Abstract
Previous microarray-based studies of acute respiratory distress syndrome (ARDS) were performed using various models to mimic disease pathogenesis. The complexity of the pathophysiologic response to direct or indirect lung injury in ARDS is difficult to reconstruct in experimental conditions. Thus, direct analysis of ARDS patient blood may provide valuable information. We investigated genome-wide gene expression profiles in paired whole blood samples from patients with ARDS (n = 8) during the acute stage (within 3 d of diagnosis) and recovery stage of ARDS (around ICU discharge). Among 126 differentially expressed genes, peptidase inhibitor 3 (PI3, encoding elafin, a potent neutrophil elastase inhibitor) had the largest fold-change (−3-fold changes, acute stage/recovery stage) in expression, indicating down-regulation during the acute stage of ARDS. We further examined plasma PI3 levels in 40 patients with ARDS and 23 at-risk control subjects from the same cohort. There was a coincidence of the microarray findings of lower PI3 gene expression with the lower plasma PI3 during the acute-stage. The plasma PI3 levels were statistically significant different among pre-diagnosis, day of diagnosis, and post-diagnosis groups (ANOVA, P = 0.001), with a trend of decreasing from pre- to post-diagnosis group. The time course of plasma PI3 decrease is well correlated with the course of early ARDS development (Pearson correlation coefficient: −0.52, P = 0.0006). Considering that PI3 can covalently binding to extracellular matrix in lung, circulating PI3 may provide a useful clinical marker for monitoring the early development of ARDS and may have implications for ARDS treatment.
Keywords: gene expression profiling, transcriptomics, acute respiratory distress syndrome, elafin, lung disease
CLINICAL RELEVANCE
This study suggests a possible protective role of elafin (PI3) in acute respiratory distress syndrome (ARDS). Circulating PI3 may provide a useful clinical marker for monitoring the early development of ARDS and may have implications for ARDS treatment.
The acute respiratory distress syndrome (ARDS) is characterized by intense inflammatory responses to direct or indirect lung injury exposures, resulting in diffuse alveolar damage and severe, life-threatening hypoxia (1–4). Risk factors for the development of ARDS include conditions commonly observed in critically ill patients, such as sepsis, trauma, pneumonia, burns, and massive transfusions of packed red blood cells. In addition, emerging viral diseases, such as the coronavirus known to cause severe acute respiratory syndrome (SARS) and the H5N1 avian influenza virus, have become important causes of ARDS in humans, with the potential for pandemic spread (5–7). These events further highlight the need for additional research to improve understanding of the pathogenesis of ARDS with the ultimate goal of developing specific treatments.
Genome-wide gene expression profiling using microarray technology has been applied successfully to the study of human disease pathogenesis. Examples include the discovery of new cancer subtypes with different prognosis and response to therapies (8), as well as the generation of new hypotheses of disease pathogenesis in asthma, pulmonary fibrosis, and ARDS (9–11). However, given the inherent difficulties of collecting RNA samples from patients with ARDS, previous microarray-based studies of ARDS were performed using various models to mimic the pathogenesis of acute lung injury (ALI), such as cell culture, animal models, ex vivo pulmonary tissue analysis, and healthy human volunteers (12). Although similar patterns of differential expression were reported among independent studies of ARDS disease models, there is no direct evidence supporting these findings in actual disease states. The complexity of the pathophysiologic response to direct or indirect lung injury in ARDS is difficult to reconstruct in single experimental stimulus studies. Microarray expression analyses on samples from patients with ARDS may provide valuable information about the complex inflammatory response in ARDS.
Previously, we have published evidence that a paired-sampling study design can detect gene expression changes in the total RNA of whole blood from individuals occupationally exposed to inhalable particulates, even though the particulate-induced changes were small compared with person-to-person variations (13). A follow-up study confirmed these findings, and suggested that particulate-induced effects on gene expression profiles are transient, with most diminishing within 18 hours after exposure, and exhibiting a dose–response pattern (unpublished data). Based on our previous experience in assessing global gene expression profiling in human blood samples, we applied a strategy of using microarray analysis to explore potential candidate genes to identify potential downstream protein products that can be used as biomarkers for ARDS. We started from an exploratory study of genome-wide gene expression in whole blood samples from patients with ARDS, with the hypothesis that there were ARDS-related differential gene expression between the acute-stage and recovery-stage of ARDS. The results of gene expression profiling were further investigated in the plasma samples from the same cohort. Since whole blood sample is a complex composition of heterogeneous cell types and its composition can be varied under different pathophysiological conditions, it is possible that the different percentages of certain subtype of blood cells between the acute stage and the recovery stage of ARDS could solely result in the observed differential expression of whole blood expression profiles. However, it is also possible that nondifferential expression of whole blood could be the result of compound effects of different percentage of blood cell subtypes and truly differential expression in certain subtypes. Although truly differential expression in subtype cells in the later situation could be revealed by adjusting cell subtypes, the overall (global) differential expression in whole blood is more meaningful for ARDS biomarker exploration.
MATERIALS AND METHODS
Study Subjects
This study was conducted within the ongoing Molecular Epidemiology of ARDS project at the Massachusetts General Hospital (MGH) and Harvard School of Public Health, both in Boston, Massachusetts. The study was started in 1999, and was approved by the Human Subjects Committees of both institutions. Study subjects were recruited from patients admitted to one of the four adult intensive care units (ICU) at MGH as described previously (14, 15). Eligible subjects were patients admitted to the ICU with at least one risk factor for the development of ARDS: (1) sepsis, (2) septic shock, (3) trauma, (4) pneumonia, (5) aspiration, or (6) massive transfusion of packed red blood cells (PRBC: defined as > 8 units of PRBC during the 24 h before admission). Patients were followed prospectively during ICU for the development of ARDS. Patients who developed ARDS, defined by the American European Consensus Committee (AECC) criteria (16), were identified as ARDS cases. Control subjects were identified as at-risk patients who did not meet criteria for ARDS during their stay in the ICU and had no prior history of ARDS. Baseline clinical information, vital signs, and laboratory testing results in the first 24 hours of ICU admission were collected for calculation of the Acute Physiological and Chronic Health Evaluation (APACHE) III score (17). In addition to a blood collection for DNA extraction during ICU stay, plasma samples were also collected for long-term storage. Based on the original protocol, two plasma samples were collected from each recruited subject, with the first samples collected during the first 24 hours of ICU admission and the second sample collected 3 days after the first collection. A written informed consent was obtained from each subject or an appropriate proxy of the patient.
RNA Sample Preparation and Microarray Hybridization
Previous evidence showed that a paired-sampling study design could minimize the biological variability among individuals and compare more precisely the gene expression profiles of different exposure states, especially under the circumstance that the expected expression variations were less than interpersonal variations (13). We applied a similar paired-sampling study design in this exploratory microarray study on blood samples, with two blood samples collected from each patient with ARDS. Given the situations of uncertain timing of ARDS development in at-risk ICU patients, extreme difficulties of obtaining full collaboration from patients and medical staff during ICU, and variable courses of disease progression, it was not only extremely difficult to collect samples at exactly two arbitrarily set time points, but also practically impossible to fractionate blood cells in time without introducing extra ex vivo variations. Therefore, for the hypothesis generating purpose, we compared the difference in gene expression of whole blood total RNA samples between the acute stage of ARDS (collected within 3 d of ARDS diagnosis) and the recovery stage (collected within the 6-d period between 3 d before and 3 d after ICU discharge), even though there were large time variations between paired sample collections.
Protocols for sample collection and processing, whole blood total RNA extraction, and quality assessment were described previously (13). RNA samples were hybridized to Affymetrix Human Genome U133A GeneChips (Affymetrix, Santa Clara, CA) at the Microarray Core Facility of the Dana-Farber Cancer Institute (Boston, MA). The paired RNA samples collected from each subject were processed together in one batch of microarray analysis to minimize inherent variations. Since the gene expression profile of the whole blood total RNA is the compound effects of hemoglobin RNA and the various subsets of white blood cells, we used DNA-Chip Analyzer 2006 (dChip, http://www.dchip.org/) software, which applied an invariant set of genes for normalization and calculation of expression values across all microarrays, to normalize raw microarray signals with the assumption that a subset of genes had constant expression among all subtypes of cell (18). The Detection Calls of a gene (Present Calls or Absent Calls) in an RNA sample was carried out by Affymetrix MAS 5.0 software using the one-sided Wilcoxon's signed-ranked algorithm (19).
We obtained high-quality microarray data of 22 paired RNA samples on 11 subjects. Although we only focused on recruiting patients with ARDS for microarray analysis, two subjects, who were misclassified originally as patients with ARDS, did not meet the AECC criteria for ARDS while in ICU and therefore were excluded from the paired microarray analysis. In addition, one ARDS case was excluded since the first sample was collected 2 days before the diagnosis of ARDS, and we attempted to standardize for time of collection. ARDS-related gene expression changes were examined among the eight remaining pairs of RNA samples. Baseline characteristics for these subjects are shown in Table 1.
TABLE 1.
PATIENT CHARACTERISTICS FOR SUBJECTS WITH RNA SAMPLES FOR MICROARRAY ANALYSIS
| Patient # | Group | Age | Sex | Etiology of ARDS | APACHE III* | Outcome | Days between Samples† |
|---|---|---|---|---|---|---|---|
| 1 | ARDS | 57 | Female | Septic Shock | 79 | Alive | 3 |
| 2 | ARDS | 62 | Female | Pneumonia/Septic Shock | 70 | Alive | 21 |
| 3 | ARDS | 20 | Female | Pneumonia/Septic Shock | 71 | Alive | 14 |
| 4 | ARDS | 83 | Male | Septic Shock | 119 | Died | 20 |
| 5 | ARDS | 79 | Male | Septic Shock | 99 | Died | 2 |
| 6 | ARDS | 28 | Male | Septic Shock | 117 | Alive | 5 |
| 7 | ARDS | 35 | Male | Septic Shock | 93 | Alive | 16 |
| 8 | ARDS | 29 | Male | Pneumonia/Septic Shock | 55 | Alive | 21 |
Definition of abbreviations: APACHE III, the Acute Physiological and Chronic Health Evaluation III score; ARDS, acute respiratory distress syndrome.
APACHE III score was calculated for each patient within 24 hours of ICU admission.
Days between the collections of two RNA samples.
Microarray Data Analysis
Out of a total of 22,215 probe sets on Affymetrix U133A microarray, a subset of 6,772 probe sets with Present Calls in over 50% of tested arrays was used in microarray data analysis. Genes with altered expression between two time points were identified by initially screening for average fold changes of paired samples larger than 1.2 or less than −1.2, and then tested genes using a one-sided paired t test with a P value < 0.05 as the cutoff for significance. Functional clustering analyses of the identified genes were carried out on the annotations defined by the Gene Ontology Consortium (GO, http://www.geneontology.org/) (20), using a non-redundant gene list for Affymetrix U133A microarray in the analysis (downloaded from Affymetrix website on March 7, 2007). GeneNotes software (http://combio.cs.brandeis.edu/GeneNotes/index.htm) was used to identify GO biological process significantly enriched with the altered genes, with Bonferroni correction (21).
Quantitative Real-Time PCR Analysis Using TaqMan Assays
To validate the microarray data, TaqMan (Applied Biosystems, Foster City, CA) quantitative real-time reverse transcriptase polymerase chain reaction (RT-PCR) was performed on available paired RNA samples from six subjects. TaqMan Probes, available as “Assay on Demand,” were used in the analyses of the expression levels of five target genes, including CYP4F3 (Hs00168521_m1), HPGD (Hs00168359_m1), IL8 (Hs00174103_m1), MMP9 (Hs00234579_m1), and PI3 (Hs00160066_m1), as well as three endogenous control genes (18S, Hs99999901_s1; ACTB, Hs99999903_m1; and RPLP0, Hs99999902_m1), according to the manufacturer-suggested procedures. Briefly, cDNA was first synthesized from approximately 2 μg RNA in a 100-μl reaction volume, using the High Capacity cDNA Archive kit (Applied Biosystems). Quantitative RT-PCR was then performed on synthesized cDNA in triplicates on an ABI PRISM 7900HT Sequence Detection System (Applied Biosystems). Of three simultaneously detected internal control genes, RPLP0 showed the lowest level of intra-assay variations, and was chosen to normalize RT-PCR results. Relative expression levels between paired RNA samples were determined using the ΔΔCt method with the value of the first RNA sample (acute stage of ARDS) of each pair as the calibrator (22).
Analysis of PI3 (Pre-Elafin) and Matrix Metalloproteinase-9 Levels in Plasma
Since each study subject of Molecular Epidemiology Study of ARDS was prospectively followed for the development of ARDS from the ICU admission and plasma samples were collected at the first few days of ICU stay, over 95% of plasma samples from patients with ARDS were collected during the period of ± 5 days of ARDS diagnosis. This unique feature of the study design enabled us to conduct a nested study of comparing plasma PI3 (pre-elafin, fully functional precursor of elafin) levels among three ARDS sample groups, including the pre-diagnosis group (5-d period before ARDS diagnosis), day-of-diagnosis group (within 24 h of diagnosis), and post-diagnosis group (3-d period after diagnosis), in addition to comparison between ARDS cases and at-risk control subjects. A total of 292 subjects, including 126 patients with ARDS and 166 at-risk control subjects, had plasma samples available for the analysis. We selected 40 patients with ARDS, including all 12 subjects of the pre-diagnosis group and 10 subjects of day-of-diagnosis group, as well as 18 subjects randomly selected from the post-diagnosis group. Twenty-three at-risk control subjects were randomly selected, who had paired plasma samples with the first sample collected during the first 24 hours of ICU admission and a second sample collected 3 days later. Plasma samples were stored at −80°C until analysis. Plasma PI3 and matrix metalloproteinase (MMP)-9 levels were quantified in duplicate using Human pre-ELAFIN/SKALP ELISA Test Kit (Cell Sciences, Canton, MA) and Human MMP-9 Immunoassay Kit (Millipore, Billerica, MA), according to the manufacturer's recommended protocol.
Statistical Analysis
The baseline characteristics between groups were compared using chi-square tests for categorical variables and by Student's t test for normally distributed continuous variables. The correlations between microarray expression data and quantitative RT-PCR data were estimated by Pearson correlation test. Since plasma PI3 and MMP-9 levels had skewed distributions, the natural log transformed data were used in the analyses. Plasma PI3 and MMP-9 levels among three ARDS sample groups and one control sample group were compared using ANOVA analysis, and comparisons between individual sample groups were conducted using Student's t test with a P value cut-off at 0.0083 (Bonferroni correction for multiple comparisons). The relationship of ARDS plasma PI3 levels and the sampling date relative to ARDS diagnosis was investigated by stepwise multivariable linear regression analyses to define associated covariates, with the sampling date relative to ARDS diagnosis forced in as covariate in all models. Candidate covariates included age, sex, APACHE III score on ICU admission, history of steroid use, pre-deposit clinical complications (including sepsis, septic shock, diabetes, chronic liver disease, pneumonia, and trauma), and laboratory testing results on ICU admission (including Hematocrit, WBC counts, platelet counts, serum sodium, serum potassium, serum urea nitrogen, serum creatinine, serum glucose, serum albumin, serum bilirubin, and serum bicarbonate). The criterion for covariate selection was P < 0.05. All statistical analyses were performed using the SAS statistical software package (version 9.1; SAS Inc., Cary, NC).
RESULTS
In this study, we compared the difference of gene expression of whole blood total RNA samples between the acute stage of ARDS (collected within 3 d of ARDS diagnosis) and the recovery-stage (collected within the 6-d period between 3 d before and 3 d after ICU discharge) on eight pairs of microarrays, with the median period between two sample collections of 15 days (range, 2–21 d; Table 1). Comparisons of gene expression were performed using a straightforward approach of fold-change ranking plus a P value cutoff (<0.05), which was proved to be a more reliable ranking criterion for gene selection in previous studies (23, 24). From a subset of 1,123 probe sets with an average paired fold-change larger than 1.2, we identified 126 genes (136 probe sets) with significantly altered gene expression. Compared to the recovery stage of ARDS, 70% (n = 88) of genes were expressed at lower levels and 30% (n = 38) were expressed at higher levels during the acute stage, suggesting that many genes were suppressed during the acute stage of ARDS (see Table E3 in the online supplement for complete list of genes). We also found a subset of 27 genes with large changes of expression (>1.5 fold-change) as shown in Table 2, as compared with previous microarray analyses of whole blood paired RNA samples (13). Of the 27 genes listed with a greater than 1.5-fold change in expression, gene PI3, IL8, and HGPD had two probe sets that showed significant ARDS-related variations.
TABLE 2.
GENES WITH > 1.5 FOLD-CHANGE IN EXPRESSION IN PERIPHERAL BLOOD BETWEEN THE ACUTE-STAGE AND RECOVERY-STAGE OF ARDS
| Symbol | Probe Set | Gene Name | Gene ID | Fold Change* |
|---|---|---|---|---|
| PI3 | 203691_at | Peptidase inhibitor 3, skin-derived (SKALP) | 5266 | −2.98 |
| 41469_at | −2.65 | |||
| IL8 | 202859_x_at | Interleukin 8 | 3576 | −2.93 |
| 205592_at | −1.79 | |||
| Unknown | 211781_x_at | Unknown (gb:BC006164.1) | −2.22 | |
| MME | 203435_s_at | Membrane metallo-endopeptidase (neutral endopeptidase, enkephalinase) | 4311 | −2.12 |
| PTGS2 | 204748_at | Prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) | 5743 | −2.03 |
| SGK | 201739_at | Serum/glucocorticoid regulated kinase | 6446 | −2.03 |
| BNIP3L | 221478_at | BCL2/adenovirus E1B 19kDa interacting protein 3-like | 665 | −1.84 |
| POLB | 203616_at | Polymerase (DNA directed), beta | 5423 | −1.83 |
| STAT1 | 200887_s_at | Signal transducer and activator of transcription 1, 91kDa | 6772 | −1.78 |
| FGL2 | 204834_at | Fibrinogen-like 2 | 10875 | −1.68 |
| GPR177 | 221958_s_at | G protein–coupled receptor 177 | 79971 | −1.58 |
| PIGF | 205077_s_at | Phosphatidylinositol glycan anchor biosynthesis, class F | 5281 | −1.56 |
| CLEC7A | 221698_s_at | C-type lectin domain family 7, member A | 64581 | −1.56 |
| C14orf159 | 218298_s_at | Chromosome 14 open reading frame 159 | 80017 | −1.56 |
| TBCC | 202495_at | Tubulin folding cofactor C | 6903 | −1.56 |
| LY75 | 205668_at | Lymphocyte antigen 75 | 4065 | −1.55 |
| BTRC | 216091_s_at | Beta-transducin repeat containing | 8945 | −1.55 |
| DUSP6 | 208892_s_at | Dual specificity phosphatase 6 | 1848 | −1.52 |
| HIP2 | 202346_at | Huntingtin interacting protein 2 | 3093 | 1.55 |
| OSBPL1A | 208158_s_at | Oxysterol binding protein-like 1A | 114876 | 1.58 |
| STXBP2 | 209367_at | Syntaxin binding protein 2 | 6813 | 1.62 |
| IDI1 | 204615_x_at | Isopentenyl-diphosphate delta isomerase 1 | 3422 | 1.58 |
| 208881_x_at | 1.65 | |||
| HSPA1A / HSPA1B | 200800_s_at | Heat shock 70-kD protein 1A / heat shock 70-kD protein 1B | 3303/3304 | 1.66 |
| GALNT2 | 217788_s_at | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2 (GalNAc-T2) | 2590 | 1.83 |
| PDGFC | 218718_at | Platelet-derived growth factor C | 56034 | 2.08 |
| HPGD | 203914_x_at | Hydroxyprostaglandin dehydrogenase 15-(NAD) | 3248 | 2.00 |
| 203913_s_at | 2.29 | |||
| GADD45A | 203725_at | Growth arrest and DNA-damage-inducible, alpha | 1647 | 2.38 |
Definition of abbreviation: ARDS, acute respiratory distress syndrome.
A negative number of fold change means the gene was down-regulated during the acute stage of ARDS, and a positive number of fold change means the gene was up-regulated during the acute stage.
The biological functions that may be associated with the genes demonstrating ARDS-related expression changes was next investigated by GO Biological Process categories and by limited literature mining in the Medline database. The GO analysis focused on GO terms with over-representation of genes that were differentially expressed more frequently than expected by chance. We found that all GO terms, which tested significantly after Bonferroni correction (P < 0.0002) for multiple comparison, were closely interconnected on GO structure under two general GO categories: Response to Stimulus (GO: 50896) and Death (GO: 16265), as listed in Table 3. In addition, a more specific GO category of Prostaglandin Metabolism (GO: 6693) was marginally significant after Bonferroni correction (P = 0.0004), with three genes (CD74, HGPD, and PTGS2) associated with ARDS-related expression changes. A total of 44 genes were identified within three categories of GO terms: Response to Stimulus (GO: 50896), Death (GO: 16265), and Prostaglandin Metabolism (GO: 6693). In addition, the list of 126 altered genes was compared with the results of MedGene literature mining in Medline database for top-ranked genes linked to ARDS, as well as ARDS-related MeSH terms, including “neutrophil,” “leukotriene,” and “prostaglandin” (25). We identified three genes associated with ARDS and an additional 27 genes associated with relevant MeSH terms (Table E4), of which 20 genes were also found in the significant GO categories. Furthermore, using MedGene literature mining of the 126 genes with altered expression, 8 genes were found directly linked to the development of ARDS, and all were located in the GO term of Response to Stimulus (GO: 50896) (Table E3).
TABLE 3.
GENE ONTOLOGY CATEGORIES WITH ENRICHMENT OF 126 DIFFERENTIALLY-EXPRESSED GENES
| Gene Ontology ID | Gene Ontology Name | P Value | No. of Genes in List | No. of Genes in Array |
|---|---|---|---|---|
| 50896 | Response to stimulus | 1.52E-06 | 36 | 1672 |
| 6950 | Response to stress | 1.14E-06 | 25 | 901 |
| 9607 | Response to biotic stimulus | 6.69E-06 | 22 | 802 |
| 6952 | Defense response | 3.77E-05 | 20 | 766 |
| 51707 | Response to other organism | 5.11E-05 | 15 | 478 |
| 6955 | Immune response | 2.93E-05 | 19 | 690 |
| 9613 | Response to pest, pathogen or parasite | 4.02E-05 | 15 | 468 |
| 16265 | Death | 3.20E-07 | 19 | 506 |
| 8219 | Cell death | 3.01E-07 | 19 | 504 |
| 12501 | Programmed cell death | 6.49E-07 | 18 | 478 |
| 6915 | Apoptosis | 6.10E-07 | 18 | 476 |
| 43067 | Regulation of programmed cell death | 5.83E-05 | 12 | 321 |
| 42981 | Regulation of apoptosis | 5.49E-05 | 12 | 319 |
All gene ontology biological processes were significantly enriched in the lists of 126 genes identified by paired t test (P < 0.05) after multiple comparison adjustment (Bonferroni correction).
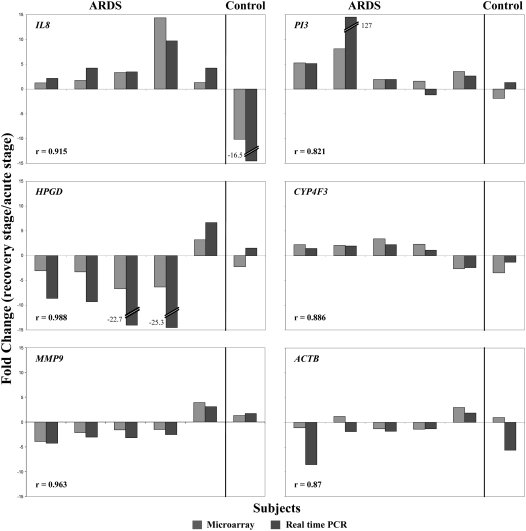
Microarray results were validated by analyzing five selected genes using quantitative RT-PCR on six subjects with enough paired RNA samples available after microarray analysis. Three genes, the down-regulated PI3 and IL8 genes and up-regulated HPGD gene at the acute stage of ARDS, were chosen based on their large differential expression between acute stage and recovery stage of ARDS, and being identified repeatedly in Medline literature mining. In addition, a gene with marginal expression change (CYP4F3, −1.5-fold change, P = 0.08; encoding enzyme involved in the process of inactivating and degrading leukotriene B4) and a gene with no change (MMP9, 1.0 fold-change, P = 1.0; encoding a matrix metalloproteinase) were chosen as reference genes. As shown in Figure 1, the microarray and quantitative RT-PCR results showed good agreement in fold-changes between two time points (correlation coefficients: 0.82–0.99). For all tested genes, there were no statistically significant differences between microarray and RT-PCR measured fold-changes by paired t test (P range 0.052–0.803).
Figure 1.
Comparison of the expression fold-changes of selected genes measured by microarray and quantitative RT-PCR. Of 11 pairs of RNA samples used in microarray analysis, only 6 pairs had enough RNA for quantitative RT-PCR analysis, including 5 patients with acute respiratory distress syndrome (ARDS) and 1 control subject. Gene PI3, IL8, and HPGD were chosen as having large differential expression between acute stage and stable stage of ARDS, as well as being identified repeatedly in Medline literature mining. Gene CYP4F3 with marginal expression change and gene MMP9 with no change were chosen as reference genes in validation. The results of gene ACTB was also illustrated, which was originally selected as endogenous control. The y-axis displays the fold change of the recovery stage to the acute stage of ARDS.
Among 126 identified genes, the PI3 gene had the largest fold-change in expression between the acute stage and the recovery stage of ARDS (−2.98 fold-changes). This gene encodes a neutrophil elastase inhibitor, peptidase inhibitor 3 (PI3), formally called elafin. PI3 is one of two low-molecular-weight protease inhibitors of the anti-leukoprotease family, which was previous reported to be localized to the injury sites (26). In addition to antiproteinase activity, PI3 proteins also demonstrate antimicrobial and anti-inflammatory activities (27). Compared with the recovery stage of ARDS, the PI3 gene was expressed at a 2.98-fold change lower level in peripheral blood during the acute stage of ARDS (Table 2), suggesting that PI3 gene product could play important roles in the development of ARDS.
We investigated further the role of PI3 during the early development of ARDS by investigating plasma levels among three ARDS sample groups, the pre-diagnosis group, day-of-diagnosis group, and post-diagnosis group, as well as between ARDS sample groups and at-risk control subjects. Demographic characterization of these patients is shown in Tables E1 and E2. Most of the descriptive characteristics were not significantly different between ARDS cases and control subjects, except that ARDS cases were more frequently received packed red blood cell transfusion (P = 0.009). In addition, for both patients with ARDS and at-risk control subjects, there was no statistically significant difference in baseline characteristics between the selected subjects and the rest of the study population (data not shown).
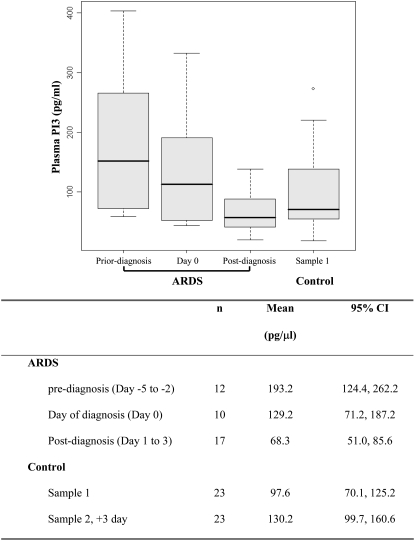
The plasma PI3 levels were statistically significant different among three ARDS sample groups (ANOVA, P = 0.001) and among four sample groups, including the first sample of control subjects (ANOVA, P = 0.003). There was a trend of decreasing plasma PI3 levels from pre-diagnosis group to post-diagnosis group, as shown in Figure 2. The post-diagnosis group had the lowest plasma PI3 level (mean, 68.3 pg/ml; 95% confidence interval [CI], 51–85.6 pg/ml), which was statistically significantly lower than the pre-diagnosis group (mean, 193.2 pg/ml; 95% CI, 124.4–262.2 pg/ml; P = 0.0009) and not significantly different from the day-of-diagnosis group (mean, 129.2 pg/ml; 95% CI, 71.2–187.2 pg/ml) after adjusting for multiple comparisons (P = 0.05). Plasma PI3 levels in the post-diagnosis group were not significantly different from those of at-risk control subjects collected during the first 24 hours of ICU admission (P = 0.19). Among three ARDS groups, the pre-diagnosis group had the highest plasma PI3 level, but was not significantly higher than the first samples of the control subjects collected during the first 24 hours of ICU admission (mean, 97.6 pg/ml; 95% CI, 70.1–125.2 pg/ml) after adjusting for multiple comparisons (P = 0.01).
Figure 2.
Levels of pre-elafin (PI3) in plasma of patients with ARDS and critically ill patients who did not develop ARDS (control subjects). Each ARDS case provides on one plasma sample. Based the date of sample collection relative to the ARDS diagnosis date, including pre-diagnosis group (Day −5 to Day −1), day of diagnosis group (Day 0), and post-diagnosis group (Day 1 to Day 3). Sample 1 of control was collected during the first 2 days of ICU admission. There was no statistically significant difference in baseline characteristics between patients with ARDS and control subjects, except that patients with ARDS more frequently received transfusion (P = 0.009). Significant results of t test: ARDS pre-diagnosis versus post-diagnosis, P = 0.0009.
We also tested the trend of plasma PI3 decrease across the ARDS development, as measured by sampling dates relative to ARDS diagnosis (Pearson correlation coefficient, −0.52; P = 0.0006), using stepwise multivariable linear regression models. Besides sampling date relative to ARDS diagnosis, pneumonia (partial R2, 0.102; P = 0.011) and lowest level of hematocrit on ICU admission (partial R2, 0.143; P = 0.006) were significantly related to plasma PI3 levels. Without forcing in any covariate, the stepwise multivariable linear regression analysis picked the same three covariates. This regression model explained 54.5% of the inter-individual variability in plasma PI3 levels, with sampling date relative to ARDS diagnosis alone explaining 30.1% of the total variance (mean regression slope: fold change of plasma PI3/day: 0.897; 95% CI, 0.834–0.967; P = 0.005). Furthermore, when extra covariates, including age, sex, APACHE III score on ICU admission, history of steroid use, sepsis, septic shock, diabetes, chronic liver disease, and trauma, were added, the model remained significant (P = 0.002) with R2 increased to 0.60. In contrast, no statistically significant difference existed in plasma levels of MMP-9 among three ARDS sample groups (ANOVA, P = 0.36) and among four sample groups including the first sample of controls (ANOVA, P = 0.54), as expected based on the lack of change (1.0-fold change, P = 1.0) in the microarray analysis (Figure E1). In addition, in the at-risk control subjects we found a statistically significant increase of plasma PI3 levels (mean fold change of plasma PI3 level, 1.53; 95% CI, 1.28–1.78; P = 0.0007) 3 days after the first 24 hours of ICU admission (Figure E2).
DISCUSSION
Using a microarray-based global gene profiling of whole blood total RNA, we identified 126 genes with altered expression between the acute stage and recovery stage in subjects with ARDS. Based on Gene Ontology annotations, the ARDS-related gene expression changes were clustered in GO biological processes related to Response to Stimulus (GO: 50896) and Death (GO: 16265). Significant subordinate GO terms of Death (GO: 16265) included Cell Death (GO: 8219), Programmed Cell Death (GO: 12501), Apoptosis (GO: 6915), Regulation of Programmed Cell Death (GO: 43067), and Regulation of Apoptosis (GO: 42981). This clustering of differentially expressed genes related to the regulation of apoptosis would support previous observations of reduced apoptosis in alveolar neutrophils isolated from patients with ARDS (28).
Among 126 identified genes, we found that gene PI3 had the largest differential expression in peripheral blood of patients with ARDS. Expression of PI3 gene was suppressed at the early, acute stage of ARDS compared with significantly higher levels during the recovery stage of ARDS. PI3 gene encodes protein peptidase inhibitor 3 with two isoforms, including a 95–amino acid molecule named pre-elafin (also known as trappin-2) and a 58–amino acid molecule called elafin, which was produced by proteolytic cleavage of pre-elafin. Both pre-elafin and elafin have two functional domains, including the C-terminal domain containing the antiproteinase active site and the N-terminal domain containing motifs of transglutaminase substrate. The transglutaminase substrate motifs, five motifs in pre-elafin molecules and one motif in elafin, enables covalent binding of proteinase inhibitors to the extracellular matrix (ECM) proteins (29). PI3 protein expression is induced by inflammation-initiating cytokines and localized to the site of inflammatory response, including airways, skin, and other mucosal surfaces (30–32). However, in this study, we found that the expression of PI3 gene in peripheral blood was differentially expressed between the acute stage and the recovery stage of ARDS.
Importantly, the microarray findings of lower PI3 gene expression during the acute stage of ARDS was well correlated with the results of the ELISA assay measurements of plasma PI3 levels, which found PI3 was expressed at the lowest level in plasma during the acute stage, compared with plasma PI3 levels pre-diagnosis and the day of ARDS diagnosis. Moreover, the time course of plasma PI3 decrease was also well correlated with the course of early ARDS development, as the plasma sampling date relative to ARDS diagnosis not only shows a moderate negative correlation with plasma PI3 level (Pearson correlation coefficient: −0.52, P = 0.0006), but also has the largest contribution in explaining the plasma PI3 variance (partial R2, 30.1%) in the multivariable linear regression model. Furthermore, there seems to be a pre-onset increase of the plasma PI3 level in the pre-diagnosis ARDS group (mean 193.2 pg/ml, 95% C: 124.4–262.2 pg/ml; P = 0.01), as compared with at-risk controls within the first 24-hour ICU admission (mean 97.6 pg/ml, 95% C: 70.1–125.2 pg/ml), even though it was not reached significantly level under multiple comparison adjustment. In contrast, there was a significant but moderate increase of plasma PI3 levels during a 3-day period after ICU admission in at-risk control subjects (mean fold change of plasma PI3 level: 1.53; 95% CI, 1.28–1.78; P = 0.0007; Figure E2). Taken together, these observations suggest that PI3 levels in plasma could be used as a biomarker for monitoring the development and progress of ARDS. Similarly, an earlier study reported that the serum levels of elafin could be used to monitor the disease activity of psoriasis, an inflammatory skin and joint disease (33). The serum elafin levels were correlated with the clinical course of psoriasis and disease severity score (Psoriasis Area Severity Index), and a decrease in serum elafin levels was associated with response to cyclosporine A treatment.
Neutrophils play a crucial role in the initiation and propagation of ARDS (3). Considerable evidence exists for the role of neutrophil-derived proteinases in the pathogenesis of ARDS, including neutrophil elastase and collagenase (34–36). A local imbalance between proteinases and their physiological inhibitors results in pulmonary parenchyma damage by leakage of a protein-rich fluid into the interstitium and alveolar spaces. Major pulmonary proteinase inhibitors include α1-proteinase inhibitor (α1-PI), secretory leukocyte proteinase inhibitor (SLPI), and elafin (PI3) (32, 37). Unlike high-molecular-weight α1-PI, which is mainly produced by the liver and reaches the lung via passive diffusion, SLPI and PI3 are low-molecular-weight inhibitors and are produced locally at neutrophil infiltration site in the lung (30). PI3 and SLPI are important antiproteinases in the lung in both health and disease (38), and also demonstrate multiple biological functions, such as antibacterial activity, anti-inflammatory activity, priming of innate immunity, tissue remodeling and cellular differentiation, and augmentation of antiviral adaptive immunity (39).
Contrary to extensive molecular characterization of PI3, there are limited studies directly investigating the potential protective effects of PI3 in acute lung injury (ALI) and ARDS. In a murine model of lung injury mediated by Pseudomonas aeruginosa, intratracheal administration of human elafin encoded on an adenovirus vector showed significant protection against lung injury by reduced protein concentrations in BAL fluid, increased elimination of bacteria from the airways, and decreased incidence of hematogenous bacterial dissemination (40). In another study by Tremblay and coworkers, recombinant human pre-elafin exhibited a significant protective effect against human neutrophil elastase (HNE) induced acute lung injury in hamsters in a dose-dependent manner (41). In contrast to pre-elafin, elafin did not show such a protective effect within the same study. Elafin was the proteolytic product of pre-elafin, containing less transglutaminase substrate motifs (42). Both pre-elafin and elafin could be cross-linked to ECM protein, catalyzed by transglutaminase, and still exert anti-proteinase function (43). However, additional transglutaminase substrate motifs contained in pre-elafin might allow stronger binding locally to inflammatory sites and show stronger protection effects than elafin.
Currently, there is only one report of measuring the BAL levels of PI3 and SLPI, as well as α1-PI, in human population (44). Although significant increases of PI3 were observed among patients with ARDS and at-risk patient without ARDS compared with healthy individuals, there was no significant difference of PI3 between patients with ARDS and at-risk patient without ARDS. In contrast, both SLPI and α1-PI demonstrated significant increases between two patient groups. Furthermore, almost all of the detectable PI3 was associated with high-molecular-weight proteins, as revealed by Western blot analysis. We believe that the difference between PI3 and SLPI in BAL could be partially explained by the unique structure of transglutaminase substrate motifs contained in PI3 protein, which allows covalent binding of PI3 to ECM proteins (29, 45). As most of PI3 is anchored to lung parenchyma and exerts its biological functions locally (44), it may be impossible to assess accurately the protective role of PI3 in ALI or ARDS in BAL fluid or in lung tissue.
We also found that gene IL8 had a large differential expression in peripheral blood of patients with ARDS, similar to the pattern of gene PI3. IL-8 is a potent neutrophil chemoattractant, which has been associated with ARDS in a number of previous studies. Markedly increased IL-8 level was consistently found in BAL fluid from patients with ARDS, with strong correlation to the numbers of BAL neutrophils, suggesting that IL-8 played a major role in promoting lung damage (1, 46–49). Persistent increase of BAL IL-8 level was also associated with poor clinic prognosis (50, 51). Unlike previous studies, we found that IL8 gene expression was suppressed at the early, acute stage of ARDS, as compared with the recovery stage in this study. However, most of the previous studies mainly focused on the measurement of IL-8 levels in BAL fluid, with fewer reports including IL-8 measurements in peripheral blood in patients with ARDS. Compared with healthy control subjects, Callister and colleagues reported significant higher levels of plasma IL-8 in patients with mixed ALI/ARDS (P < 0.0001) (46). When focusing on the early stage of ARDS development, Goodman and coworkers observed only a marginal significant increase of plasma IL-8 level in six patients (P = 0.05) within 24 hours of post-traumatic ARDS, with a wide concentration range, than those of healthy control subjects (52). In another study with serum IL-8 measurement in the first 24 hours of ARDS, there was no significant difference between ARDS and ALI, in contrast to significantly elevated IL-8 levels in BAL fluid in ARDS (49). The same study also reported concentrations of IL-8 in both BAL fluid and pulmonary epithelial lining fluid were significantly higher than serum IL-8 levels in all ARDS and ALI control subjects, indicating that there was no significant correlations between pulmonary and blood IL-8 levels. Considering that many factors associated with underlying causes of ARDS could affect the plasma IL-8 levels, we did not further investigate plasma IL-8 level in the current study.
In this study, we were able to uniquely investigate the decrease of plasma PI3 along with the early ARDS development, specifically, from pre- to post-ARDS diagnosis. It is worth noting the drawback that the available ARDS plasma samples represented different patients, instead of ideal longitudinal samples from each patient. However, the observed decrease of plasma PI3 is not likely to be solely associated with ARDS risk factors, such as sepsis or trauma, as the regression model remained significant after adjusting for important clinic covariates of ARDS, and there were no significant differences of these risk factors between patients with ARDS and at-risk control subjects in the studied population. In addition, we only observed the coincidence of the lower PI3 gene expression and the lower plasma PI3 level during the acute stage of ARDS; additional studies are needed to address whether the variance of plasma PI3 level is merely driven by the expression change of PI3 gene in blood cells or reflects different PI3 levels in lung, or both. In conclusion, this study is the first report of the decrease of PI3 in the peripheral blood is associated with the early ARDS development, and suggests that circulating PI3 may provide a useful clinical marker for monitoring the early development of ARDS and may have significant implications for ARDS treatment.
Supplementary Material
Acknowledgments
The authors thank Michelle Gong, Weiling Zhang, Kelly McCoy, Thomas McCabe, Marcia Chertok, and Julia Shin for patient recruitment for the Molecular Epidemiology of ARDS project; Andrea Shafer and Lia Shimada for research support; Ian James for laboratory expertise; and Janna Frelich and Lucille Pothier for data management.
This work was supported by National Institutes of Health grants HL60710 and ES00002 (to D.C.C.).
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2007-0354OC on January 18, 2008
Conflict of Interest Statement: None of the authors has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Goodman RB, Strieter RM, Martin DP, Steinberg KP, Milberg JA, Maunder RJ, Kunkel SL, Walz A, Hudson LD, Martin TR. Inflammatory cytokines in patients with persistence of the acute respiratory distress syndrome. Am J Respir Crit Care Med 1996;154:602–611. [DOI] [PubMed] [Google Scholar]
- 2.Holter JF, Weiland JE, Pacht ER, Gadek JE, Davis WB. Protein permeability in the adult respiratory distress syndrome: loss of size selectivity of the alveolar epithelium. J Clin Invest 1986;78:1513–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weiland JE, Davis WB, Holter JF, Mohammed JR, Dorinsky PM, Gadek JE. Lung neutrophils in the adult respiratory distress syndrome: clinical and pathophysiologic significance. Am Rev Respir Dis 1986;133:218–225. [DOI] [PubMed] [Google Scholar]
- 4.Strieter RM, Kunkel SL, Keane MP, Standiford TJ. Chemokines in lung injury: Thomas A. Neff Lecture. Chest 1999;116:103S–110S. [DOI] [PubMed] [Google Scholar]
- 5.Brundage JF. Interactions between influenza and bacterial respiratory pathogens: implications for pandemic preparedness. Lancet Infect Dis 2006;6:303–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen CY, Lee CH, Liu CY, Wang JH, Wang LM, Perng RP. Clinical features and outcomes of severe acute respiratory syndrome and predictive factors for acute respiratory distress syndrome. J Chin Med Assoc 2005;68:4–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Looney MR. Newly recognized causes of acute lung injury: transfusion of blood products, severe acute respiratory syndrome, and avian influenza. Clin Chest Med 2006;27:591–600. (abstract viii). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science 1999;286:531–537. [DOI] [PubMed] [Google Scholar]
- 9.Izuhara K, Saito H. Microarray-based identification of novel biomarkers in asthma. Allergol Int 2006;55:361–367. [DOI] [PubMed] [Google Scholar]
- 10.Selman M, Pardo A, Barrera L, Estrada A, Watson SR, Wilson K, Aziz N, Kaminski N, Zlotnik A. Gene expression profiles distinguish idiopathic pulmonary fibrosis from hypersensitivity pneumonitis. Am J Respir Crit Care Med 2006;173:188–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wurfel MM. Microarray-based analysis of ventilator-induced lung injury. Proc Am Thorac Soc 2007;4:77–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coldren CD, Nick JA, Poch KR, Woolum MD, Fouty BW, O'Brien JM, Gruber MP, Zamora MR, Svetkauskaite D, Richter DA, et al. Functional and genomic changes induced by alveolar transmigration in human neutrophils. Am J Physiol Lung Cell Mol Physiol 2006;291:L1267–L1276. [DOI] [PubMed] [Google Scholar]
- 13.Wang Z, Neuburg D, Li C, Su L, Kim JY, Chen JC, Christiani DC. Global gene expression profiling in whole-blood samples from individuals exposed to metal fumes. Environ Health Perspect 2005;113:233–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gong MN, Zhou W, Williams PL, Thompson BT, Pothier L, Boyce P, Christiani DC. -308GA and TNFB polymorphisms in acute respiratory distress syndrome. Eur Respir J 2005;26:382–389. [DOI] [PubMed] [Google Scholar]
- 15.Gong MN, Wei Z, Xu LL, Miller DP, Thompson BT, Christiani DC. Polymorphism in the surfactant protein-B gene, gender, and the risk of direct pulmonary injury and ARDS. Chest 2004;125:203–211. [DOI] [PubMed] [Google Scholar]
- 16.Bernard GR, Artigas A, Brigham KL, Carlet J, Falke K, Hudson L, Lamy M, Legall JR, Morris A, Spragg R. The American-European Consensus Conference on ARDS. Definitions, mechanisms, relevant outcomes, and clinical trial coordination. Am J Respir Crit Care Med 1994;149:818–824. [DOI] [PubMed] [Google Scholar]
- 17.Knaus WA, Wagner DP, Draper EA, Zimmerman JE, Bergner M, Bastos PG, Sirio CA, Murphy DJ, Lotring T, Damiano A, et al. The APACHE III prognostic system. Risk prediction of hospital mortality for critically ill hospitalized adults. Chest 1991;100:1619–1636. [DOI] [PubMed] [Google Scholar]
- 18.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA 2001;98:31–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu WM, Mei R, Di X, Ryder TB, Hubbell E, Dee S, Webster TA, Harrington CA, Ho MH, Baid J, et al. Analysis of high density expression microarrays with signed-rank call algorithms. Bioinformatics 2002;18:1593–1599. [DOI] [PubMed] [Google Scholar]
- 20.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000;25:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hong P, Wong WH. GeneNotes–a novel information management software for biologists. BMC Bioinformatics 2005;6:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001;25:402–408. [DOI] [PubMed] [Google Scholar]
- 23.Shi L, Reid LH, Jones WD, Shippy R, Warrington JA, Baker SC, Collins PJ, de Longueville F, Kawasaki ES, Lee KY, et al. The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat Biotechnol 2006;24:1151–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo L, Lobenhofer EK, Wang C, Shippy R, Harris SC, Zhang L, Mei N, Chen T, Herman D, Goodsaid FM, et al. Rat toxicogenomic study reveals analytical consistency across microarray platforms. Nat Biotechnol 2006;24:1162–1169. [DOI] [PubMed] [Google Scholar]
- 25.Hu Y, Hines LM, Weng H, Zuo D, Rivera M, Richardson A, LaBaer J. Analysis of genomic and proteomic data using advanced literature mining. J Proteome Res 2003;2:405–412. [DOI] [PubMed] [Google Scholar]
- 26.Sallenave JM. The role of secretory leukocyte proteinase inhibitor and elafin (elastase-specific inhibitor/skin-derived antileukoprotease) as alarm antiproteinases in inflammatory lung disease. Respir Res 2000;1:87–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roghanian A, Williams SE, Sheldrake TA, Brown TI, Oberheim K, Xing Z, Howie SE, Sallenave JM. The antimicrobial/elastase inhibitor elafin regulates lung dendritic cells and adaptive immunity. Am J Respir Cell Mol Biol 2006;34:634–642. [DOI] [PubMed] [Google Scholar]
- 28.Matute-Bello G, Liles WC, Radella F II, Steinberg KP, Ruzinski JT, Jonas M, Chi EY, Hudson LD, Martin TR. Neutrophil apoptosis in the acute respiratory distress syndrome. Am J Respir Crit Care Med 1997;156:1969–1977. [DOI] [PubMed] [Google Scholar]
- 29.Schalkwijk J, Wiedow O, Hirose S. The trappin gene family: proteins defined by an N-terminal transglutaminase substrate domain and a C-terminal four-disulphide core. Biochem J 1999;340:569–577. [PMC free article] [PubMed] [Google Scholar]
- 30.Sallenave JM, Shulmann J, Crossley J, Jordana M, Gauldie J. Regulation of secretory leukocyte proteinase inhibitor (SLPI) and elastase-specific inhibitor (ESI/elafin) in human airway epithelial cells by cytokines and neutrophilic enzymes. Am J Respir Cell Mol Biol 1994;11:733–741. [DOI] [PubMed] [Google Scholar]
- 31.Alkemade JA, Molhuizen HO, Ponec M, Kempenaar JA, Zeeuwen PL, de Jongh GJ, van Vlijmen-Willems IM, van Erp PE, van de Kerkhof PC, Schalkwijk J. SKALP/elafin is an inducible proteinase inhibitor in human epidermal keratinocytes. J Cell Sci 1994;107:2335–2342. [DOI] [PubMed] [Google Scholar]
- 32.Pfundt R, van Ruissen F, van Vlijmen-Willems IM, Alkemade HA, Zeeuwen PL, Jap PH, Dijkman H, Fransen J, Croes H, van Erp PE, et al. Constitutive and inducible expression of SKALP/elafin provides anti-elastase defense in human epithelia. J Clin Invest 1996;98:1389–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alkemade HA, de Jongh GJ, Arnold WP, van de Kerkhof PC, Schalkwijk J. Levels of skin-derived antileukoproteinase (SKALP)/elafin in serum correlate with disease activity during treatment of severe psoriasis with cyclosporin A. J Invest Dermatol 1995;104:189–193. [DOI] [PubMed] [Google Scholar]
- 34.Lee CT, Fein AM, Lippmann M, Holtzman H, Kimbel P, Weinbaum G. Elastolytic activity in pulmonary lavage fluid from patients with adult respiratory-distress syndrome. N Engl J Med 1981;304:192–196. [DOI] [PubMed] [Google Scholar]
- 35.McGuire WW, Spragg RG, Cohen AB, Cochrane CG. Studies on the pathogenesis of the adult respiratory distress syndrome. J Clin Invest 1982;69:543–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Christner P, Fein A, Goldberg S, Lippmann M, Abrams W, Weinbaum G. Collagenase in the lower respiratory tract of patients with adult respiratory distress syndrome. Am Rev Respir Dis 1985;131:690–695. [DOI] [PubMed] [Google Scholar]
- 37.Kramps JA, Rudolphus A, Stolk J, Willems LN, Dijkman JH. Role of antileukoprotease in the human lung. Ann N Y Acad Sci 1991;624:97–108. [DOI] [PubMed] [Google Scholar]
- 38.Tremblay GM, Sallenave JM, Israel-Assayag E, Cormier Y, Gauldie J. Elafin/elastase-specific inhibitor in bronchoalveolar lavage of normal subjects and farmer's lung. Am J Respir Crit Care Med 1996;154:1092–1098. [DOI] [PubMed] [Google Scholar]
- 39.Williams SE, Brown TI, Roghanian A, Sallenave JM. SLPI and elafin: one glove, many fingers. Clin Sci (Lond) 2006;110:21–35. [DOI] [PubMed] [Google Scholar]
- 40.Simpson AJ, Wallace WA, Marsden ME, Govan JR, Porteous DJ, Haslett C, Sallenave JM. Adenoviral augmentation of elafin protects the lung against acute injury mediated by activated neutrophils and bacterial infection. J Immunol 2001;167:1778–1786. [DOI] [PubMed] [Google Scholar]
- 41.Tremblay GM, Vachon E, Larouche C, Bourbonnais Y. Inhibition of human neutrophil elastase-induced acute lung injury in hamsters by recombinant human pre-elafin (trappin-2). Chest 2002;121:582–588. [DOI] [PubMed] [Google Scholar]
- 42.Guyot N, Zani ML, Berger P, Dallet-Choisy S, Moreau T. Proteolytic susceptibility of the serine protease inhibitor trappin-2 (pre-elafin): evidence for tryptase-mediated generation of elafin. Biol Chem 2005;386:391–399. [DOI] [PubMed] [Google Scholar]
- 43.Guyot N, Zani ML, Maurel MC, Dallet-Choisy S, Moreau T. Elafin and its precursor trappin-2 still inhibit neutrophil serine proteinases when they are covalently bound to extracellular matrix proteins by tissue transglutaminase. Biochemistry 2005;44:15610–15618. [DOI] [PubMed] [Google Scholar]
- 44.Sallenave JM, Donnelly SC, Grant IS, Robertson C, Gauldie J, Haslett C. Secretory leukocyte proteinase inhibitor is preferentially increased in patients with acute respiratory distress syndrome. Eur Respir J 1999;13:1029–1036. [DOI] [PubMed] [Google Scholar]
- 45.Nara K, Ito S, Ito T, Suzuki Y, Ghoneim MA, Tachibana S, Hirose S. Elastase inhibitor elafin is a new type of proteinase inhibitor which has a transglutaminase-mediated anchoring sequence termed “cementoin.” J Biochem (Tokyo) 1994;115:441–448. [DOI] [PubMed] [Google Scholar]
- 46.Callister ME, Burke-Gaffney A, Quinlan GJ, Nicholson AG, Florio R, Nakamura H, Yodoi J, Evans TW. Extracellular thioredoxin levels are increased in patients with acute lung injury. Thorax 2006;61:521–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miller EJ, Cohen AB, Nagao S, Griffith D, Maunder RJ, Martin TR, Weiner-Kronish JP, Sticherling M, Christophers E, Matthay MA. Elevated levels of NAP-1/interleukin-8 are present in the airspaces of patients with the adult respiratory distress syndrome and are associated with increased mortality. Am Rev Respir Dis 1992;146:427–432. [DOI] [PubMed] [Google Scholar]
- 48.Donnelly SC, Strieter RM, Kunkel SL, Walz A, Robertson CR, Carter DC, Grant IS, Pollok AJ, Haslett C. Interleukin-8 and development of adult respiratory distress syndrome in at-risk patient groups. Lancet 1993;341:643–647. [DOI] [PubMed] [Google Scholar]
- 49.Wiedermann FJ, Mayr AJ, Kaneider NC, Fuchs D, Mutz NJ, Schobersberger W. Alveolar granulocyte colony-stimulating factor and alpha-chemokines in relation to serum levels, pulmonary neutrophilia, and severity of lung injury in ARDS. Chest 2004;125:212–219. [DOI] [PubMed] [Google Scholar]
- 50.Meduri GU, Kohler G, Headley S, Tolley E, Stentz F, Postlethwaite A. Inflammatory cytokines in the BAL of patients with ARDS: persistent elevation over time predicts poor outcome. Chest 1995;108:1303–1314. [DOI] [PubMed] [Google Scholar]
- 51.Ikuta N, Taniguchi H, Kondoh Y, Takagi K, Hayakawa T. Sustained high levels of circulatory interleukin-8 are associated with a poor outcome in patients with adult respiratory distress syndrome. Intern Med 1996;35:855–860. [DOI] [PubMed] [Google Scholar]
- 52.Goodman ER, Kleinstein E, Fusco AM, Quinlan DP, Lavery R, Livingston DH, Deitch EA, Hauser CJ. Role of interleukin 8 in the genesis of acute respiratory distress syndrome through an effect on neutrophil apoptosis. Arch Surg 1998;133:1234–1239. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.