Abstract
Post-translational sulfation of tyrosines affects the affinity and binding of at least some chemokine receptors to their ligand(s) and has been hypothesized to be a feature in all chemokine receptors. This binding initiates downstream signaling cascades. By this mechanism, tyrosine sulfation can influence the cells involved in acute and chronic events of cellular immunity. These events include leukocyte trafficking and airway inflammation important in asthma and chronic obstructive pulmonary disease (COPD). We are using computational methods to convert the poorly defined hypothesis of more widespread sulfation of chemokine receptors to more specific assessments of how closely the sequence environment of each tyrosine residue resembles the sequence environment of tyrosine residues proven to be sulfated. Thus, we provide specific and readily tested hypotheses about the tyrosine residues in all of the chemokine receptors. Tyrosine sulfation was predicted with high scores in the N-terminus domain of 13 out of 18 human chemokine receptor proteins using a position-specific scoring matrix, which was determined to be 94.2% accurate based on Receiver Operating Characteristic analysis. The remaining chemokine receptors have sites exhibiting features of tyrosine sulfation. These putative sites demonstrate clustering in a manner consistent with known tyrosine sulfation sites and conservation both within the chemokine receptor family and across mammalian species. Human chemokine receptors important in asthma and COPD, such as CXCR1, CXCR2, CXCR3, CXCR4, CCR1, CCR2, CCR3, CCR4, CCR5, and CCR8, contain at least one known or predicted tyrosine sulfation site. Recognition that tyrosine sulfation is found in most clinically relevant chemokine receptors could help the development of specific receptor-ligand antagonists to modulate events important in airway diseases.
Keywords: tyrosine sulfation, post-translational modification, chemokine receptors, asthma, chronic obstructive pulmonary disease
CLINICAL RELEVANCE
We have predicted tyrosine sulfation in 13 chemokine receptors. Knowledge of this modification would enable more specific antagonists to be modeled for targeting asthma, chronic obstructive pulmonary disease, and other similar inflammatory lung diseases.
Our understanding of the events that initiate and promote chronic inflammation in asthma and chronic obstructive pulmonary disease (COPD) is changing with the recognition that chemokines are central to their pathogenesis (1). Evidence from animal models and humans indicates that leukocytes and mononuclear cells play a crucial role in mediating the inflammation that underlies asthma and COPD, in which adaptive immune responses become and remain abnormal. Chemokine receptors on mononuclear cells and leukocytes mediate inflammatory cell trafficking, activation, and differentiation, but how chemokine receptors interact with chemokines in vivo is not completely known (2, 3). Human chemokine receptors are a family of 18 seven-transmembrane (7TM) G protein–coupled receptors that bind to a diverse group of chemokine ligands (2). Unlike most 7TM receptors, chemokine receptors have a broad and overlapping specificity for their chemokine ligands and are expressed on various leukocytes (4). Leukocytes respond to specific chemokines, determined by which chemokine receptor(s) they express on their cell membranes.
Chemokine receptors important in asthma include those that are known to be sulfated (CCR2, CCR5, CCR8, CXCR3, and CXCR4) and those not yet known to be sulfated (CCR1, CCR3, CCR4, CXCR1, and CXCR2) (1, 3). Knowledge of sulfation in these chemokine receptors may elucidate unrecognized chemokine and chemokine receptor interactions leading to the development of additional receptor agonist/antagonists. Finding experimental evidence for tyrosine sulfation would support the hypothesis that tyrosine sulfation plays an important role in the function of these critical proteins (5, 6). Here we computationally examine the sequence data to see if it is likely that such experimental evidence can ultimately be found and determine at which tyrosine residues such evidence is most likely to be found. This converts the general hypothesis of tyrosine sulfation of these proteins into a more specific and readily testable prediction about specific residues.
Tyrosine sulfation is a selective post-translational modification of proteins involving the addition of a negatively charged sulfate group to an exposed tyrosine residue, yielding tyrosyl O-sulfate. Tyrosyl protein sulfotransferases (TPSTs), which catalyze this reaction, are localized in the trans-Golgi (7). Huttner and Baeuerle estimated that sulfation of tyrosines occurs in approximately 1% of all tyrosines in eukaryotic proteins (7), based on Drosophila data available 25 years ago. Only 70 proteins are currently known to contain sulfated tyrosines, and 89 individual sulfated tyrosines have been experimentally identified. From these tyrosine sites, it has been observed that negatively charged, acidic residues frequently flank sulfated tyrosines, and that sulfated tyrosines are often grouped in well-defined clusters. The major determinants of tyrosine sulfation, then, are the presence of negatively charged acidic amino acid residues and secondary and tertiary protein structure that likely promotes sulfation by exposing adjacent tyrosines to TPSTs (8).
Sulfated tyrosines have been experimentally confirmed in only five human chemokine receptors: CXCR3, CXCR4, CCR2b, CCR5, and CX3CR1 (5, 9–12), and one murine receptor, CCR8 (13). Sulfated tyrosines in each of the five human chemokine receptors were shown to be in the N-terminus, a protein domain critical for high binding affinity (12, 14–17). Sulfated tyrosines and their flanking acidic amino acid residues are required for optimal chemokine binding in CXCR4, CCR2b, and CCR5. After mutagenesis of either of those key residues, all of the receptors demonstrated a significant decrease in binding (16, 18, 19). Because of the correlation between sulfated tyrosines and chemokine receptor binding, we analyzed all chemokine receptors with our newest position-specific scoring matrix (PSSM), which includes the data for the experimentally confirmed chemokine receptor tyrosine sulfation sites, to predict the existence of tyrosine sulfation sites in chemokine receptors. By making the hypothesis of sulfation more specific and more readily tested for each chemokine receptor, these predictions may elucidate chemokine receptor–ligand interactions relevant to inflammatory response and disease.
MATERIALS AND METHODS
Definitions of Chemokine Receptors
Chemokine receptors are defined according to the standards set by the Nomenclature Committee of the International Union of Pharmacology (NC-IUPHAR), which state that chemokine receptors are proteins that both bind to established chemokines and produce intracellular signaling (4).
Definition and Extraction of a Tyrosine Sulfation Site
All protein sequences were obtained from the Swiss-Prot release 54.2 of September 11, 2007. Tyrosine sites were then extracted from the sequences using a program that screens sequences for the five amino acids immediately upstream and downstream of a tyrosine. Hence, each tyrosine site consists of the candidate tyrosine and 10 flanking amino acids, five on either side of the tyrosine.
The Position-Specific-Scoring Matrix
The construction of the p89n438 PSSM (with 89 positive sites and 438 negative sites for sulfation) has been previously described in detail (8). The p89n438 PSSM assigns unique log-odds scores for every possible amino acid in each position of the defined sulfation site relative to the central tyrosine. The score is the log of the ratio of the fraction of a particular amino acid observed in the 89 experimentally confirmed sulfation site to the fraction of the same amino acid observed at the same position in the 438 experimentally confirmed nonsulfated sites. The resulting cumulative scores represent the likelihood of sulfation in the examined sites based on their similarities and differences with known sulfated and nonsulfated sites (8). The Receiver Operating Characteristic (ROC) analysis established that the PSSM distinguishes between sulfated and nonsulfated tyrosines with 94.2% accuracy.
The cutoff score for a predicted tyrosine sulfation site has been set at 2.5 for the p89n438 PSSM because this score minimizes the total number of false positive and false negative predictions. Predicted tyrosine sulfation sites with scores 2.5 and above are referred to as “sites with high scores”; predicted tyrosine sulfation site with scores between 1.48 and 2.5 are referred to as “sites with intermediate scores.” These sites with the lower scores are included because they are in the range of scores for some known tyrosine sulfation sites. Any site with a score greater than zero indicates that the tyrosine site is more similar to sulfated sites than to nonsulfated sites.
Creation of Multiple Protein Sequence Alignments
Chemokine receptor protein sequences were truncated in such a way to include the N-termini and the beginning of the first transmembrane helix before they were converted to PIR format and submitted to the program, ProbCons (20), which generated a highly optimized alignment. The ProbCons program goes through several steps to create the best possible final alignment (20) and has been found to be better at this than the alternative programs available (21).
RESULTS
Prediction of Tyrosine Sulfation in Chemokine Receptors
Sixteen chemokine receptors of the 18 receptors tested have a total of 29 predicted tyrosine sulfation sites, which include both high-scoring (2.5 and above) and intermediate scoring (1.48–2.5) sites (Table 1). Of these, nine receptors had sites with only high scores, four receptors had sites with both high and intermediate scores, and three receptors had only intermediate-scoring predicted sites. All chemokine receptors, even CXCR5 and CCR7 (not included in Table 1), have sites with scores greater than 0, which indicates that these are more likely to be sulfated than nonsulfated. The PSSM scores of known tyrosine sulfation sites in human chemokine receptors ranged from 2.51 to 8.97, with a mean of 5.15 ± 2.54 SD. Thus, in these receptors all of the known sites had high scores. Scores for all 22 predicted tyrosine sulfation sites with high scores covered a range from 2.51 to 8.97 with a mean of 4.87 ± 2.16 SD. In the set of predicted sites, there were tyrosines with intermediate scores below 2.5, such as tyrosine (Y) 22 in CCR4, which scored 1.48. However, because of additional evidence we believe that Y22 is a strong candidate for sulfation as it aligns with, and is thus postulated to be homologous with, five other tyrosines known to be sulfated in human and mouse chemokine receptors (Figure 1). In addition to receiving high PSSM scores, two pairs of predicted tyrosine sulfation sites in two distinct chemokine receptors CXCR3 and CCR8 were within five residues of each other, consistent with a clustering pattern observed in known tyrosine sulfated proteins such as CCR5 (Table 1). All predicted sites were located in extracellular domains.
TABLE 1.
CHEMOKINE RECEPTORS PREDICTED TO BE TYROSINE SULFATED
| Common Name | Swiss-Prot Name | Accession # | Tyrosine # | Sequence | Score |
|---|---|---|---|---|---|
| CXCR1 | CXCR1_HUMAN | P25024 | 27 | padedYspcml | 4.13 |
| CXCR2 | CXCR2_HUMAN | P25025 | 197 | vspacYedmgn | 1.80 |
| CXCR3 | CXCR3_HUMAN | P49682 | 27*† | nfsssYdygen | 2.94 |
| 29*† | sssydYgenes | 7.97 | |||
| CXCR4 | CXCR4_HUMAN | P30991 | 12 | ytsdnYteemg | 5.06 |
| 21* | mgsgdYdsmke | 7.72 | |||
| CXCR6 | CXCR6_HUMAN | O00574 | 6† | maehdYhedyg | 8.19 |
| 10† | dyhedYgfssf | 5.59 | |||
| CCR1 | CCR1_HUMAN | P32246 | 10‡ | nttedYdttte | 5.13 |
| 18‡ | ttefdYgdatp | 7.31 | |||
| CCR2b | CCR2_HUMAN | P41597 | 26*† | ttffdYdygap | 7.55 |
| 28†‡ | ffdydYgapch | 3.55 | |||
| CCR3§ | CCR3_HUMAN | P51677 | 17 | gttsyYddvgl | 3.45 |
| 172 | pefifYeteel | 2.01 | |||
| CCR4 | CCR4_HUMAN | P51679 | 22 | snyylYesipk | 1.48 |
| 284 | eryldYaiqat | 2.51 | |||
| CCR5 | CCR5_HUMAN | P51681 | 3* | …mdYqvssp | 2.51 |
| 10*† | vsspiYdinyy | 2.59 | |||
| 14*† | iydinYytsep | 2.78 | |||
| 15*† | ydinyYtsepc | 3.36 | |||
| 187 | fpysqYqfwkn | 2.42 | |||
| CCR6 | CCR6_HUMAN | P51684 | 18 | dssedYfvsvn | 1.72 |
| CCR8§‖ | CCR8_HUMAN | P51685 | 15†‡ | ttvtdYyypdi | 3.74 |
| 17†‡ | vtdyyYpdifs | 2.87 | |||
| CCR9 | CCR9_HUMAN | P51686 | 17 | nmaddYgsest | 5.60 |
| 28 | ssmedYvnfnf | 2.25 | |||
| CCR10 | CCR10_HUMAN | P46092 | 22 | deedaYsaepl | 3.58 |
| CX3CR1 | CX3C1_HUMAN | P49238 | 14* | tenfeYddlae | 8.97 |
| XCR1 | XCR1_HUMAN | P46094 | 14 | sttffYydlqs | 2.32 |
Tyrosines that have a demonstrated presence of O-sulfate on one or more tyrosines. The scores of these tyrosines, whose sites are incorporated into the PSSM, are used as a standard for comparison.
Predicted tyrosine sulfation sites that exhibit clustering.
N-terminal peptides have been shown to be sulfated by TPST1 and TPST2 in a cell-free system (44)
Protein has been previously shown to be tyrosine sulfated through fusion with heavy chain of IgG1 (5).
Murine CCR8 is known to be sulfated at tyrosines 14 and 15.
Figure 1.
Conservation of tyrosine sulfation sites in human chemokine receptors. Sulfation sites with scores of 2.5 or higher are in black. Sites with intermediate scores between 1.5 and 2.5 are in gray. The following tyrosines are known to be sulfated: Y12 and Y21 of CXCR4; Y26 of CCR2; Y3, Y10, Y14, and Y15 of CCR5; and Y14 of CX3CR1. CCR8 is known to be tyrosine sulfated in the mouse, probably in both Y14 and Y15.
Conservation of Tyrosine Sulfation Sites
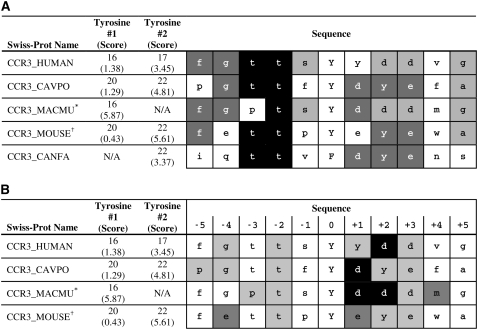
Predicted tyrosine sulfation sites were well conserved across multiple receptor types in humans and other animal species. Sulfation sites were divided into sites with high scores (2.5 or higher) and intermediate scores (1.5–2.5) in an alignment between all human chemokine receptors (Figure 1). Notably, the alignment revealed that many tyrosines predicted to be sulfated were in a region localized to the N-terminus of their receptor. Across mammalian species, predicted tyrosine sulfation sites were frequently completely conserved (data not shown). The example of CCR3 sites in Figure 2A demonstrates the lesser degree of conservation frequently observed in regions with more than one tyrosine in close proximity. Figure 2B shows the contribution of each amino acid at each position to the cumulative score for the central, lower-scoring tyrosine site. Except for the CCR3_MACMU example, which has only one tyrosine and a high score, the other sites score poorly because of the amino acids at positions at Y-1, Y-3, Y-5, Y+4, and Y+5.
Figure 2.
Conservation of Sulfation in CCR3. Most predicted tyrosine sulfation sites in chemokine receptors are highly or completely conserved across animal species. The predicted Ys along with the glutamic and aspartic acid residues form a negative patch on the receptors. *CCR3_MACFA and CCR3_CERAE have the same sequence as CCR3_MACMU and therefore are not included as a separate sequence. †CCR3_RAT is the same as the CCR3_MOUSE sequence. (A) % Identity for amino acids in column at various positions in relation to the Y. Black boxes, 80% identify or above; darkly shaded boxes, 60% or above; lightly shaded boxes, 40% or above. Amino acids with less than 40% identity are not shaded. (B) Contribution of each amino acid around the central tyrosine to the cumulative score for the whole site (Y ± 5 residues). Black boxes, +2 or above; darkly shaded boxes, dfd between +1 and +2; lightly shaded boxes, dfd between 0 and +1. Amino acids with 0 or negative contributions are not shaded.
DISCUSSION
Chemokine receptors are expressed by leukocytes, mononuclear cells, airway epithelial cells, and smooth muscle cells of the lower respiratory tract. Leukocytes and mononuclear cells play a crucial role in mediating the inflammation that underlies asthma and COPD, in which adaptive immune responses become and remain abnormal. The discovery of the chemokine CCL11/eotaxin and its chemokine receptor CCR3 ushered in a new appreciation for the role of chemokines and their receptors in asthma (22). CCR3 activation recruits key inflammatory cells such as the eosinophil into the airway in both lung diseases. CXCR3, as a mediator of chemotaxis for mast cells, is also a critical receptor in asthma (23). Furthermore, CCR4- and CCR8-mediated recruitment of Th2 cells may be relevant in the development of asthma (1, 24, 25). CXCR1 and CXCR2 are neutrophil receptors for CXC chemokines (e.g., CXCL8/IL-8) important in COPD (25) and perhaps in adult-onset asthma, in which neutrophils are more damaging.
Our prediction is that tyrosine sulfation in chemokine receptors in asthma and COPD is more prevalent than currently known. In addition to the 5 chemokine receptors with known sulfated tyrosines, we predict that 11 other chemokine receptors contain sulfated tyrosines, including 3 receptors with scores between 1.5 and 2.5. Some of the predicted sites are clustered and most are highly conserved across diverse mammalian species. Both of these qualitative attributes are characteristic of known tyrosine sulfation sites and corroborate our predictions. All but three of the predicted tyrosine sites are located in the extracellular N-terminal domain, where chemokine receptor–ligand interactions occur in vivo. The other three predicted sites are located in the second extracellular loop, although experimental evidence does not support sulfated tyrosines in this loop of CCR5, the site of one of our predictions (5).
Recent studies indicate that the two sulfotransferases have overlapping but distinct enzymatic profiles and substrate specificities. For instance, the TPSTs have different optimum pH and Mn2+ concentration (26). TPST-2 is also stimulated by Mg2+ ions, whereas TPST-1 is not, and expression patterns of the two isoenzymes are significantly different across tissues. These data strongly suggest that the substrate specificities of the two TPSTs are not completely redundant. Indeed, the two strains of TPST knockout mice have separate mutant phenotypes: TPST-1−/− mice have moderately reduced body weight and fecundity, while TPST-2−/− mice have significantly slower growth rates and impaired male fertility compared with wild-type (27). A mutation in TPST2, leading to a loss of TPST2 activity, causes the lack of sulfation and signaling in TSHR, which results in hypothyroidism in mice (28). Because the PSSM was designed to compare only amino acid sequences, it was not expected to incorporate additional structural features that might affect enzyme substrate specificity.
There are, however, several limitations inherent in the application of the PSSM to chemokine amino acid sequences. First, the PSSM is constructed from 89 known sites. Previous estimates by Moore of the prevalence of sulfation found that as many as 7% of all mouse proteins may be tyrosine sulfated (27). If this estimate is correct, the 41 proteins with known sulfation sites (from the total 70 sulfated proteins published) used in the PSSM would represent only a very small sample of the actual total number of proteins. Another limitation is the selection bias toward sites with acidic residues in the PSSM. Of the known sites, 44% have aspartate and 17% have glutamate at the −1 position in relation to the tyrosine (Y-1). A few known tyrosine sulfation sites, in peptides such as enkephalin and phyllokinin, with only basic residues and no acidic residues, are not included in the PSSM because there may be a second, unknown mechanism of recognition for these basic sites. Since different Km and Vmax (dissociation constant and maximum velocity, respectively) values in TPST-1 and TPST-2 imply that these two enzymes have different requirements for the recognition of a substrate (26), one of these TPSTs may sulfate basic sites better than the other.
Another concern with our prediction of tyrosine sulfation sites is the fact that one site in some tyrosine rich regions may score much higher than the site near by. For example, some tyrosine sites in the CCR3 alignment (Figure 2A) have low scores. Even with lower scores, these sites show highly conservative replacement of amino acids with the preservation of their physical and chemical properties. Because the PSSM does not take into account a nearby sulfated tyrosine, a tyrosine close to this sulfated tyrosine may have a higher probability of sulfation than the prediction score indicates.
The presence of an acidic residue at the Y-1 position is common in the known sites, as 61% of these sites have either glutamic acid or aspartic acid in this position. Not only do these acidic residues increase the frequency of tyrosine sulfation, but they may also increase binding of a sulfated protein to its ligand. In experiments with gastrin in the cell line HIT, mutating the Y-1 alanine to aspartate increases tyrosine sulfation from 60 to 100% (29). In a nuclear magnetic resonance study (NMR) of CXCR1 peptide complexed to CXCL1/IL-8, the acidic residues just N-terminal to the predicted Y27 interact with lysine and arginine residues in the chemokine. These interactions would be expected to increase receptor-ligand binding affinity (30).
The conservation of tyrosine sulfation sites, though imperfect, suggests some fundamental role for tyrosine sulfation that is evolutionarily favorable. The lack of definite patterns in the tyrosine sites of different human chemokine receptors results in large gaps in the alignment shown in Figure 1. Nevertheless, the predicted sulfated tyrosines are generally conserved in the position that corresponds to Y18 of CXCR1. Though the negative charge of a sulfated tyrosine may simply enhance chemokine receptor–ligand interactions, the conservation of a localized tyrosine sulfation site suggests that this region is vital to chemokine receptor function.
The alignment of a single chemokine receptor, CCR3, from five mammalian species (Figure 2) shows better conservation of sites than the alignment of all the human chemokine receptors (Figure 1), as expected. In this alignment, Y16 from human CCR3 is conserved in all sequences except the dog. Notice that in every case the predicted tyrosine, as well as flanking glutamic and aspartic acid residues, would form a negative patch complementing the basic residues in the chemokines to which they bind. In addition to the sulfated tyrosine, the amino acid sequence of the tyrosine sulfation site probably influences chemokine receptor specificity, since most chemokine receptors can bind several different chemokines.
Mounting evidence suggests that tyrosine sulfation of chemokine receptors plays a vital role in chemokine receptor-ligand binding and signaling. In CCR3, the predicted tyrosine sulfation sites are located in the N-terminus and the second extracellular loop, the protein domains critical for receptor binding and activation (31). Furthermore, NMR observations indicate that predicted tyrosines Y27 and Y17 in N-terminal fragments of CXCR1 and CCR3 bind with hydrophobic interactions to their respective ligands, CXCL8/IL-8 and CCL11/eotaxin (30, 32) If these tyrosines are sulfated and act similarly to those in known sulfated proteins, electrostatic interactions (salt-bridges) and H-bonding would be expected between sulfonate and the chemokine ligands. For example Y21 of CXCR4 peptide, the only sulfated chemokine receptor whose specific binding is known, participates in an electrostatic interaction with a site in CXCL12/SDF-1 including an arginine (33). In a model of sulfated CXCR1 based on the NMR structure above, Y27 and an adjacent aspartate residue participate in H-bonds with lysines in the ligand CXCL8/IL-8 (data not shown). In each case, acidic residues surrounding the tyrosines structurally complement a hydrophobic groove bordered by basic residues on the chemokine ligands (34). Thus, experimental evidence where tyrosine sulfation is known to occur shows a strong relationship between tyrosine sulfation and ligand binding, though in vitro experiments with CCR2b and CX3CR1 suggest that sulfation may also be important for intracellular signaling (10, 11). The pattern and location of the tyrosine residues that we predict to be sulfated in the chemokine receptors suggest that the predicted sites may play a role in increasing binding to their ligands similar to the known sulfation sites (see Table 2). This role, in mediating the interaction and binding with ligands, is consistent with the role of tyrosine sulfation in other biological systems. Tyrosine sulfation mediates protein–protein interactions and binding, in such processes as coagulation, bacterial lysis, and anti-coagulation (27).
TABLE 2.
SPECIFIC EFFECTS OF TYROSINE SULFATION
| Effect | Chemokine Receptor | Details | Reference |
|---|---|---|---|
| Increased binding of chemokines | CMV US28 | Viral receptor CMV US28: Increased binding to CCL3*, CCL4†, CCL5‡, CX3CL1§ | (45) |
| Increased binding of chemokines | CXCR3 | Receptor CXCR3: Increased binding to CXCL9‖, CXCL10**, CXCL11†† | (12) |
| CXCR4 | Receptor CXCR4: Increased binding to CXCL12‡‡ | (9) | |
| CCR2b | Receptor CCR2b: Increased binding to CCL2§§ | (10) | |
| CCR5 | Receptor CCR5: Increased binding to CCL3*, CCL4†, and CCL5‡ | (5,46) | |
| Increased binding to chemokine/ adhesion molecules | CX3CR1 | Receptor CX3CR1: Increased firm adhesion to CX3CL1§ | (11) |
| CXCR3 | Receptor CXCR3: Increases chemotaxis after binding to CXCL9‖, CXCL10**, and CXCL11†† | (12) | |
| Increased signaling/ Downstream events | CCR2b | Receptor CCR2b: Increased Ca++ influx and chemotaxis after binding to CCL2§§ | (10) |
| CX3CR1 | Receptor CX3CR1: Increased Ca++ influx after binding to CXC3L1§ | (11) |
CCL3/MIP-1α
CCL4/MIP-1β
CCL5/RANTES.
CX3CL1/Fractalkine.
CXCL9/MIG.
CXCL10/IP-10.
CXCL11/I-TAC.
CXCL12/SDF-1.
CCL2/MCP-1.
The role of tyrosine sulfation in the pathogenesis of diseases such as AIDS is becoming increasingly well documented. Tyrosine sulfation is critical for HIV-1 infection of T cells via the major coreceptor CCR5, though it appears to be dispensable for the lesser coreceptor CXCR4 (5, 9). While it has been hypothesized that all chemokine-like coreceptors for HIV-1 are tyrosine sulfated (5), the exact mechanism for viral entry via sulfated chemokine receptors is not completely understood. Under the prevailing theory, HIV mimics the electrostatic interactions that facilitate chemokine ligand-receptor binding. Thus, tyrosine sulfation of HIV coreceptors would produce patches of negatively charged residues. Negatively charged patches would then enhance receptor binding to the HIV envelope glycoprotein (gp120), which is known to possess a conserved arginine R-298 critical for viral infection (35). Similar mechanisms may be used by anti-gp120 antibodies to neutralize HIV infection. These antibodies demonstrate tyrosyl O-sulfate or have residue compositions that are highly favorable for tyrosine sulfation (6). In select antibodies, X-ray crystallography showed that sulfated tyrosines come into direct contact and bind with gp120, suggesting that tyrosine sulfation is critical for antigen recognition and viral binding (36). A tyrosine-sulfated peptide derived from the heavy-chain of an HIV-1–neutralizing antibody binds gp120 and inhibits HIV-1 infection (37). An area of future translational clinical research with great potential would involve targeting tyrosyl O-sulfate on HIV coreceptors, especially since our PSSM predictions indicate that HIV coreceptors CCR3, CCR8, CXCR6, D6, and DEZ are tyrosine sulfated, similar to those N-terminal sites in CCR5 (data not shown).
In the pathobiology of asthma, Th2 cells, mast cells, eosinophils, and epithelial cells all appear to be important in causing persistent airway inflammation. Some human chemokine receptors expressed on these cells are confirmed to be tyrosine sulfated, such as CCR2, CXCR3, CXCR4, and CX3CR1, while some are predicted but not confirmed, such as CCR1, CCR3, CCR4, and CCR8 (12, 24, 38). Mast cells in particular play a crucial role in asthma. Airway hyperresponsiveness is strongly correlated to the number of mast cells in smooth muscle of the lung (23). The recent discovery that CXCR3, expressed by a subset of human lung mast cells (39), undergoes tyrosine sulfation is significant (12) because the tyrosine sulfation is necessary for the binding of three chemokines—CXCL9 (MIG), CXCL10 (IL-10), and CXCL11 (I-TAC)—as well as internalization of the receptor and for chemotaxis. When Y27 and Y29 are mutated to alanine, no chemotaxis is observed when the chemokines are added to the medium. Mast cell progenitors express chemokine receptors CCR3, CCR5, CCR2, and CXCR4, though only CXCR4 is necessary for migration from the bone marrow (40). Recently, Sutcliffe and coworkers showed that CCR3 and CCR1 were necessary for migration to the airway smooth muscles (41). Similar to CXCR3, predicted tyrosine sulfation sites in CCR1, CCR3, and CCR4 are expected to increase binding to their respective ligands and enhance migration of mast cells (12).
A significant portion of the pharmaceutical research and development modulating inflammation in asthma and COPD focuses on chemokine receptor antagonists (25, 42). The modulation of specific chemokine ligand-receptor networks, necessary for leukocyte trafficking and downstream inflammatory events, is important for the control of asthma and COPD (25). Antagonizing chemokines/chemokine receptors is complicated by their overlapping function and binding specificities, although several promising small-molecule antagonists for chemokine receptors such as CCR5 and CXCR4 are in phase 2 or 3 trials (43). The predictions presented above open the possibility that tyrosine sulfation may be even more widespread among the chemokine receptors than has been previously recognized. Consideration of these sulfated tyrosines during the structural design of specific receptor antagonists may lead to more precise targeting of chemokine receptors and effective drug therapy for asthma, COPD, and similar inflammatory lung diseases.
Acknowledgments
Michelle Louie and Henry Lin have our gratitude for their insight and editorial assistance during the early stages of this research. The authors especially thank Linda Tran, who offered many creative comments during the preparation of the manuscript, and Martha Teeter for her model of CXCR1 peptide complexed with IL-8.
This research was supported by the National Center for Research Resources (RR06009) and the National Genome Research Institute (HG00015).
Originally Published in Press as DOI:10.1165/rcmb.2007-0118OC on January 24, 2008
Conflict of Interest Statement: None of the authors has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Panina-Bordignon P, D'Ambrosio D. Chemokines and their receptors in asthma and chronic obstructive pulmonary disease. Curr Opin Pulm Med 2003;9:104–110. [DOI] [PubMed] [Google Scholar]
- 2.Olson TS, Ley K. Chemokines and chemokine receptors in leukocyte trafficking. Am J Physiol Regul Integr Comp Physiol 2002;283:R7–R28. [DOI] [PubMed] [Google Scholar]
- 3.Smit JJ, Lukacs NW. A closer look at chemokines and their role in asthmatic responses. Eur J Pharmacol 2006;533:277–288. [DOI] [PubMed] [Google Scholar]
- 4.Murphy PM, Baggiolini M, Charo IF, Hebert CA, Horuk R, Matsushima K, Miller LH, Oppenheim JJ, Power CA. International Union of Pharmacology: XXII. Nomenclature for chemokine receptors. Pharmacol Rev 2000;52:145–176. [PubMed] [Google Scholar]
- 5.Farzan M, Mirzabekov T, Kolchinsky P, Wyatt R, Cayabyab M, Gerard NP, Gerard C, Sodroski J, Choe H. Tyrosine sulfation of the amino terminus of CCR5 facilitates HIV-1 entry. Cell 1999;96:667–676. [DOI] [PubMed] [Google Scholar]
- 6.Choe H, Li W, Wright PL, Vasilieva N, Venturi M, Huang CC, Grundner C, Dorfman T, Zwick MB, Wang L, et al. Tyrosine sulfation of human antibodies contributes to recognition of the CCR5 binding region of HIV-1 gp120. Cell 2003;114:161–170. [DOI] [PubMed] [Google Scholar]
- 7.Huttner W, Baeuerle P. Protein sulfation on tyrosine. Mol Cell Biol 1988;6:97–140. [Google Scholar]
- 8.Nicholas HB Jr, Chan SS, Rosenquist GL. Reevaluation of the determinants of tyrosine sulfation. Endocrine 1999;11:285–292. [DOI] [PubMed] [Google Scholar]
- 9.Farzan M, Babcock GJ, Vasilieva N, Wright PL, Kiprilov E, Mirzabekov T, Choe H. The role of post-translational modifications of the CXCR4 amino terminus in stromal-derived factor 1 alpha association and hiv-1 entry. J Biol Chem 2002;277:29484–29489. [DOI] [PubMed] [Google Scholar]
- 10.Preobrazhensky AA, Dragan S, Kawano T, Gavrilin MA, Gulina IV, Chakravarty L, Kolattukudy PE. Monocyte chemotactic protein-1 receptor CCR2B is a glycoprotein that has tyrosine sulfation in a conserved extracellular n-terminal region. J Immunol 2000;165:5295–5303. [DOI] [PubMed] [Google Scholar]
- 11.Fong AM, Alam SM, Imai T, Haribabu B, Patel DD. CX3CR1 tyrosine sulfation enhances fractalkine-induced cell adhesion. J Biol Chem 2002;277:19418–19423. [DOI] [PubMed] [Google Scholar]
- 12.Colvin RA, Campanella GS, Manice LA, Luster AD. CXCR3 requires tyrosine sulfation for ligand binding and a second extracellular loop arginine residue for ligand-induced chemotaxis. Mol Cell Biol 2006;26:5838–5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gutierrez J, Kremer L, Zaballos A, Goya I, Martinez AC, Marquez G. Analysis of post-translational CCR8 modifications and their influence on receptor activity. J Biol Chem 2004;279:14726–14733. [DOI] [PubMed] [Google Scholar]
- 14.Doranz BJ, Orsini MJ, Turner JD, Hoffman TL, Berson JF, Hoxie JA, Peiper SC, Brass LF, Doms RW. Identification of CXCR4 domains that support coreceptor and chemokine receptor functions. J Virol 1999;73:2752–2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Monteclaro FS, Charo IF. The amino-terminal extracellular domain of the MCP-1 receptor, but not the Rantes/MIP-1alpha receptor, confers chemokine selectivity: evidence for a two-step mechanism for MCP-1 receptor activation. J Biol Chem 1996;271:19084–19092. [DOI] [PubMed] [Google Scholar]
- 16.Blanpain C, Doranz BJ, Vakili J, Rucker J, Govaerts C, Baik SS, Lorthioir O, Migeotte I, Libert F, Baleux F, et al. Multiple charged and aromatic residues in CCR5 amino-terminal domain are involved in high affinity binding of both chemokines and HIV-1 env protein. J Biol Chem 1999;274:34719–34727. [DOI] [PubMed] [Google Scholar]
- 17.Mizoue LS, Bazan JF, Johnson EC, Handel TM. Solution structure and dynamics of the CX3C chemokine domain of fractalkine and its interaction with an N-terminal fragment of CX3CR1. Biochemistry 1999;38:1402–1414. [DOI] [PubMed] [Google Scholar]
- 18.Brelot A, Heveker N, Montes M, Alizon M. Identification of residues of CXCR4 critical for human immunodeficiency virus coreceptor and chemokine receptor activities. J Biol Chem 2000;275:23736–23744. [DOI] [PubMed] [Google Scholar]
- 19.Hemmerich S, Paavola C, Bloom A, Bhakta S, Freedman R, Grunberger D, Krstenansky J, Lee S, McCarley D, Mulkins M, et al. Identification of residues in the monocyte chemotactic protein-1 that contact the MCP-1 receptor, CCR2. Biochemistry 1999;38:13013–13025. [DOI] [PubMed] [Google Scholar]
- 20.Do CB, Mahabhashyam MS, Brudno M, Batzoglou S. Probcons: probabilistic consistency-based multiple sequence alignment. Genome Res 2005;15:330–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wallace IM, O'Sullivan O, Higgins DG, Notredame C. M-coffee: combining multiple sequence alignment methods with t-coffee. Nucleic Acids Res 2006;34:1692–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ponath PD, Qin S, Post TW, Wang J, Wu L, Gerard NP, Newman W, Gerard C, Mackay CR. Molecular cloning and characterization of a human eotaxin receptor expressed selectively on eosinophils. J Exp Med 1996;183:2437–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brightling CE, Bradding P, Symon FA, Holgate ST, Wardlaw AJ, Pavord ID. Mast-cell infiltration of airway smooth muscle in asthma. N Engl J Med 2002;346:1699–1705. [DOI] [PubMed] [Google Scholar]
- 24.Bisset LR, Schmid-Grendelmeier P. Chemokines and their receptors in the pathogenesis of allergic asthma: progress and perspective. Curr Opin Pulm Med 2005;11:35–42. [DOI] [PubMed] [Google Scholar]
- 25.Charo IF, Ransohoff RM. The many roles of chemokines and chemokine receptors in inflammation. N Engl J Med 2006;354:610–621. [DOI] [PubMed] [Google Scholar]
- 26.Mishiro E, Sakakibara Y, Liu MC, Suiko M. Differential enzymatic characteristics and tissue-specific expression of human TPST-1 and TPST-2. J Biochem (Tokyo) 2006;140:731–737. [DOI] [PubMed] [Google Scholar]
- 27.Moore KL. The biology and enzymology of protein tyrosine O-sulfation. J Biol Chem 2003;278:24243–24246. [DOI] [PubMed] [Google Scholar]
- 28.Sasaki N, Hosoda Y, Nagata A, Ding M, Cheng JM, Miyamoto T, Okano S, Asano A, Miyoshi I, Agui T. A mutation in TPST2 encoding tyrosylprotein sulfotransferase causes dwarfism associated with hypothyroidism. Mol Endocrinol 2007;24:24. [DOI] [PubMed] [Google Scholar]
- 29.Bundgaard JR, Vuust J, Rehfeld JF. Tyrosine O-sulfation promotes proteolytic processing of progastrin. EMBO J 1995;14:3073–3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Skelton NJ, Quan C, Reilly D, Lowman H. Structure of a CXC chemokine-receptor fragment in complex with interleukin-8. Structure 1999;7:157–168. [DOI] [PubMed] [Google Scholar]
- 31.Pease JE, Wang J, Ponath PD, Murphy PM. The N-terminal extracellular segments of the chemokine receptors CCR1 and CCR3 are determinants for MIP-1alpha and eotaxin binding, respectively, but a second domain is essential for efficient receptor activation. J Biol Chem 1998;273:19972–19976. [DOI] [PubMed] [Google Scholar]
- 32.Ye J, Kohli LL, Stone MJ. Characterization of binding between the chemokine eotaxin and peptides derived from the chemokine receptor CCR3. J Biol Chem 2000;275:27250–27257. [DOI] [PubMed] [Google Scholar]
- 33.Veldkamp CT, Seibert C, Peterson FC, Sakmar TP, Volkman BF. Recognition of a CXCR4 sulfotyrosine by the chemokine stromal cell-derived factor-1alpha (SDF-1alpha/CXCL12). J Mol Biol 2006;359:1400–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blanpain C, Doranz BJ, Bondue A, Govaerts C, De Leener A, Vassart G, Doms RW, Proudfoot A, Parmentier M. The core domain of chemokines binds CCR5 extracellular domains while their amino terminus interacts with the transmembrane helix bundle. J Biol Chem 2003;278:5179–5187. [DOI] [PubMed] [Google Scholar]
- 35.Wang WK, Dudek T, Zhao YJ, Brumblay HG, Essex M, Lee TH. Ccr5 coreceptor utilization involves a highly conserved arginine residue of HIV type 1 gp120. Proc Natl Acad Sci USA 1998;95:5740–5745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang CC, Venturi M, Majeed S, Moore MJ, Phogat S, Zhang MY, Dimitrov DS, Hendrickson WA, Robinson J, Sodroski J, et al. Structural basis of tyrosine sulfation and VH-gene usage in antibodies that recognize the HIV type 1 coreceptor-binding site on gp120. Proc Natl Acad Sci USA 2004;101:2706–2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dorfman T, Moore MJ, Guth AC, Choe H, Farzan M. A tyrosine-sulfated peptide derived from the heavy-chain CDR3 region of an HIV-1-neutralizing antibody binds gp120 and inhibits HIV-1 infection. J Biol Chem 2006;281:28529–28535. [DOI] [PubMed] [Google Scholar]
- 38.Barnes PJ. Drugs for asthma. Br J Pharmacol 2006;147:S297–S303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brightling CE, Ammit AJ, Kaur D, Black JL, Wardlaw AJ, Hughes JM, Bradding P. The CXCL10/CXCR3 axis mediates human lung mast cell migration to asthmatic airway smooth muscle. Am J Respir Crit Care Med 2005;171:1103–1108. [DOI] [PubMed] [Google Scholar]
- 40.Lin TJ, Issekutz TB, Marshall JS. Human mast cells transmigrate through human umbilical vein endothelial monolayers and selectively produce IL-8 in response to stromal cell-derived factor-1 alpha. J Immunol 2000;165:211–220. [DOI] [PubMed] [Google Scholar]
- 41.Sutcliffe A, Kaur D, Page S, Woodman L, Armour CL, Baraket M, Bradding P, Hughes JM, Brightling CE. Mast cell migration to Th2 stimulated airway smooth muscle from asthmatics. Thorax 2006;61:657–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Elsner J, Escher SE, Forssmann U. Chemokine receptor antagonists: a novel therapeutic approach in allergic diseases. Allergy 2004;59:1243–1258. [DOI] [PubMed] [Google Scholar]
- 43.Allen SJ, Crown SE, Handel TM. Chemokine: receptor structure, interactions, and antagonism. Annu Rev Immunol 2007;25:787–820. [DOI] [PubMed] [Google Scholar]
- 44.Yu Y, Hoffhines AJ, Moore KL, Leary JA. Determination of the sites of tyrosine O-sulfation in peptides and proteins. Nat Methods 2007;10:10. [DOI] [PubMed] [Google Scholar]
- 45.Casarosa P, Waldhoer M, LiWang PJ, Vischer HF, Kledal T, Timmerman H, Schwartz TW, Smit MJ, Leurs R. CC and CX3C chemokines differentially interact with the N terminus of the human cytomegalovirus-encoded US28 receptor. J Biol Chem 2005;280:3275–3285. [DOI] [PubMed] [Google Scholar]
- 46.Bannert N, Craig S, Farzan M, Sogah D, Santo NV, Choe H, Sodroski J. Sialylated O-glycans and sulfated tyrosines in the NH2-terminal domain of cc chemokine receptor 5 contribute to high affinity binding of chemokines. J Exp Med 2001;194:1661–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]