Abstract
Both transcriptional activation and repression have essential functions in maintaining proper spatial and temporal control of gene expression. Although Wnt signalling is often associated with gene activation, we have identified several directly repressed targets of Wnt signalling in Drosophila. Here, we explore how individual Wnt target genes are specified for signal-induced activation or repression. Similar to activation, repression required binding of Armadillo (Arm) to the N terminus of TCF. However, TCF/Arm mediated repression by binding to DNA motifs that are markedly different from typical TCF-binding sites. Conversion of the novel motifs to standard TCF-binding sites reversed the mode of regulation, resulting in Wnt-mediated activation instead of repression. A mutant form of Arm defective in activation was still functional for repression, indicating that distinct domains of the protein are required for each activity. This study suggests that the sequence of TCF-binding sites allosterically regulates the TCF/Arm complex to effect either transcriptional activation or repression.
Keywords: allosteric, Armadillo, β-catenin, TCF, Wingless
Introduction
Much of the cell–cell communication that occurs during animal development is controlled by a small number of signalling pathways that influence cell-fate decisions primarily through transcriptional regulation (Barolo and Posakony, 2002). Spatial and temporal specificity of gene expression are conferred by cis-acting regulatory elements that recruit various combinations of transcription factors. Although transcription factor-binding sites are typically considered passive docking sites, variations in some binding site sequences have been shown to alter transcription factor cofactor requirements (Leung et al, 2004; Luecke and Yamamoto, 2005) or its ability to activate or repress transcription (Scully et al, 2000). These examples demonstrate that some DNA sequences can have an active function in gene regulation by allosterically regulating the activity of bound transcription factors.
The Wnt/β-catenin (hereafter called Wnt) signalling pathway is one of the cell–cell communication pathways that influences numerous aspects of animal development (Cadigan and Nusse, 1997; Logan and Nusse, 2004). The signalling mechanism revolves around the stability of β-catenin (β-cat), called Armadillo (Arm) in flies. In the absence of Wnt signalling, β-cat/Arm is targeted for degradation by a complex that includes Axin, adenomatous polyposis coli (APC), and glycogen synthase kinase-3 (GSK3) (Kikuchi et al, 2006). Activation of Wnt signalling inhibits β-cat/Arm degradation, allowing it to accumulate in the cytoplasm and translocate to the nucleus where it interacts with members of the TCF/Lef1 (TCF) family of DNA-binding proteins to regulate gene transcription (Parker et al, 2007).
The TCF family of sequence-specific DNA-binding proteins binds DNA as monomers through a conserved HMG domain (Giese et al, 1991). In the absence of β-cat/Arm, TCF binds co-repressors such as Groucho to prevent gene activation (Cavallo et al, 1998). Upon activation of the pathway, nuclear β-cat/Arm binds to TCF and activates transcription by displacing Groucho and recruiting co-activators such as p300/CBP, Legless, Pygopus, and Hyrax (Stadeli et al, 2006; Parker et al, 2007). A variety of integrated Wnt reporters containing multiple TCF-binding sites upstream of generic promoters are upregulated upon activation of Wnt signalling in many species, leading to the general view that Wnt signalling activates transcription through TCF (Barolo, 2006).
Genetic analysis of Drosophila development (Cadigan et al, 1998, 2002; Payre et al, 1999) and microarray analyses in mammalian cells (van de Wetering et al, 2002; Willert et al, 2002; Jung and Kim, 2005; Naishiro et al, 2005) have established that Wnt signalling can also repress the transcription of many genes. Very little is known about the mechanism of signal-activated repression by the Wnt pathway, or the means by which individual genes are specified for repression or activation. This is in part because only a handful of repressed genes have been studied in enough detail to establish that they are directly repressed by the pathway, as opposed to indirectly repressed through a Wnt-activated repressor protein. In fact, the only genes shown to be directly repressed by Wnt signalling in a manner that depends on TCF and β-cat/Arm are stripe in the Drosophila epidermis (Piepenburg et al, 2000), Decapentaplegic (Dpp) in the fly leg imaginal disc (Theisen et al, 2007), and CDH1 (E-cadherin) in mouse keratinocytes (Jamora et al, 2003). Repression required traditional TCF-binding sites at these targets, and it remains a mystery why these genes are repressed by Wnt/β-cat signalling rather than activated.
Here, we address the question of how individual genes are specified for signal-induced repression by the Wnt pathway. We present several new targets of direct Wnt repression in Drosophila, along with a detailed analysis of how one of these targets, Ugt36Bc, is specified for transcriptional repression instead of activation by Wnt/Arm signalling. We found that allosteric effects of DNA sequence on TCF/Arm activity can determine whether a Wnt target is activated or repressed by signalling.
Results
Wnt signalling activates and represses transcription in a gene-specific manner
Transcriptional targets of the Wnt pathway were identified using microarray analysis with Affymetrix Drosophila Genome Array 2.0 genechips. Drosophila Kc167 (Kc) cells were used in these studies as they contain an intact Wnt pathway (Fang et al, 2006; Li et al, 2007). To minimize artefacts, two different comparisons were analysed on the genechips, addition of Wingless-conditioned media (Wg-CM) versus control-conditioned media (Ctrl-CM), and RNAi knockdown of Axin versus control (Ctrl) RNAi knockdown. Wg is a fly Wnt ligand, and Axin RNAi allows Arm to accumulate even in the absence of Wg (data not shown). Genes that responded similarly to Wg-CM and Axin RNAi were further characterized using quantitative RT–PCR to measure the magnitude of Wnt regulation.
RNAi knockdown of Axin resulted in robust activation of two previously identified direct Wnt targets, naked cuticle (nkd) and CG6234 (Fang et al, 2006) and simultaneous 5- to 10-fold repression of Ugt36Bc, Peroxidasin (Pxn), Tiggrin (Tig), and Ugt58Fa (Figure 1A). Wg-CM treatment resulted in qualitatively similar responses, although the amplitude of regulation was lower for all targets (Figure 1B). We have previously established that nkd and CG6234 require both TCF and Arm for signal-induced activation by Wnt (Fang et al, 2006). RNAi knockdown of Axin with simultaneous knock down of TCF or Arm showed that repression of Ugt36Bc, Pxn, Tig, and Ugt58Fa also required both TCF and Arm (Figure 1C), implicating the standard Wnt pathway in repression.
Figure 1.
Wnt signalling both activates and represses transcription in Kc cells. (A) qRT–PCR analysis of gene expression in Kc cells grown in the presence of dsRNA-targeting Axin (Wnt ON) or control sequences (Wnt OFF) for 7 days. Wnt signalling activated transcription of nkd and CG6234, whereas repressing the transcription of four novel Wnt targets, Ugt36Bc, Pxn, Tig, and Ugt58Fa. (B) qRT–PCR analysis of gene expression in Kc cells exposed to Wg-conditioned media (Wg-CM) or control-conditioned media (Ctrl-CM) for 6 h. (C) Both TCF and Arm were required for Wnt-mediated repression. Kc cells were treated as in (A), except that dsRNA targeting TCF or Arm was added in combination with control or Axin dsRNA (10 ng/ml each). (D) TCF has opposite functions at Wnt-repressed and Wnt-activated genes in the absence of signalling. RNAi-mediated knockdown of TCF lowered expression of all four repressed Wnt targets, but de-repressed expression of the Wnt-activated targets nkd and CG6234.
In addition to mediating Arm-dependent transcriptional activation, TCF also silences many Wnt targets in the absence of Wnt signalling (Parker et al, 2007), including nkd and CG6234 in Kc cells (Figure 1D; Fang et al, 2006). Conversely, TCF appears to activate transcription of Ugt36Bc, Pxn, Tig, and Ugt58Fa in the absence of Wnt signalling (Figure 1D). In other words, the repressed genes behave exactly opposite of activated genes; the repressed targets are activated by TCF in the absence of signalling and repressed in an Arm- and TCF-dependent manner upon pathway activation.
Wnt signalling represses gene expression in embryonic haemocytes
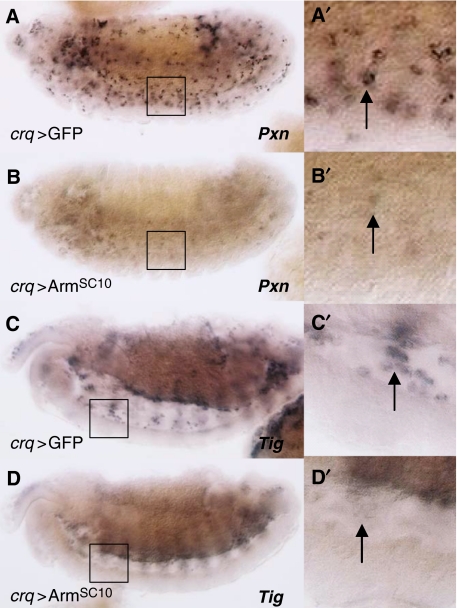
Kc cells are derived from embryonic haemocytes (Goto et al, 2001), and both Pxn and Tig encode extracellular matrix proteins that are expressed in embryonic haemocytes (Fogerty et al, 1994; Nelson et al, 1994; Alfonso and Jones, 2002). To confirm that Wnt signalling also repressed these genes in vivo, we expressed a constitutively active form of Arm (ArmSC10) (Pai et al, 1997) in haemocytes using the crq-Gal4 driver (Olofsson and Page, 2005). The crq-Gal4 driver is expressed in haemocytes from late stage 11 onward (data not shown). Ectopic Wnt signalling repressed both Pxn and Tig expression in haemocytes (Figure 2A–D).
Figure 2.
Wnt signalling represses transcription in Drosophila haemocytes. DIC images ( × 10 magnification) of stage 13 (A, B) or 14 (C, D) embryos following in situ hybridization with the indicated cRNA probes. P[UAS-GFP], P[crq79-Gal4] (crq>GFP) control embryos are compared with P[UAS-ArmSC10], P[crq79-Gal4] (crq>ArmSC10) embryos in which Wnt signalling has been activated in haemocytes. Both Pxn (compare A to B) and Tig (compare C to D) transcript levels are reduced by Wnt signalling. Insets (A′–D′) show × 40 magnification to highlight individual haemocytes migrating along the ventral boundary of the embryo. Anterior is left and dorsal is up in all images.
Ugt36Bc and Ugt58Fa encode members of the UDP glycosyltransferase superfamily of enzymes that are thought to exert an effect in detoxification (Luque and O'Reilly, 2002). Despite a previous report indicating expression of Ugt36Bc in a subset of haemocytes (Tomancak et al, 2002), we were unable to discern a reproducible expression pattern in embryos with two distinct probes (data not shown). Nonetheless, the results with Pxn and Tig demonstrate that Wnt-mediated transcriptional repression occurs in Drosophila haemocytes and suggest that Kc cells provide a suitable model in which to explore the mechanism of Wnt-mediated transcriptional repression.
Wnt signalling directly represses Ugt36Bc transcription
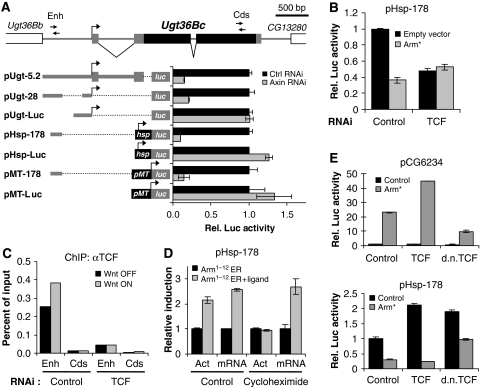
To determine if the repressed targets were directly regulated by TCF/Arm, we attempted to localize the Wnt response elements (WREs) responsible for repression. We tested several fragments of genomic DNA near the repressed genes, but only one fragment recapitulated Wnt regulation in a reporter gene assay, a 1.5-kb sequence surrounding the Ugt36Bc transcription start site (TSS). Deletion analysis identified a 178-bp fragment between −632 and −455 bp relative to the Ugt36Bc TSS as the minimal WRE (Figure 3A). This WRE conferred transcriptional activity in the absence of Wnt signalling and Wnt-induced repression to a variety of core promoters, including the endogenous Ugt36Bc promoter, the Hsp70 promoter, and the metallothionein promoter (Figure 3A). The 178 bp WRE worked equally well at a variety of distances from the endogenous promoter and in either orientation relative to the Hsp70 promoter (data not shown). Similar to the endogenous Ugt36Bc gene, the minimal WRE also required TCF for full activity in the absence of Wnt signalling, and for Wnt-mediated repression (Figure 3B).
Figure 3.
A minimal Ugt36Bc enhancer is directly repressed by Wnt signalling. A cartoon of the Ugt36Bc genomic locus is shown. Black rectangles represent Ugt36Bc coding sequences, grey rectangles represent untranslated regions, and white rectangles represent the boundaries of the upstream and downstream genes. Arrows represent the regions probed by PCR in ChIP experiments. Enh shows the WRE and Cds shows coding sequence near the 3′ end. (A) A minimal 178 bp WRE in the 5′ intergenic region of Ugt36Bc was repressed by Wnt signalling in the context of the endogenous Ugt36Bc promoter (pUgt-28), the Hsp70Bb minimal promoter (pHsp-178), and the metalothionein promoter (pMT-178). Reporters lacking the 178 bp WRE (pUgt-Luc, pHsp-Luc, and pMT-Luc) were not repressed. Luciferase activity in the Wnt OFF state was normalized to 1.0 for each reporter. (B) pHsp-178 (25 ng) was transiently transfected with pAc5.1/V5-His (empty vector) or pAcArm* (100 ng of each) into Kc cells pretreated with dsRNA targeting control sequences or TCF for 4 days. Luciferase and β-gal activities were measured 1 day after transfection. Arm* is a constitutively active form of Arm. pHsp-178 was repressed by Arm* in a TCF-dependent manner, and required TCF for full expression in the Wnt OFF state. (C) Kc cells treated with dsRNA targeting β-lactamase (Ctrl) or TCF were subjected to ChIP with TCF antibodies in the Wnt OFF and Wnt ON states (Ctrl and Axin RNAi, respectively). TCF binds the endogenous Ugt36Bc WRE (Enh) in both the absence and presence of Wnt signalling. TCF RNAi greatly reduced TCF binding at the WRE. Control sequences near the 3′ end of the Ugt36Bc coding region (Cds) were not bound by TCF. The experiment shown is representative of three independent experiments showing similar results. (D) Kc cells transfected with pAcArm1–12ER (500 ng) and pHsp178 (100 ng) were grown for 19 h before being treated for 6 h with 10 μg/ml cycloheximide and/or 400 ng/ml 17-β-estradiol (ligand). In the absence of cycloheximide, ligand activated pHsp-178 two-fold, as measured by luciferase mRNA levels and luciferase activity. In the presence of cycloheximide, ligand still activated transcription luciferase mRNA, but protein synthesis was inhibited as indicated by the lack of increased luciferase activity. ‘Act' refers to luciferase activity, ‘mRNA' refers to luciferase mRNA levels as measured by qRT–PCR. (E) Arm works through TCF to repress the 178 bp WRE. Kc cells were transfected with 50 ng pCG6234 or pHsp-178 reporters, 50 ng control or Arm* expression vector, and 200 ng control, wild-type TCF, or dominant-negative TCF (dnTCF) expression vectors. dnTCF partially disrupted Arm* activation of pCG6234 (top panel) and Arm* repression of pHsp-178 (lower panel). Wild-type TCF enhanced the Wnt responsiveness of both reporters.
The optimal in vitro-binding site for Drosophila TCF has been defined as CCTTTGATCTT (van de Wetering et al, 1997) and we have previously generated a position-weighted consensus sequence resembling SCTTTGWW (S=G/C; W=A/T), which was successfully used to identify genuine TCF-binding sites in nkd and CG6234 (Fang et al, 2006). No sequences matching these TCF sites are found within the minimal Ugt36Bc WRE. However, chromatin immunoprecipitation (ChIP) demonstrated that TCF strongly bound the chromosomal region of Ugt36Bc containing the WRE in Kc cells (Figure 3C). TCF showed greater than 10-fold enrichment at the Ugt36Bc WRE locus relative to coding sequences near the 3′ end of the gene. TCF bound the Ugt36Bc WRE locus both in the absence and presence of Wnt signalling, consistent with its roles in the absence and presence of Wnt signaling. TCF binding was strongly reduced when TCF protein was knocked down by RNAi, confirming the specificity of the ChIP analysis (Figure 3C).
The non-traditional nature of Ugt36Bc regulation by Wnt signalling led us to explore whether Arm was also directly involved. Our attempts to locate Arm at the Ugt36Bc locus through ChIP with a monoclonal Arm antibody have been inconclusive (data not shown). As an alternative, we tested the ability of an Arm fusion protein to regulate Ugt36Bc in the absence of protein synthesis, to rule out an indirect effect of Arm. The mammalian oestrogen receptor ligand-binding domain contains a ligand-dependent transcriptional activation domain (ER) (Webster et al, 1988). This ER domain was fused to the central portion of Arm (containing Arm repeats 1–12) to generate Arm1–12ER. When expressed in Kc cells without ligand, Arm1–12ER repressed transcription of the Ugt36Bc reporter approximately five-fold, the same extent as constitutively active Arm (Arm*; data not shown). Addition of ligand to cells that contain Arm1–12ER caused a two-fold increase in Ugt36Bc reporter-derived luciferase mRNA levels and protein activity (Figure 3D). When cells containing Arm1–12ER were treated with 10 μg/ml cycloheximide prior to ligand addition, the ligand still activated transcription of luciferase mRNA, even though protein synthesis is shut off as indicated by the lack of increased luciferase activity (Figure 3D). These results confirm that Arm is directly recruited to the Ugt36Bc WRE.
To ensure that TCF and Arm work together to repress Ugt36Bc, the effect of a dominant-negative form of TCF (dnTCF) (van de Wetering et al, 1997), which no longer binds Arm was examined. dnTCF inhibits Wnt signalling in a variety of contexts (van de Wetering et al, 1997; Cadigan et al, 1998) and it partially inhibits Arm* activation of a CG6234 reporter in Kc cells (Figure 3E, top panel). Interestingly, both wild-type TCF and dnTCF activated the Ugt36Bc reporter in the absence of Wnt signalling (Figure 3E, bottom panel). However, TCF enhanced Arm*-mediated repression of pHsp-178, whereas dnTCF inhibited this repression (Figure 3E, bottom panel). Together with the TCF ChIP and Arm1–12ER results, these data confirm that Arm works through TCF to directly repress transcription of the Ugt36Bc WRE.
Wnt signalling represses Ugt36Bc through novel TCF-binding sites
Although the Ugt36Bc WRE lacks traditional TCF-binding sites, the TCF ChIP data suggest that it is bound by TCF (Figure 3C). Our attempts to use sequence conservation among various Drosophila species to identify important DNA sequences were not helpful in that no particularly well-conserved DNA sequences stood out in the alignment generated by the University of California Santa Cruz Genome Browser (Supplementary Figure S1; Kent et al, 2002). Instead, we tested whether TCF directly recognized DNA sequences in the minimal WRE using DNaseI protection assays with the HMG domain of TCF fused to glutathione-S-transferase (GST–HMG) (Lee and Frasch, 2000; Knirr and Frasch, 2001). A novel protocol that employed fluorescently labelled probes and utilized automated DNA sequencing was used (see Materials and methods; Yindeeyoungyeon and Schell, 2000). Three regions within the 178 bp WRE were highly protected from DNaseI digestion by GST–HMG (Figure 4A shows representative data for the TCF site 2 footprint). No protection from DNaseI was observed with GST alone (data not shown). All three protected sequences contained a similar stretch of six nucleotides conforming to the consensus AGAWAW, with additional flanking sequences also protected in two of the three footprints (Figure 4B).
Figure 4.
Wnt-mediated repression of Ugt36Bc requires novel TCF-binding sites. (A) DNaseI protection assays were used to identify TCF-binding sites in the 178 bp WRE (see Materials and methods). Blue traces show the protection pattern in the presence GST and green traces show the protection pattern in the presence of GST–HMG. Top panel: wild-type Ugt-178 probes. Bottom panel: Ugt-178 probes with mutated TCF-binding sites. The lower green signal relative to blue signal in the boxed region indicates protection of this sequence by the HMG domain. Binding is sequence specific as protection is abolished when the sequence is mutated (bottom panel). The chromatogram region shown corresponds to the TCF site 2 region and is representative of the other two TCF-binding sites. (B) Summary of DNaseI protection assays showing the sequence and location of the three strongest GST–HMG footprints within the 178 bp WRE. Underlined regions were mutated to disrupt TCF-binding sites as described below. Bold sequences generated the loose consensus AGAWAW (W=A or T). (C) GST and GST–HMG binding to an oligo containing TCF-binding site 2 (WT) or an oligo containing a mutated site (Mut.). (D) TCF-binding sites to the 178 bp WRE were probed using TCF ChIP in Kc cell lines containing stably integrated wild-type (pMT-178) or TCF site mutant (pMT-178*123) reporters. Cloned WREs were distinguished from the endogenous Ugt36Bc WRE using PCR primers specific for vector-derived sequences adjacent to the WRE. (E) Luciferase assay in transiently transfected Kc cells. Mutation of any individual TCF site within pHsp-178 partially reduced WRE activity in the absence of Wnt signalling, and mutation of all three TCF-binding sites (pHsp-178*123) lowered activity to background levels. All activities are expressed relative to pHsp-Luc. (F) The role of the TCF-binding sites in repression was measured using the metallothionein promoter to provide activity to mutant reporters in the absence of Wnt signalling. Here, 50 ng of control, wild-type, and TCF site mutant reporters was transiently transfected with 150 ng control or Arm* expression vectors. Here, 50 μM CuSO4 was added to cells 24 h before harvesting to enhance reporter expression. Uninduced expression levels were normalized to 1.0 for each reporter. Arm* repressed the wild-type WRE (pMT-178), but not a reporter in which all three TCF sites were mutated (pMT-178*123). (G) Site-directed mutagenesis was used to convert all three footprinted sites in the pMT-178 reporter to sequences that match the traditional TCF-binding site consensus (CCTTTGATCTT) to generate pMT-178*TCF Swap. These mutations reduced the activity in Ctrl RNAi-treated cells to enhancer-less vector levels. More importantly, the TCF site swap caused a 15-fold activation by Axin RNAi.
The binding of TCF to the protected regions was confirmed in two ways. GST–HMG was able to bind to an oligonucleotide containing the TCF site 2 footprint (Figure 4C). In addition, the ability of TCF to bind the Ugt36Bc WRE in the context of chromatin was assayed by performing TCF ChIP on polyclonal stable cell lines containing either the wild-type pMT-178 reporter or a version of that reporter in which all three footprinted TCF sites were mutated (pMT-178*123). We found that TCF strongly bound the cloned wild-type WRE, and mutation of the TCF-binding sites reduced TCF binding to the background levels observed with TCF RNAi treatment (Figure 4D).
To assess the functional role of the novel TCF-binding sites, they were mutated individually and in combination in the context of the pHsp-178 reporter. Mutation of any individual TCF-binding site resulted in a partial loss of enhancer activity in the absence of Wnt signalling (Figure 4E). Mutation of all three TCF-binding sites reduced reporter activity in the absence of Wnt signalling to levels seen with the enhancer-less vector. These results are consistent with our previous observations that TCF activates Ugt36Bc in the absence of Wnt stimulation (Figures 1D, 3B and E).
As the novel TCF-binding sites were required for activity of the Ugt36Bc WRE in the absence of Wnt, we could not assess their role in Wnt-mediated repression using the pHsp-178 reporter. To circumvent this problem, the Hsp70 promoter was replaced with the metal-inducible metallothionein promoter to generate pMT-178 (Figure 3A). Luciferase expression from this reporter is enhanced by adding CuSO4 to the growth media (data not shown). When pMT-178 was transfected into cells grown in the presence of 50 mM CuSO4, this reporter was highly expressed in the absence of Wnt signalling and repressed approximately three-fold by Wnt signalling (Figure 4F), similar to regulation of stably integrated reporters (data not shown). Mutation of all three TCF sites (pMT-178*123) greatly abrogated the Wnt-mediated repression (Figure 4F).
TCF-binding site sequence specifies activation or repression of the Ugt36Bc WRE
The fact that the TCF-binding sites required for repression differ so substantially from traditional TCF-binding sites suggested that they might determine the response to Wnt signalling. If this were the case, then changing the novel TCF sites in pMT-178 to traditional TCF sites (CCTTTGATCTT) should cause the WRE to behave similarly to a traditional WRE, that is, have low activity in the absence of Wnt and be activated by pathway stimulation. When traditional TCF sites replaced the three footprinted TCF sites in pMT-178, Wnt signalling activated transcription of the reporter approximately 15-fold (Figure 4G). In addition, transcription in the absence of Wnt signalling dropped to background levels, showing that DNA sequence influences TCF activity in both the absence and presence of Wnt signalling. The simplest interpretation of the data is that the DNA sequence to which TCF binds has an important function in determining whether the gene will be activated or repressed by Wnt signalling.
The activation and repression domains of arm are physically separate
β-cat/Arm consists of an N-terminal regulatory domain, a central domain containing 12 Arm repeats, and a C-terminal domain (Daniels et al, 2001). Both the N- and C-terminal halves of β-cat/Arm possess the ability to activate transcription (Hsu et al, 1998; Fang et al, 2006). Activation by the N-terminal half requires a region in the first Arm repeat that binds to Legless (Hoffmans and Basler, 2004). The C-terminal half binds several co-activators, including p300/CBP that binds Arm repeat 10 to the C terminus (Daniels and Weis, 2002) and Parafibromin/Hyrax that binds Arm repeat 12 to the C terminus (Daniels and Weis, 2002; Mosimann et al, 2006). If Arm uses distinct domains for transcriptional activation and repression, then introducing mutations into co-activator-binding sites might compromise activation without affecting repression. To this end, two point mutations known to disrupt Legless binding (E170A and E172A) (Hoffmans and Basler, 2004) were introduced into a truncated version of Arm* lacking the C-terminal domain to generate an allele of Arm known as DisArmed.
As predicted, DisArmed showed a dramatic reduction in the ability to activate several Wnt reporter genes, including reporters for nkd (Li et al, 2007), notum (Stadeli and Basler, 2005), and CG6234 (Fang et al, 2006) (Figure 5A). Amazingly, DisArmed retained the ability to repress pHsp-178 nearly as well as Arm*, showing that Arm contains distinct activation and repression domains (Figure 5A).
Figure 5.
Arm uses distinct domains for transcriptional activation and repression. (A) Kc cells were transiently transfected for 3 days with 150 ng empty vector, Arm*, or DisArmed expression vectors along with the 50 ng of the indicated reporter. DisArmed was severely compromised for transcriptional activation of all reporters, but repressed pHsp-178 nearly as well as Arm*. (B) Pxn, Tig, and Ugt36Bc are directly repressed by Wnt signalling. Kc cells were transiently transfected with 200 ng pAc-, pAcArm*, or pAcDisArmed along with 100 ng pAcIL2α expression vector. At 3 days after transfection, cells were isolated on αCD25 magnetic beads and used for real-time PCR or western blotting. (C) Western blots of whole-cell extract from cells used in (B). Arm* and DisArmed were expressed at approximately the same level, as measured by using antibodies against an N-terminal portion of Arm (top panel) and the C-terminal V5 tag (bottom panel).
As DisArmed is severely compromised for activation, genes directly repressed by Wnt signalling should be equally repressed by DisArmed and Arm*, whereas genes indirectly repressed through a Wnt-activated repressor protein should not be repressed by DisArmed. The ability of Arm* and DisArmed to repress the endogenous transcript levels of Ugt36Bc, Pxn, Tiggrin, and Ugt58Fa was measured in populations of Kc cells enriched for successful transfection (see Materials and methods for details). Pxn, Tig, and Ugt36Bc were all repressed by DisArmed nearly as well as they were by Arm* (Figure 5B). Ugt58Fa on the other hand, was not repressed by DisArmed (data not shown), suggesting that the repression observed in Figure 1 is indirect, and providing a simple proof of principle that DisArmed can distinguish between some directly and indirectly repressed genes. Note that endogenous nkd is not activated by DisArmed, consistent with the reporter genes (Figure 5B). Western blots of whole-cell lysate from transfected cells showed that Arm* and DisArmed were expressed at similar levels (Figure 5C).
To extend these studies, we also generated transgenic flies expressing DisArmed or Arm*. When expressed in haemocytes under the control of the crq-Gal4 driver, DisArmed repressed the transcription of Pxn and Tig just as well as Arm* (Figure 6). We anticipate that DisArmed will provide a useful reagent for dissecting the role of direct Wnt-mediated repression in a variety of developmental contexts.
Figure 6.
DisArmed represses transcription in vivo. DIC images ( × 10 magnification) of stage 13 (A–C) or 14 (D–F) embryos following in situ hybridization with the indicated probes. P[UAS-GFP], P[crq79-Gal4] (crq>GFP) control embryos are compared with P[UAS-Arm*], P[crq79-Gal4] (crq>Arm*) and P[UAS-DisArmed], P[crq79-Gal4] (crq>DisArmed) embryos. DisArmed represses Pxn and Tig expression in haemocytes as efficiently as Arm*. Insets (A′–F′) show × 40 magnification to highlight individual haemocytes migrating along the ventral boundary of the embryo. Anterior is left and dorsal is up in all images.
Discussion
TCF is a programmable transcriptional switch
The transcription factors that mediate signalling by the Wnt, Notch, Hh, and nuclear receptor pathways are often referred to as ‘transcriptional switches' because the same protein mediates transcriptional repression in the absence of ligand and transcriptional activation when the pathway is stimulated (Barolo and Posakony, 2002). For many Wnt targets, TCF represses transcription in the absence of signalling and switches to an activator when complexed with β-cat/Arm (Stadeli et al, 2006; Parker et al, 2007). Here, we identified several direct Wnt targets that were instead activated by TCF in the absence of signalling and repressed by TCF/Arm upon pathway activation (Figures 1, 2 and 3). Our detailed analysis of the WRE controlling transcription of one of these, Ugt36Bc, revealed novel TCF-binding sites that were essential for both basal activation and Wnt-mediated repression (Figure 4A–F). Replacing the novel TCF-binding sites with traditional TCF-binding sites caused the WRE to behave similarly to a traditional Wnt target, that is, it was not active in the absence of Wnt signalling and was activated upon pathway stimulation (Figure 4G). Thus, TCF is more aptly described as a ‘programmable transcription switch', in which the DNA sequence to which TCF binds determines whether flipping the switch turns gene expression on or off (Figure 7).
Figure 7.
Model for allosteric regulation of TCF and arm by DNA. Our study suggests that TCF can bind two different classes of DNA sequences. When TCF is bound to traditional consensus sites resembling CCTTTGATCTT, it represses transcription in the absence of signalling. Upon Wnt stimulation, TCF/Arm activates transcription through Arm-associated co-activators. When TCF is bound to the sequences containing AGAWAW in Ugt36Bc, possibly with the aid of cofactors, it activates transcription in the absence of signalling and Wnt stimulation results in Arm-dependent transcriptional repression. The different shapes of TCF and Arm at activated and repressed genes represent potential structural differences in the complex when bound to the different classes of DNA sequence.
Many transcription factors change conformation upon binding DNA (reviewed in Lefstin and Yamamoto, 1998). However, there are only a few other examples of transcription factors whose activity is different when bound to different DNA sequences. These include the fly Smad proteins Mad and Medea and the mouse POU domain protein Pit-1, where the spacing of half sites influences whether these proteins activate or repress transcription (Scully et al, 2000; Pyrowolakis et al, 2004). Another example is NF-κB, where subtle changes in the sequence of its binding sites (1 or 2 bp) alter gene-specific co-activator requirements or determine whether a NF-κB target can be repressed by the glucocorticoid receptor (Leung et al, 2004; Luecke and Yamamoto, 2005).
The TCF mechanism described here is similar to thyroid hormone receptor (TR). TR represses numerous genes in the absence of ligand, and binding of thyroid hormone (T3) switches the receptor into a transcriptional activator (Glass and Rosenfeld, 2000). In contrast, several other thyroid responsive elements (nTREs) show the opposite response, that is, TR activates expression in the absence of hormone and this activation is blocked by T3 treatment (Saatcioglu et al, 1993; Lazar, 2003; Nygard et al, 2003, 2006). The TR-binding site sequence is at least partially responsible for this alteration, as conversion of the nTRE to the consensus TRE compromised the ability of TR to activate transcription in the absence of ligand (Saatcioglu et al, 1993).
The mechanism described for allosteric regulation of TCF by DNA is distinct from the mechanisms described above in a few ways. First, the proteins discussed above bind DNA as homo- or heterodimers. The POU domain transcription factor Pit-1 is the one exception, and it utilizes the bipartite POU domain to bind two half-sites separated by 4 or 6 bp (Scully et al, 2000). To our knowledge, TCF is unique in that it binds as a monomer to a single contiguous DNA sequence. Also, unlike the other cases, there is a dramatic difference in the sequence of TCF-binding sites found in the repressed target Ugt36Bc (AGAWAW) and those found in activated targets (SCTTTGWW). Interestingly, in none of the examples above was substituting one type of binding site for another able to completely reverse regulation in both the absence and presence of signalling as we have shown for the Ugt36Bc WRE.
The AGAWAW sequences are clearly necessary for the Wnt regulation of the Ugt36Bc WRE but are they also sufficient? A reporter that contained three tandem copies of TCF site 2 in the pMT-Luc reporter was not repressed by Wnt signalling, nor could three copies of TCF site 2 impart activation to hsp70-based reporter constructs (data not shown). Given that the TCF HMG domain binds preferentially to sites containing the traditional CCTTTGAT motif in vitro (van de Wetering et al, 1997; Hallikas et al, 2006; Atcha et al, 2007), perhaps additional cofactors are required to facilitate TCF binding to AGAWAW-type sites (Figure 7).
Repression of Ugt36Bc is distinct from other Wnt-repressed targets
Prior to these studies, only three other genes had been shown to be directly repressed by TCF and β-cat/Arm. In all three cases, Wnt-mediated repression required traditional TCF-binding sites. For Dpp and CDH1, it is not clear why Arm/β-cat represses transcription when recruited to TCF at traditional binding sites, although the transcriptional repressors Brinker and Snail contribute to the repression of each gene, respectively (Jamora et al, 2003; Theisen et al, 2007). Repression of the stripe gene also required two traditional TCF sites, but these sites partially overlap binding sites for Cubitus Interruptis (Ci), an essential activator of stripe (Piepenburg et al, 2000). This suggests a mechanism in which Arm binding to TCF occludes Ci from the stripe WRE. In our study, the results from the TCF-binding site swap experiment (Figure 4F) argue against the possibility that Arm represses Ugt36Bc transcription by displacing an essential activator, as the activator should be displaced regardless of the DNA sequence used to recruit TCF and Arm.
Wnt signalling and haemocyte development/function
We have shown that Wnt signalling can repress the transcription of several genes expressed in haemocytes. However, we did not observe any effect of ectopic Wnt signalling or loss-of-function Wnt mutations on the number, class (plasmatocytes or crystal cells), or migration pattern of embryonic haemocytes (data not shown). However, it has been reported that Wnt signalling can inhibit lamellocyte (a specific type of haemocyte) formation in fly larva (Zettervall et al, 2004). In addition, Tig is a major component of larval haemolymph clots (Scherfer et al, 2004), which are important in preventing loss of haemolymph and wound healing. This suggests that the Wnt pathway and perhaps Wg-mediated transcriptional repression may have a function in haemocyte development and/or function.
DisArmed provides a tool to probe direct repression by Wnt signalling
Three of the four repressed genes identified by our microarray analyses were directly repressed by TCF/Arm (Figure 5B). This raises the possibility that Wnt-mediated repression may be more common than currently appreciated. Searching for traditional TCF-binding sites near a gene's promoter is common first step in screening for direct targets. However, the TCF-binding sites identified in the Ugt36Bc enhancer differ significantly from the traditional TCF-binding sites and would not be identified in such searches. Currently, the AGAWAW consensus does not contain enough information to be useful in genome-wide searches for additional repressed Wnt targets, as such sequences are found very frequently throughout the genome. For example, 110 such motifs are found within 2 kb of the Pxn gene and 26 motifs within the Tig gene (including 2 kb on either side of each genes' UTRs). A more detailed understanding of the sequences that specify Wnt-mediated repression should facilitate the identification of additional repressed targets using this approach.
DisArmed also provides a valuable reagent to help identify additional direct targets of Wnt repression. The mutations or deletions in known co-activator-binding sites of Arm severely compromise transcriptional activation without significantly affecting repression of Ugt36Bc, Tig, and Pxn (Figures 5 and 6). These mutations demonstrate that the activation and repression domains of Arm are distinct. They also allow one to probe directness of repressed Wnt targets without prior identification of the WRE and without the use of protein synthesis inhibitors. In theory, a target that is repressed by both Arm and DisArmed would be a good candidate for direct repression by Wnt signalling, as is the case for Pxn and Tig in haemocytes (Figure 6). The applicability of this approach to a broad range of Wnt repressed targets awaits further analysis.
Materials and methods
Plasmids
All protein expression vectors used in Kc cells were generated by cloning the ORF and a 5′ Kozak sequence into pAc5.1/v5-His-A (Invitrogen). pAcArm* expresses a stabilized form of Arm (T52A/S56A) fused to C-terminal V5 epitope and 6 × His tags. pAcDisArmed was generated by introducing additional nucleotide substitutions that code for D170A/D172A into pAcArm* using the QuickChange II kit (Strategene) and subcloning all but the C-terminal domain following Arm repeat 12 in frame with C-terminal V5 and 6 × His tags. pAc-TCF expresses full-length untagged TCF. pAc-dnTCF is similar, except that the codons for amino acids 2–33 are missing from the TCF coding region. pAcArm1–12ER expresses Arm repeats 1–12 fused to the wild-type version of the ligand-binding domain of mouse ERα without additional tags.
All luciferase reporter vectors are derivatives of pGL2 (Promega). pUgt5.2 contains the genomic region extending from −730 to +780 relative to the annotated TSS of Ugt36Bc cloned into the KpnI/HindIII sites of pUASluc (Fang et al, 2006). pUgt-28 contains the −730 to −430 and −150 to −1 sequences cloned into the XmaI/KpnI and KpnI/HindIII sites of pUASluc, respectively. pUgt-Luc (control) contains only the −279 to −1 sequence cloned into the KpnI/HindIII sites of pUASluc. pHsp-Luc (control) contains the −44 to +203 region of Drosophila Hsp70Bb cloned into the XhoI/HindIII sites of pGL2-Basic (Promega). It also contains 83 bp of spacer sequence between the 5′ end of the Hsp70Bb promoter and the 3′ end of cloned WREs. pHsp-178 contains the −630 to −456 sequences cloned into the XmaI/KpnI of pHsp-Luc. pMT-Luc and pMT-178 were generated by replacing the Hsp70Bb promoter with the metallothionein promoter from pMT/Rpr/V5-His (Zachariou et al, 2003) in pHsp-Luc and pHsp-178, respectively. QuickChange II (Stratagene) was used to mutate the TCF sites of pMT-178*123 and to generate pMT-178*TCF Swap. Primer sequences used for cloning available upon request. All plasmids were sequenced at the University of Michigan DNA Sequencing Core to confirm integrity.
Drosophila cell culture, RNAi knockdown, and Wg-CM treatment and qRT–PCR
Kc cells were routinely cultured in Schneider's Drosophila media (Invitrogen) supplemented with 5% FBS at room temp. For RNAi-mediated gene knockdown, cells were seeded at 1 × 106 cells/ml in growth media supplemented with 10 μg/ml dsRNA for 4 days, diluted to 1 × 106 cells/ml, and grown with the same concentration of dsRNA for 3 more days for gene expression experiments or 2 more days for ChIP experiments. When multiple genes were targeted by RNAi, the final concentration of all dsRNA species was not more than 20 μg/ml. dsRNA constructs targeting two different regions of Arm, Axin, and TCF all showed similar effects, and control dsRNA sequences targeting either, β-lactamase, or eGFP also showed similar results. See Supplementary Table S1 for primer sequences.
Wg-CM was prepared as described previously (Fang et al, 2006). Wg-CM (400 μl) was added to 1 ml of Kc cells at 2 × 106 cells/ml for 6 h to induce Wnt signalling.
Kc cell lines with stably integrated copies of pMT-Luc, pMT-178, or pMT-178*123 were generated by co-transfecting 2 μg of reporter and 2 μg pCoPuro (Iwaki et al, 2003). Cells were maintained in growth media supplemented with 7 μg/ml puromycin and showed greater than 90% viability after 6 weeks.
qRT–PCR was performed as previously described (Fang et al, 2006) except that real-time PCR reactions were carried out in standard PCR buffer supplemented with 25% DMSO, SYBR Green, 10 mM fluoroscein, and Platinum Taq (Invitrogen) in an iCycler real-time PCR detection system (Bio-Rad). Gene expression among different samples was normalized to tubulin56D levels. Primers used for PCR are shown in Supplementary Table S1.
Transient transfections and reporter gene assays
Transient transfections were carried out as previously described (Fang et al, 2006). Reporter assays used 25–100 ng of luciferase reporter DNA as indicated in figure legends and 5 ng pAc5.1-LacZ (Invitrogen). Here, 50–200 ng of pAcArm*, pAcDisArmed, pAcArm1–12ER, pAc-dnTCF, or pAc-TCF were co-transfected with the reporter genes as indicated in figure legends. pAc5.1-V5/His-A vector was used to equalize DNA content between samples and as a negative control for expression vectors. Luciferase and β-galactosidase activities were measured 3 days after transfection unless otherwise noted. Luciferase activity was normalized to β-galactosidase activity from pAc5.1-LacZ to control for difference in transfection efficiency among samples.
Purification of transfected cells was accomplished as described by Ogawa et al (2002) with the following modifications. Cells were co-transfected with 100 ng of pAc-IL2α along with 200 ng pAcArm*, pAcDisArmed, or pAc5.1-V5His-A. At 3 days after transfection, cells were collected by low-speed centrifugation, washed once with growth media, and incubated with 5 μl packed αCD25 magnetic beads (Dynabeads), and washed again with 500 μl fresh growth media at room temperature for 20 min. Half the beads were resuspended in protein loading buffer for western blot analysis using monoclonal antibodies to Arm (Riggleman et al, 1990) or V5 (Invitrogen), and total RNA was isolated from the remaining cells using Trizol reagent.
ChIP
ChIP was performed essentially as described by the Upstate ChIP Assay Kit protocol (Upstate Biotechnology), with the previously described modifications (Fang et al, 2006). DNA was fragmented using a Branson600 sonicator to an average size of 200–1000 bp. Whole-cell extract from 3 × 106 crosslinked cells and 15 μl polyclonal TCF antisera (Fang et al, 2006) were used in each ChIP experiment. Precipitated DNA was measured with qPCR using the primers listed in Supplementary Table S1.
Statistical analysis
Unless otherwise noted, every experiment using cell culture was repeated at least twice, usually three or four times, with two or more replicates in each independent experiment. All error bars represent standard deviations of at least two replicates in a representative experiment.
Fluorescent DNA footprinting
Footprinting was performed as described (Yindeeyoungyeon and Schell, 2000) with modifications. In brief, DNA sequences corresponding to the Ugt36Bc minimal WRE plus an additional 50–150 bp on both sides were PCR amplified using one primer with a 5′-fluorescent tag (FAM or HEX) and one unlabelled primer to generate dsDNA probes with a single fluorescent label. Fluorescent probes were used in lieu of radioactively labelled probes in standard DNaseI protection assays (see Promega Core Footprinting System protocol for salt and buffer concentrations). We used 500 nM to10 μM purified GST–HMG (Lee and Frasch, 2000) with 12 nM HEX-labelled probes (green) and similar concentrations of GST alone with 12 nM FAM-labelled probes (blue) in 50 μl digestion reactions. After digestion, the blue and green probes were combined, purified by phenol/chloroform extraction, concentrated by ethanol precipitation, and analysed by the University of Michigan DNA sequencing core. Peaks in the chromatograms were assigned to specific nucleotides using NED-labelled ddATP in a manual sequencing reaction, which was run in an adjacent lane in the automated sequencing gel. The DNA sequences identified as novel TCF-binding sites in the Ugt36Bc WRE were reproducibly protected at in at least two separate experiments for each of three independent probe preparations. A detailed description of this method will be reported elsewhere.
Electrophoretic mobility shift assay
Electrophoretic mobility shift assays (EMSAs) were performed using the Lightshift Chemiluminescent EMSA Kit (Pierce) in combination with the Chemiluminescent Nucleic Acid Detection Module (Pierce). Glycerol (4.35%), magnesium chloride (5 mM), poly (dI–dC) (50 ng/μl), and NP-40 (0.05%) were included in the binding reaction. Wild-type (5′-Bio-CTATACAGATAAAAACATTT) and mutant (5′-Bio-CTAGAAATAGACACAAAGTT) probes were present at 20 nM final concentration. GST and GST–HMG were purified from Escherichia coli and present at 250 ng/μl final concentration in the binding reaction.
Drosophila genetics
The P[crq79-Gal4] driver and P[UAS-ArmSC10] have been previously described (Pai et al, 1997; Olofsson and Page, 2005). P[UAS-Arm*] and P[UAS-DisArmed] were generated by cloning the Arm* or DisArmed ORFs described above into pUAST (Brand and Perrimon, 1993). Transgenic flies were generated by BestGene Inc. (Chino Hills, CA).
In situ hybridization
Drosophila embryos were prepared for in situ hybridization or immunostaining as previously described (Lin et al, 2004). In situ probes were generated by PCR amplification of cDNA templates (see Supplementary Table S1 for primers). In situ hybridization was performed at 55°C using Dig-UTP RNA probes at 100 ng/ml. All embryos were photographed on an Nikon Elipse 800 microscope using DIC optics.
Supplementary Material
Supplementary Table S1
Supplementary Figure S1
Acknowledgments
We are very grateful to Birgitta Olofsson for crq79-Gal4 flies, as well as David van Mater, Eric Fearon, Pascal Meier, Manfred Frasch, and Francis Castellino for DNA plasmids. Special thanks to Roel Nusse for the pTub-Wg cells and to Dan Bochar for help with the purification of transfected cells. We thank the Bloomington Stock Center and the Hybridoma Bank for providing fly stocks and antibodies. We are very grateful to Scott Barolo, Eric Fearon, David Parker and members of the Cadigan lab for many discussions. TAB was supported by post-doctoral fellowships from the University of Michigan Cancer Center and the American Heart Association (0425723Z). Special thanks also to Gyorgyi Csankovszki for providing support to TAB during the completion of this study. This research was supported by NIH grant RO1 CA95869 and a grant from the University of Michigan Cancer Center to KMC.
References
- Atcha FA, Syed A, Wu B, Hoverter NP, Yokoyama NN, Ting JH, Munguia JE, Mangalam HJ, Marsh JL, Waterman ML (2007) A unique DNA binding domain converts T-cell factors into strong Wnt effectors. Mol Cell Biol 27: 8352–8363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfonso TB, Jones BW (2002) gcm2 promotes glial cell differentiation and is required with glial cells missing for macrophage development in Drosophila. Dev Biol 248: 369–383 [DOI] [PubMed] [Google Scholar]
- Barolo S (2006) Transgenic Wnt/TCF pathway reporters: all you need is Lef? Oncogene 25: 7505–7511 [DOI] [PubMed] [Google Scholar]
- Barolo S, Posakony JW (2002) Three habits of highly effective signaling pathways: principles of transcriptional control by developmental cell signaling. Genes Dev 16: 1167–1181 [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415 [DOI] [PubMed] [Google Scholar]
- Cadigan KM (2002) Regulating morphogen gradients in the Drosophila wing. Semin Cell Dev Biol 13: 83–90 [DOI] [PubMed] [Google Scholar]
- Cadigan KM, Fish MP, Rulifson EJ, Nusse R (1998) Wingless repression of Drosophila frizzled 2 expression shapes the Wingless morphogen gradient in the wing. Cell 93: 767–777 [DOI] [PubMed] [Google Scholar]
- Cadigan KM, Nusse R (1997) Wnt signaling: a common theme in animal development. Genes Dev 11: 3286–3305 [DOI] [PubMed] [Google Scholar]
- Cavallo RA, Cox RT, Moline MM, Roose J, Polevoy GA, Clevers H, Peifer M, Bejsovec A (1998) Drosophila Tcf and Groucho interact to repress Wingless signalling activity. Nature 395: 604–608 [DOI] [PubMed] [Google Scholar]
- Daniels DL, Eklof Spink K, Weis WI (2001) Beta-catenin: molecular plasticity and drug design. Trends Biochem Sci 26: 672–678 [DOI] [PubMed] [Google Scholar]
- Daniels DL, Weis WI (2002) ICAT inhibits beta-catenin binding to Tcf/Lef-family transcription factors and the general coactivator p300 using independent structural modules. Mol Cell 10: 573–584 [DOI] [PubMed] [Google Scholar]
- Fang M, Li J, Blauwkamp T, Bhambhani C, Campbell N, Cadigan KM (2006) C-Terminal-binding protein directly activates and represses Wnt transcriptional targets in Drosophila. EMBO J 25: 2735–2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogerty FJ, Fessler LI, Bunch TA, Yaron Y, Parker CG, Nelson RE, Brower DL, Gullberg D, Fessler JH (1994) Tiggrin, a novel Drosophila extracellular matrix protein that functions as a ligand for Drosophila alpha PS2 beta PS integrins. Development 120: 1747–1758 [DOI] [PubMed] [Google Scholar]
- Giese K, Amsterdam A, Grosschedl R (1991) DNA-binding properties of the HMG domain of the lymphoid-specific transcriptional regulator LEF-1. Genes Dev 5: 2567–2578 [DOI] [PubMed] [Google Scholar]
- Glass CK, Rosenfeld MG (2000) The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev 14: 121–141 [PubMed] [Google Scholar]
- Goto A, Kumagai T, Kumagai C, Hirose J, Narita H, Mori H, Kadowaki T, Beck K, Kitagawa Y (2001) A Drosophila haemocyte-specific protein, hemolectin, similar to human von Willebrand factor. Biochem J 359: 99–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallikas O, Palin K, Sinjushina N, Rautiainen R, Partanen J, Ukkonen E, Taipale J (2006) Genome-wide prediction of mammalian enhancers based on analysis of transcription-factor binding affinity. Cell 124: 47–59 [DOI] [PubMed] [Google Scholar]
- Hoffmans R, Basler K (2004) Identification and in vivo role of the Armadillo–Legless interaction. Development 131: 4393–4400 [DOI] [PubMed] [Google Scholar]
- Hsu SC, Galceran J, Grosschedl R (1998) Modulation of transcriptional regulation by LEF-1 in response to Wnt-1 signaling and association with beta-catenin. Mol Cell Biol 18: 4807–4818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwaki T, Figuera M, Ploplis VA, Castellino FJ (2003) Rapid selection of Drosophila S2 cells with the puromycin resistance gene. Biotechniques 35: 482–484 [DOI] [PubMed] [Google Scholar]
- Jamora C, DasGupta R, Kocieniewski P, Fuchs E (2003) Links between signal transduction, transcription and adhesion in epithelial bud development. Nature 422: 317–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung HC, Kim K (2005) Identification of MYCBP as a beta-catenin/LEF-1 target using DNA microarray analysis. Life Sci 77: 1249–1262 [DOI] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D (2002) The human genome browser at UCSC. Genome Res 12: 996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi A, Kishida S, Yamamoto H (2006) Regulation of Wnt signaling by protein–protein interaction and post-translational modifications. Exp Mol Med 38: 1–10 [DOI] [PubMed] [Google Scholar]
- Knirr S, Frasch M (2001) Molecular integration of inductive and mesoderm-intrinsic inputs governs even-skipped enhancer activity in a subset of pericardial and dorsal muscle progenitors. Dev Biol 238: 13–26 [DOI] [PubMed] [Google Scholar]
- Lazar MA (2003) Thyroid hormone action: a binding contract. J Clin Invest 112: 497–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HH, Frasch M (2000) Wingless effects mesoderm patterning and ectoderm segmentation events via induction of its downstream target sloppy paired. Development 127: 5497–5508 [DOI] [PubMed] [Google Scholar]
- Lefstin JA, Yamamoto KR (1998) Allosteric effects of DNA on transcriptional regulators. Nature 392: 885–888 [DOI] [PubMed] [Google Scholar]
- Leung TH, Hoffmann A, Baltimore D (2004) One nucleotide in a kappaB site can determine cofactor specificity for NF-kappaB dimers. Cell 118: 453–464 [DOI] [PubMed] [Google Scholar]
- Li J, Sutter C, Parker DS, Blauwkamp T, Fang M, Cadigan KM (2007) CBP/p300 are bimodal regulators of Wnt signaling. EMBO J 26: 2284–2294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin HV, Rogulja A, Cadigan KM (2004) Wingless eliminates ommatidia from the edge of the developing eye through activation of apoptosis. Development 131: 2409–2418 [DOI] [PubMed] [Google Scholar]
- Logan CY, Nusse R (2004) The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol 20: 781–810 [DOI] [PubMed] [Google Scholar]
- Luecke HF, Yamamoto KR (2005) The glucocorticoid receptor blocks P-TEFb recruitment by NFkappaB to effect promoter-specific transcriptional repression. Genes Dev 19: 1116–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luque T, O'Reilly DR (2002) Functional and phylogenetic analyses of a putative Drosophila melanogaster UDP-glycosyltransferase gene. Insect Biochem Mol Biol 32: 1597–1604 [DOI] [PubMed] [Google Scholar]
- Mosimann C, Hausmann G, Basler K (2006) Parafibromin/Hyrax activates Wnt/Wg target gene transcription by direct association with beta-catenin/Armadillo. Cell 125: 327–341 [DOI] [PubMed] [Google Scholar]
- Naishiro Y, Yamada T, Idogawa M, Honda K, Takada M, Kondo T, Imai K, Hirohashi S (2005) Morphological and transcriptional responses of untransformed intestinal epithelial cells to an oncogenic beta-catenin protein. Oncogene 24: 3141–3153 [DOI] [PubMed] [Google Scholar]
- Nelson RE, Fessler LI, Takagi Y, Blumberg B, Keene DR, Olson PF, Parker CG, Fessler JH (1994) Peroxidasin: a novel enzyme-matrix protein of Drosophila development. EMBO J 13: 3438–3447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nygard M, Becker N, Demeneix B, Pettersson K, Bondesson M (2006) Thyroid hormone-mediated negative transcriptional regulation of Necdin expression. J Mol Endocrinol 36: 517–530 [DOI] [PubMed] [Google Scholar]
- Nygard M, Wahlstrom GM, Gustafsson MV, Tokumoto YM, Bondesson M (2003) Hormone-dependent repression of the E2F-1 gene by thyroid hormone receptors. Mol Endocrinol 17: 79–92 [DOI] [PubMed] [Google Scholar]
- Ogawa H, Ishiguro K, Gaubatz S, Livingston DM, Nakatani Y (2002) A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science 296: 1132–1136 [DOI] [PubMed] [Google Scholar]
- Olofsson B, Page DT (2005) Condensation of the central nervous system in embryonic Drosophila is inhibited by blocking hemocyte migration or neural activity. Dev Biol 279: 233–243 [DOI] [PubMed] [Google Scholar]
- Pai LM, Orsulic S, Bejsovec A, Peifer M (1997) Negative regulation of Armadillo, a Wingless effector in Drosophila. Development 124: 2255–2266 [DOI] [PubMed] [Google Scholar]
- Parker DS, Blauwkamp T, Cadigan KM (2007) Wnt/β-catenin-mediated transcriptional regulation. In Wnt Signaling in Embryonic Development, Sokol S (ed), Advances in Developmental Biology, Wassarman PM (ed), Vol. 17, pp. 1–61. San Diego: Elsevier [Google Scholar]
- Payre F, Vincent A, Carreno S (1999) ovo/svb integrates Wingless and DER pathways to control epidermis differentiation. Nature 400: 271–275 [DOI] [PubMed] [Google Scholar]
- Piepenburg O, Vorbruggen G, Jackle H (2000) Drosophila segment borders result from unilateral repression of hedgehog activity by wingless signaling. Mol Cell 6: 203–209 [PubMed] [Google Scholar]
- Pyrowolakis G, Hartmann B, Muller B, Basler K, Affolter M (2004) A simple molecular complex mediates widespread BMP-induced repression during Drosophila development. Dev Cell 7: 229–240 [DOI] [PubMed] [Google Scholar]
- Riggleman B, Schedl P, Wieschaus E (1990) Spatial expression of the Drosophila segment polarity gene armadillo is posttranscriptionally regulated by wingless. Cell 63: 549–560 [DOI] [PubMed] [Google Scholar]
- Saatcioglu F, Deng T, Karin M (1993) A novel cis element mediating ligand-independent activation by c-ErbA: implications for hormonal regulation. Cell 75: 1095–1105 [DOI] [PubMed] [Google Scholar]
- Scherfer C, Karlsson C, Loseva O, Bidla G, Goto A, Havemann J, Dushay MS, Theopold U (2004) Isolation and characterization of hemolymph clotting factors in Drosophila melanogaster by a pullout method. Curr Biol 14: 625–629 [DOI] [PubMed] [Google Scholar]
- Scully KM, Jacobson EM, Jepsen K, Lunyak V, Viadiu H, Carriere C, Rose DW, Hooshmand F, Aggarwal AK, Rosenfeld MG (2000) Allosteric effects of Pit-1 DNA sites on long-term repression in cell type specification. Science 290: 1127–1131 [DOI] [PubMed] [Google Scholar]
- Stadeli R, Basler K (2005) Dissecting nuclear Wingless signalling: recruitment of the transcriptional co-activator Pygopus by a chain of adaptor proteins. Mech Dev 122: 1171–1182 [DOI] [PubMed] [Google Scholar]
- Stadeli R, Hoffmans R, Basler K (2006) Transcription under the control of nuclear Arm/beta-catenin. Curr Biol 16: R378–R385 [DOI] [PubMed] [Google Scholar]
- Theisen H, Syed A, Nguyen BT, Lukacsovich T, Purcell J, Srivastava GP, Iron D, Gaudenz K, Nie Q, Wan FY, Waterman ML, Marsh JL (2007) Wingless directly represses DPP morphogen expression via an armadillo/TCF/Brinker complex. PLoS ONE 2007: e142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomancak P, Beaton A, Weiszmann R, Kwan E, Shu S, Lewis SE, Richards S, Ashburner M, Hartenstein V, Celniker SE, Rubin GM (2002) Systematic determination of patterns of gene expression during Drosophila embryogenesis. Genome Biol 3: RESEARCH0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Wetering M, Cavallo R, Dooijes D, van Beest M, van Es J, Loureiro J, Ypma A, Hursh D, Jones T, Bejsovec A, Peifer M, Mortin M, Clevers H (1997) Armadillo coactivates transcription driven by the product of the Drosophila segment polarity gene dTCF. Cell 88: 789–799 [DOI] [PubMed] [Google Scholar]
- van de Wetering M, Sancho E, Verweij C, de Lau W, Oving I, Hurlstone A, van der Horn K, Batlle E, Coudreuse D, Haramis AP, Tjon-Pon-Fong M, Moerer P, van den Born M, Soete G, Pals S, Eilers M, Medema R, Clevers H (2002) The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell 111: 241–250 [DOI] [PubMed] [Google Scholar]
- Webster NJ, Green S, Jin JR, Chambon P (1988) The hormone-binding domains of the estrogen and glucocorticoid receptors contain an inducible transcription activation function. Cell 54: 199–207 [DOI] [PubMed] [Google Scholar]
- Willert J, Epping M, Pollack JR, Brown PO, Nusse R (2002) A transcriptional response to Wnt protein in human embryonic carcinoma cells. BMC Dev Biol 2: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yindeeyoungyeon W, Schell MA (2000) Footprinting with an automated capillary DNA sequencer. Biotechniques 29: 1034–1038 [DOI] [PubMed] [Google Scholar]
- Zachariou A, Tenev T, Goyal L, Agapite J, Steller H, Meier P (2003) IAP-antagonists exhibit non-redundant modes of action through differential DIAP1 binding. EMBO J 22: 6642–6652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zettervall CJ, Anderl I, Williams MJ, Palmer R, Kurucz E, Ando I, Hultmark D (2004) A directed screen for genes involved in Drosophila blood cell activation. Proc Natl Acad Sci USA 101: 14192–14197 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table S1
Supplementary Figure S1