Abstract
We describe a new method, which identifies protein fragments for soluble expression in Escherichia coli from a randomly fragmented gene library. Inhibition of E. coli dihydrofolate reductase (DHFR) by trimethoprim (TMP) prevents growth, but this can be relieved by murine DHFR (mDHFR). Bacterial strains expressing mDHFR fusions with the soluble proteins green fluroscent protein (GFP) or EphB2 (SAM domain) displayed markedly increased growth rates with TMP compared to strains expressing insoluble EphB2 (TK domain) or ketosteroid isomerase (KSI). Therefore, mDHFR is affected by the solubility of fusion partners and can act as a reporter of soluble protein expression. Random fragment libraries of the transcription factor Fli1 were generated by deoxyuridine incorporation and endonuclease V cleavage. The fragments were cloned upstream of mDHFR and TMP resistant clones expressing soluble protein were identified. These were found to cluster around the DNA binding ETS domain. A selected Fli1 fragment was expressed independently of mDHFR and was judged to be correctly folded by various biophysical methods including NMR. Soluble fragments of the cell-surface receptor Pecam1 were also identified. This genetic selection method was shown to generate expression clones useful for both structural studies and antibody generation and does not require a priori knowledge of domain architecture.
INTRODUCTION
Expression of mammalian proteins in Escherichia coli often results in protein misfolding with protein degradation and inclusion body formation. This may be because prokaryotic expression systems lack the necessary chaperones, natural binding partners and ability to perform the post-translational modifications required for correct folding of a eukaryotic protein. The addition of solubility enhancing tags can improve expression, but this is dependent on the properties of the protein target and precipitation can occur upon tag removal (1,2). A strategy employed by many laboratories when attempting to express a large multi-domain protein for structural or functional studies, including antibody production, is truncation to produce smaller single domains that are easier to express in a soluble form in E. coli. This requires the domains to be annotated onto the linear polypeptide sequence and primers designed to the domain boundaries to amplify specific domains for cloning into expression vectors. However, the precise domain boundaries can vary depending on the prediction method used. Although methods based on sequence alignment are good at highlighting the core consensus motif, they are less good at predicting the fold edges because these regions can contain more sequence variability. For example, the domains predicted from the Pfam database (3) are often smaller than the structural domain boundaries. Attempted expression of a construct based on the Pfam boundaries would result in premature truncation of N- or C-terminal α-helixes or β-sheets required to complete the domain folding unit giving an unstable construct with poor expression yield. It is possible, in a ‘primer pair walking’ strategy, to design primers spaced at a progressively greater distances from the core Pfam domain, clone the PCR products into expression vectors and screen for soluble expression. However, this approach is laborious, costly and limited by requiring a mapped protein domain.
A possible solution to this problem is to take a combinatorial approach by preparing a library of gene fragments and select those that express well. This requires efficient methods for gene fragmentation and selection for soluble protein expression. In this work, we describe both an improved method for random library generation and a novel genetic selection of soluble protein expression involving murine dihydrofolate reductase (mDHFR). Random insert libraries were generated by first amplifying the gene by PCR in the presence of dUTP followed by cleavage with Endonuclease V in the presence of MnCl2, which promotes double stranded DNA cleavage at the second and third phosphodiester bonds 3′ of the site of uracil incorporation (4).
DHFR is an essential enzyme for the survival of E. coli and converts dihydrofolate into tetrahydrofolate, which can then be converted to tetrahydrofolate co-factors used in one-carbon transfer reactions for the de novo synthesis of purines, thymidylic acid and certain amino acids. Trimethoprim (TMP) is a potent inhibitor of bacterial DHFR but not mDHFR, allowing selection for functional mDHFR by plating the library on minimal expression plates containing TMP and IPTG for protein induction. Only transformants expressing functional mDHFR confer TMP resistance and are able to grow on the selection plates. mDHFR was previously shown not to perturb the folding of a set of N-terminal fusion proteins (1), which together with its monomeric state makes it an ideal reporter. We show here that expression of functionally active DHFR is dependent on the folding state of a variety of upstream control fusion proteins. The selection process was further validated by producing a library of the transcription factor Fli1. Screening selected for the ETS (erythroblast transformation specific) domain which was soluble when expressed in isolation (with a hexahistidine tag). This protein was judged to be folded when 15N labelled and examined by 2D NMR.
A library of random DNA fragments was also generated of the type 1 integral membrane receptor Pecam1. Selection identified a number of extracellular and intracellular protein expression constructs. A cytoplasmic construct was expressed with a hexahistidine tag and although not folded as judged by 1D and 2D NMR, this construct was used successfully to produce antibodies in a phage display selection that gave a specific membrane staining to an endothelial cell line. Previously, rationally designed constructs to this receptor failed E. coli expression. This illustrates that this novel genetic selection method will be useful for discovery of expression constructs for both structural work and monoclonal antibody production for functional studies.
METHODS
Materials
Oligonucleotides were synthesized by Sigma-Genosys (Haverhill, UK). Restriction enzymes and Endonuclease V were from New England Biolabs (Hitchin, UK). The vectors pENTR1A, pDEST17 and Gateway LR clonase were from Invitrogen (Paisley, UK). Plasmid, gel extraction and PCR purification kits were purchased from Qiagen (Crawley, UK). All other chemicals including antibiotics unless stated were from Sigma-Aldrich (Gillingham, UK).
Preparation of uracil containing templates, Endonuclease V digestion and dA tailing of random fragmented DNA libraries
The uracil-containing Fli1 and Pecam1 genes were prepared with PCR mixtures, which contained 10 mM Tris–HCl (pH 8.0), 50 mM KCl, 1.5 mM MgCl2 and 0.2 mM each of dATP, dGTP, dCTP and 0.2 mM of dTTP/dUTP mixture, 0.25 μM of each forward and reverse primers, 10 ng of each template plasmid and 1.25U of Taq polymerase (Sigma-Aldrich) in a final volume of 50 μl. PCR reaction conditions were: 95°C for 2 min, followed by 30 cycles of 94°C for 30 s, 54°C for 30 s, and extension at 72°C for 3.5 min for Fli1 and 5 min for Pecam1 and a final extension at 72°C for 7 min. Amplified DNA was purified using a PCR purification column (Qiagen) and eluted with 50 μl of ultra-pure water. These samples were diluted in 2× Endonuclease V digestion buffer [20 mM HEPES–KOH (pH 7.4), 100 mM NaCl, 1 mM MnCl2]. Two units of Endonuclease V were added to the diluted DNA sample (3 μg) and incubated at 37°C for 12 h followed by 95°C for 10 min. Digested fragments were purified using PCR purification columns and eluted in 50 μl of 10 mM TE (pH 8.0) for subsequent reactions.
Smaller fragments (<100 bp) were removed using microspin 400 columns (GE Healthcare, Little Chalfont, UK) according to the manufacturer's instructions and then concentrated using microcon centrifugal filter devices (Millipore, Watford, UK). One microgram of fragmented DNA was blunt-ended in a reaction containing 50 mM Tris–HCl (pH 8.0), 5 mM MgCl2, 0.1 mg/ml nuclease-free BSA, 0.2 mM dNTP mix, 1 mM DTT and 1.5 units of T4 DNA polymerase (NEB) in a total volume of 25 μl and incubated at 11°C for 20 min followed by 75°C for 10 min. Following the flushing reaction, the entire mixture was used in the dA tailing step. A total of 8.5 μl of 10× dA tailing buffer (100 mM Tris–HCl, pH 9.0, 0.5M KCl, 0.1% gelatin, 1% Triton X-100), 0.5 μl (1.25U) of Tth polymerase (Novagen, Nottingham, UK) was added in a final volume of 85 μl and incubated at 70°C for 15 min. The dA-tailed samples were purified using PCR purification spin columns and eluted in 30 μl of 10 mM TE (pH 8.0), digested with 5U of Dpn1 enzyme at 37°C for 4 h, to remove methylated plasmid DNA and purified by PCR purification spin columns to give a final insert concentrations of between 15 and 20 ng/µl.
Construction of entry and expression vectors
The T-vector was constructed by digestion of pENTR1A with DraI and EcoRV, agarose gel purification and the dT tailing carried out as previously described (5). The pDEST17-MCS was constructed by whole plasmid PCR as described previously (1) using forward primer 5′ CATATGGGTACCTAATGAGTTTGATCCGGCTGCTAACAAAGCCCGAAAGGAAG 3′, reverse primer 5′ ATGCATCACTTCGTGCACCACTTTGTACAAGAAAGCTGAACG 3′ and template pDEST17 to insert a DraIII site downstream of the attR2 sequence. The pRLP101 was constructed by ligation of the 1791 bp fragment, generated by XbaI/DraIII digestion of pDEST17-MCS, with the 6190 bp XbaI/DraIII vector back-bone from pDEST-C102 (1).
Library production and selection
T-tailed entry plasmid (50 ng) was ligated with the dA tailed insert (40 ng) at a 1:3 molar ratio (6), purified using PCR purification spin columns and 1 µl used to electroporate 20 µl of E. coli DH5α-E electrocompetent cells (Invitrogen). Four electroporations were carried out for each fragmented gene library and plated onto LB kanamycin (100 µg/ml) plates. The colonies from plates were scraped, resuspended in 20 ml LB media and a total of 10 OD600 units spun down and miniprep plasmid DNA prepared (Qiagen). Purified DNA was used in LR Gateway recombinations (1) with pRLP101 expression vector. The reactions were purified by PCR spin column, eluted with 30 µl of ultra-pure water and 1 µl of the eluate was used to transform E.coli DH5α-E electrocompetent cells as described earlier. Two electroporations were carried out for each gene library and the average library size was 1 × 106. DNA was purified as described earlier and 1 µl of miniprep DNA was used to transform E. coli Ultra BL21(DE3) cells (Edge BioSystems, Gaithersburg, USA). Transformations were plated onto minimal media agar plates (1× M9 salt, 0.002% glucose, 1 mM MgSO4 and 100 µg/ml ampicillin). Plates used in selection contained 75 µg/ml TMP and 100 µM IPTG in addition to the above described recipe. All the plates were incubated at 30°C for either 18 h (non-selective) or 36 h (selective +TMP + IPTG).
Expression screening
Colonies were picked and inoculated into 500 µl of non-inducing P0.5G minimal media (7) in a 96 deep well block and these were shaken at 800 r.p.m. (3 mm orbital throw) overnight at 37°C. The overnight cultures (1.25 μl) were used to inoculate 1.25 ml of ZYP-5052 auto-induction media (7) in a 96 deep well block, which was grown at 37°C, shaken at 800 r.p.m. for 4 h followed by 20°C for 18 h. Total and soluble protein analysis was essentially as described previously (1) except that solubility screening filter plates (Novagen) were employed.
Ligation-independent cloning and protein expression with 15N labelling
Selected fragments from Fli1 and Pecam1 were PCR amplified with primers containing forward linker 5′ GGCGGTGGTGGCGGCATG 3′ and reverse linker 5′ CAGTTCTTCTCCTTTGCGCCCCTA 3′ to clone into to pLIC.B3 [gift from Rosalind Kim, Lawrence Berkeley National Laboratory, USA (8)]. Ligation independent cloning was carried out as previously described (http://www.strgen.org/protocols/). The resultant constructs with N-terminal hexahistidine tags were expressed at the 2 l scale in P-5052 media (7) for 15N isotope incorporation.
Protein purification, analytical size exclusion chromatography and nuclear magnetic resonance (NMR)
Purification of His-tagged protein fragments and size exclusion chromatography were performed as detailed in the Supplementary Methods section. 1H and 1H-15N HSQC NMR data were collected at 25°C on a Bruker DRX 500 spectrometer equipped with 5 mm triple resonance H/C/N/z-gradient probe on 15N labelled 50 and 100 µM samples in 20 mM Na phosphate buffer (pH 6.0), 300 mM NaCl, 10% D2O. A spin-echo sequence was used to collect 1H spectra and a 15N HSQC with water flip-back and a Watergate sequence was used to collect the 2D spectra. Spectra were processed with Azara software (written by Wayne Boucher, University of Cambridge) and analysed with relevant tools.
Generation of single chain variable fragments (ScFv) and immunocytochemistry
ScFv were selected by antibody phage display panning, expressed and purified as previously described (9–11). Hemangioendothelioma (EOMA) endothelial cell line was grown in chamber slides until 70% confluent and fixed with 1% paraformaldehyde (see Supplementary Methods). The slides were incubated with ScFv at 5 µg/ml concentration overnight, washed, followed by the addition anti-FLAG biotin antibody (2.5 µg/ml) and streptavidin-peroxidase (Molecular Probes, Glasgow, UK). Slides were then incubated in tyramide-AF647 followed by 4′,6-diamidino-2-phenylindole (DAPI) (Molecular Probes). Slides were mounted with Prolong gold antifade mountant (Molecular Probes) and dried overnight before viewing under the microscope. Slides were viewed using Olympus BX51 microscope, with an EXFO X-cite 120 fluorescence illuminator. Images were capture at 20× objective under a plain field lenses and Olympus F-view II cooled black & white camera. Cell B imagine software was used to create composite images.
RESULTS
mDHFR acts as a protein solubility reporter
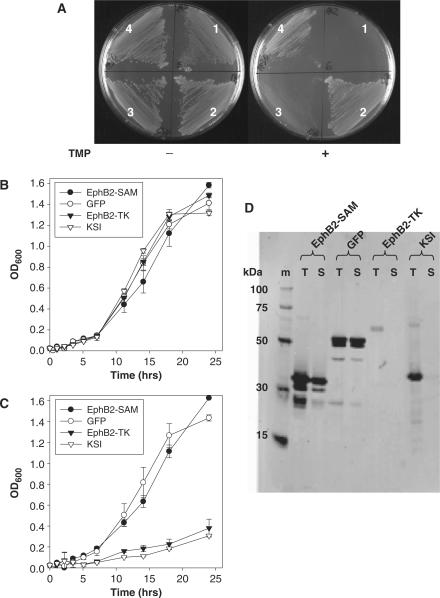
To test if mDHFR could be used as a genetic selection reporter of soluble protein expression, a number of E. coli BL21(DE3) expression clones were streaked onto minimal plates containing ampicillin and IPTG in the presence or absence of TMP. TMP is a potent inhibitor of bacterial DHFR, but does not effectively inhibit murine or human DHFR. Therefore, only bacteria expressing TMP resistant (TMPR) mammalian DHFR will enable growth in the presence of TMP. The strains hosted the expression plasmid pDEST-C102-Dhfr, described previously (1), designed to express cloned open reading frames (ORFs) with a C-terminal mDHFR fusion protein. The clones expressed the tyrosine kinase or SAM domain of the murine EphB2 receptor (EphB2-TK or EphB2-SAM, respectively), the murine transcription factor Fos and Aequorea victoria green fluorescent protein (GFP) mutant (4) engineered for soluble expression in E. coli (12). As shown in Figure 1A, all bacterial strains grew well when streaked onto expression plates in the absence of TMP, but in the presence of TMP only the strains expressing the known soluble proteins EphB2-SAM and GFP (1) grew well (plate segments 2 and 4), whereas the insoluble proteins EphB2-TK and Fos either failed to grow or grew very slowly after a 30 h incubation at 30°C (plate segments 1 and 3, Figure 1). The growth results illustrated in Figure 1A provide evidence that mDHFR can act as a dominant genetic reporter of soluble protein expression. Upstream protein fusions to DHFR that are prone to misfolding and inclusion body formation can perturb the folding and therefore enzymatic activity of DHFR. Additional evidence for the ability of DHFR to act as a solubility reporter came from liquid culture experiments in minimal media containing IPTG and ampicillin, where the growth rates of strains expressing the soluble EphB2-SAM and GFP or insoluble EphB2-TK or ketosteroid isomerase (KSI) were compared in the absence or presence of TMP. As shown in Figure 1B, all expression strains grew equally well in the absence of TMP, but in the presence of TMP (Figure 1C) the strains expressing low or undetectable levels of soluble protein (Figure 1D) displayed markedly reduced growth kinetics.
Figure 1.
Murine dihydrofolate reductase (mDHFR) can act as a protein solubility reporter. (A) Escherichia coli BL21(DE3) expressing mDHFR fused to tyrosine kinase (1) or SAM domain (2) of the murine EphB2 receptor, c-Fos (3) or GFP (4) were streaked onto minimal agar plates containing ampicillin and IPTG in the presence (+) or absence (−) of trimethoprim. (B) and (C) Growth rate studies of E. coli BL21(DE3) expressing various N-termimal fusion proteins to mDHFR in the absence (B) or presence (C) of trimethoprim. (D) SDS–PAGE western blots for total (T) and soluble (S) expression of EphB2-SAM, GFP, EphB2-TK and KSI expressed as mDHFR fusions. Marker lane (m) = His-tag ladder (Qiagen).
Construction of fragmented libraries
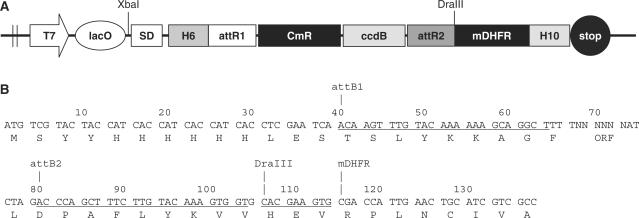
The DHFR reporter vector pRLP101 (Figure 2) is essentially the same as the previously described expression plasmid pDEST-C102-Dhfr (1), except that a Shine–Dalgarno sequence and start codon were inserted downstream of the lac operator (lacO). The presence of the lacO and plasmid encoded lac repressor (lacI) was found to be important in the selection experiments to reduce the effects of protein over-expression toxicity caused by high rates of transcript synthesis (data not shown). GATEWAY vectors were used because they allow the flexibility of shuffling inserts between multiple expression vectors and reporter systems without the need to use PCR amplification or restriction enzyme cloning (13,14).
Figure 2.
Map of the mDHFR reporter vector. (A) Schematic depiction of pRLP101. T7, T7 RNA polymerase promoter; SD, Shine–Dalgarno; lacO, lac operator; attR1, attR2, att recombination sites; CmR, chloramphenicol resistance gene; ccdB, bacterial toxin gene (inhibitor of DNA gyrase); mDHFR, murine dihydrofolate reductase; H6 or H10, hexahistidine or decahistidine; ORF, open reading frame. (B) DNA sequence of the final expression construct (see Materials and methods section). The sequence covers from the start codon to the first eight amino acids of mDHFR. The ORF is cloned at position 68–73 bp.
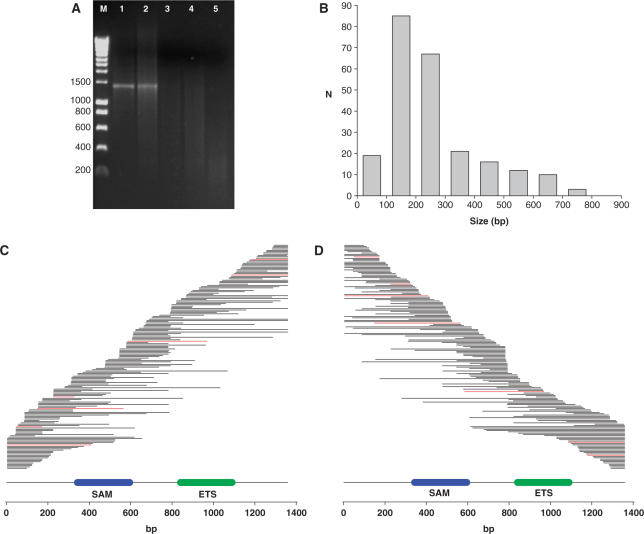
We adapted the method of Miyazaki (4), originally used for random DNA fragment generation prior to DNA shuffling mutagenesis, for creation of a library of gene fragments to be used for protein solubility selections. The proto-oncogene Fli1 (Friend leukemia integration 1 transcription factor, UNIPROT accession P26323) was chosen as the initial target because previous studies showed that even when expressed with the N-terminal solubility enhancing fusions maltose binding-protein (MBP) and thioredoxin (Trx), extensive proteolytic cleavage of the full-length product occurred (1) resulting in very poor yields during attempted purification. The Fli1 ORF was amplified with gene-specific primers in the presence of different ratios of dTTP and dUTP and the product was digested with E. coli Endonuclease V, a repair enzyme that recognizes deoxyinosine and deoxyuridine and cleaves at the second and third phosphodiester bonds 3′ to the mismatch with a 95% efficiency for the second bond and a 5% efficiency for the third bond leaving a nick with 3′-hydroxyl and 5′-phosphate (15). Endonuclease V cleavage in the presence of manganese (II) promotes double strand cleavage (K. Miyazaki, personal communication). As shown in Figure 3A, Endonuclease V does not cleave the Fli1 ORF amplified without dUTP (lane 1), but as the percentage dUTP employed in the PCR increased progressively greater fragmentation occurred (lanes 2–5). The fragments generated by Endonuclease V cleavage of PCR product amplified with 50% and 75% dUTP (lanes 3 and 4) were end repaired with T4 DNA polymerase and 5′ A-tailed with Tth polymerase. The inserts were cloned into the T-tailed DraI/EcoRV cut pENTR1A vector (see Materials and methods section) to give a total library size of 9.8 × 105. To check library diversity 240 clones were picked and DNA sequenced. A size distribution was achieved in the range from 0 to 800 bp (Figure 3B) centred at 100–200 bp. The sequences of the non-redundant Fli1 inserts were aligned to the full-length reference sequence and ordered from their 5′-end (Figure 3C) or 3′-end (Figure 3D), to check library diversity. Inserts in frame at the 5′ and 3′-ends and the correct orientation are coloured red. Steps in the fragment distribution were observed indicating some Endonuclease V cleavage ‘hot-spots’, such as the 3′ 780–792 bp region. This sequence was characterized by being relatively AT rich (67%) and was flanked by two 12 bp GC-rich regions. Diversity could be increased by also preparing a deoxyinosine doped PCR product performing Endonuclease V cleavage and combining this with the library generated from the deoxyuridine incorporated PCR product. This was not performed in this report because the library was considered to have sufficient diversity, with good coverage of the two annotated domains to proceed with selection experiments. Coverage of the entire gene was achieved and no region of the gene appeared to dominate the library.
Figure 3.
Generation of a library of Fli1 fragments. (A) Full-length Fli1 gene was PCR amplified in the presence of different dTTP/dUTP ratios, digested with endonuclease V and analysed by agarose gel (1.6%) electrophoresis. Lane M, DNA standard (HyperLadder1;BIOLINE marker); lane 1, 0.2 mM dTTP/0 mM dUTP; lane 2, 0.15 mM dTTP/0.05 mM dUTP; lane 3, 0.1 mM dTTP/0.1 mM dUTP; lane 4, 0.05 mM dTTP/0.15 mM dUTP; lane 5, 0 mM dTTP/0.2 mM dUTP. (B) Histogram showing the size distribution frequency (N) of 240 randomly picked Fli1 library entry plasmids. (C) Coverage plot of a non-redundant set of 173 sequence confirmed Fli1 entry plasmids arrayed against the 1359 bp Fli1 sequence (lower line) with annotated SAM domain (blue, 343–595 bp) and ETS domains (green, 841–1090 bp) sorted from their 5′ end. Red lines are inserts in frame at the 5′ and 3′ junctions and the correct orientation. (D) as (C) except the clones are ordered from their 3′ end.
Selection of soluble Fli1 fragments
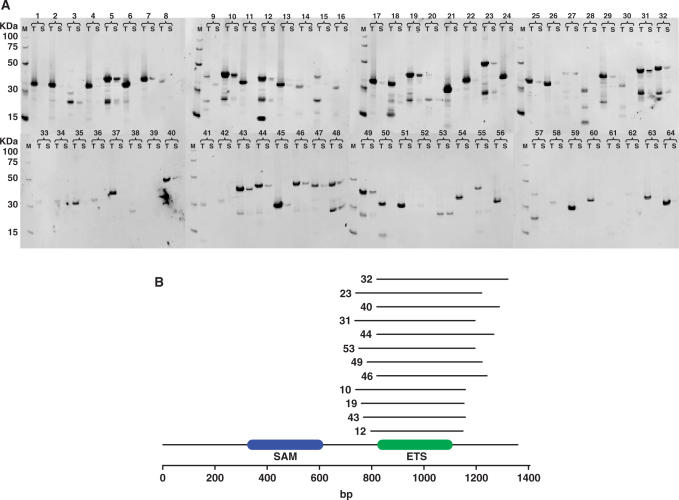
The Fli1 fragment library, cloned into pENTR1A, was recombined with pRLP101 to give a library of expression clones. The library was plated onto minimal agar plates containing ampicillin and IPTG in the presence or absence of TMP. Comparing the selective (+TMP) with the non-selective (−TMP) plates there was an average 66-fold reduction in the number of transformants, indicating that selection was taking place. Growth on the TMP plates was dependent on plasmid induced protein expression because no growth was observed on the TMP plates in the absence of IPTG. A total of 64 TMP resistant transformants were picked into minimal media, expressed using auto-induction medium [see Materials and methods section (7)], analysed for soluble protein expression (16) (Figure 4A) and sequenced (Supplementary Table 1). Thirty percent (19/64) of the TMP-resistant transformants gave soluble expression and of those sequence confirmed 75% (12/16) were cloned in the forward orientation and in frame at both the 5′ and 3′ junctions (Supplementary Table 1). The aligned in frame soluble expression hits (Figure 4B) gave a striking consensus relative to the random nature of the initial starting library (Figure 3C and D) homing on the DNA binding ETS domain. The equivalent human Fli1 ETS domain is the only region of this protein to have been expressed in E. coli and crystallized (17). The solved domain consisted of aa276–373 and it is interesting to note the difference with the annotated Pfam (3) ETS coordinates at aa280–363, being 14 amino acids smaller than the structural domain. In this study, the successful selection illustrates that the method can be used to rapidly identify folded regions of a larger protein that are capable of soluble expression at levels that are sufficient for structural determination.
Figure 4.
Selection of soluble fragments of Fli1. (A) Western blot results for total (T) and soluble (S) expression of all 64 Fli1 expression clones after selection for growth on trimethoprim plates. (B) Schematic diagram for soluble Fli1 hits aligned to the reference sequence with annotated SAM (blue) and ETS (green) SMART (45) domains. The coordinates of the expression clones are contained in Supplementary Table 1. Clone numbers correspond between Figure 4A and B, Supplementary Table 1.
Selection of soluble Pecam1 fragments
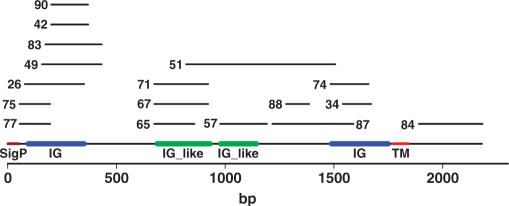
The method was tested on a larger, more complex protein: murine platelet endothelial cell adhesion molecule (Pecam1 or Cd31, Uniprot accession number Q08481). Pecam1 is a type 1 integral membrane cell adhesion molecule expressed on platelets and at endothelial cell intercellular junctions. A Pecam1 fragment library was generated in pENTR1A, as for Fli1, cloned into pRLP101 and 96 TMP-resistant transformants were picked, screened for soluble expression and DNA sequenced (Supplementary Table 2). Twenty-eight percent (27/96) of the TMP-resistant clones gave soluble protein expression and of those DNA sequenced 65% (17/26) were in the correct orientation and in frame at the 5′ and 3′ junctions. The soluble protein expression hits covered several extracellular regions of the protein including all the individual Ig domains and one was located in the intracellular cytoplasmic tail region (#84, Figure 5).
Figure 5.
Schematic diagram for soluble Pecam1 hits aligned to the reference sequence with annotated Ig (blue) and Ig-like (green) SMART (45) domains, signal sequence (brown) and transmembrane domain (red). The coordinates of the expression clones are contained in Supplementary Table 2.
Biophysical characterization of the soluble Fli1 and Pecam1 fragments
To test if the regions of Fli1 and Pecam1 identified from the mDHFR screen were correctly folded in the absence of the mDHFR fusion, selected constructs were sub-cloned into a His tag expression vector, expressed and affinity purified. Fli1 clone #12 was chosen as this was the smallest selected fragment (266–383/452aa) covering the annotated Pfam ETS domain (280–363aa). Fli1 clone #10 (247–386aa) was selected as an example of a larger fragment containing the ETS domain with good levels of soluble expression (Figure 4A). Pecam1 clones #26, #71, #57, #74 and #84 were picked for further characterization because these covered all the individual SMART annotated extracellular Ig and Ig-like domains and the intracellular domain. Expression in 50 ml of auto-induction media (7) and affinity purification was successful for the Fli1 clones and Pecam1 intracellular clone #84, but the selected Pecam1 extracellular domains failed to give sufficient material for biophysical characterization.
Filtration or centrifugation is commonly used to separate the soluble protein fraction from inclusion bodies, but this soluble fraction can contain aggregates which may not be natively folded (18,19). For this reason, size exclusion chromatography (SEC) was used to determine the aggregation state of the His-tagged Fli1 and Pecam1 fragments. Table 1 shows the expected molecular weight, the calculated molecular weight from the SEC calibration curves and the inferred aggregation state. Fli1 constructs 10 and 12 (Table 1) covering the ETS domain with different flanking lengths gave calculated molecular weights consistent with them having a monomeric state, although the shorter fragment 12 gave the best agreement between the experimentally determined and theoretical molecular weights. The Pecam1 fragment most closely approximated to an apparent trimeric state.
Table 1.
Apparent aggregation state of selected Fli1 and Pecam1 protein fragments
| Gene | Construct | Fragment (aa) | Ve (ml) | KAV | MWcalc (kDa) | MWtheor (kDa) | Mwcalc/MWtheor |
|---|---|---|---|---|---|---|---|
| Fli1 | 10 | 247–386 | 1.29 | 0.304 | 24.9 | 17.3 | 1.44 |
| Fli1 | 12 | 266–383 | 1.39 | 0.366 | 15.5 | 14.8 | 1.05 |
| Pecam1 | 44 | 631–727 | 1.19 | 0.242 | 39.9 | 14.3 | 2.79 |
Comparison of calculated molecular weights (MWcalc) from SEC data and theoretical molecular weights (MWtheor) of His-tagged proteins.
Fli1 fragment 12 containing the DNA binding ETS domain and Pecam1 fragment 84 covering the receptor cytoplasmic domain were expressed at a 2 l scale in minimal auto-induction media for 15N labelling of proteins [see Materials and methods section (7)] and purified by affinity, ion-exchange and SEC to give purified yields of 0.5 and 3.0 mg of labelled protein, respectively for NMR studies.
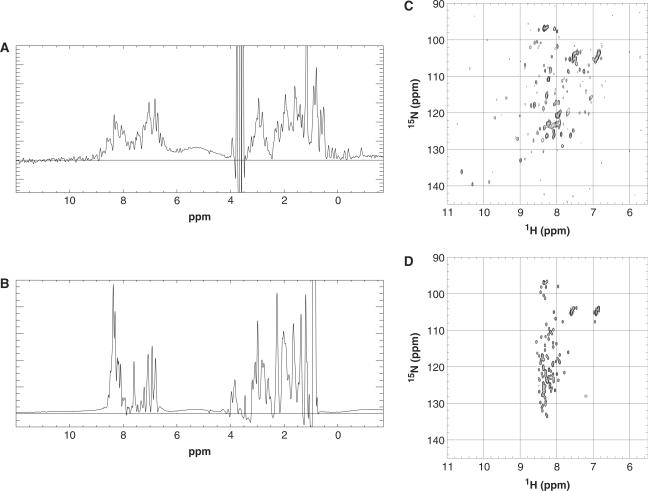
The proteins were first analysed by 1D 1H NMR. Methyl group proton chemical shift signals in the region of 1.0 to −1.0 p.p.m. disperse when a methyl group is placed in the core of the protein and can be indicative of correct protein folding (20,21). The Fli1 fragment clearly shows some well-dispersed peaks in the region of 1 to −1 p.p.m. (Figure 6A), whereas the Pecam1 fragment does not (Figure 6B). 1H-15N HSQC (Heteronuclear Single Quantum Coherence) NMR spectra (21,22) showed well dispersed and sharp resonances for the Fli1 construct (Figure 6C), but this was not apparent for the Pecam1 cytoplasmic domain (Figure 6D). The most intense and sharp signals in the centre of the Fli1 HSQC spectrum belongs to the disordered His-tag. All of the resonances in the spectrum of Pecam1, a domain of similar size, are at least as sharp, indicating a disordered and well soluble protein. Pecam1 does not form larger aggregates, which would markedly widen the resonances. Taken together, the 1D NMR and 2D NMR data are consistent with the monomeric Fli1 12 construct being folded, whereas the Pecam1 84 fragment is disordered rather than trimeric, as indicated by size exclusion. Disordered proteins are known to occupy a larger volume than compact globular proteins under native conditions.
Figure 6.
Biophysical analysis of selected soluble domains. 1D proton NMR spectra of Fli1 10 (A) fragment and Pecam1 84 fragment. (B) 2D 15N-HSQC spectra results of Fli1 10 (C) fragment and Pecam1 84 fragment (D).
Utility of selected fragments in antibody generation
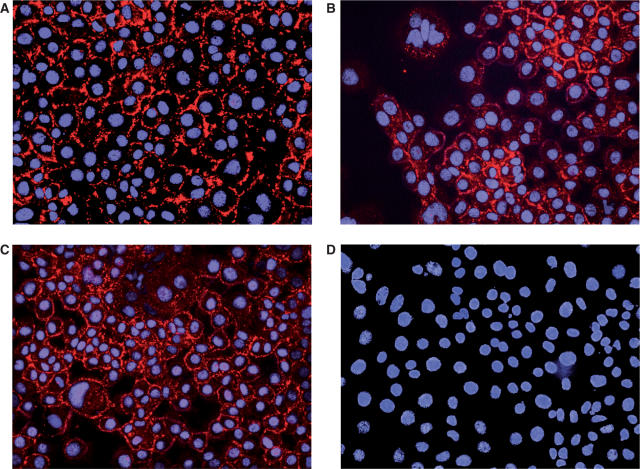
Although the Pecam1 cytoplasmic domain construct #84 was judged to be unfolded by the NMR screens, we were interested to investigate if this soluble, disordered protein could be used to produce specific antibodies capable of recognizing the full-length Pecam1 cell surface receptor in cultured cells. The his-tagged construct was used to generate specific single chain (scFv) antibodies by antibody phage display selection (10). ScFv were sub-cloned into pSANG14-3F, which expresses scFv–alkaline phosphatase fusions (9), and expressed in auto-induction media (7). These were affinity purified and ranked for binding to Pecam1 #84 as described previously (11). From a panel of 96 selected scFv clones, 36 possessed unique DNA sequences (11) and specifically recognized Pecam1 #84 indicating that the selection was successful. The antibody clone and ELISA specificity data are available via the AtlasDB (11) web-site (http://www.sanger.ac.uk/cgi-bin/teams/team86/AtlasDB.pl). Clones ant699_808_B04 and ant699_808_G08 were chosen for immunocytochemistry (ICC) analysis because they gave the highest ELISA signals for binding to the purified His-tagged Pecam1 intracellular #84 protein. A scFv (ant65_d05) specific for the receptor Jagged-1 (11) was used as a positive control as a known marker that is co-expressed on endothelial cells with Pecam1 (23). The anti-Jagged-1 scFv, and anti-Pecam1 scFv clones ant699_808_B04 and ant699_808_G08 all gave a characteristic membrane staining pattern when incubated with fixed mouse EOMA cells (Figure 7A–C, respectively) and detected via the FLAG tag (see Materials and methods section). A scFv specific to Rab9b (ant308_180_G02), which is expressed specifically in brain and spinal cord (http://cgap.nci.nih.gov/SAGE) was included as a negative control and this gave no staining of the EOMA cells (Figure 7D).
Figure 7.
ScFv giving membrane staining to the EOMA endothelial cell-line. Immunocytochemistry results for single chain Fv (in red) raised against Jagged-1 (A), Pecam1 #84 fragment, clone ant699_808_B04 (B) or clone ant699_808_G08 (C) and Rab9B. (D) Nuclei were stained with DAPI (blue) as described in Materials and methods section.
DISCUSSION
This work combines an efficient method of DNA fragmentation with a genetic screen, employing a C-terminal mDHFR fusion, for the identification of protein constructs capable of soluble expression in E. coli. Production of a pool of random DNA fragments by Endonuclease V has some advantages over the alternate methods (24). The physical methods of sonication (14) or shearing (25) produce the most random libraries due to their lack of dependence on sequence, but DNA damage can occur, perhaps due to free radical generation during cavitation (26). DNAse (27) or exonuclease digestion (28–30) requires that the reaction conditions must be determined empirically on a gene by gene basis. Also, the exonuclease methods may not allow sampling of all the available internal sequence space when making successive N- or C-terminal deletions. Random PCR (31,32) can result in biases in the preferential amplification of GC-rich regions resulting in a non-random library. PCR with a dTTP/dUTP mix (33) followed by first uracil-DNA glycosylase addition and next cleavage at the resulting abasic sites with Endonuclease IV has the advantage that no enzyme titrations or time courses are required. Cleavage is driven to completion with the insert size library being solely determined by the ratio of dTTP/dUTP ratio used in the original insert PCR. The fragmentation method described in this report is similar to the method of Reich et al. (33), but in this method cleavage is effected in a single enzyme step.
We describe mDHFR as a new genetic reporter of soluble expression and provided evidence to support this with some known soluble and insoluble proteins. The principle of this technique is that only soluble N-terminal fusions allow the correct folding of the downstream reporter protein, whereas misfolded N-terminal fusions perturb the folding and therefore activity of mDHFR. Alternate reporters of soluble protein expression can be separated into those that lead to a colour (34) or fluorescence (12,35,36) read-out allowing one to pick transformants displaying the correct phenotype, and those that act as dominant genetic markers (37–39) where, in the presence of an appropriate antibiotic, a growth advantage is conferred on clones that express soluble protein. A non-fusion reporter system has been used where β-galactosidase was cloned downstream of the chromosomal ibpAB promoter, which is upregulated in response to an accumulation of unfolded protein (40). However, larger library sizes can be screened more rapidly with the dominant genetic markers (41). This is particularly useful for random insert cloning where there is a 1 in 18 chance of an insert being in the correct orientation and in frame at both the 5′ and 3′ junctions meaning that larger libraries must be sampled to screen sufficient correctly cloned inserts for soluble expression. Chloramphenicol acetyl transferase has been used as a genetic marker (38), but this is active as a trimeric protein and may lead to higher order aggregate protein fusions. Selection based on protein secretion to the bacterial periplasm and fusion to β-lactamase (BlaC) is a very promising system (37), although one needs to be careful to eliminate false positive clones that occur through passive BlaC leakage from cells and secretion is limited to proteins of less than 90 kDa in molecular mass. Recently, over-expression of E. coli DHFR has been used as a reporter of soluble protein expression, but this is a highly soluble protein and may increase the solubility of the upstream fusion, although the authors were successful selecting for mutants of an acetyltransferase (39) with improved properties. The advantage of mDHFR as a genetic reporter is that it is monomeric and has been shown previously (1) not to significantly perturb the folding of N-terminal fusion proteins. Also, DHFR is not a common plasmid marker in E. coli unlike β-lactamase, chloramphenicol acetyl transferase or kanamycin resistance making it compatible with a wider range of bacterial expression systems.
The method was validated using the transcription factor Fli1, which cannot be expressed as a full length protein in E. coli (1). From a pool of random DNA fragments, mDHFR selection successfully identified soluble expression clones of the ETS domain of murine Fli1, judged to be folded by 1D and 2D NMR. Indeed, this was the only region of the human orthologue to have its structure solved (17). Some false positive TMPR clones were identified which did not express soluble protein as judged by a filter plate screen (16). The reason for this could be that initial expression of these constructs as DHFR fusions resulted in soluble expression allowing growth on TMP plates, but as the levels of over-expressed proteins increased in the cell during induction, this gave rise to inclusion body formation. Alternatively, low levels of proteolysis between the fusion protein and mDHFR may release sufficient soluble and active mDHFR to allow growth on TMP plates despite the misfolding and aggregation of the full-length fusion protein. Also, some reverse orientation insertions were observed and here it is likely that internal initiation from an upstream in frame start codon occurred. It might be possible to reduce the false positive rate by mutating mDHFR to make it more sensitive to misfolding of an N-terminal fusion partner. However, despite the occurrence of these false positive TMPR clones, sufficient enrichment of soluble expression clones occurred to be useful in subsequent studies.
A comparison between the Fli1 and Pecam1 selection results indicates that the success rate of this selection method is likely to be target specific and might favour proteins normally expressed in the nucleus or cytoplasm compared with extracellular proteins. Several soluble expression fragments of the more complex target platelet endothelial cell adhesion molecule (Pecam1) were identified. When expressed as His-tagged proteins, only the cytoplasmic domain construct was capable of high levels of soluble expression. This may reflect the difficulty of expressing ectodomains of mammalian cell surface receptors, which are naturally glycosylated and contain disulphide bonds in vivo and may require a mammalian expression system (42) or expression with specific fusion partners and E. coli strains (43) with co-expression of chaperones for optimal expression. The possibility of false negative clones cannot be ruled out for slow-folding fusions. If a fragment is still in the process of folding when the mDHFR polypeptide emerges from the translating ribosome, this could interfere with the folding of the reporter. The observation that the Pecam1 extracellular fragments appeared soluble in the filter plate screen (16) but failed attempted purification could be that the filter plate screen fails to differentiate between monomeric or dimeric protein and higher order aggregates, which appear an soluble inclusion bodies (18). Alternatively, the proteins were initially expressed correctly folded, but they aggregated during purification, perhaps due to their lack of hydrophilic glycosylation. The Pecam1 selection illustrates that rigorous validation of TMPR expression clones should be performed including solubility screening and purification. The Pecam1 extracellular selected fragments could be useful for suggesting constructs to be tested in alternate prokaryotic (43) or eukaryotic expression systems assuming that they were initially expressed solubly, but aggregated over time due to not being in the correct environment.
It was previously found that some mammalian proteins expressed in E. coli as MBP fusions failed to work in phage—antibody display selections (11). They may have been prone to soluble inclusion body formation, which could have sterically hindered the antigen's accessibility for single chain antibody selection. The selected Pecam1 fragment was shown not to be a soluble higher order aggregate, but most likely as a disordered monomeric species, as judged from the NMR data, and did not require to be expressed as a MBP fusion for high level soluble expression. The selected cytoplasmic domain of Pecam1 is likely to be an example of a protein existing in a natively unfolded state (44) because this region is phosphorylated and during receptor signalling recruits the protein-tyrosine phosphatases SHP-1 and SHP-2 (39) and therefore requires flexibility for function. The selected Pecam1 cytoplasmic domain consisted of 631–727aa, whereas a rationally designed construct to express the cytoplasmic domain of the receptor, consisting of 620–727aa could only be expressed as a MBP fusion and this failed antibody phage display selection [see Schofield et al. (11), ant238]; presumably, because it gave rise to soluble inclusion bodies. This illustrates the power of screening versus design where a difference of a few amino acids can make the difference between success and failure.
CONCLUSIONS
We provide a method combining random DNA fragmentation by Endonuclease V together with genetic selection using mDHFR for the identification of protein fragments capable of soluble expression in E. coli. This is the first study to combine random fragmentation with genetic selection to identify soluble protein expression constructs. The methods will be particularly useful in cases where the attempted expression of the full-length protein has failed or where there is insufficient domain annotation for rational design.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We would like to thank Dr William Howat (Cancer Research Institute, Cambridge, UK) for help with immunocytochemistry. This work was funded by the Wellcome Trust. Funding to pay the Open Access publication charges for this article was provided by the Wellcome Trust.
Conflict of interest statement. None declared.
REFERENCES
- 1.Dyson MR, Shadbolt SP, Vincent K, Perera R, McCafferty J. Production of soluble mammalian proteins in Escherichia coli: identification of protein features that correlate with successful expression. BMC Biotechnol. 2004;4:32. doi: 10.1186/1472-6750-4-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Esposito D, Chatterjee DK. Enhancement of soluble protein expression through the use of fusion tags. Curr. Opin. Biotech. 2006;17:353–358. doi: 10.1016/j.copbio.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 3.Finn RD, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, Durbin R, et al. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miyazaki K. Random DNA fragmentation with endonuclease V: application to DNA shuffling. Nucleic Acids Res. 2002;30 doi: 10.1093/nar/gnf139. e139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marchuk D, Drumm M, Saulino A, Collins FS. Construction of T-vectors, a rapid and general system for direct cloning of unmodified PCR products. Nucleic Acids Res. 1991;19 doi: 10.1093/nar/19.5.1154. 1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sambrook J, Russell D. Molecular Cloning: A Laboratory Manual. 3rd. Cold Spring Harbor Laboratory Press, Woodbury, USA; 2001. [Google Scholar]
- 7.Studier FW. Protein production by auto-induction in high-density shaking cultures. Protein Expres. Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 8.Kim R. Protein expression in Escherichia coli, Chapter 2. In: Dyson MR, Durocher Y, editors. Expression Systems. Bloxham, Oxfordshire: Scion Publishing Ltd.; 2007. pp. 13–28. [Google Scholar]
- 9.Martin C, Rojas G, Mitchell J, Vincent K, Wu J, McCafferty J, Schofield D. A simple vector system to improve performance and utilisation of recombinant antibodies. BMC Biotechnol. 2006;6:46. doi: 10.1186/1472-6750-6-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCafferty J, Griffiths AD, Winter G, Chiswell DJ. Phage antibodies: filamentous phage displaying antibody variable domains. Nature. 1990;348:552–554. doi: 10.1038/348552a0. [DOI] [PubMed] [Google Scholar]
- 11.Schofield DJ, Pope AR, Clementel V, Buckell J, Chapple SD, Clarke KF, Conquer JS, Crofts AM, Crowther SR, Dyson MR, et al. Application of phage display to high throughput antibody generation and characterisation. Genome Biol. 2007;8:R254. doi: 10.1186/gb-2007-8-11-r254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Waldo GS, Standish BM, Berendzen J, Terwilliger TC. Rapid protein-folding assay using green fluorescent protein. Nat. Biotechnol. 1999;17:691–695. doi: 10.1038/10904. [DOI] [PubMed] [Google Scholar]
- 13.Hartley JL, Temple GF, Brasch MA. DNA cloning using in vitro site-specific recombination. Genome Res. 2000;10:1788–1795. doi: 10.1101/gr.143000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakayama M, Ohara O. A system using convertible vectors for screening soluble recombinant proteins produced in Escherichia coli from randomly fragmented cDNAs. Biochem. Bioph. Res. Commun. 2003;312:825–830. doi: 10.1016/j.bbrc.2003.10.193. [DOI] [PubMed] [Google Scholar]
- 15.Yao M, Kow YW. Further characterization of Escherichia coli endonuclease V. Mechanism of recognition for deoxyinosine, deoxyuridine and base mismatches in DNA. J. Biol. Chem. 1997;272:30774–30779. doi: 10.1074/jbc.272.49.30774. [DOI] [PubMed] [Google Scholar]
- 16.Knaust RKC, Nordlund P. Screening for soluble expression of recombinant proteins in a 96-well format. Anal. Biochem. 2001;297:79–85. doi: 10.1006/abio.2001.5331. [DOI] [PubMed] [Google Scholar]
- 17.Liang H, Mao X, Olejniczak ET, Nettesheim DG, Yu L, Meadows RP, Thompson CB, Fesik SW. Solution structure of the ets domain of Fli-1 when bound to DNA. Nat. Struct. Biol. 1994;1:871–875. doi: 10.1038/nsb1294-871. [DOI] [PubMed] [Google Scholar]
- 18.Schrodel A, de Marco A. Characterization of the aggregates formed during recombinant protein expression in bacteria. BMC Biochem. 2005;6:10. doi: 10.1186/1471-2091-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zanier K, Nomine Y, Charbonnier S, Ruhlmann C, Schultz P, Schweizer J, Trave G. Formation of well-defined soluble aggregates upon fusion to MBP is a generic property of E6 proteins from various human papillomavirus species. Protein Expres. Purif. 2007;51:59–70. doi: 10.1016/j.pep.2006.07.029. [DOI] [PubMed] [Google Scholar]
- 20.Rehm T, Huber R, Holak TA. Application of NMR in structural proteomics: screening for proteins amenable to structural analysis. Structure. 2002;10:1613–1618. doi: 10.1016/s0969-2126(02)00894-8. [DOI] [PubMed] [Google Scholar]
- 21.Scheich C, Leitner D, Sievert V, Leidert M, Schlegel B, Simon B, Letunic I, Bussow K, Diehl A. Fast identification of folded human protein domains expressed in E. coli suitable for structural analysis. BMC Struct. Biol. 2004;4:4. doi: 10.1186/1472-6807-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woestenenk EA, Hammarstrom M, Hard T, Berglund H. Screening methods to determine biophysical properties of proteins in structural genomics. Anal. Biochem. 2003;318:71–79. doi: 10.1016/s0003-2697(03)00162-3. [DOI] [PubMed] [Google Scholar]
- 23.Villa N, Walker L, Lindsell CE, Gasson J, Iruela-Arispe ML, Weinmaster G. Vascular expression of Notch pathway receptors and ligands is restricted to arterial vessels. Mech. Develop. 2001;108:161–164. doi: 10.1016/s0925-4773(01)00469-5. [DOI] [PubMed] [Google Scholar]
- 24.Prodromou C, Savva R, Driscoll PC. DNA fragmentation-based combinatorial approaches to soluble protein expression: Part I. Generating DNA fragment libraries. Drug Discov. Today. 2007;12:931–938. doi: 10.1016/j.drudis.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 25.Christ D, Winter G. Identification of protein domains by shotgun proteolysis. J. Mol. Biol. 2006;358:364–371. doi: 10.1016/j.jmb.2006.01.057. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook J, Russell DW. Fragmentation of DNA by sonication. Cold Spring Harbor Protocols. 2006 doi: 10.1101/pdb.prot4538. 2006, pdb.prot4538. [DOI] [PubMed] [Google Scholar]
- 27.Cochrane D, Webster C, Masih G, McCafferty J. Identification of natural ligands for SH2 domains from a phage display cDNA library. J. Mol. Biol. 2000;297:89–97. doi: 10.1006/jmbi.2000.3561. [DOI] [PubMed] [Google Scholar]
- 28.King DA, Hall BE, Iwamoto MA, Win KZ, Chang JF, Ellenberger T. Domain structure and protein interactions of the silent information regulator Sir3 revealed by screening a nested deletion library of protein fragments. J. Biol. Chem. 2006;281:20107–20119. doi: 10.1074/jbc.M512588200. [DOI] [PubMed] [Google Scholar]
- 29.Tarendeau F, Boudet J, Guilligay D, Mas PJ, Bougault CM, Boulo S, Baudin F, Ruigrok RWH, Daigle N, Ellenberg J, et al. Structure and nuclear import function of the C-terminal domain of influenza virus polymerase PB2 subunit. Nat. Struct. Mol. Biol. 2007;14:229–233. doi: 10.1038/nsmb1212. [DOI] [PubMed] [Google Scholar]
- 30.Cornvik T, Dahlroth SL, Magnusdottir A, Flodin S, Engvall B, Lieu V, Ekberg M, Nordlund P. An efficient and generic strategy for producing soluble human proteins and domains in E. coli by screening construct libraries. Proteins. 2006;65:266–273. doi: 10.1002/prot.21090. [DOI] [PubMed] [Google Scholar]
- 31.Jacobs SA, Podell ER, Wuttke DS, Cech TR. Soluble domains of telomerase reverse transcriptase identified by high-throughput screening. Protein Sci. 2005;14:2051–2058. doi: 10.1110/ps.051532105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kawasaki M, Inagaki F. Random PCR-based screening for soluble domains using green fluorescent protein. Biochem. Biophys. Res. Commun. 2001;280:842–844. doi: 10.1006/bbrc.2000.4229. [DOI] [PubMed] [Google Scholar]
- 33.Reich S, Puckey LH, Cheetham CL, Harris R, Ali AAE, Bhattacharyya U, Maclagan K, Powell KA, Prodromou C, Pearl LH, et al. Combinatorial domain hunting: an effective approach for the identification of soluble protein domains adaptable to high-throughput applications. Protein Sci. 2006;15:2356–2365. doi: 10.1110/ps.062082606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wigley WC, Stidham RD, Smith NM, Hunt JF, Thomas PJ. Protein solubility and folding monitored in vivo by structural complementation of a genetic marker protein. Nat. Biotechnol. 2001;19:131–136. doi: 10.1038/84389. [DOI] [PubMed] [Google Scholar]
- 35.Hedhammar M, Stenvall M, Lonneborg R, Nord O, Sjolin O, Brismar H, Uhlen M, Ottosson J, Hober S. A novel flow cytometry-based method for analysis of expression levels in Escherichia coli, giving information about precipitated and soluble protein. J. Biotechnol. 2005;119:133–146. doi: 10.1016/j.jbiotec.2005.03.024. [DOI] [PubMed] [Google Scholar]
- 36.Cabantous S, Terwilliger TC, Waldo GS. Protein tagging and detection with engineered self-assembling fragments of green fluorescent protein. Nat. Biotech. 2005;23:102–107. doi: 10.1038/nbt1044. [DOI] [PubMed] [Google Scholar]
- 37.Fisher AC, Kim W, Delisa MP. Genetic selection for protein solubility enabled by the folding quality control feature of the twin-arginine translocation pathway. Protein Sci. 2006;15:449–458. doi: 10.1110/ps.051902606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maxwell KL, Mittermaier AK, Forman-Kay JD, Davidson AR. A simple in vivo assay for increased protein solubility. Protein Sci. 1999;8:1908–1911. doi: 10.1110/ps.8.9.1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu J-W, Boucher Y, Stokes HW, Ollis DL. Improving protein solubility: the use of the Escherichia coli dihydrofolate reductase gene as a fusion reporter. Protein Expres. Purif. 2006;47:258–263. doi: 10.1016/j.pep.2005.11.019. [DOI] [PubMed] [Google Scholar]
- 40.Lesley SA, Graziano J, Cho CY, Knuth MW, Klock HE. Gene expression response to misfolded protein as a screen for soluble recombinant protein. Protein Eng. 2002;15:153–160. doi: 10.1093/protein/15.2.153. [DOI] [PubMed] [Google Scholar]
- 41.Hart DJ, Tarendeau F. Combinatorial library approaches for improving soluble protein expression in Escherichia coli. Acta Crystallogr. D Biol. Crystallogr. 2006;62:19–26. doi: 10.1107/S0907444905036097. [DOI] [PubMed] [Google Scholar]
- 42.Chapple S, Crofts A, Shadbolt SP, McCafferty J, Dyson MR. Multiplexed expression and screening for recombinant protein production in mammalian cells. BMC Biotechnol. 2006;6:49. doi: 10.1186/1472-6750-6-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bessette PH, Aslund F, Beckwith J, Georgiou G. Efficient folding of proteins with multiple disulfide bonds in the Escherichia coli cytoplasm. Proc. Natl Acad. Sci. 1999;96:13703–13708. doi: 10.1073/pnas.96.24.13703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fink AL. Natively unfolded proteins. Curr. Opin. Struct. Biol. 2005;15:35–41. doi: 10.1016/j.sbi.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 45.Letunic I, Copley RR, Schmidt S, Ciccarelli FD, Doerks T, Schultz J, Ponting CP, Bork P. SMART 4.0: towards genomic data integration. Nucleic. Acids Res. 2004;32:D142–D144. doi: 10.1093/nar/gkh088. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.