are all classified as the C4-type; however, the XPA zinc fingers do not share the conserved glycine residues that follow the CX2C motif as in UvrA. Regardless, it has been shown that mutation or deletion of the XPA zinc finger leads to a UV-sensitive phenotype (37, 38). Amino acids 98–219 are the central domain of XPA, which includes its zinc finger and is defined as its minimal DNA binding domain (39). Chemical shift perturbation experiments using the 15N-labeled minimal DNA binding domain of XPA revealed that a basic cleft adjacent to the zinc finger was responsible for DNA binding. Because of the observation that the zinc finger did not directly interact with the DNA, but mutations of it caused a UV-sensitive phenotype, the authors proposed that the XPA zinc finger is required to support the structure of the DNA binding domain through hydrophobic contacts. In addition, they speculated that loss of the zinc finger structure would lead to unfolding and conformational distortions in the DNA binding domain. We suggest that a similar mechanism may be at work within UvrA. The UvrA C-terminal zinc finger may not directly interact with DNA, but it may be important for structural integrity by bringing the ABC transporter motifs into juxtaposition.
The majority of proteins in the ABC ATPase super family mediate the transport of small molecules. These transport type-ABC ATPases are known to undergo dramatic protein-protein rearrangements following ligand binding. The crystal structure of MalK has been equated with tweezers-like motion (40, 41). Therefore, it is not unreasonable to suggest that UvrA might undergo dramatic structural changes upon DNA binding, which could involve the C-terminal zinc finger as well. There is evidence that the N- and C-terminal domains of UvrA may need to interact to achieve specific DNA binding because when the N- and C-terminal domains of UvrA were separated, each isolated domain possessed DNA binding but lacked specific DNA binding (7). This latter result is consistent with the work presented here, that the C-terminal domain while showing tight DNA binding, shows no ability for damage discrimination.
Two other regions of UvrA have been evaluated for their involvement in DNA binding, a helix-turn-helix motif in the N terminus, and a glycine-rich region in the C terminus (15, 42, 43). We have modeled the three-dimensional structure of the Bca UvrA C-terminal ABC domains based on the monomeric HisP structure ((25), PDB code 1B0U) and the dimeric Rad 50 structure ((44), PDB code 1F2U), Fig. 11. It is clear that the N-terminal helix-turn-helix motif, previously identified as being important for DNA binding by UvrA, is in fact part of the ABC ATPase fold and furthermore not predicted to lie on the surface of the protein (3).
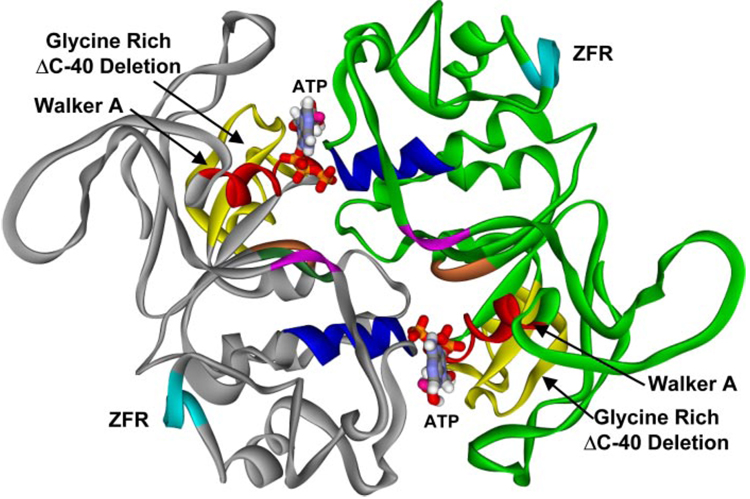
FIGURE 11. Structural model of the B. caldotenax UvrA dimer.
A model of the UvrA dimer was created based on the structural similarity between the ABC ATPase domains of UvrA and Rad50 (PDB codes: 1F2T, 1F2U). The dimer consists of two monomers, one in gray and the other in green. The ABC ATPase motifs: the Walker A (red), signature sequence (blue), Q-loop (purple), Walker B (dark green), and His loop (orange) are shown. The zinc finger region, containing 88 amino acids, has been deleted in the model, and six alanines were substituted (pale blue, ZFR). The position of the glycine-rich ΔC-40 deletion is depicted in yellow.
We speculate that the deletion of the C-terminal glycine-rich region produces a non-functional protein due to the loss of amino acids supporting the Walker A residues, the essential ATP binding motif of the ABC ATPase. Consequently, this deletion may disrupt the ABC ATPase dimer interface and could explain why both of the UvrA proteins with the deleted glycine-rich region had to be purified by refolding the protein from the insoluble fraction of cell extracts (42, 43). In Fig. 11, the amino acids highlighted in yellow represent the corresponding glycine-rich C-40 deletion previously described in E. coli (42). Based on our structural modeling of UvrA we predict that both regions, the helix-turn-helix motif and the glycine-rich sequences, have an indirect effect on DNA binding because of altered ATPase function or structure and therefore are not directly responsible for DNA binding by UvrA. Additional site-directed mutagenesis and protein-DNA cross-linking will be required to elucidate the true DNA binding domains within UvrA.
Finally, our working model suggests that the ZnG UvrA protein fails to distinguish damaged from non-damaged DNA because of a regulatory defect controlling damage discrimination. Assuming that the on rates of DNA binding are diffusion limited, then the ZnG UvrA protein must have a slower off rate from non-damaged DNA. Experiments deriving the on and off rates are beyond the scope of this present study and are currently in progress using fluorescence spectroscopic approaches.
Acknowledgments
We thank Dr. Leroy Worth, Dr. William Beard, and Dr. Caroline Kisker for critically reading the manuscript and Dr. Robert Petrovich and Lori Edwards of the NIEHS protein core facility for their assistance with the production of Wt UvrA.
Footnotes
This research was supported by the Intramural Research Program of the NIEHS, National Institutes of Health.
The abbreviations used are: NER, nucleotide excision repair; ABC ATPase, ATP-binding cassette ATPase; CABC, UvrA C-terminal ABC ATPase domain; dsDNA, double-stranded DNA; EMSA, electrophoretic mobility shift assay; GST, glutathione S-transferase; SC DNA, supercoiled DNA; UV DNA, UV-irradiated DNA; IPTG, isopropyl-1-thio-β-d-galactopyranoside; DTT, dithiothreitol; Wt, wild type; Bca, Bacillus caldotenax; RMS, root mean-square; PDB, Protein Data Bank.
D. L. Croteau, unpublished observations.
REFERENCES
- 1.Van Houten B, Croteau DL, DellaVecchia MJ, Wang H, Kisker C. Mutat. Res. 2005;577:92–117. doi: 10.1016/j.mrfmmm.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 2.Truglio JJ, Croteau DL, Van Houten B, Kisker C. Chem. Rev. 2006;106:233–252. doi: 10.1021/cr040471u. [DOI] [PubMed] [Google Scholar]
- 3.Croteau DL, Dellavecchia MJ, Skorvaga M, Van Houten B. In: DNA Damage Recognition. Wolfram S, Kow YW, Doetsch PW, editors. Boca Raton, FL: Taylor & Francis; 2005. pp. 111–138. [Google Scholar]
- 4.Reardon JT, Sancar A. Cell Cycle. 2004;3:141–144. [PubMed] [Google Scholar]
- 5.Doolittle RF, Johnson MS, Husain I, Van Houten B, Thomas DC, Sancar A. Nature. 1986;323:451–453. doi: 10.1038/323451a0. [DOI] [PubMed] [Google Scholar]
- 6.Navaratnam S, Myles GM, Strange RW, Sancar A. J. Biol. Chem. 1989;264:16067–16071. [PubMed] [Google Scholar]
- 7.Myles GM, Sancar A. Biochemistry. 1991;30:3834–3840. doi: 10.1021/bi00230a005. [DOI] [PubMed] [Google Scholar]
- 8.Orren DK, Sancar A. Proc. Natl. Acad. Sci. U. S. A. 1989;86:5237–5241. doi: 10.1073/pnas.86.14.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mazur S, Grossman L. Biochemistry. 1991;30:4432–4443. doi: 10.1021/bi00232a009. [DOI] [PubMed] [Google Scholar]
- 10.Reardon JT, Nichols AF, Keeney S, Smith CA, Taylor JS, Linn S, Sancar A. J. Biol. Chem. 1993;268:21301–21308. [PubMed] [Google Scholar]
- 11.Thiagalingam S, Grossman L. J. Biol. Chem. 1993;268:18382–18389. [PubMed] [Google Scholar]
- 12.Berg JM, Shi Y. Science. 1996;271:1081–1085. doi: 10.1126/science.271.5252.1081. [DOI] [PubMed] [Google Scholar]
- 13.Matthews JM, Sunde M. IUBMB Life. 2002;54:351–355. doi: 10.1080/15216540216035. [DOI] [PubMed] [Google Scholar]
- 14.Visse R, de Ruijter M, Ubbink M, Brandsma JA, van de Putte P. Mutat. Res. 1993;294:263–274. doi: 10.1016/0921-8777(93)90009-6. [DOI] [PubMed] [Google Scholar]
- 15.Wang J, Grossman L. J. Biol. Chem. 1993;268:5323–5331. [PubMed] [Google Scholar]
- 16.Wang J, Mueller KL, Grossman L. J. Biol. Chem. 1994;269:10771–10775. [PubMed] [Google Scholar]
- 17.Schofield MJ, Lilley DM, White MF. Biochemistry. 1998;37:7733–7740. doi: 10.1021/bi980399s. [DOI] [PubMed] [Google Scholar]
- 18.Caplan AJ, Douglas MG. J. Cell Biol. 1991;114:609–621. doi: 10.1083/jcb.114.4.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kimura Y, Yahara I, Lindquist S. Science. 1995;268:1362–1365. doi: 10.1126/science.7761857. [DOI] [PubMed] [Google Scholar]
- 20.Ziegelhoffer T, Lopez-Buesa P, Craig EA. J. Biol. Chem. 1995;270:10412–10419. doi: 10.1074/jbc.270.18.10412. [DOI] [PubMed] [Google Scholar]
- 21.Li J, Qian X, Sha B. Structure (Camb.) 2003;11:1475–1483. doi: 10.1016/j.str.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 22.Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, Khanna A, Marshall M, Moxon S, Sonnhammer EL, Studholme DJ, Yeats C, Eddy SR. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Diederichs K, Diez J, Greller G, Muller C, Breed J, Schnell C, Vonrhein C, Boos W, Welte W. EMBO J. 2000;19:5951–5961. doi: 10.1093/emboj/19.22.5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith PC, Karpowich N, Millen L, Moody JE, Rosen J, Thomas PJ, Hunt JF. Mol. Cell. 2002;10:139–149. doi: 10.1016/s1097-2765(02)00576-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hung LW, Wang IX, Nikaido K, Liu PQ, Ames GF, Kim SH. Nature. 1998;396:703–707. doi: 10.1038/25393. [DOI] [PubMed] [Google Scholar]
- 26.DellaVecchia MJ, Croteau DL, Skorvaga M, Dezhurov SV, Lavrik OI, Van Houten B. J. Biol. Chem. 2004;279:45245–45256. doi: 10.1074/jbc.M408659200. [DOI] [PubMed] [Google Scholar]
- 27.Truglio JJ, Croteau DL, Skorvaga M, DellaVecchia MJ, Theis K, Mandavilli BS, Van Houten B, Kisker C. EMBO J. 2004;23:2498–2509. doi: 10.1038/sj.emboj.7600263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Skorvaga M, Theis K, Mandavilli BS, Kisker C, Van Houten B. J. Biol. Chem. 2002;277:1553–1559. doi: 10.1074/jbc.M108847200. [DOI] [PubMed] [Google Scholar]
- 29.Lin JJ, Phillips AM, Hearst JE, Sancar A. J. Biol. Chem. 1992;267:17693–17700. [PubMed] [Google Scholar]
- 30.Moolenaar GF, Visse R, Ortiz-Buysse M, Goosen N, van de Putte P. J. Mol. Biol. 1994;240:294–307. doi: 10.1006/jmbi.1994.1447. [DOI] [PubMed] [Google Scholar]
- 31.Moolenaar GF, Hoglund L, Goosen N. EMBO J. 2001;20:6140–6149. doi: 10.1093/emboj/20.21.6140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park SM, Jung HY, Chung KC, Rhim H, Park JH, Kim J. Biochemistry. 2002;41:4137–4146. doi: 10.1021/bi015961k. [DOI] [PubMed] [Google Scholar]
- 33.Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Annu. Rev. Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 34.Myles GM, Hearst JE, Sancar A. Biochemistry. 1991;30:3824–3834. doi: 10.1021/bi00230a004. [DOI] [PubMed] [Google Scholar]
- 35.Thiagalingam S, Grossman L. J. Biol. Chem. 1991;266:11395–11403. [PubMed] [Google Scholar]
- 36.Batty DP, Wood RD. Gene (Amst.) 2000;241:193–204. doi: 10.1016/s0378-1119(99)00489-8. [DOI] [PubMed] [Google Scholar]
- 37.Miyamoto I, Miura N, Niwa H, Miyazaki J, Tanaka K. J. Biol. Chem. 1992;267:12182–12187. [PubMed] [Google Scholar]
- 38.Asahina H, Kuraoka I, Shirakawa M, Morita EH, Miura N, Miyamoto I, Ohtsuka E, Okada Y, Tanaka K. Mutat. Res. 1994;315:229–237. doi: 10.1016/0921-8777(94)90034-5. [DOI] [PubMed] [Google Scholar]
- 39.Kuraoka I, Morita EH, Saijo M, Matsuda T, Morikawa K, Shirakawa M, Tanaka K. Mutat. Res. 1996;362:87–95. doi: 10.1016/0921-8777(95)00038-0. [DOI] [PubMed] [Google Scholar]
- 40.Chen J, Lu G, Lin J, Davidson AL, Quiocho FA. Mol. Cell. 2003;12:651–661. doi: 10.1016/j.molcel.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 41.Lu G, Westbrooks JM, Davidson AL, Chen J. Proc. Natl. Acad. Sci. U. S. A. 2005;102:17969–17974. doi: 10.1073/pnas.0506039102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Claassen LA, Grossman L. J. Biol. Chem. 1991;266:11388–11394. [PubMed] [Google Scholar]
- 43.Kulkarni AS, Khalap N, Joshi VP. Indian J. Exp. Biol. 2006;44:7–13. [PubMed] [Google Scholar]
- 44.Hopfner KP, Karcher A, Shin DS, Craig L, Arthur LM, Carney JP, Tainer JA. Cell. 2000;101:789–800. doi: 10.1016/s0092-8674(00)80890-9. [DOI] [PubMed] [Google Scholar]
- 45.Gattiker A, Michoud K, Rivoire C, Auchincloss AH, Coudert E, Lima T, Kersey P, Pagni M, Sigrist CJ, Lachaize C, Veuthey AL, Gasteiger E, Bairoch A. Comput. Biol. Chem. 2003;27:49–58. doi: 10.1016/s1476-9271(02)00094-4. [DOI] [PubMed] [Google Scholar]