Abstract
Nucleotide excision repair (NER) is responsible for the recognition and removal of numerous structurally unrelated DNA lesions. In prokaryotes, the proteins UvrA, UvrB and UvrC orchestrate the recognition and excision of aberrant lesions from DNA. Despite the progress we have made in understanding the NER pathway it remains unclear how the UvrA dimer interacts with DNA to facilitate DNA damage recognition. The purpose of this study was to define amino acids residues in UvrA that provide binding energy to DNA. Based on conservation among ~300 UvrA sequences and 3D-modeling, two positively charged residues, Lys680 and Arg691, were predicted to be important for DNA binding. Mutagenesis and biochemical analysis of Bacillus caldontenax UvrA variant proteins containing site directed mutations at these residues demonstrate that Lys680 and Arg691 make a significant contribution toward the DNA binding affinity of UvrA. Replacing these side chains with alanine or negatively charged residues decreased UvrA binding 3–37-fold. Survival studies indicated that these mutant proteins complemented a WP2 uvrA− strain of bacteria 10% to 100% of WT UvrA levels. Further analysis by DNase I footprinting of the double UvrA mutant revealed that the UvrA DNA binding defects caused a slower rate of transfer of DNA to UvrB. Consequently, the mutants initiated the oligonucleotide incision assay nearly as well as WT UvrA thus explaining the observed mild phenotype in the survival assay. Based on our findings we propose a model of how UvrA binds to DNA.
1. Introduction
Damage recognition in the nucleotide excision repair pathway is an ATP-dependent multi-step process, for recent reviews see [1–3]. While several models have been proposed for prokaryotic damage recognition, most suggest that UvrA, as part of an UvrA2B or UvrA2B2 complex [4], first scans DNA for altered base-pair conformations. Upon encountering an altered DNA structure UvrA delivers UvrB to the DNA by providing two important functions: 1) UvrA bends the DNA at the site of the damage and helps to open the two strands, and 2) UvrA relieves UvrB’s domain 4 autoinhibitory domain thereby activating UvrB's cryptic ATPase allowing transfer of the damaged DNA to UvrB [5–7]. UvrB then searches the DNA for the exact site of the damaged nucleotide using aromatic side-chains along a β-hairpin motif that is inserted between the two strands of DNA [8–12]. During this verification step, UvrB takes possession of the DNA causing UvrA to dissociate. The formation of a stable UvrB·DNA complex increases repair specificity by effectively separating damage recognition from the endonuclease events. In the second phase of the reaction, UvrB recruits UvrC and holds the DNA in the proper geometry, possibly by ATP binding and/or hydrolysis [13,14], to facilitate cutting by UvrC. UvrC contains two nuclease centers and each site is responsible for a single strand cut [15]. The first cut is mediated by the GIY-YIG type-nuclease in the N-terminus of UvrC [16]. It produces a single strand break four to five nucleotides 3′ to the DNA lesion. The second incision is mediated by an RNase H-like nuclease in the C-terminus of UvrC [17,18]. This endonuclease cleaves eight nucleotides 5′ to the lesion. UvrD and Pol I are responsible for the release of the 12-nucleotide fragment containing the DNA lesion and re-synthesis of the DNA [19–21]. DNA ligase seals the nick to finalize the reaction [21].
UvrA is a 105 kD protein with multiple domains and structural motifs [22]. Most notably, it contains two ATP binding cassette-type (ABC) ATPase domains separated by a flexible protease sensitive hinge region. Like all ABC ATPases, Bacillus caldotenax UvrA (Bca UvrA) has the conserved ABC ATPase elements such as: the Walker A, Q-loop, signature sequence, Walker B and His-loop (Fig. 1a). While most ABC ATPase domains are contiguous, UvrA has sequences between the ABC ATPase Walker A and signature sequences. A C4-type zinc finger is present within each insertion segment. Previously, it was thought that UvrA interacts with DNA via the zinc finger at the C-terminus of the protein [23,24]. Recently, we observed increased DNA binding when amino acids within this zinc finger were deleted [25]. Thus, the C-terminus zinc finger of UvrA is not required for DNA binding. However, these studies also revealed that the zinc finger is required for efficient damage recognition in the context of large amounts of non-damaged DNA and/or subsequent processing by UvrB. Based on these findings it was reasoned that the true DNA binding domain resides in another region of the protein.
Fig. 1. Region of UvrA under investigation.
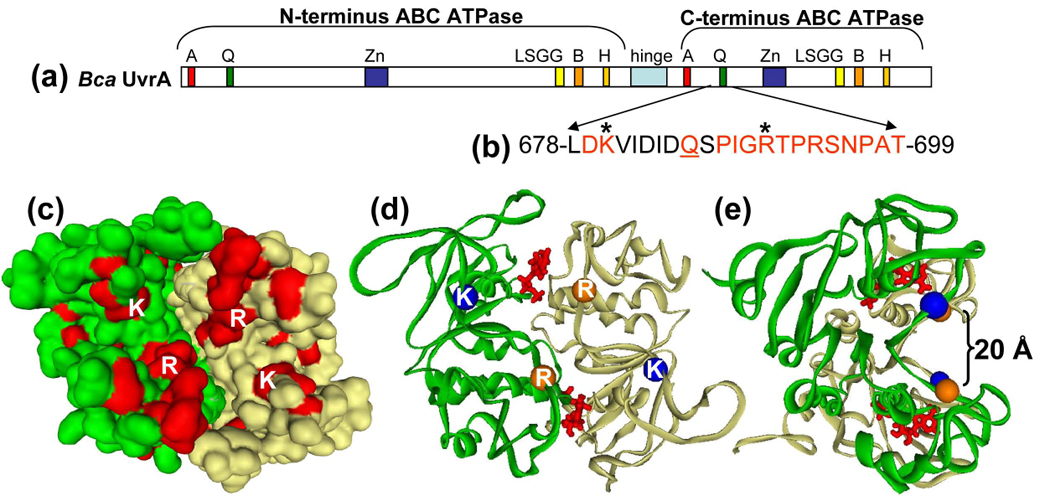
Panel a, Linear sequence of Bca UvrA. The important features of UvrA are denoted by the boxed areas: Walker A, A; Q-loop, Q; zinc finger, Zn; signature sequence, LSGG; Walker B, B; Histidine-loop, H and hinge region. Panel b, The highly conserved sequence under investigation is shown by the red amino acids. The Q-loop glutamine is underlined and the asterisks denote amino acids Lys680 and Arg691. Panel c, An alignment of 30 UvrA sequences was constructed and the conserved amino acids, excluding the ABC ATPase motifs, were painted onto the 3D model of UvrA's C-terminal dimer. Lys680 (K) and Arg691 (R) are depicted on the surface. This model lacks the zinc finger domain. Panel d, Ribbon diagram showing the α carbons of Lys680 (blue spheres) and Arg691 (orange spheres). ATP is shown in red at the dimer interface. Panel e, Rotation of the model in panel c to show that Lys680 and Arg691 are predicted to lie on the walls of a concave cleft on the surface of the UvrA dimer.
In this study, we sought to define amino acids that contribute to DNA binding by UvrA. We identified two amino acid positions within Bca UvrA that are critical for DNA binding, Lys680 and Arg691. Several Bca UvrA mutants, K680A, K680E, R691A, R691D and a double mutant K680A/R691A were created to evaluate the functional significance of these residues. Variant UvrA proteins were expressed in E. coli, purified and characterized. Our results demonstrate that Lys680 and Arg691 are important for DNA binding by Bca UvrA, with the double alanine mutant having reduced affinity for DNA, 37-fold, relative to WT UvrA. However, this protein is still competent at initiating the early steps of NER. Our results show that a decrease in UvrA's relative DNA binding affinity has a minor adverse effect on the DNA damage recognition properties of the UvrA2B complex or on the overall incision reaction mediated by UvrA, UvrB and UvrC. Our data support a model in which damage recognition is achieved through the combined action of UvrA in conjunction with UvrB in the context of the UvrA2B complex.
2. Experimental Procedures
2.1 Cloning
All mutants were created using site specific mutagenesis primers, pTYB1-Bca UvrA plasmid, Pfu turbo DNA polymerase (Stratagene) and Dpn I (New England Biolabs). The sense primer sequences were as follows: K680A, 5′ CGG GCT GGA GCA TCT CGA TGC AGT CAT TGA CAT CGA CCA GTC GC 3′; K680E, 5′ CGG GCT GGA GCA TCT CGA TGA AGT CAT TGA CAT CGA CCA GTC GC 3′; R691A, 5′ CGA CCA GTC GCC GAT CGG CGC CAC GCC ACG CTC GAA CCC G 3′; R691D, 5′ CGA CCA GTC GCC GAT CGG CGA CAC GCC ACG CTC GAA CCC G 3′. The double mutant was created by PCR amplification of the pTYB1-R691A Bca UvrA vector with the K680A primers. The inserts of all vectors were sequenced to confirm no additional mutations were introduced during PCR amplification.
2.2 Expression and Purification
All proteins were expressed in BL21(DE3)RIL cells (New England Biolabs) and purified using the T7 IMPACT system (New England Biolabs) by standard procedures. UvrA and UvrB were maintained at −20°C in storage buffer (50 mM Tris-HCl, pH 7.5, 500 mM KCl, 0.1 mM EDTA and 50% glycerol) until use. Thermotoga maritima UvrC was stored at −20°C in UvrC storage buffer (50 mM Tris-HCl, pH. 8.5, 500 mM KCl, 0.1 mM EDTA and 50% glycerol).
2.3 UV Survival Assay
Survival assays following UV exposure were conducted as described previously [25]. Briefly, E. coli WP2 uvrA− trp− cells (Mol Tox Inc.) were transfected with a vector that expresses the T7 polymerase upon IPTG exposure, pT7pol26, and a vector that expresses either WT UvrA or a UvrA mutant. Individual colonies were selected and grown to an A600 ~ 1. The cell culture was diluted 2-fold and the proteins were induced by the addition of IPTG (0.1 mM) for 1 h at 37 °C. In duplicate, three serial dilutions of each sample culture (100 µl) were spread onto LB plates containing appropriate antibiotics. The plates were irradiated with an 8W germicidal lamp and the dose was calculated based on measurements from a 245 nm UVX Radiometer (UVP Inc.) Serial dilutions of nonirradiated cultures were plated to determine the plating efficiency of each transformant. The number of colonies obtained after 20 h of incubation at 37 °C was recorded and the percent survival was calculated from the plating efficiency of the nonirradiated controls. Two independent experiments were performed for each sample. The mean survival of the experiments is plotted as a function of UV fluence.
2.4 DNA Substrates
All DNA substrates were synthesized and PAGE purified by Sigma-Genosys. The "damaged" DNA oligonucleotide, F2650, consisted of a 50-mer with a centrally located fluorescein adduct thymine (FldT). The F2650 sequence was: 5′-GAC TAC GTA CTG TTA CGG CTC CAT C(FldT)C TAC CGC AAT CAG GCC AGA TCT GC-3′. The complementary oligonucleotide, NDB, sequence contained a dA opposite the lesion. The damaged DNA, F2650, was 5′ end labeled using Optikinase (USB Corp.) and (γ-32P) ATP (3000 Ci/mmol, GE Healthcare) according to standard procedures. The reactions were terminated by the addition of EDTA (20 mM) and the enzyme was heat denatured by incubation for 10 min at 65 °C. Unincorporated radioactive nucleotides were removed by gel filtration chromatography (Biospin-6, Bio-Rad). The labeled oligonucleotide was annealed with equimolar amounts of the complementary oligonucleotide, NDB. The double stranded character of the duplex was analyzed on a native 10% polyacrylamide gel.
2.5 Electrophoretic Mobility Shift Assay (EMSA)
For the UvrA EMSAs, binding reactions were performed with duplexed 50-mer DNA oligonucleotides containing a site specific lesion at position 26 (F2650/NDB, 2nM) and increasing UvrA2 protein concentrations (7.5 - 125 nM) in 20 µl reaction buffer (50 mM Tris-HCl, pH7.5, 100 mM KCl, 10 mM MgCl2, 1 mM ATP, 5 mM DTT and 1 µM bovine serum albumin) for 15 min at 55 °C. The reactions were loaded onto a 3.5% native polyacrylamide gels (29:1, acrylamide:bis ratio) containing 1 mM ATP and 10 mM MgCl2. The buffer contained 44.5 mM Tris pH 8.3, 44.5 mM boric acid, 1 mM EDTA, 1 mM ATP and 10 mM MgCl2. Electrophoresis was carried out for 1 h at 100 V at 4 °C. The gels were dried and exposed to a phosphorimager screen. Data are reported as the percent bound versus protein concentration. Binding isotherms were fitted by nonlinear regression analysis to the equation Fb = ((1+KaP + KaD) - ((1 + KaP KaD)2 - (4DKa2P))1/2)/2DTKa; where Fb is the fraction bound; P is the protein concentration ; DT is the total DNA concentration; Ka = 1/Kd(app); Kd(app) is the apparent dissociation constant using Kaleidagraph and the method of Schofield [26].
For the UvrA·UvrB·DNA EMSAs, the proteins were preheated to 65 °C for 10 min prior to initiation of the reactions. Binding reactions were performed with F2650/NDB duplex (2 nM), UvrA (20 nM) and WT UvrB (100 nM) in 20 µl of reaction buffer (50 mM Tris-HCl pH 7.5, 100 mM KCl, 10 mM MgCl2, 1 mM ATP, 5 mM DTT and 1 µM BSA) for 15 min at 55 °C. In those reactions containing competitor DNA, 309 ng of pUC19 DNA (New England Biolabs) was added prior to addition of the oligonucleotide. The reactions were loaded onto a 3.5% native polyacrylamide gel (29:1, acrylamide:bis) and subjected to electrophoresis at 100 V for 1 h at 4 °C. The gels and buffers contained 44.5 mM Tris, 44.5 mM boric acid and 1 mM EDTA, 1 mM ATP and 10 mM MgCl2. The gels were dried and exposed to a phosphorimager screen. The percent of DNA bound in the DNA·protein complexes was calculated based on the total radioactivity in the lane. The percentage bound is reported as the mean ± standard deviation of three independent experiments.
2.6 ATP Hydrolysis Assay
The ATPase assays were performed as previously described [25]. The conversion of ATP to ADP by the UvrAB system was monitored using a coupled enzyme assay system consisting of pyruvate kinase and lactic dehydrogenase to couple the hydrolysis of ATP to the oxidation of NADH (ε340nm = 6220 M−1 cm−1). ATP (Roche) was added to a final concentration of 1 mM in a 100 µl reaction mixture containing 50 mM Tris-HCl pH 7.5, 55 mM KCl, 4 mM MgCl2, 1 mM DTT, 12.6 units/ml L-lactic dehydrogenase (Sigma), 10 units/ml pyruvate kinase (Sigma), 2 mM phosphoenol pyruvate (Roche), 0.15 mM NADH (Roche), 50 nM Bca UvrA (WT or mutants) and 100 nM Bca UvrB. The individual proteins were preheated to 65 °C for 10 min prior to initiation of the reactions. Each protein was assayed in the absence of DNA as well as in the presence of UV-irradiated pUC19 DNA (UV DNA, 1 µg). The rate of hydrolysis was calculated from the linear change in absorbance at 340 nm and 55 °C using a Beckman DU-640 spectrophotometer. The data are reported as the mean rate (M/min) ± the standard deviation of the mean, n = 3.
2.7 Incision Assay
Incision assays were performed as described previously [25]. Separately, the UvrA, UvrB and UvrC proteins were heated to 65 °C for 10 min prior to initiation of the reactions. The 5’ end labeled duplex DNA containing a site specific fluorescein adducted thymine at position 26 (2 nM, F2650/NDB) was treated with UvrABC (20 nM WT or mutant UvrA, 100 nM Bca UvrB and 50 nM Tma UvrC) in 20 µl of UvrABC buffer (50 mM Tris-HCl pH 7.5, 50 mM KCl, 10 mM MgCl2, 1 mM ATP and 5 mM DTT) at 55 °C for the indicated time. For those reactions containing competitor DNA, 309 ng pUC19 DNA (New England Biolabs) was added. The reactions were terminated by addition of EDTA (20 mM). Ten percent of the reaction was removed, denatured with formamide and heated to 85 °C for 5 min. Incision products were resolved on a 10% denaturing polyacrylamide gel and electrophoresis was performed at 325 V in Tris-Borate-EDTA buffer (89 mM Tris, 89 mM boric acid and 2 mM EDTA) for 40 mins. Gels were dried and exposed to a phosphorimager screen (Molecular Dynamics overnight. The percent of the DNA incised was calculated using the Molecular Dynamics software, ImageQuant and based on band intensities within each lane. The percentage of DNA incised is reported as the mean ± the standard deviation of the mean, n = 3.
2.8 DNase I Footprinting
The UvrA and UvrB proteins were heated to 65 °C for 10 min. The DNA binding reactions contained 2 nM F2650/NDB duplex in 50 mM Tris-HCl, pH 7.5, 50 mM KCl, 10 MgCl2, 1 mM ATP, 10 mM DTT, 100 nM bovine serum albumin and the indicated amounts of UvrA and UvrB in a total volume of 10 µL. After incubation at 37 °C for the indicated amount of time, DNase I (1 µL of 0.03 U/µL in 50 mM CaCl2 and 100 nM bovine serum albumin, New England BioLabs) was added and after digestion for 30 sec at room temperature the reactions were stopped by the addition of sarkosyl (0.5%) and EDTA (15 mM). The reactions were rapidly frozen on dry ice and an equal volume of formamide plus dyes were added. The samples were heated for 15 min at 85 °C followed by quick cooling. A portion of each sample (2–5 µL) was applied to a 12% polyacrylamide sequencing gel and electrophoresed for 2 h at 65 W. The gel was dried and exposed to a phosphorimager screen. Quantitation of the individual DNase I sensitive bands was performed using the ImageQuant software (GE Healthcare). Two bands were selected for analysis which correspond to DNase I incision between nucleotides G17 -P- G18 (p1) and between T13-P-T14 (p2). The percent of initial value was calculated by dividing the band intensities for p1 or p2 by the initial band intensities for p1 or p2 observed upon DNase I digestion in the absence of added proteins. Data are reported as the percent bound versus protein concentration. Binding isotherms were fitted by nonlinear regression analysis to the equation Fb = ((1+KaP + KaD) - ((1 + KaP KaD)2 - (4DKa2P))1/2)/2DTKa; where Fb is the fraction bound; P is the protein concentration ; DT is the total DNA concentration; Ka = 1/Kd(app); Kd(app) is the apparent dissociation constant using Kaleidagraph and the method of Schofield [26]. The average Kds for WT and KRAA UvrA were determined by the analysis of these two bands from three separate experiments.
In order to determine the initial rate of Uvr(A)B loading, the intensity of the band p1 was monitored over time. The initial rate of Uvr(A)B binding were obtained by fitting the mean of three experiments ± S.D. to an exponential. For WT UvrA, the best fit was to a double exponential in which the initial rate represented an amplitude of 70%. In order to calculate the rate of Uvr(A)B loading (nmoles/min) for WT UvrA the first order term was multiplied by 70% then by 40 nmoles (the total DNA substrate in the reaction), to yield a rate of 27.8 nmoles/min. The rate of (KRAA UvrA)UvrB loading fit a single exponential and was 8.1 nmoles/min.
2.9 Structural Model for the C-terminus dimer of Bca UvrA (residues 603–935) bound to DNA
A model for the C-terminal domain of UvrA residues 603–713 and 801–935 was created as described in our previous publication; Model A [25]. This model lacked 88 amino acids which included the zinc finger motif. In order to create a complete structural model, residues 714–740 and 766–800 were modeled into Model A using the “loop search” command of the package Sybyl 7.2 (Tripos, Inc) [27]. Six Ala residues were used to connect the region between residues 740 and 766 during the loop search process. UvrA's C-terminus zinc finger domain (residues 740–766) was separately modeled using the X-ray crystal structure of the zinc finger domain of an Hsp40 protein (PDB entry Ydj1: for more detail see [28]). After removing the six connecting Ala residues from Model A, the modeled zinc finger domain residues were docked in using Sybyl 7.2 (Tripos, Inc) [27]. The structure was energy minimized in vacuum using the Sander module of Amber 9.0 [29] with the standard amber force field (parm99.dat). The X-ray crystal structure of the Rad50 dimer (PDB codes: 1F2U 1F2T) was used as a model template to construct the C-terminal domains of the UvrA dimer unit. The superimposition of UvrA's ABC ATPase motifs was aligned with the conserved backbone atoms of Rad50's ABC ABC ATPase motifs creating model 2 structure of the UvrA C-terminus dimer model. The resultant structure was energy minimized and a DNA segment (ACGT)7 was created and docked into the energy minimized structure of the C-terminal UvrA dimer, model 2. The protein-DNA complex was solvated in a box of water with box boundaries extending at least 15Å away from the closest protein or DNA atom to the box boundary. The final model system containing the C-terminal UvrA dimer, two ATP molecules, the DNA segment, two Zn+2 ions bound to the zinc finger domains and 42078 solvent water molecules (total of 138878 atoms) was then subjected to an energy minimization. The minimization used the Sander module of Amber 9.0 with the standard Amber force field (param99.dat) followed by a short constraint molecular dynamics simulation (a 100 ps temperature ramping and 500 ps constant volume-constant temperature (300K) simulation with 2 kcal/mol position constraints used in heavy atom positions). The final system was energy re-minimized.
3. RESULTS
3.1 Selection of the site directed mutagenesis targets
Historically, it was thought that UvrA interacted with DNA via its zinc finger in the C-terminus of the protein [23,24]. However, we have recently shown that this zinc finger is not required for DNA binding by UvrA [25]. A multiple sequence alignment of 30 UvrA sequences was created to identify potential DNA binding sites within the C-terminal domain of UvrA. From this alignment, several conserved sequence patches were revealed. We did not consider those conserved sequences which represented motifs of the ABC ATPase or the zinc finger. This analysis left two segments of conservation. One region, adjacent to the ABC ATPase Q-loop, contained several conserved positively charged residues, amino acids 678–699, (Fig. 1b and c). The second site was a polyglycine region. We elected to pursue the region containing the highly conserved positively charged amino acids because DNA binding by UvrA is salt sensitive [30].
There are 318 UvrA sequences deposited in HOGENOM database [31]. Of these sequences, 35 or 11% of the UvrA proteins did not possess either a Lys or Arg at position 680 and 691. However, upon closer inspection, a majority of these protein sequences were in organisms that had more than one UvrA protein sequence. In 32 out of the 35 outlier cases, the organism possessed another UvrA protein sequence which did code for either a Lys or Arg at position 680 and 691. In two halophilic archaeon UvrA sequences, Lys680 was replaced by Thr and in Oceanobaccillus iheyensis Lys680 was replaced with an Ala. A lysine or arginine is found at position 691 in the remaining 286 sequences. Previously, we reported a theoretical 3D-model for the C-terminus dimer of UvrA [25]. Using this model, it was determined that this region of interest would lay on the surface of the UvrA dimer and furthermore that Lys680 (blue) and Arg691 (orange) were approximately 20 Å apart on the walls of a shallow concave depression, (Fig. 1d and e). Therefore, we elected to create mutations at these sites that would neutralize these charged amino acids (K680A, R691A), alter the charge (K680E, R691D) or neutralize both charged amino acids (K680A/R691A).
3.2 Relative DNA Binding Affinities of WT and UvrA mutants
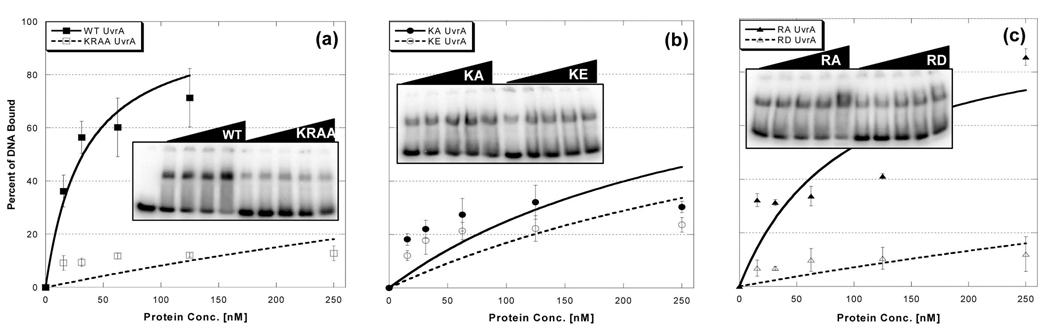
The protein variants were soluble and readily purified. Each protein was subjected to electrophoretic mobility shift analysis to determine its relative binding affinity to a damaged DNA duplex. As can be seen in Fig. 2, the UvrA proteins were tested at a variety of protein concentrations and the estimated apparent equilibrium dissociation constant, Kd, was calculated as described in the methods section, see Table I. All protein variants exhibited altered dsDNA binding as compared to WT UvrA. The double mutant, K680A/R691A bound the dsDNA oligonucleotide with the lowest affinity (Fig. 2a). It displays a 37-fold reduction in dsDNA affinity relative to WT UvrA. As expected, the two alanine-substituted proteins, K680A and R691A, displayed greater affinity than either charge switch mutants, K680E and R691D (Fig. 2b and c). These results support our proposal that Lys680 and Arg691 are indeed critical amino acids for dsDNA binding by UvrA as a dimer.
Fig. 2. Damaged DNA binding profiles of UvrA in the presence of ATP and magnesium.
EMSA was used to monitor the DNA binding properties of the proteins. Increasing amounts of the indicated UvrA protein were incubated with 2 nM F2650/NDB duplex DNA in reaction buffer containing ATP (1 mM) and MgCl2 (10 mM) for 15 min at 37 °C, see Experimental Procedures for additional details. The reactions were separated on 3.5% polyacrylaminde native gels in the presence of ATP (1 mM) and MgCl2 (10 mM). Panels a–c, Quantitative analysis of EMSA gels with representative gels as inserts. The data are reported as the mean ± S.D., n=3. Binding isotherms were fitted by nonlinear regression analysis using Kaleidagraph and the method of Schofield [26].
Table I.
Apparent Dissociation Constants a (Kd) for Specific DNA Binding
| Fold Change | ||
|---|---|---|
| Kd(app), nM ± standard deviation | Relative to WT | |
| WT | 15. 4 ± 2.1 | - |
| K680A R691A | 572 ± 89 | 37 |
| K680A | 153 ± 37 | 10 |
| K680E | 251 ± 60 | 16 |
| R691A | 44.9 ± 0.9 | 2.9 |
| R691D | 271 ± 79 | 18 |
Samples containing different amounts of the indicated protein were incubated with damaged (F2650/NDB) duplex (2 nM) in the presence of ATP (1 mM) and MgCl2 (10 mM) and fractioned by electrophoresis as in Fig. 3.
The apparent dissociation constant reported represents the mean of three independent determinations with associated error. The errors for the relative affinities were derived from the standard deviation of the three independent fits. The curve fitting was created by Kaleidegraph® using the method of Schofield [26].
Attempts to develop another independent DNA binding method using a solution based fluorescence anisotropy assay were complicated by what appears as multiple aggregation states of UvrA. We tested WT UvrA and KRAA UvrA at various KCl concentrations to assess each protein's DNA binding capacity over a wide range of salt concentrations (50, 100, 150 and 200 mM KCl). The anisotropy signal varied as a function of salt, with low KCl (50 mM) yielding high anisotropy values in the presence of DNA (data not shown). Higher salt yielded lower anisotropy values even at concentrations of DNA and UvrA which were well above the apparent Kd. Higher order aggregates of UvrA are also observed with lower KCl (50 mM KCl) concentrations in the EMSA analysis which were observed as slowly migrating bands (data not shown).
3.3 In vivo Complementation and Survival
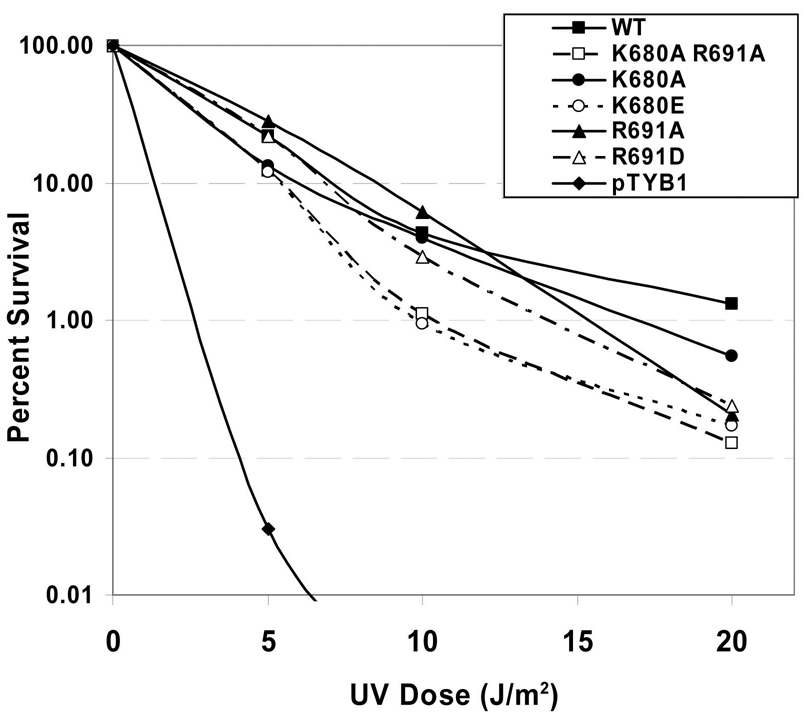
In vivo complementation and UV survival studies were conducted in E.coli WP2 trp−, uvrA− cells as previously described [25]. As can be seen in Fig. 3, each of the mutant proteins provided a significant level of UV protection relative to the sample which received the empty vector, pTYB1. At 10 J/m2, WT UvrA survival was 4.3% as compared to 0.93% for K680E and 1.1% for K680A/R691A. The K680E and K680A/R691A UvrA expressing cells are approximately 4-fold more sensitive to UV than cells complemented with WT UvrA.
Fig. 3. UV survival of E. coli WP2 uvrA− strain transformed with UvrA variants.
WP2 (trp−, uvrA−) cells were transformed with pT7pol26, a plasmid encoding an IPTG-inducible T7 polymerase and pTYB1 or pTYB1-uvrA vector. After individual colonies were selected and grown to an A600 of ~1, the cell cultures were diluted 2-fold, and the proteins were induced with IPTG (0.1 mM) for 1h at 37 °C. Serial dilutions of each sample of culture (100 µl) were spread onto a nutrient rich-media and UV-irradiated with a 254 nM germicidal light source. The numbers of colonies visible after 20 h of growth at 37 °C were recorded and the fraction of cells surviving after each dose of UV was calculated based on the plating efficiency of the nonirradiated controls. The mean of two or three independent experiments is reported. Transformants contained pT7pol26 and pTYB1 (diamonds), pTYB1-WT uvrA (solid squares), pTYB1-K680A/R691A uvrA (open squares), pTYB1-K680A uvrA (solid circle), pTYB1-K680E uvrA (open circle), pTYB1-R691A uvrA (solid triangles) or pTYB1-R691D uvrA (open triangles).
3.4 Electrophorectic Mobility Shift Assays in the Presence of Competitor DNA
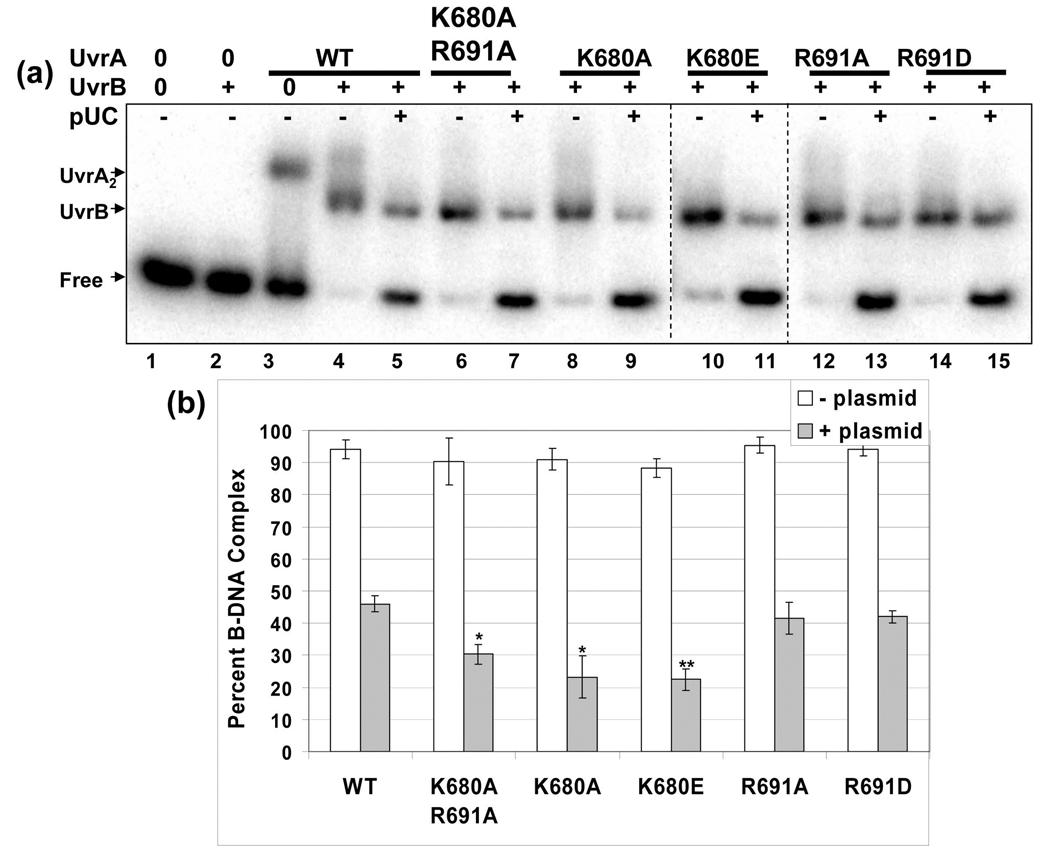
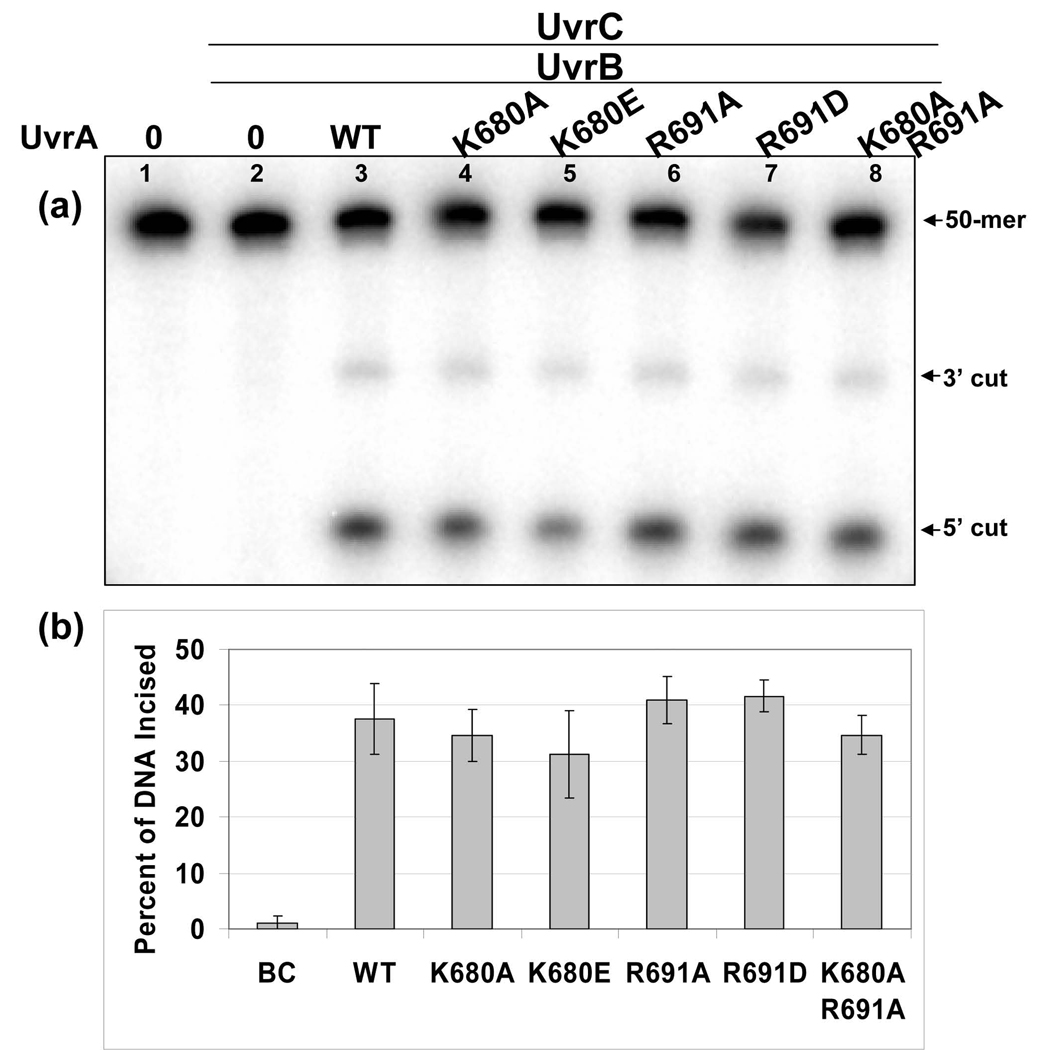
Since the survival studies did not recapitulate the large differences observed for the DNA binding affinities of the UvrA mutants, we sought to examine the next step in damage recognition, namely the ability of these mutant UvrA proteins to cooperate with UvrB to form a productive UvrB-DNA complex. In order to more faithfully duplicate the in vivo conditions we tested the formation of the UvrB·DNA complex by EMSA analysis in the presence and absence of competitor DNA. Previously, we demonstrated that the zinc finger deletion mutant of UvrA did not reveal its true defect in damage recognition until competitor DNA was added to the incision reactions [25]. As can be seen in Fig. 4, each of the UvrA proteins delivered UvrB onto DNA such that it could form a stable UvrB·DNA complex in the absence of competitor DNA. Note that UvrB does not have appreciable affinity for duplex DNA in the absence of UvrA (Fig. 4a, lane 2). Upon addition of excess plasmid DNA equivalent to about one lesion per 12 kb of undamaged DNA, the amount of UvrB loaded onto the radiolabeled oligonucleotide is inhibited. Under these reaction conditions, WT UvrA transferred 46% of the DNA to UvrB, a reduction of 49 % relative to WT UvrA without competitor pUC19 DNA. The Lys680 mutants, K680A and K680E, transferred 23% and 22% of the DNA onto UvrB. Therefore, K680A and K680E UvrA mediated transfer was down two-fold compared to WT UvrA. The double alanine mutant, K680A/R691A, also showed a significant reduction. It transferred 30% of the DNA to UvrB. Thus, the K680A/R691A UvrA protein transferred 66% as much as WT UvrA. These results suggest that while the individual UvrA mutants have different relative affinities for DNA, the dual action of the UvrA2B complex on damaged DNA can effectively complement the UvrA DNA binding defects and drive the reaction forward by moving the DNA into a stable UvrB·DNA complex.
Fig. 4. Lys680 mutants transfer less UvrB onto the damaged oligonucleotide than the other UvrA variants.
Panel a, The ability of the individual UvrA proteins to load WT UvrB onto the radiolabeled fluorescein adducted oligonucleotide was monitored by EMSA. The UvrA proteins (20 nM) were incubated with UvrB (100 nM) with or without competitor DNA (pUC19) for 15 min at 55 °C. The protein-DNA complexes were separated on 3.5% native polyacrylamide gels containing ATP (1 mM) and MgCl2 (10 mM). Dashed lines indicate merged gels run at the same time. pUC is the plasmid, pUC19. Panel b. Analysis of the EMSAs reporting the percentage of DNA in the B·DNA complex. White and filled bars represent B·DNA complexes produced in the absence and presence of competitor DNA, respectively. The data are reported as the mean ± S.D., n=3. Paired Student's T-tests were performed between WT and the variants and (*) indicates that the probability was less than 0.05 while (**) reflects a probability less than 0.01.
3.5 ATPase Activity
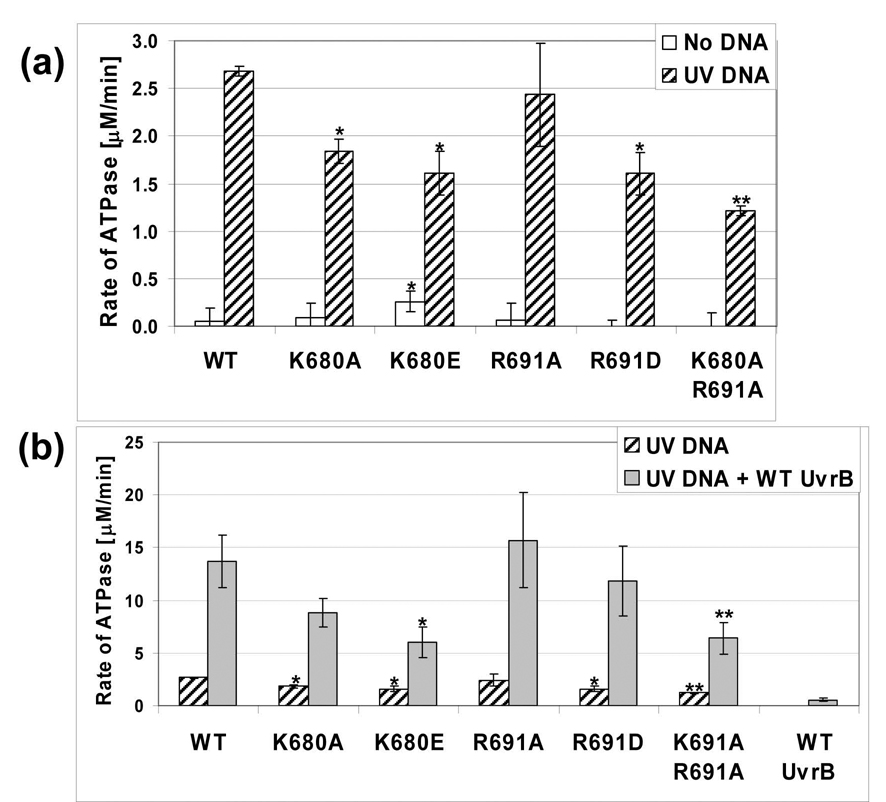
The binding and hydrolysis of nucleotides by UvrA is known to modulate its DNA binding capacity. Currently, it is thought that bound ATP facilitates DNA binding and that hydrolysis promotes dissociation of UvrA from DNA [32,33]. Each UvrA variant was evaluated for ATPase activity in the absence or presence of UV irradiated DNA to determine if the amino acid substitutions altered their ability to hydrolyze ATP. Bca UvrA is a DNA-stimulated ATPase. Therefore, if a mutant cannot interact with DNA as effectively as WT UvrA then a decrease in ATPase activity is expected. As can be seen in Fig. 5a, each of the UvrA variant proteins exhibited a DNA-stimulated ATPase phenotype. The K680E and R691D mutants displayed comparable activity, 60% as active as WT UvrA while the double mutant, K680A/R691A, exhibited the lowest rate of ATPase activity, only 45% as active as WT UvrA. The trends in the reduction of the DNA stimulated-ATPase activity of the UvrA variants correlates with their relative dsDNA binding affinities.
Fig. 5. ATPase activity of the UvrA variants.
Panel a, The conversion of ATP to ADP by the UvrA proteins (50 nM) in the presence or absence of UV irradiated DNA (1 µg) was monitored using a coupled enzyme assay system consisting of pyruvate kinase and lactate dehydrogenase, which links the hydrolysis of ATP to the oxidation of NADH (see Experimental Procedures). White bars indicate activity in the absence of DNA while hatched bars designate the presence of UV irradiated DNA. Panel b, UvrA·UvrB ATPase activity. Hatched bars indicate the level of ATPase activity of the UvrA proteins (50 nM) with UV DNA while the filled bars represent the activity of UvrA (50 nM) and UvrB (100 nM) in the presence of UV DNA. The data are reported as the mean ± S.D, n=3. Paired Student's T-tests were performed between WT and the variants and (*) indicates that the probability was less than 0.05 while (**) reflects a probability less than 0.01.
UvrB also has an ATPase center and we have recently shown that domain 4 of UvrB acts as an autoinhibitory domain for both DNA binding and ATPase activity [7]. These activities are stimulated by the action of UvrA on UvrB through direct protein-protein contacts. Thus, UvrA in the presence of damaged DNA acts to "unlock" UvrB’s inherent DNA binding and ATPase functions [7,34]. As shown in Fig. 5b, upon mixing WT UvrA, WT UvrB and UV irradiated DNA there is a 5.1-fold increase in the rate of ATPase activity above WT UvrA and UV DNA. UvrA and UvrB are proposed to interact via the N-terminal domain of UvrA [35,36] and the mutations described here are exclusively in the C-terminal domain, we therefore hypothesized that the UvrA variant proteins should interact with UvrB normally and elicit UvrB's cryptic ATPase activity. As can be seen in Fig. 5b, the ATPase of UvrB is activated by all of the UvrA variants. The R691A and R691D mutants show a 6.5- and 7.4-fold increase, K680A/R691A a 5.3-fold increase, K680A a 4.8-fold increase and K680E a 3.7-fold increase in ATPase activity. These relative fold increases in ATPase activity are very similar to that produced by WT UvrA, 5.1-fold. From these results, we can conclude that the UvrA variants are properly folded, are able to interact with UvrB and activate UvrB's cyrptic ATPase center.
3.6 Oligonucleotide Incision Assay
Oligonucleotide incision assays were conducted to determine whether the UvrA protein DNA binding variants would initiate the NER reaction as well as WT UvrA. The incision of a 32P-end labeled oligonucleotide containing a site specific fluorescein adducted thymine, F2650/NDB, duplex was monitored using denaturing polyacrylamide gel electrophoresis. Using our standard incision conditions, 2 nM duplex F2650/NDB, 20 nM Bca UvrA, 100 nM Bca UvrB and 50 nM Tma UvrC, we did not observe a difference between the variant UvrA proteins after 5 minutes of incubation (Fig. 6). Incision assays are optimized for 50 mM KCl but we also conducted experiments using 100 mM KCl. At this salt concentration there were no statistically significant difference between the mutants and WT UvrA, (data not shown). In addition, all of the mutant UvrA proteins, except KRAA UvrA, functioned as well as WT UvrA in the NER reaction in the presence of excess competitor DNA, data not shown. KRAA UvrA displayed a modest reduction in the extent of incision, 30% verses 44% for WT UvrA. Thus, these UvrA DNA binding mutants in the presence of UvrB were able to support damage recognition, subsequent UvrB loading, and UvrC mediated incisions.
Fig. 6. All UvrA variants are capable of initiating the NER reaction.
Panel a, Incision of a 5′ end-labeled substrate (F2650/NDB) was monitored by denaturing polyacrylamide gel electrophoresis. The fluorescein adducted 50-bp duplex (F2650/NDB, 2 nM) was incubated with UvrB (100 nM), UvrC (50 nM) and the indicated UvrA protein (20 nM), for 5 min at 55 °C in reaction buffer. The reactions were terminated by the addition of EDTA (20 mM) and the incision products were analyzed on a 10% denaturing polyacrylamide gel. Panel b, Graphic representation of the extent of incision for the reactions containing the various UvrA proteins. Data are reported as the mean ± S.D., n=3.
3.7 Rates of loading of UvrB Using DNase I Footprinting
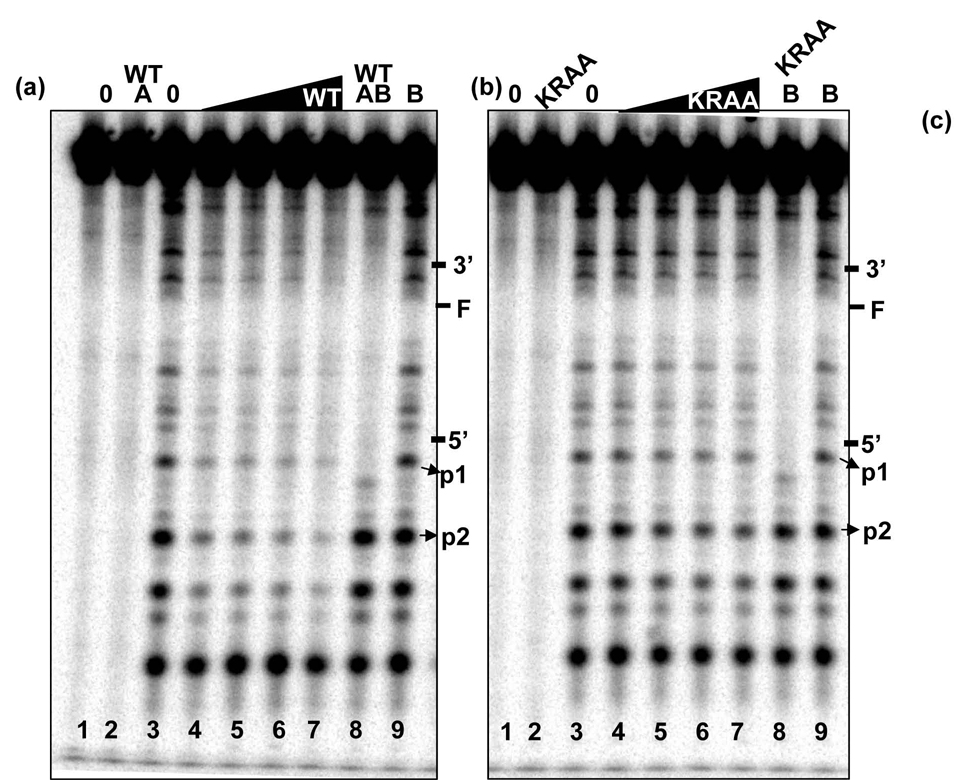
DNase I footprinting was conducted using the 50-mer fluorescein containing oligonucleotide. We sought to find an explanation for the apparent discrepancies between the observed DNA binding defects and efficient incision mediated by the mutants. All of the DNase I sensitive sites we observed are consistent with that of the unmodified oligonucleotide, data not shown. In addition, all of the DNase I digestion samples were conducted under single hit kinetics. Upon increasing amounts of WT UvrA, we observed a general loss of DNase I sensitivity throughout the oligonucleotide except for the most 5′ band, Fig. 7. We compared the UvrA DNase I footprint of WT and the KRAA UvrA proteins, Fig. 7 panels a and b. Band intensities for p1 and p2 (see Materials and Methods) were used to generate the DNA binding isotherms graphed in panel c. The apparent equilibrium dissociation binding constants were for WT UvrA for p1 and p2 were 11.8 ± 4.6 nM, respectively, and 88 ± 15.6 nM, respectively, for KRAA UvrA. Consistent with our EMSA data, the KRAA UvrA mutant displayed weaker DNA binding affinity as visualized by the persistent DNase I sensitivity, Fig. 7 lanes 4–7.
Fig. 7. DNase I Footprinting.
DNase I footprint of the WT and KRAA UvrA proteins bound to the 50 bp F2650/NBD duplex with the 32P on the 5′ end of the damaged strand. The indicated proteins were incubated with 2 nM duplex in the presence of 50 mM Tris-HCl, pH 7.5, 50 mM KCl, 10 MgCl2, 1 mM ATP, 10 mM DTT, 100 nM bovine serum albumin for 15 min at 37 °C then DNase I treated and processed as described in the methods. Panel a, WT UvrA DNase I footprint. Panel b, KRAA UvrA DNase I footprint. Both gels are loaded in the same order. The lanes contained the following: lane 1, no protein and no DNase I; lane 2, UvrA2, 10 nM and no DNase I; lane 3, no protein plus DNase I; lanes 4–7, increasing concentrations of UvrA2, 10 – 80 nM, plus DNase I; lane 8, UvrA2, 10 nM, WT UvrB, 100 nM, plus DNase I; lane 9, WT UvrB, 100 nM, plus DNase I. The position of the adduct and the 3′ and 5′ incision sites are noted on the left-hand side of the gels. The bands that were quantified for the graph in panel c are denoted by the arrows, p1 and p2. Panel c, graphic representation of the band intensities of p1 and p2 relative to the band intensity observed in lane 3, DNase I digestion in the absence of proteins. The average of 3 independent experiments was plotted with the standard deviation at each point. Binding isotherms were fitted by nonlinear regression analysis using Kaleidagraph and the method of Schofield [26]. For more details see the Materials and Methods.
Using this technique we were also able to detect the appearance of a discrete UvrB footprint after incubation with WT and the KRAA UvrA proteins, Fig 7a and b, lane 8. Note that UvrB alone does not generate a footprint on the double-stranded F2650/NDB, Fig 7a and b, lane 9. As can be seen from the gel, several bands near the bottom of the gel reappear upon UvrB binding. This was expected because the size of the DNA contact region of UvrB is smaller than UvrA's footprint [4,14,37,38]. The UvrB-footprint begins 3-nucleotides 5′ to the 5′ incision site and appears to extend almost to the same region covered by UvrA. It is clear from the gels that Bca UvrB binds to the DNA is such a way that it protects both incision sites from DNase I digestion.
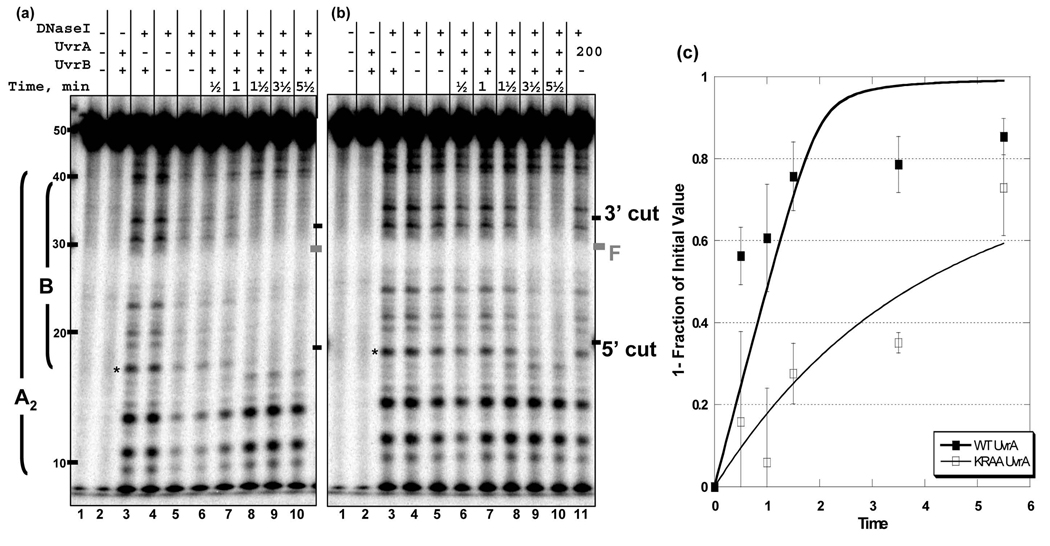
In order to gain insight into the apparent defect of the KRAA UvrA mutant, we monitored the rate of UvrB loading using a brief DNase I digestion (30 sec). Careful analysis of the DNase I footprinting at the earliest time point reveals the first footprint visualized is that of an UvrA footprint. Based on previous work and results presented here, the UvrA and transient UvrAB footprint appear to be identical. However, for the KRAA mutant there is a slight, but reproducible, apparent increase in the affinity of the UvrAB-complex for DNA, relative to the KRAA UvrA's affinity without UvrB, compare Fig. 8b lanes 3 verses 4. This was also true for the WT UvrA, but it is not as clear due to the higher affinity of WT UvrA for DNA in the absence of UvrB, compare Fig. 8a, lanes 3 and 4. Nonetheless, the rate of disappearance of band, p1, the last band protected by UvrB, was plotted and it showed a significant delay in DNase I sensitivity between WT and the KRAA UvrA, Fig. 8 panel c. The initial rates of Uvr(A)B (WT or KRAA UvrA) binding were obtained by fitting these data to an exponential in which p1's band intensities were converted to nmoles of substrate protected per min. These data revealed a biphasic reaction for the rate of (WT UvrA)B complex formation with an initial rate of 27.8 nmols of DNA bound per min. and a slower rate constant of 1.5 nmols per min. In contrast, the rate of formation of the Uvr(A)B complex mediated by KRAA UvrA mutant fit a single exponential with a rate constant of 8.1 nmols per min, a 3.4-fold reduction relative to WT UvrA's initial rate. These experiments indicate that the addition of UvrB to the KRAA UvrA mutant, complements the relatively weak DNA binding, such that UvrB can be loaded, albeit at a reduced rate.
Fig. 8. Rate of transfer of DNA from UvrA to UvrB.
DNase I footprinting of Bca UvrA and Bca UvrB-DNA complexes. Reaction mixtures contained UvrA2 (10 nM), ± 100 nM UvrB, 2 nM F2650/NDB duplex with the radiolabel on the damaged strand. The reactions were incubated at 37 °C then DNase I was added for 30 sec at RT. Then the samples were processed as described in the methods. The time indicated in the figure includes the time for DNase I digestion. Panel a, Gel image showing the transition of the WT UvrA footprint to WT UvrB footprint. The right side of the gel contains a graphic representation of the DNase I footprints observed. Panel b, Gel image showing the transition of the KRAA UvrA footprint to the WT UvrB footprint. The position of the adduct and the 3′ and 5′ incision sites are noted on the left-hand side of the gels. Asterisks indicate the position of the band used to quantify the gels, p1. Panel c, Graphic representation of the band intensities of p1 relative to the band intensity observed in lane 4, DNase I digestion in the absence of proteins. The average of 3 independent experiments was plotted with the standard deviation at each point. Data were fit to a single or double exponential using Excel.
4. Discussion
Understanding how UvrA interacts with DNA is fundamental to defining the mechanistic details of the NER pathway. While we have made great strides towards understanding both UvrB and UvrC due to recent crystal structures [16,18,39–45], there is limited knowledge of UvrA’s structure and how it initiates the NER reaction. We have begun to define UvrA's functions by deletion and site directed mutagenesis. Here we demonstrate that Lys680 and Arg691 make significant contributions to the binding affinity of UvrA for DNA. However, the altered DNA binding properties of the mutant UvrA proteins were complemented upon adding UvrB, suggesting that the affinity of the UvrA2B complex is more important determinant for damage recognition with the substrate used in this study.
Based on this work, we propose that DNA lies across the UvrA C-terminus dimer interface and that Lys680 and Arg691 make a significant contribution towards the affinity of UvrA for damaged DNA (Fig. 9). This model predicts that the UvrA dimer created by the C-terminus would interact with approximately 17 bp of DNA. This is approximately half of the reported DNase I-detected DNA footprint of ~33 bp of UvrA on a psoralen-containing substrate [37] and on a fluorescein modified thymine-containing substrate (this study). However, DNase I footprinting experiments with a GST-C-terminus UvrA fusion did not yield a discrete footprint due to a lack of damage specificity (data not shown). By defining the amino acids that are important for UvrA's interaction with DNA, we can begin to dissect how UvrA interacts with DNA and mediates efficient transfer of DNA to UvrB. Interestingly, our current data confirms and extends previous observations from this laboratory that the relative DNA binding affinity of UvrA did not correlate with incision efficiency [46]. In this present study, we have monitored the time course of UvrAB binding and the transition to the UvrB·DNA complex through the use of DNase I footprinting. The initial footprint observed upon WT Uvr(A)B binding was consistent with that of UvrA's footprint followed by the formation of an UvrB footprint.
Fig. 9. Proposed structural model of the C-terminus ABC ATPase dimer of Bca UvrA with DNA.
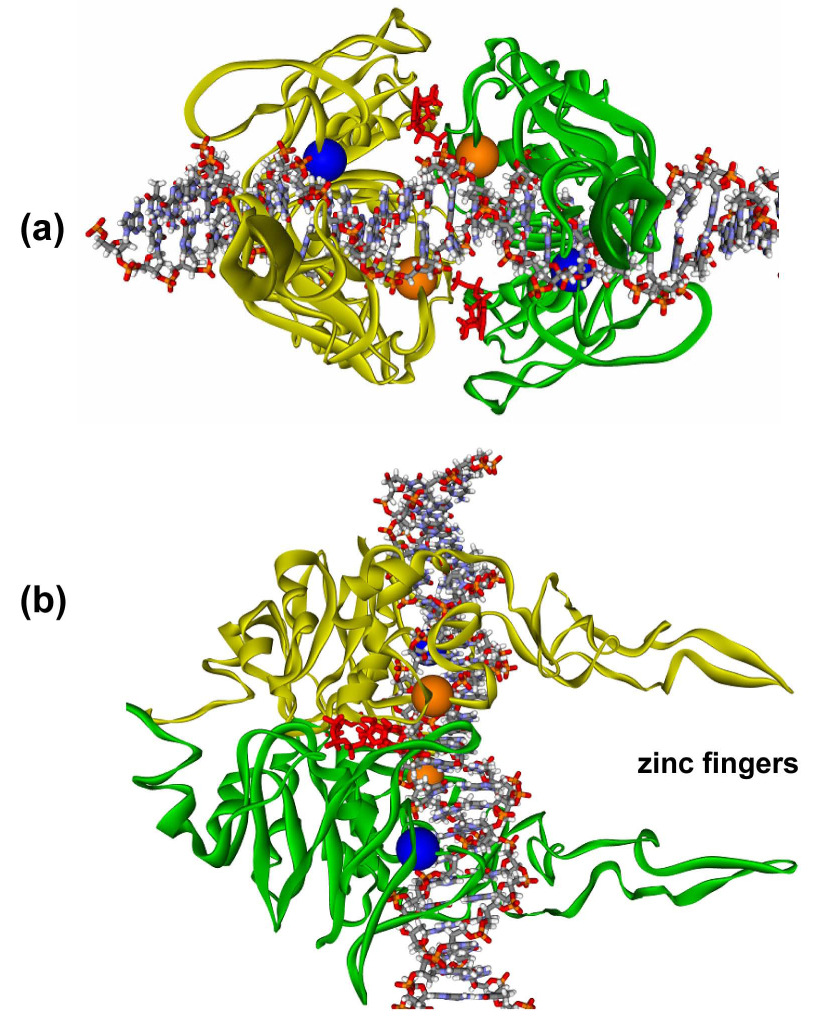
The model was constructed as described in the Experimental Procedures section. The model predicts that the DNA lies across the C-terminus ABC ATPase dimer of UvrA such that the DNA interacts with Lys680 and Arg691. In this model, approximately 17 base pairs of DNA would be in contact with the dimer. The zinc finger projections extend away from the DNA binding cleft. Panel a, View of the C-terminus UvrA dimer looking down the axis of the zinc fingers. Panel b, Side view showing the DNA laying across the dimer interface and the zinc fingers projecting out to the right. The two ATP molecules are depicted in red at the dimer interface. The α carbons of Lys680 (blue spheres) and Arg691 (orange spheres) are shown in CPK style.
In addition, using Biacore analysis, we recently calculated the affinity of UvrA2 for UvrB to be in the ~ 10 nM range (Y. Peng and B. Van Houten unpublished observations). This together with the finding that the KRAA UvrA variant showed a 37-fold loss of DNA binding (Fig. 2a) and yet was efficient at initiating the NER reaction (Figs. 6) supports our proposal that damage recognition is achieved by the cooperative action of both UvrA and UvrB. While additional research is required to more precisely define the surface contacts between UvrA and DNA, this study clearly indicates that this region of UvrA is important for DNA binding.
Our theoretical C-terminus ATPase UvrA dimer model is derived from Rad50 and the Rad 50 dimer has been described as a two-lobed structure [47]. Based on our model, the modifications we have made to the protein would lie along the interlobe connection. This region contains the Q-loop, an element of the C-terminus ABC ATPase. If our mutations had altered the relative position of the Q-loop, this would be expected to have a large effect on the ATPase function of the C-terminus, and might be expected to disrupt DNA binding. While this is a possibility, our data do not support this conclusion. Myles et al. demonstrated that when the C-terminus Walker A ATPase motif was mutated, GKS to GAS, the mutant proteins failed to load UvrB [48]. If the C-terminus ATPase had been disrupted in our proteins then the interaction of these mutants with UvrB should have been disrupted and this is not what we observed by EMSA in the absence of competitor, Fig. 4, or by the ATPase assay in the presence of UvrA, UvrB and damaged DNA, Fig. 5b. The fold-increase in ATPase activity upon adding UvrB for all the proteins is roughly the same, which suggests that the mutant UvrA proteins interact well with UvrB.
One of UvrA's functions during NER is that of an allosteric activator. We have recently presented evidence that domain 4 of UvrB acts as an autoinhibitory domain [7] and this autoinhibition is relieved by UvrA. Each of the UvrA variants described in this present study effectively interacted with UvrB as judged by their ability to activate UvrB's cryptic ATPase activity (Fig. 5b). A second important role of UvrA is to facilitate DNA opening to allow insertion of UvrB’s β-hairpin through the two DNA strands. Since strand opening is energetically linked to bending, we believe UvrA helps promote DNA opening by bending the DNA by about 60 degrees (H. Wang, D. Erie, and B. Van Houten, unpublished observation). While the KRAA UvrA variant showed a 37-fold reduction in its binding affinity for DNA by EMSA (Fig. 4), and weak DNA binding by the persistent DNase I sensitivity (Fig. 7–Fig. 8), the KRAA mutant was capable of allosterically activating UvrB and loading it onto damage DNA.
Additional research is required to fully understand how UvrB rescues these UvrA DNA binding mutants because it is not merely because UvrB structurally stabilized the mutant or altered the monomer-dimer equilibrium of the UvrA variant proteins. If that were the case then we should have seen a greater increase in the DNA binding affinity of the KRAA UvrA2B complexes with DNA. Currently, we are exploring single molecule studies of the UvrA and UvrB proteins to visualize the DNA damage recognition process. This type of analysis will yield clues of how UvrB rescues the KRAA variant protein. In lieu of such analysis, we are left with the conclusion that the DNA binding mutants of UvrA are rescued by their ability to interact with and activate UvrB’s high-affinity damage-specific DNA binding activity which drives the reaction forward into the UvrB-DNA complex. This UvrB-DNA complex is then recognized by UvrC and the dual single strand incisions occurs.
The results presented in this paper are similar to those reported for mammalian NER system. XPA is one of the damage recognition proteins involved in mammalian NER. The NMR structure of its DNA binding domain is known [49,50] and has recently been subjected to mutagenesis [51]. By monitoring luciferase expression following UV host cell reactivation in XPA deficient cells, the investigators assessed how various XPA DNA binding site mutants could initiate repair. Their results indicated that single substitutions of positively charged residues within the DNA binding domain of XPA did not adversely affect in vivo repair capacity even though DNA binding to linear dsDNA had been disrupted. This result is very similar to what is observed with the UvrA mutants. These observations combined with our present results reinforces the idea that damage recognition, during NER, occurs through the association of a multi-component system in which each constituent provides appreciable binding energy such that specificity for damaged DNA over non-damaged is only achieved through the cooperation of the entire ensemble of proteins.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
We would like to thank Leroy Worth, William Beard, Wade Lehmann, and Caroline Kisker for critically reading the manuscript. We would also like to thank Hong Wang and Ye Peng for insightful comments. This research was supported by the Intramural Research Program at NIEHS, NIH.
The abbreviations used are: NER, nucleotide excision repair; ABC ATPase, ATP binding cassette ATPase; dsDNA, double-stranded DNA; EMSA, electrophoretic mobility shift assay; UV DNA, UV irradiated DNA, Bca, Bacillus caldotenax.
REFERENCES
- 1.Croteau DL, Dellavecchia MJ, Skorvaga M, Van Houten B. Damage recognition by the bacterial nucleotide excision repair machinery. In: Wolfram S, Kow YW, Doetsch PW, editors. DNA Damage Recognition. Boca Raton, FL: Taylor & Francis; 2005. pp. 111–138. [Google Scholar]
- 2.Van Houten B, Croteau DL, DellaVecchia MJ, Wang H, Kisker C. 'Close-fitting sleeves': DNA damage recognition by the UvrABC nuclease system. Mutat Res. 2005;577:92–117. doi: 10.1016/j.mrfmmm.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 3.Truglio JJ, Croteau DL, Van Houten B, Kisker C. Prokaryotic Nucleotide Excision Repair: The UvrABC System. Chem Rev. 2006;106:233–252. doi: 10.1021/cr040471u. [DOI] [PubMed] [Google Scholar]
- 4.Moolenaar GF, Schut M, Goosen N. Binding of the UvrB dimer to non-damaged and damaged DNA: residues Y92 and Y93 influence the stability of both subunits. DNA Repair (Amst) 2005;4:699–713. doi: 10.1016/j.dnarep.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 5.Orren DK, Sancar A. Formation and enzymatic properties of the UvrB.DNA complex. J Biol Chem. 1990;265:15796–15803. [PubMed] [Google Scholar]
- 6.Sancar A, Hearst JE. Molecular matchmakers. Science. 1993;259:1415–1420. doi: 10.1126/science.8451638. [DOI] [PubMed] [Google Scholar]
- 7.Wang H, DellaVecchia MJ, Skorvaga M, Croteau DL, Erie DA, Van Houten B. UvrB domain 4: an autoinhibitory gate for regulation of DNA binding and ATPase activity. J Biol Chem. 2006;281:15227–15237. doi: 10.1074/jbc.M601476200. [DOI] [PubMed] [Google Scholar]
- 8.Moolenaar GF, Monaco V, van der Marel GA, van Boom JH, Visse R, Goosen N. The effect of the DNA flanking the lesion on formation of the UvrB-DNA preincision complex. Mechanism for the UvrA-mediated loading of UvrB onto a DNA damaged site. J Biol Chem. 2000;275:8038–8043. doi: 10.1074/jbc.275.11.8038. [DOI] [PubMed] [Google Scholar]
- 9.Moolenaar GF, Moorman C, Goosen N. Role of the Escherichia coli nucleotide excision repair proteins in DNA replication. J Bacteriol. 2000;182:5706–5714. doi: 10.1128/jb.182.20.5706-5714.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moolenaar GF, Hoglund L, Goosen N. Clue to damage recognition by UvrB: residues in the beta-hairpin structure prevent binding to non-damaged DNA. EMBO J. 2001;20:6140–6149. doi: 10.1093/emboj/20.21.6140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Skorvaga M, Theis K, Mandavilli BS, Kisker C, Van Houten B. The beta -hairpin motif of UvrB is essential for DNA binding, damage processing, and UvrC-mediated incisions. J Biol Chem. 2002;277:1553–1559. doi: 10.1074/jbc.M108847200. [DOI] [PubMed] [Google Scholar]
- 12.Verhoeven EE, Wyman C, Moolenaar GF, Goosen N. The presence of two UvrB subunits in the UvrAB complex ensures damage detection in both DNA strands. EMBO J. 2002;21:4196–4205. doi: 10.1093/emboj/cdf396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moolenaar GF, Franken KL, Dijkstra DM, Thomas-Oates JE, Visse R, van de Putte P, Goosen N. The C-terminal region of the UvrB protein of Escherichia coli contains an important determinant for UvrC binding to the preincision complex but not the catalytic site for 3′-incision. J Biol Chem. 1995;270:30508–30515. doi: 10.1074/jbc.270.51.30508. [DOI] [PubMed] [Google Scholar]
- 14.Moolenaar GF, Herron MF, Monaco V, van der Marel GA, van Boom JH, Visse R, Goosen N. The role of ATP binding and hydrolysis by UvrB during nucleotide excision repair. J Biol Chem. 2000;275:8044–8050. doi: 10.1074/jbc.275.11.8044. [DOI] [PubMed] [Google Scholar]
- 15.Verhoeven EE, van Kesteren M, Moolenaar GF, Visse R, Goosen N. Catalytic sites for 3′ and 5′ incision of Escherichia coli nucleotide excision repair are both located in UvrC. J Biol Chem. 2000;275:5120–5123. doi: 10.1074/jbc.275.7.5120. [DOI] [PubMed] [Google Scholar]
- 16.Truglio JJ, Rhau B, Croteau DL, Wang L, Skorvaga M, Karakas E, DellaVecchia MJ, Wang H, Van Houten B, Kisker C. Structural Insights into the first incision reaction during nucleotide excision repair. EMBO J. 2005;24:885–894. doi: 10.1038/sj.emboj.7600568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin JJ, Sancar A. Active site of (A)BC excinuclease. I. Evidence for 5′ incision by UvrC through a catalytic site involving Asp399, Asp438, Asp466, and His538 residues. J Biol Chem. 1992;267:17688–17692. [PubMed] [Google Scholar]
- 18.Karakas E, Truglio JJ, Croteau D, Rhau B, Wang L, Van Houten B, Kisker C. Structure of the C-terminal half of UvrC reveals an RNase H endonuclease domain with an Argonaute-like catalytic triad. Embo J. 2007;26:613–622. doi: 10.1038/sj.emboj.7601497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Husain I, Van Houten B, Thomas DC, Abdel-Monem M, Sancar A. Effect of DNA polymerase I and DNA helicase II on the turnover rate of UvrABC excision nuclease. Proc Natl Acad Sci U S A. 1985;82:6774–6778. doi: 10.1073/pnas.82.20.6774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumura K, Sekiguchi M, Steinum AL, Seeberg E. Stimulation of the UvrABC enzyme-catalyzed repair reactions by the UvrD protein (DNA helicase II) Nucleic Acids Res. 1985;13:1483–1492. doi: 10.1093/nar/13.5.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caron PR, Kushner SR, Grossman L. Involvement of helicase II (uvrD gene product) and DNA polymerase I in excision mediated by the uvrABC protein complex. Proc Natl Acad Sci U S A. 1985;82:4925–4929. doi: 10.1073/pnas.82.15.4925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Husain I, Van Houten B, Thomas DC, Sancar A. Sequences of Escherichia coli uvrA gene and protein reveal two potential ATP binding sites. J Biol Chem. 1986;261:4895–4901. [PubMed] [Google Scholar]
- 23.Visse R, de Ruijter M, Ubbink M, Brandsma JA, van de Putte P. The first zinc-binding domain of UvrA is not essential for UvrABC-mediated DNA excision repair. Mutat Res. 1993;294:263–274. doi: 10.1016/0921-8777(93)90009-6. [DOI] [PubMed] [Google Scholar]
- 24.Wang J, Mueller KL, Grossman L. A mutational study of the C-terminal zinc-finger motif of the Escherichia coli UvrA protein. J Biol Chem. 1994;269:10771–10775. [PubMed] [Google Scholar]
- 25.Croteau DL, Dellavecchia MJ, Wang H, Bienstock RJ, Melton MA, Van Houten B. The C-terminal zinc finger of UvrA does not bind DNA directly, but regulates damage-specific DNA binding. J Biol Chem. 2006;281:26370–26381. doi: 10.1074/jbc.M603093200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schofield MJ, Lilley DM, White MF. Dissection of the sequence specificity of the Holliday junction endonuclease CCE1. Biochemistry. 1998;37:7733–7740. doi: 10.1021/bi980399s. [DOI] [PubMed] [Google Scholar]
- 27.SYBYL Tripos Inc. St. Louis, Missouri: [Google Scholar]
- 28.Li J, Qian X, Sha B. The crystal structure of the yeast Hsp40 Ydj1 complexed with its peptide substrate. Structure (Camb) 2003;11:1475–1483. doi: 10.1016/j.str.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 29.Case DA, Darden T, Cheatham TE, 3rd, Simmerling C, Wang RE, Duke R, Luo R, Merz KM, Jr, Pearlman DA, Crowley M, Walker RC, Zhang W, Wang B, Hayik S, Roitberg A, Seabra G, Wong KF, Paesani F, Wu X, Brozell S, Tsui V, Gohlke H, Yang L, Tan C, Mongan J, Hornak V, Cui G, Beroza P, Mathews DH, Schafmeister C, Ross WS, Kollman PA. Amber 9. San Francisco: University of California; 2006. [Google Scholar]
- 30.Mazur S, Grossman L. Dimerization of Echerichia colli UvrA and Its Binding to Undamaged and Ultraviolet Light Damaged DNA. Biochemistry. 1991;30:4432–4443. doi: 10.1021/bi00232a009. [DOI] [PubMed] [Google Scholar]
- 31.HOGENOM UvrA. http://pbil.univ-lyon1.fr/cgi-bin/acnuc-link-ac2aln?db=Hogenprot&query=P0A698".
- 32.Myles GM, Sancar A. Isolation and characterization of functional domains of UvrA. Biochemistry. 1991;30:3834–3840. doi: 10.1021/bi00230a005. [DOI] [PubMed] [Google Scholar]
- 33.Thiagalingam S, Grossman L. Both ATPase sites of Escherichia coli UvrA have functional roles in nucleotide excision repair. J Biol Chem. 1991;266:11395–11403. [PubMed] [Google Scholar]
- 34.Caron PR, Grossman L. Involvement of a cryptic ATPase activity of UvrB and its proteolysis product, UvrB* in DNA repair. Nucleic Acids Res. 1988;16:10891–10902. doi: 10.1093/nar/16.22.10891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Claassen LA, Ahn B, Koo HS, Grossman L. Construction of deletion mutants of the Escherichia coli UvrA protein and their purification from inclusion bodies. J Biol Chem. 1991;266:11380–11387. [PubMed] [Google Scholar]
- 36.Claassen LA, Grossman L. Deletion mutagenesis of the Escherichia coli UvrA protein localizes domains for DNA binding, damage recognition, and protein-protein interactions. J Biol Chem. 1991;266:11388–11394. [PubMed] [Google Scholar]
- 37.Van Houten B, Gamper H, Sancar A, Hearst JE. DNase I footprint of ABC excinuclease. J Biol Chem. 1987;262:13180–13187. [PubMed] [Google Scholar]
- 38.Voigt JM, Van Houten B, Sancar A, Topal MD. Repair of O6-methylguanine by ABC excinuclease of Escherichia coli in vitro. J Biol Chem. 1989;264:5172–5176. [PubMed] [Google Scholar]
- 39.Theis K, Chen PJ, Skorvaga M, Van Houten B, Kisker C. Crystal structure of UvrB, a DNA helicase adapted for nucleotide excision repair. EMBO J. 1999;18:6899–6907. doi: 10.1093/emboj/18.24.6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakagawa N, Sugahara M, Masui R, Kato R, Fukuyama K, Kuramitsu S. Crystal structure of Thermus thermophilus HB8 UvrB protein, a key enzyme of nucleotide excision repair. J Biochem (Tokyo) 1999;126:986–990. doi: 10.1093/oxfordjournals.jbchem.a022566. [DOI] [PubMed] [Google Scholar]
- 41.Machius M, Henry L, Palnitkar M, Deisenhofer J. Crystal structure of the DNA nucleotide excision repair enzyme UvrB from Thermus thermophilus. Proc Natl Acad Sci U S A. 1999;96:11717–11722. doi: 10.1073/pnas.96.21.11717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Waters TR, Eryilmaz J, Geddes S, Barrett TE. Damage detection by the UvrABC pathway: crystal structure of UvrB bound to fluorescein-adducted DNA. FEBS Lett. 2006;580:6423–6427. doi: 10.1016/j.febslet.2006.10.051. [DOI] [PubMed] [Google Scholar]
- 43.Eryilmaz J, Ceschini S, Ryan J, Geddes S, Waters TR, Barrett TE. Structural insights into the cryptic DNA-dependent ATPase activity of UvrB. J Mol Biol. 2006;357:62–72. doi: 10.1016/j.jmb.2005.12.059. [DOI] [PubMed] [Google Scholar]
- 44.Truglio JJ, Croteau DL, Skorvaga M, DellaVecchia MJ, Theis K, Mandavilli BS, Van Houten B, Kisker C. Interactions between UvrA and UvrB: the role of UvrB's domain 2 in nucleotide excision repair. EMBO J. 2004;23:2498–2509. doi: 10.1038/sj.emboj.7600263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Truglio JJ, Karakas E, Rhau B, Wang H, DellaVecchia MJ, Van Houten B, Kisker C. Structural basis for DNA recognition and processing by UvrB. Nat Struct Mol Biol. 2006;13:360–364. doi: 10.1038/nsmb1072. [DOI] [PubMed] [Google Scholar]
- 46.Snowden A, Van Houten B. Initiation of the UvrABC nuclease cleavage reaction. Efficiency of incision is not correlated with UvrA binding affinity. J Mol Biol. 1991;220:19–33. doi: 10.1016/0022-2836(91)90378-j. [DOI] [PubMed] [Google Scholar]
- 47.Hopfner KP, Karcher A, Shin DS, Craig L, Arthur LM, Carney JP, Tainer JA. Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell. 2000;101:789–800. doi: 10.1016/s0092-8674(00)80890-9. [DOI] [PubMed] [Google Scholar]
- 48.Myles GM, Hearst JE, Sancar A. Site-specific mutagenesis of conserved residues within Walker A and B sequences of Escherichia coli UvrA protein. Biochemistry. 1991;30:3824–3834. doi: 10.1021/bi00230a004. [DOI] [PubMed] [Google Scholar]
- 49.Ikegami T, Kuraoka I, Saijo M, Kodo N, Kyogoku Y, Morikawa K, Tanaka K, Shirakawa M. Solution structure of the DNA- and RPA-binding domain of the human repair factor XPA. Nat Struct Biol. 1998;5:701–706. doi: 10.1038/1400. [DOI] [PubMed] [Google Scholar]
- 50.Buchko GW, Daughdrill GW, de Lorimier R, Rao BK, Isern NG, Lingbeck JM, Taylor JS, Wold MS, Gochin M, Spicer LD, Lowry DF, Kennedy MA. Interactions of human nucleotide excision repair protein XPA with DNA and RPA70 Delta C327: chemical shift mapping and 15N NMR relaxation studies. Biochemistry. 1999;38:15116–15128. doi: 10.1021/bi991755p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Camenisch U, Dip R, Schumacher SB, Schuler B, Naegeli H. Recognition of helical kinks by xeroderma pigmentosum group A protein triggers DNA excision repair. Nat Struct Mol Biol. 2006;13:278–284. doi: 10.1038/nsmb1061. [DOI] [PubMed] [Google Scholar]