Abstract
S100A2 is generally found expressed in the epidermis and was recently shown to play a crucial role in the differentiation of keratinocytes. Also known as CaN19, S100A2 was identified as a potential tumor suppressor. Expression of S100A2 is upregulated by p53. The proteins p63 and p73 are related to p53 and are expressed as several splice variants with partially overlapping tasks but also functions different from p53. It had been shown that p63 proteins with mutations in their DNA-binding domain cause severe phenotypes in man as autosomal dominantly inherited disease including EEC, AEC, SHFM, LMS and ADULT syndromes. Here we show that S100A2 is a transcriptional target of p63/p73 family members, particularly the p63 splice variant TAp63γ. The regulation is mediated by a novel transcriptional element in the S100A2 promoter which is bound by TAp63γ but not by p53. Mutant p63 proteins derived from EEC and ADULT syndrome patients cannot activate S100A2 transcription whereas SHFM-related mutants still can stimulate the S100A2 promoter. Consistent with a function in tumor suppression S100A2 expression is stimulated upon DNA damage. After doxorubicin treatment p63γ proteins are recruited to the S100A2 promoter in vivo. This may indicate a function of the p63-dependent S100A2 regulation in tumor suppression.
INTRODUCTION
S100A2 was first isolated from bovine lung tissue (1) and is generally found expressed in the epidermis (2). Recently, S100A2 was shown to play a crucial role during differentiation when its expression silenced by siRNA-mediated mRNA knockdown resulted in decreased expression of two keratinocyte differentiation markers (3). In contrast to other S100 family members, S100A2 is located in the cell nucleus (4,5) and is involved in keratinocyte response to oxidative stress (6).
S100A2 is a member of the S100 family representing the largest family within the EF-hand proteins. These proteins are characterized by two distinct EF-hand motifs (7) flanking a central hinge region and act as Ca2+ signaling or Ca2+ buffering proteins. In addition to Ca2+ many S100 family members display high affinity also towards Zn2+ and Cu2+ ions. The cDNA of S100A2 codes for a 10.7-kDa protein which can form homodimers in living cells (8) with affinity to Ca2+ and Zn2+ ions (9). Dimerization of S100 proteins appears to be important for their biological function.
Members of the S100 family show a large diversity in structure and function. They are involved in the regulation of contraction, motility, cell growth, differentiation, cell cycle progression, transcription and secretion. In contrast to other EF-hand proteins, S100 proteins have so far been found only in vertebrates and consequently form a phylogenetically young group (10). Genes of most group members are clustered in region 1q21 of human chromosome 1 which is known as epidermal differentiation complex EDC (11). An analogous cluster is found on chromosome 3 in mice. Usually the highly conserved gene structure consists of three exons of which the first exon is noncoding. Furthermore, S100 proteins are expressed in a cell and tissue-specific manner (12) implying that the relatively large number of family members is not due to redundancy (13).
Interestingly, S100A2—then named CaN19—was identified as a potential tumor suppressor gene by differential expression in normal versus tumor-derived human mammary epithelial cells (14). It was shown that S100A2 expression is markedly downregulated in several tumor tissues (15–18). Methylation of the promoter mediates S100A2 repression during breast cancer progression (19). Furthermore, it was shown that the tumor suppressor p53 activates transcription of S100A2 (20). In addition, S100A2 overexpression was found in gastric cancer (21), ovarian cancer (22), lymphoma (23), head and neck squamous cell carcinoma (24,25) and early-stage nonsmall cell lung cancer (26). It was concluded that overexpression of S100A2 may be an early tumorigenic event (21). Another finding is that S100A2 modulates transcriptional activity of p53 due to protein-protein interaction (27). Recently it was shown that S100A2 could exert its antitumor activity by repression of cyclooxygenase-2 expression (28).
Many biological processes like apoptosis and development are critically regulated by members of the p53 family (29). p63 and p73 are a group of proteins that, like p53, are transcription factors which activate target genes through sequence-specific DNA binding (30–33). The high level of amino acid sequence similarity within the p53 family, particularly in the DNA-binding domain, allows transactivation of common target genes. However, members of the p53 family are not entirely functionally redundant. While p53 is known as a classical tumor suppressor, p63 seems to be capable of enforcing some of the tumor suppressive mechanisms that p53 also mediates (34). Furthermore, p63 plays an important role in development. Mutations in the p63 DNA-binding domain cause severe phenotypes in man as autosomal dominantly inherited syndromes. This family of disorders includes the EEC, AEC, SHFM, LMS and ADULT syndromes. They are characterized by combination of ectrodactyly, ectodermal dysplasia and facial clefting (35–41). Recently, it was shown that p63 is able to bind DNA elements which differ from classical p53 consensus (42,43). This implies that p53 and p63 can regulate different target genes.
Here we identify S100A2 as a novel transcriptional target of the p63 isoform TAp63γ. We show that mutants of p63 which are responsible for the human EEC syndrome fail to activate S100A2 transcription. In addition, we find that recruitment of p63γ proteins to the S100A2 promoter correlates with increased expression of S100A2 following DNA damage.
MATERIALS AND METHODS
Cell culture, transfections and luciferase assays
SaOS-2 cells obtained from DSMZ (Braunschweig, Germany) were cultured and transfected in 24-well plates as described previously (44). DLD-1 cells stably transfected with various p53 family-expressing plasmids were cultured as described previously (44). DLD-1 cells express only inactive mutant p53 protein (45) and display a single nucleotide exchange in one p63 allele resulting in a replacement of proline at position 279 in the TA*p63 protein by histidine (46). The p73 gene in DLD-1 cells is wild type (47).
Transient transfections using expression plasmids for wild-type and mutant p53 family members (25 ng) were carried out according to the manufacturer's instructions using Fugene 6 (Roche, Mannheim, Germany) with 400 ng of the plasmid carrying the human S100A2 promoter and 25 ng pRL-null vector (Promega, Mannheim, Germany) per assay. The total amount of transfected DNA was held constant. Luciferase assays were carried out as reported earlier (48).
HepG2 and p53-negative Hep3B cells (49) were also obtained from DSMZ and cultured as described (46). As a DNA-damaging agent, doxorubicin was employed at 0.2 ng/ml.
RNA extraction and real-time RT–PCR
Extraction of total RNA was performed employing the RNeasy Protect Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Cells carrying p53 family transgenes used for RNA preparations were described earlier (44). Real-time RT–PCR mRNA quantification including calculations have been described (50–52). Specific primers for human S100A2 5′-GCG ACA AGT TCA AGC TGA GT-3′; 5′-CAC CTG CTG GTC ACT GTT CT-3′ (GenBank accession number NM_005978) were used at 1 µM on 50 ng total RNA template in the QuantiTect SYBR Green RT–PCR mix (Qiagen, Hilden, Germany) employing a LightCycler instrument (Roche).
Generation of polyclonal antibodies raised against human S100A2
Full-length cDNA of S100A2 prepared from human blood RNA where amplified using the primer 5′-AAA CCA TGG GCA GTT CTC TGG AGC AG 3′ and 5′-TTT CTC GAG GGG TCG GTC TGG GCA GC-3′. The fragment was cloned into the NcoI/XhoI site of the pTriEx-2 expression vector (Novagen, Madison, WI, USA). The plasmid was used to transform Escherichia coli BL21 cells. Cells were grown to a density of OD600 nm of 0.5 and expression of recombinant protein was induced by adding 1 mM isopropyl thio-β-d-galactopyranoside (IPTG) to the culture for 4 h. Cells were harvested by centrifugation and cell lysates were used for Ni–His-affinity chromatography purification. The eluate was further purified using preparative SDS–PAGE. Bands carrying S100A2 were cut out from the gel, ground in a mortar in liquid nitrogen and suspended in PBS. For immunization the antigen suspension was mixed with an equal volume of the adjuvant TiterMax® Gold (CytRx Corporation, Los Angeles, CA, USA) and injected subcutaneously into the neck area of a rabbit. In intervals of 4 weeks antibody quality was tested by using decreasing amounts of recombinant protein in western blot analyses. Antiserum obtained after five immunizations was used for western blot analysis.
Western blot analysis
Western blots were prepared essentially as previously described (50). The polyclonal rabbit anti-human S100A2 serum was used in a 1 : 100 dilution. The blot was stripped and reprobed with a 1 : 5000 dilution of the mouse monoclonal anti-β-actin antibody (clone AC-15, Sigma, Taufkirchen, Germany). The induction of p53 was detected with the monoclonal mouse antibody DO-1 (Calbiochem, Darmstadt, Germany; 1 : 5000 dilution), whereas the induction of TAp63γ was detected using the mouse monoclonal anti-p63 antibody (clone 4A4; sc-8431 Santa Cruz Biotechnology, Santa Cruz, CA, USA) with a 1 : 250 dilution.
Cloning and mutation of human S100A2 promoter and p63 expression constructs
The wild-type human S100A2-promoter firefly-luciferase construct S100A2 short (20) was kindly provided by Dr Beat W. Schäfer (University of Zurich, Switzerland). However, most experiments were performed with a newly created longer promoter construct. To this end, the first intron of S100A2 was amplified using the primers 5′-TTT GGT ACC GCC CCA GGT TGC TTC TCT C-3′ and 5′-TTT AGA TCT TGG ATC TGT GGC TGC AGA G-3′. This fragment was cloned into the KpnI/BgIII sites of pGI3 basic vector (Promega, Mannheim, Germany). In order to obtain the S100A2 long luciferase construct the intron 1 fragment was cut out by SfiI/BgIII and inserted into the S100A2 short promoter in pGI3 that had been linearized by SfiI/HindIII digestion (GenBank accession number EU036993). Promoter mutants were created by PCR-based targeted mutagenesis on the basis of S100A2 long employing the primers site1-5′mut-fwd, 5′-GGA TAG AGG GTG CAG GCA TGT GTG GGT CGA TTC TGA AC-3′; site1-3′mut-fwd, 5′-TAG AGG GCA TGG GTC GAT GTG GGT CGA TTC TGA AC-3′; site2-mut1-fwd, 5′-GGA TT GGA TTG AGG TGG ATT TGG TTT CC-3′; site2-mut2-fwd, 5′-GGA TCA TGT TGA GGC ATG TTT GGT TTC C-3′; and the respective reverse primers.
TAp63γ mutants were created by PCR-based targeted mutagenesis using the QuikChange (Stratagene, La Jolla, CA, USA) according to the manufacturer's protocol on the basis of TAp63γ-pcDNA3.1/HisC (44) employing the primers TAp63γ-K193E-fwd, 5′-GCC ATG CCT GTC TAC GAA AAA GCT GAG CAC GTC AC-3′; TAp63γ-K194E-fwd, 5′-CAT GCC TGT CTA CAA AGA AGC TGA GCA CGT CAC GG-3′; TAp63γ-R204W-fwd 5′-GGA GGT GGT GAA GTG GTG CCC CAA CCA TG-3′; TAp63γ-R279H-fwd, 5′-GTT GTG TTG GAG GGA TGA ACC ACC GTC CAA TTT TAA TCA TTG-3′; TAp63γ-R298Q-fwd, 5′-CAA GTC CTG GGC CAA CGC TGC TTT GAG GC-3′; and the respective reverse primers. Identity of constructs was confirmed by DNA sequencing. TAp63γ-R304H was published earlier (44).
Electrophoretic mobility shift assay (EMSA)
EMSAs were carried out as previously described (53). TAp63γ was supershifted by using the goat polyclonal anti-p63γ antibody (C-18, sc-8370, Santa Cruz Biotechnology) whereas p53 was shifted by adding a monoclonal mouse anti-p53 antibody (pAb421, SA-293, Biomol, Hamburg, Germany). Probes were generated by annealing the following oligonucleotides with respective reverse oligonucleotides: S100A2 site2, fwd 5′-GGG TGG GAT CAG GTT GAG GCA GGT TTG GTT TCC TT-3′; S100A2 site2 mut-1 fwd, 5′-GGG TGG GAT TGG ATT GAG GTG GAT TTG GTT TCC TT-3′; S100A2 site2 mut-2 fwd, 5′-GGG TGG GAT CAT GTT GAG GCA TGT TTG GTT TCC TT-3′; p21 fwd, 5′-GGC CAT CAG GAA CAT GTC CCA ACA TGT TGA GCT CT-3′; mdm2 fwd, 5′-GGG CGG CCG CTG GTC AAG TTG GGA CAC GTC CGG-3′.
Chromatin immunoprecipitation (ChIP) assays
ChIPs were carried out guided by a published procedure (54) using HepG2 cells (49) or DLD-1 colorectal adenocarcinoma cells with p53 or TAp63γ as tet-off-regulated transgenes (44). Protein crosslinks were precipitated using 5 µg of a goat polyclonal anti-p63γ (C-18, sc-8370, Santa Cruz Biotechnology) or monoclonal anti-p53 antibody (DO-1, Calbiochem, Darmstadt, Germany). Samples were analyzed as described earlier (52) employing the primers IP-S100A2-site1-fwd, 5′-CAG GAC AGA ACA GGT AGA CAC TGA A-3′; IP-S100A2-site1-rev, 5′-CCT GCT GCT GCG TGT CC-3′; IP-S100A2-site2-fwd, 5′-GGT CCA GGA TGC CCA GTC-3′; and IP-S100A2-site2-rev, 5′-GAA GGA GAG CAA GGC AGC-3′.
RESULTS
Induction of S100A2 expression by members of the p53 family
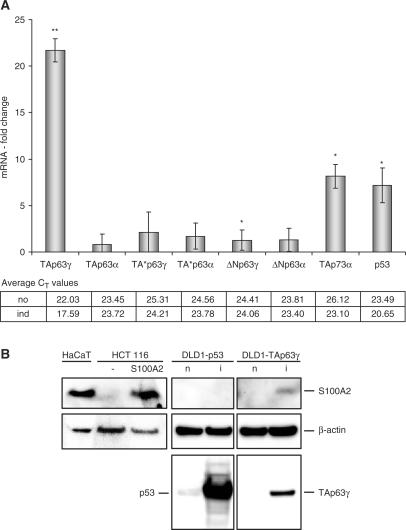
S100A2 is a TAp63γ-target gene identified earlier in a DNA-microarray screening (55). Another report had previously shown that S100A2 can be upregulated by p53 (20). With these two observations as a starting point, we were interested to test how these two and some of the other members of the p53 family control S100A2 expression and which functional implications this may have. We had earlier established a tet-off regulated expression system for the most relevant p63 splice variants using the colorectal adenocarcinoma cell line DLD-1 (44). We analyzed regulation of S100A2 comparing its mRNA expression by real-time RT–PCR before and 9 h after induction of TAp63γ, TAp63α, ΔNp63γ, ΔNp63α, TA*p63γ, TA*p63α, TAp73α or p53 transgenes (Figure 1A). We find the mRNA of S100A2 upregulated after induction of p53, TAp73α as well as TAp63γ proteins. The other p53 family proteins did not affect S100A2 expression significantly. Overexpression of TAp63γ results in a strong increase of S100A2 mRNA up to about 22-fold (Figure 1A). S100A2 induction by p53 is consistent with a previous report by Tan and coworkers (20).
Figure 1.
(A) Stimulation of S100A2 mRNA expression after selective induction of p53 family members. In RNA preparations from DLD-1 colorectal adenocarcinoma cells stably transfected with tet-off vectors expressing members of the p53 family S100A2 mRNA levels were measured. Relative mRNA levels were determined by real-time RT–PCR and changes in expression are given as induction factor comparing mRNA levels 9 h after tet-off induction to levels before induction. Averages of three experiments including standard deviations are shown. Average CT values are denoted before (no) and after (ind) tet-off induction. t-test was carried out to yield statistic significance values (**P-value ≤ 0.01; *P-value ≤ 0.05). GAPDH expression was used for standardization. (B) Stimulation of S100A2 protein expression after induction of TAp63γ. S100A2 protein was analyzed by western blot comparing expression before (n) and 9 h after (i) tet-off regulated p53 or TAp63γ expression in the colorectal adenocarcinoma cell line DLD-1. In each lane 60 µg of total cell lysate was loaded. S100A2 protein was detected with polyclonal antibodies raised against full-length human S100A2. Lysates from HCT116 cells (15 µg) transfected with an S100A2-expressing plasmid and HaCaT cells (5 µg) served as positive controls. Induction of p53 and p63 expression was analyzed by comparison of cell lysates before (n) and after induction (i) of the transgenes. Detection of β-actin served as a loading control.
More important for S100A2 function is an induction on the protein level. Since a number of commercially available antibodies were not of sufficient sensitivity, we generated polyclonal antibodies against the full-length human S100A2 protein. S100A2 protein was detected from human HaCaT keratinocytes, known to express high levels of the protein (3), and human colon cancer HCT116 cells transfected with an S100A2 expression plasmid as controls by western analysis (Figure 1B). Employing this antibody preparation, we tested S100A2 expression comparing the protein level before and 9 h after induction of the respective transgene in the DLD-1-system. A clear induction of S100A2 was observed following the induction of TAp63γ (Figure 1B). However, no S100A2 protein was detectable after induction of p53 despite a clear increase in p53 protein expression. Taken together with the observations on the mRNA level, this finding indicates that S100A2 protein expression induced by TAp63γ is stronger than the induction by p53.
TAp63γ regulates transcription of the S100A2 gene
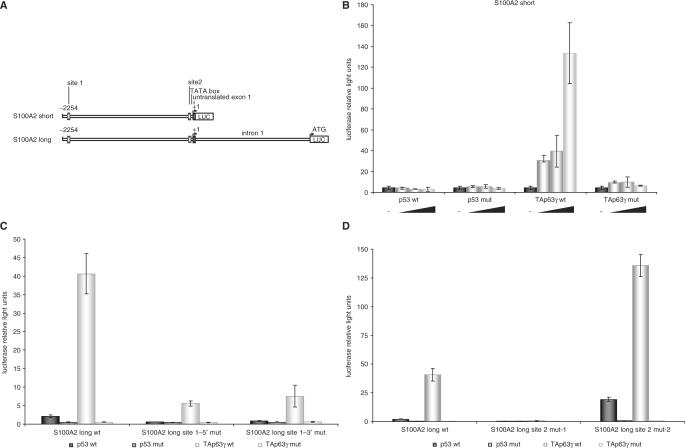
After finding that increased expression of TAp63γ leads to elevated S100A2 mRNA and protein levels, we tested if this regulation is controlled on the transcriptional level. Initially, we used in our assays an S100A2-promoter fragment-reporter construct which was kindly provided by Dr Beat W. Schäfer (University of Zurich) and had been published to be regulated by p53 (20). This construct, designated S100A2 short (Figure 2A), was transfected into SaOS-2 cells. Increasing amounts of p53- or TAp63γ-expressing plasmids were cotransfected. As controls, their DNA binding-deficient mutants were also assayed. Wild-type TAp63γ was able to activate the promoter fragment about 16-fold (Figure 2B). Surprisingly, p53 was not able to induce expression from the S100A2 short construct, leaving the question unanswered why S100A2 mRNA increases after p53 induction. Trying to explain this discrepancy we extended the S100A2 short promoter by a fragment of about 2 kb up to the translational start creating S100A2 long. This reporter construct includes the untranslated exon 1 and the first intron (Figure 2A). Activation by p53 of this promoter results in an increase of about 3.5-fold. Much more substantial is the activation by TAp63γ which enhances the expression of the reporter gene about 43-fold (Figure 2C).
Figure 2.
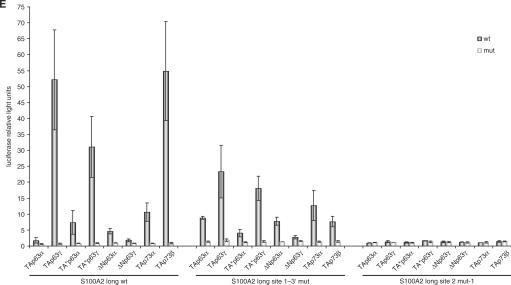
A novel site in the S100A2 promoter different from a previously recognized element mediates transcriptional activation by TAp63γ. (A) Structure of the analyzed promoter fragments of S100A2 upstream of the translational start containing previously published p53 element (site 1) and the newly identified TAp63γ-binding element (site 2). (B) The S100A2 short-reporter is activated by increasing amounts of TAp63γ but not by p53 or DNA binding-deficient mutants of TAp63γ or p53. In SaOS-2 cells 250 ng of the S100A2 short plasmid were cotransfected with increasing amounts starting with 2 ng up to 25 ng of the plasmids expressing wild-type or DNA binding-deficient mutants of TAp63γ or p53. All experiments were standardized to Renilla luciferase activity expressed from cotransfected pRL-null vector. The total amount of DNA transfected was held constant. Averages from four experiments with standard deviations are given. (C) The S100A2 long reporter is activated by TAp63γ and to a lesser extent by p53 but not by DNA binding-deficient mutants of TAp63γ and p53. Mutation of site 1 abrogates p53-mediated but not TAp63γ-mediated transactivation. In SaOS-2 cells, 400 ng reporter plasmid containing wild-type or mutated S100A2 long were cotransfected with 25 ng of the plasmids expressing wild-type or DNA binding-deficient mutants of TAp63γ or p53. Standardization was done as described above. (D) Mutations of the novel consensus element (site 2) influence TAp63γ- and p53-mediated activation of the S100A2 promoter. Transfections were done as described above. (E) Transactivation of S100A2 by several members of the p63/p73 family is mediated by the novel element 2. Plasmids expressing p63/p73 family members or their DNA-binding deficient mutants were transfected with the S100A2 long reporter construct in SaOS-2 cells. Luciferase reporter activities from three independent experiments with standard deviations are shown.
Furthermore, we analyzed the influence of other p63/p73 splice variants in the same assay. We find a strong activation of the reporter by expressing TAp73β which was not tested in our previous approaches. Also TAp73α, TA*p63γ and TA*p63α enhance transcription of S100A2 (Figure 2E). The other p53 family proteins tested only have a minor role in transcriptional activation of the S100A2 promoter.
In summary, p53 is able to activate transcription of S100A2. However, p63 and p73 proteins appear to be more potent activators of S100A2 transcription.
A novel response element mediates TAp63γ-dependent regulation of S100A2
In order to determine the responding element mediating S100A2 transcription by strong activators like TAp63γ we created some promoter mutants in sites similar to the p53 consensus (Supplementary Table 1). Initially the described p53-binding element (20) designated here as site 1 (Supplementary Table 1) was tested. In addition to this potential p53-binding site, referred to as S100A2 site 1–3′, in silico analyses revealed an additional potential consensus in this region, denoted as S100A2 site 1–5′. Mutation of these elements results in a loss of the p53-dependent activation whereas TAp63γ is still able to activate this promoter mutant (Figure 2C). We narrowed down the TAp63γ-responsive part of the S100A2 promoter by creating and testing several deletion mutants. A region of about 80-bp upstream from the untranslated exon 1 was identified to be required for TAp63γ-mediated transcription (data not shown). Comparison of this S100A2 promoter segment to the known p53-consensus sequence revealed a potential binding element consisting of two consecutive palindromes lacking any spacer region (Supplementary Table 1). This element, designated site 2, contains 4 bp which are different from the classical p53 consensus (RRRCWWGYYY) (56). Instead of the established core-binding element CWWG the S100A2 promoter shows the nucleotide sequence CAGG in each half of the palindrome. Furthermore, next to the core another 2 bp are changed in the upstream palindrome in comparison to the consensus (Supplementary Table 1). Moreover, phylogenetical footprint analyses reveal a strong conservation of this site within vertebrates (data not shown), implying a possible function for this region. To test functional importance of site 2 we mutated this element in a way that the core of the site was destroyed and used the resulting reporter in transient transfection assays (Figure 2D, site 2 mut-1). Activation of S100A2 transcription by TAp63γ is essentially lost upon mutation of site 2, identifying this element as the major site for TAp63γ-dependent transcription in the S100A2 promoter (Figure 2D). When this mutant was tested with other proteins of the p63/p73 family, we observed that the newly identified site 2 appears to be essential also for transactivation by these other splice variants (Figure 2E, site 2 mut-1). In contrast, mutation of site 1 affects transactivation of the S100A2 reporter by p63/p73 family members only to a minor extent (Figure 2E, site 1–3′ mut), emphasizing the importance of the newly identified element 2 for mediating p63/p73-dependent regulation of S100A2.
In another mutant, a bona fide p53 consensus was created by changing the guanine in position three to a thymidine in the core-binding element CAGG of site 2 (Supplementary Table 1). TAp63γ was able to activate this mutant S100A2 promoter clearly. Furthermore, this promoter construct shows a significant induction also after expression of p53 (Figure 2D, site 2 mut-2). Changing just this single base in each half of the palindrome appears sufficient to alter a site preferentially bound by p63 and p73 proteins into a functional p53 element.
Taken together, we have identified a novel promoter element which is different from the established p53 consensus mediating S100A2 transactivation selectively by p63/p73 proteins.
TAp63γ, but not p53, binds the novel S100A2 promoter element in vitro
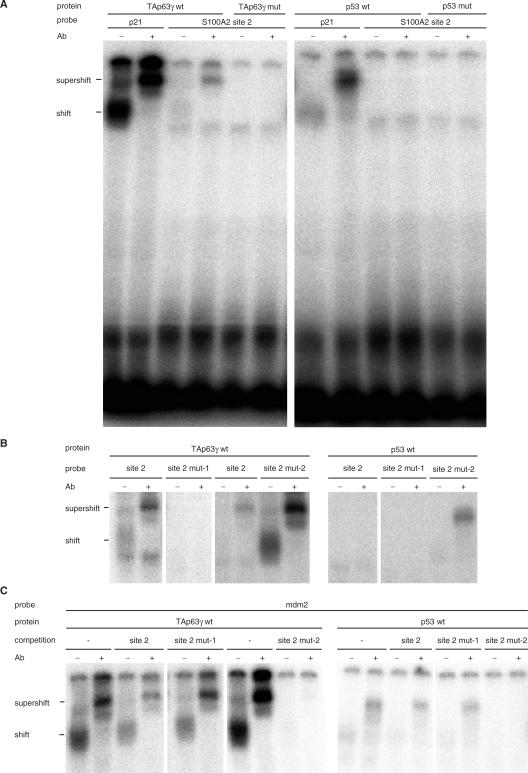
Binding of TAp63γ and p53 to the novel S100A2 promoter element site 2 was tested by EMSA employing proteins translated in vitro. An established p53-consensus element from the p21WAF1/CIP1 promoter was used as a positive control. Both, wild-type TAp63γ and p53 gave a specific signal for the control element. Addition of antibodies against p53 or p63 led to a supershifted band (Figure 3A). EMSAs yielded binding of TAp63γ to the site 2 element in the S100A2 promoter. p53 did not show significant binding to this site. Enhancement of p53 binding upon supplementing antibodies had been described (57). However, even addition of antibodies did not yield a detectable p53 binding (Figure 3A). As a negative control, DNA binding-deficient mutants of p53 or TAp63γ proteins were employed.
Figure 3.
TAp63γ is able to bind to the novel S100A2 promoter consensus element in vitro, whereas no binding of p53 is detectable. Wild-type or mutant TAp63γ or p53 were produced by translation in vitro and incubated with probes representing a p53 site from p21WAF1/CIP1 promoter as positive control or the novel site 2 from the S100A2 promoter. In some samples, specificity of detected binding was verified by adding antibody against p63γ or p53. Samples were analyzed in EMSAs. (A) Binding of TAp63γ respectively p53 to the p21WAF1/CIP1 probe or to the S100A2 promoter wild-type site 2. (B) TAp63γ or p53 binding to the S100A2 promoter wild-type site 2 or two mutants, S100A2 site 2 mut-1 and S100A2 site 2 mut-2. (C) A probe carrying a p53 site from the mdm2 promoter was employed in EMSAs. Binding to the labeled probe was competed with a 100-fold excess of unlabelled DNA of wild-type or two mutants of the S100A2 site 2.
Applying a probe representing a mutant of site 2 which does not allow TAp63γ-dependent transactivation of the reporter binds neither p63 nor p53 in an EMSA (Figure 3B, S100A2 site 2 mut-1). However, using a mutant-2 probe containing the restored p53 consensus results in an enhanced binding of wild-type TAp63γ. Consistent with our data from the reporter assays we also find binding of p53 to this DNA fragment (Figure 3B, S100A2 site 2 mut-2).
Furthermore, in competition experiments, S100A2 wild-type oligonucleotide is able to attenuate the binding of TAp63γ to a probe containing the mdm2 p53-binding site (Figure 3C S100A2 site 2). To a lesser extent, also binding of p53 to mdm2 is reduced. Mutant S100A2 oligonucleotide (S100A2 site 2 mut-1) failed to diminish these interactions, whereas oligonucleotides containing the restored p53 consensus (S100A2 site 2 mut-2) prevented binding of TAp63γ or p53 to the mdm2 probe (Figure 3C).
In conclusion, we showed a sequence-specific binding of TAp63γ to a novel S100A2 promoter element in vitro, whereas p53 was not able to bind this fragment. Changing one base in each half of the palindrome in order to create a classical p53 consensus sequence resulted in an enhanced binding by TAp63γ and a clear detectable interaction also with p53.
In vivo binding of TAp63γ and p53 to the S100A2 promoter
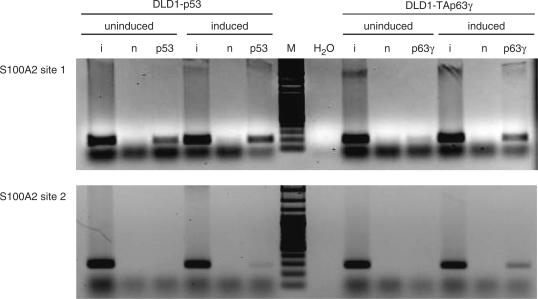
To test for binding of TAp63γ protein to the novel S100A2 promoter element in vivo we carried out ChIP analyses. Employing DLD-1 cells carrying a tet-off-regulated system expressing TAp63γ or p53 binding to two regions of the S100A2 promoter including binding site 1 or site 2 was tested. TAp63γ and p53 were shown to bind to the S100A2 promoter in vivo (Figure 4).
Figure 4.
TAp63γ is able to bind to the novel S100A2 promoter consensus element in vivo. Chromatin from DLD-1 colorectal adenocarcinoma cells stably transfected with tet-off vectors expressing members of the p53 family were cross-linked before (uninduced) and 9 h after induction (induced) of TAp63γ or p53 expression. After precipitation with antibodies against p63γ or p53, the S100A2 promoter regions containing the p53 element published earlier (site 1) or the novel element (site 2) were amplified by PCR from the precipitated DNA. Lanes are input (i), no antibody (n), water control (H2O) and DNA ladder (M).
EEC syndrome-specific mutants of TAp63γ fail to transactivate S100A2
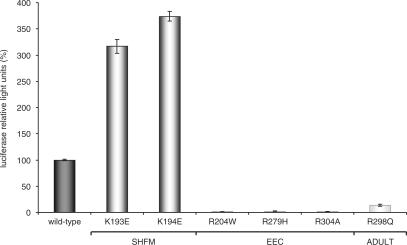
Since we found that S100A2 expression is regulated by p63 we were interested to test mutants of this protein relevant in human syndromes for their properties. Mutations such as R204, R279 and R304 found in EEC patients correspond to hotspot mutations in the DNA-binding domain of p53 observed in cancer (58). Unlike to those mutants, p63 mutations responsible for the SHFM phenotype such as K193 or K194 do not directly participate in DNA binding. In contrast to people suffering from EEC syndrome, SHFM patients do not exhibit ectodermal dysplasia and facial cleftings, which are also found in individuals with the ADULT syndrome. Several syndrome-derived p63 mutants were assayed as transcriptional activators of the S100A2 promoter: EEC syndrome, R204W, R279H, R304A; SHFM syndrome, K193E, K194E; ADULT syndrome, R298Q (Figure 5). As a control, p63 mutant proteins were tested for comparable expression by western analysis (data not shown). The reporter assays indicate that EEC syndrome-derived mutants are not able to transactivate transcription of S100A2. In contrast, p63 mutants derived from SHFM patients even appear to enhance S100A2 reporter activity. Mutations related to the ADULT syndrome are still able to stimulate some S100A2 transcription (Figure 5).
Figure 5.
TAp63γ mutants related to SHFM, EEC and ADULT human developmental syndromes show a functional difference in regulating S100A2 transcription. TAp63γ mutants originating from patients with developmental syndromes SHFM, EEC or ADULT differentially activate the S100A2 long reporter. Transfections and analyses were done as described above.
Binding of p63γ protein to the S100A2 promoter correlates with S100A2 expression after DNA damage
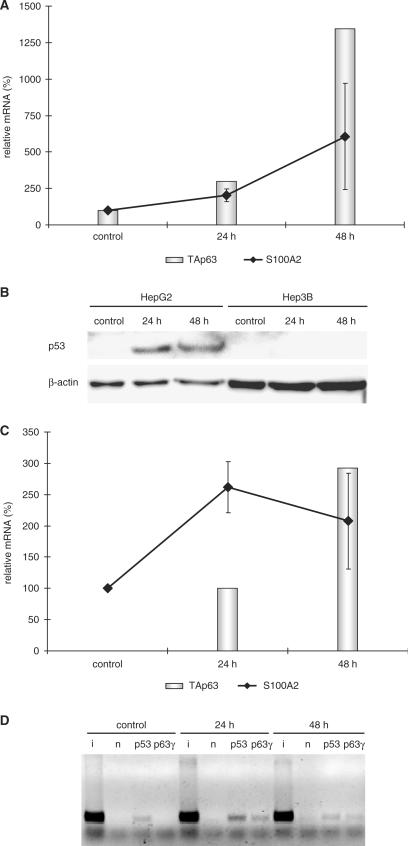
S100A2 was described as a potential tumor suppressor (14). Therefore, we tested if regulation of S100A2 expression may be connected to DNA damage through p63. It had been observed earlier that p63 expression can be induced by DNA damage (59). Consistent with these results we found enhanced expression of TAp63 mRNA upon treatment of HepG2 hepatocellular carcinoma cells with doxorubicin for 24 to 48 hours. Interestingly, in line with TAp63 expression also S100A2 mRNA expression increases (Figure 6A). Of note is that mRNA levels of ΔNp63 and TAp73 isoforms did not change significantly in this experiment (data not shown).
Figure 6.
Recruitment of p63γ protein to the S100A2 promoter may connect S100A2 expression to DNA damage. (A) TAp63γ and S100A2 mRNA expression increase after DNA damage. Real-time RT–PCR measurements of S100A2 and TAp63 mRNAs from doxorubicin-treated HepG2 cells. Relative mRNA levels were normalized to expression of GAPDH mRNA. Cells before treatment were employed as control. Control levels were set to 100%. Times of doxorubicin treatment are indicated. (B) Western blot analyses of lysates from p53-positive HepG2 and p53-negative Hep3B cells after doxorubicin-induced DNA damage. Controls contain samples taken before treatment. Doxorubicin-treated samples were analyzed after 24 h or 48 h. Detection of β-actin served as a loading control. (C) Expression of S100A2 mRNA correlates with an increase of TAp63 mRNA after doxorubicin-induced DNA damage in p53-negative Hep3B cells. Relative mRNA levels were measured by real-time RT–PCR. Expression of GAPDH was used for normalization. S100A2 mRNA from untreated control cells was set to 100%. In control samples no TAp63 mRNA was detectable. Therefore, the TAp63 mRNA measurement at 24 h was employed as the 100% reference. (D) Chromatin immunoprecipitation (ChIP) assays of p53 and p63γ proteins binding to the S100A2 promoter in HepG2 cells following DNA damage. Control and doxorubicin-treated cells were prepared as described above. Lanes are input (i), no antibody (n), p53 and p63γ antibodies.
HepG2 cells are positive for p53 protein expression. In addition to these cells we also tested the p53-negative liver cell line Hep3B for p53-protein induction after DNA damage. As expected, only in HepG2 cells p53 increases (Figure 6B). We examined if S100A2 expression also changes independently of p53. Also in Hep3B cells, S100A2 mRNA expression is induced when the p53-negative cells are treated with doxorubicin. Furthermore, the TAp63 mRNA level increases as well. However, TAp63 expression is not detectable in untreated cells, hampering a correlation between TAp63 and S100A2 expression as cause and effect (Figure 6C). Also in this cell system, levels of ΔNp63 and TAp73 mRNAs did not change (data not shown).
We wished to correlate expression of p63 proteins with binding to the S100A2 promoter in vivo by ChIP assays. However, p53-independent analysis of p63γ-protein binding was not detectable likely due to low endogenous protein amounts (data not shown). Nevertheless, in HepG2 cells we could show binding of p63γ protein to the S100A2 promoter in vivo following DNA damage. Recruitment of p63γ is not observed in untreated cells (Figure 6D). Distinct from binding of p63γ is the recruitment of p53 to the S100A2 promoter. p53 binds already to the promoter in cells before DNA damage (Figure 6D) although p53 is induced only after doxorubicin treatment (Figure 6B). In conclusion, binding of endogenous p63γ protein to the S100A2 promoter matches with enhanced mRNA expression of TAp63 isoforms after doxorubicin-induced DNA damage.
DISCUSSION
In contrast to p53, mutations of p63 are not frequently found in human cancer. Therefore, p63 is not a classical tumor suppressor despite strong structural homologies to p53 (60). Elucidating other possible p63 functions is difficult due to the presence of various p63 splice variants and their divergent expression pattern. The TAp63γ protein closely resembles p53. One reason for the similarity is its lack of an inhibitory domain present in the α variants. TAp63γ is found to be a very potent activator of p53 target genes (33) and seems to be able to enforce tumor suppressive mechanisms in which p53 is involved (34).
In addition to a possible function as a tumor suppressor, it was discovered that p63 plays a role in development (36,40). Heterozygous p63 germ line mutations cause several skin and other developmental defects in man (39,58). These observations suggest for p63 other transcriptional targets than for p53.
Here we identify S100A2 as a transcriptional target of p63. Particularly the p63 variant TAp63γ is rather active in enhancing S100A2 expression. Selective expression of TAp63γ leads to a dramatic increase in mRNA amounts (Figure 1A) and subsequently to enhanced expression of S100A2 protein (Figure 1B). A novel binding site in the S100A2 promoter is essential for TAp63γ-mediated activation (Figure 2). We demonstrated that TAp63γ binds to this element in a sequence-specific manner in vitro (Figure 3). Furthermore, the recruitment of TAp63γ to the S100A2 promoter in vivo was demonstrated (Figure 4). A recent report showed activation of S100A2 transcription by p73α and p73β (3). Interestingly, we could show that the newly identified p63-response element is the activating site also for TAp73 proteins (Figure 2E). However, an additional element relevant for part of the p73-dependent activation in the second intron of the S100A2 gene, as discussed by others, cannot be excluded (3). We did not observe a strong S100A2 repression by ΔNp63α (Figures 1 and 2E) as detected by other researchers (3). This may be caused by different experimental approaches. Lapi and coworkers employed firefly luciferase for measurement of reporter activity and β-galactosidase for normalization instead of the firefly/Renilla-luciferase combination used here. The difference in protein stability between luciferase and β-galactosidase may result in distortion of relative reporter activities. Other differences are the use of a DNA–binding-deficient mutant of ΔNp63α as a control instead of empty vector and employment of distinct cell systems (3). However, Lapi and coworkers observed under their experimental conditions that TAp63α is not able to activate the S100A2 promoter which is perfectly consistent with our findings (Figure 2E).
S100A2 is known to be a transcriptional target of p53 (20). Consistent with this we find an induction of S100A2 mRNA after expression of p53, but activation of the S100A2 promoter by p53 was substantially lower than the increase in expression after TAp63γ induction (Figure 2). Furthermore, on the protein level S100A2 increased after TAp63γ expression, whereas an equivalent protein induction was not detectable following p53 expression (Figure 1B). These findings suggest that TAp63γ is a more potent transcriptional activator than p53 also implying that regulation through p63 generally may be more relevant for S100A2 function.
We demonstrated that changing one nucleotide in the core of the newly identified binding element 2 is sufficient to differentiate between binding of p53 or TAp63γ (Figures 2D and 3B). These observations are in line with previously published results which demonstrate that p63 preferentially binds to degenerate p53 consensus elements (42,43). This mechanism appears to be important for differentiating functions of p63 versus p53. Furthermore, the evolutionary conservation of the identified binding element in S100A2 promoters from different species (data not shown) suggests an essential biological relevance of the TAp63γ-dependent regulation (61).
One function of p63 is the regulation of developmental processes. Several mutations in the p63 DNA-binding domain are responsible for a family of human syndromes characterized by a combination of ectrodactyly, ectodermal dysplasia and facial clefting (37–39,58). These observations led us to test the influence of disease-related p63 mutants on transcriptional activation of S100A2. In reporter assays, we found an even stronger transcriptional activation employing two TAp63γ mutants derived from SHFM patients compared to wild-type p63 (Figure 5). In contrast, three p63 mutants responsible for the EEC syndrome had essentially lost their activation potential on the S100A2 promoter. A TAp63γ mutant related to the ADULT syndrome yielded some residual S100A2 activation when compared to wild-type TAp63γ (Figure 5). Interestingly, these observations correlate with severity of the epidermal syndrome phenotype since SHFM patients, in contrast to patients suffering from EEC and ADULT syndromes, are characterized by a lack of epidermal dysplasia (62). Therefore, it is tempting to speculate that the ability of p63 to stimulate expression of S100A2 plays a role in developing the phenotype of these patients. Further research is required to establish a stronger link.
S100A2 was identified as a potential tumor suppressor by subtractive hybridization between normal and tumor-derived mammary epithelial cells in man (14). Consistent with these findings S100A2 expression is markedly downregulated in several tumor tissues (15–18). However, it was shown that S100A2 expression is increased in other tumors (21–26). It was discussed that overexpression is an early event in tumorigenesis. Furthermore, a physical interaction between S100A2 and p53 proteins enhances transcriptional activity of p53 which implies antitumorigenic properties of S100A2 (27). A possible interaction also between p63 and S100A2 proteins has not yet been investigated. Recently, it was demonstrated that S100A2 expression is able to diminish expression of Cox-2 protein which provides further evidence for a tumor suppressive function of S100A2 (28). Interestingly, in line with these observations we showed that expression of S100A2 increases following DNA damage (Figure 6A and C). The enhanced expression of S100A2 correlates with binding of p63γ protein to the S100A2 promoter (Figure 6D). Generally, detection of different p63 isoforms on the mRNA and protein levels is difficult. It is possible to differentiate between TAp63 and ΔNp63 mRNA variants. On the protein level we were able to observe p63γ versus total p63 protein expression. After DNA damage we exclusively find an increase in the TAp63 isoforms on the mRNA level. Combining these observations with the data from the ChIP assays suggests that TAp63γ is induced after DNA damage and subsequent binding to the S100A2 promoter mediates its regulation (Figure 6D). Supportive of this notion is the finding that p53-negative Hep3B cells also show an increase in transcription of S100A2 together with enhanced expression of the TAp63 isoforms after DNA damage is observed (Figure 6C). In conclusion, it is possible that regulation of S100A2 transcription by TAp63γ is a supporting mechanism to complement and enhance cellular response in preventing tumor transformation (34). This is consistent with earlier observations implying that TAp63γ is able to substitute partially p53 function in hepatocellular carcinomas lacking p53 expression by transactivating the maspin tumor suppressor (55).
In summary, we show that S100A2 is a novel transcriptional target of the p63 splice variant TAp63γ. The regulation is mediated by a novel and strongly conserved transcriptional element in the S100A2 promoter which is bound by TAp63γ but not by p53. Transcriptional properties of p63 mutants derived from EEC, ADULT and SHFM syndrome patients yield evidence for a role of a TAp63γ-mediated transcription of S100A2 in developmental processes. Recruitment of p63γ proteins to the S100A2 promoter following DNA damage correlates with enhanced expression of S100A2 and suggest a function in tumor suppression.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Jana Lorenz for technical assistance, Drs Bert Vogelstein and Beat W. Schäfer for generously providing cells and plasmids. G.A.M. was the recipient of a graduate fellowship provided by the Freistaat Sachsen. This work was supported by a grant from the Interdisciplinary Center for Clinical Research (IZKF) at the University of Leipzig (to K.E.). Funding to pay the Open Access publication charges for this article was provided by the IZKF.
Conflict of interest statement. None declared.
REFERENCES
- 1.Glenney JR, Kindy MS, Zokas L. Isolation of a new member of the S100 protein family – amino-acid sequence, tissue, and subcellular-distribution. J. Cell Biol. 1989;108:569–578. doi: 10.1083/jcb.108.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boni R, Burg G, Doguoglu A, Ilg EC, Schafer BW, Muller B, Heizmann CW. Immunohistochemical localization of the Ca2+ binding S100 proteins in normal human skin and melanocytic lesions. Br. J. Dermatol. 1997;137:39–43. [PubMed] [Google Scholar]
- 3.Lapi E, Iovino A, Fontemaggi G, Soliera AR, Iacovelli S, Sacchi A, Rechavi G, Givol D, Blandino G, Strano S. S100A2 gene is a direct transcriptional target of p53 homologues during keratinocyte differentiation. Oncogene. 2006;25:3628–3637. doi: 10.1038/sj.onc.1209401. [DOI] [PubMed] [Google Scholar]
- 4.Mandinova A, Atar D, Schafer BW, Spiess M, Aebi U, Heizmann CW. Distinct subcellular localization of calcium binding S100 proteins in human smooth muscle cells and their relocation in response to rises in intracellular calcium. J. Cell Sci. 1998;111:2043–2054. doi: 10.1242/jcs.111.14.2043. [DOI] [PubMed] [Google Scholar]
- 5.Mueller A, Bachi T, Hochli M, Schafer BW, Heizmann CW. Subcellular distribution of S100 proteins in tumor cells and their relocation in response to calcium activation. Histochem. Cell Biol. 1999;111:453–459. doi: 10.1007/s004180050381. [DOI] [PubMed] [Google Scholar]
- 6.Zhang T, Woods TL, Elder JT. Differential responses of S100A2 to oxidative stress and increased intracellular calcium in normal, immortalized, and malignant human keratinocytes. J. Invest. Dermatol. 2002;119:1196–1201. doi: 10.1046/j.1523-1747.2002.19520.x. [DOI] [PubMed] [Google Scholar]
- 7.Nakayama S, Kretsinger RH. Evolution of the Ef-hand family of proteins. Annu. Rev. Biophys. Biomol. Struct. 1994;23:473–507. doi: 10.1146/annurev.bb.23.060194.002353. [DOI] [PubMed] [Google Scholar]
- 8.Deshpande R, Woods TL, Fu J, Zhang T, Stoll SW, Elder JT. Biochemical characterization of S100A2 in human keratinocytes: Subcellular localization, dimerization, and oxidative cross-linking. J. Invest. Dermatol. 2000;115:477–485. doi: 10.1046/j.1523-1747.2000.00078.x. [DOI] [PubMed] [Google Scholar]
- 9.Franz C, Durussel I, Cox JA, Schafer BW, Heizmann CW. Binding of Ca2+ and Zn2+ to human nuclear S100A2 and mutant proteins. J. Biol. Chem. 1998;273:18826–18834. doi: 10.1074/jbc.273.30.18826. [DOI] [PubMed] [Google Scholar]
- 10.Marenholz I, Heimann CW, Fritz G. S100 proteins in mouse and man: from evolution to function and pathology (including an update of the nomenclature) Biochem. Biophys. Res. Commun. 2004;322:1111–1122. doi: 10.1016/j.bbrc.2004.07.096. [DOI] [PubMed] [Google Scholar]
- 11.Volz A, Korge BP, Compton JG, Ziegler A, Steinert PM, Mischke D. Physical mapping of a functional cluster of epidermal differentiation genes on chromosome-1Q21. Genomics. 1993;18:92–99. doi: 10.1006/geno.1993.1430. [DOI] [PubMed] [Google Scholar]
- 12.Zimmer DB, Cornwall EH, Landar A, Song W. The S100 protein family – history, function, and expression. Brain Res. Bull. 1995;37:417–429. doi: 10.1016/0361-9230(95)00040-2. [DOI] [PubMed] [Google Scholar]
- 13.Donato R. Intracellular and extracellular roles of s100 proteins. Microsc. Res. Tech. 2003;60:540–551. doi: 10.1002/jemt.10296. [DOI] [PubMed] [Google Scholar]
- 14.Lee SW, Tomasetto C, Sager R. Positive selection of candidate tumor-suppressor genes by subtractive hybridization. Proc. Natl Acad. Sci. USA. 1991;88:2825–2829. doi: 10.1073/pnas.88.7.2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng G, Xu XC, Youssef EM, Lotan R. Diminished expression of S100A2, a putative tumor suppressor, at early stage of human lung carcinogenesis. Cancer Res. 2001;61:7999–8004. [PubMed] [Google Scholar]
- 16.Lee SW, Tomasetto C, Swisshelm K, Keyomarsi K, Sager R. Down-regulation of a member of the S100 gene family in mammary-carcinoma cells and reexpression by azadeoxycytidine treatment. Proc. Natl Acad. Sci. USA. 1992;89:2504–2508. doi: 10.1073/pnas.89.6.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maelandsmo GM, Florenes VA, Mellingsaeter T, Hovig E, Kerbel RS, Fodstad O. Differential expression patterns of S100A2, S100A4 and S100A6 during progression of human malignant melanoma. Int. J. Cancer. 1997;74:464–469. doi: 10.1002/(sici)1097-0215(19970822)74:4<464::aid-ijc19>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 18.Zhang X, Hunt JL, Shin DM, Chen ZG. Down-regulation of S100A2 in lymph node metastases of head and neck cancer. Head Neck. 2006;29:236–243. doi: 10.1002/hed.20511. [DOI] [PubMed] [Google Scholar]
- 19.Wicki R, Franz C, Scholl FA, Heizmann CW, Schafer BW. Repression of the candidate tumor suppressor gene S100A2 in breast cancer is mediated by site specific hypermethylation. Cell Calcium. 1997;22:243–254. doi: 10.1016/s0143-4160(97)90063-4. [DOI] [PubMed] [Google Scholar]
- 20.Tan M, Heizmann CW, Guan K, Schafer BW, Sun Y. Transcriptional activation of the human S100A2 promoter by wild-type p53. FEBS Lett. 1999;445:265–268. doi: 10.1016/s0014-5793(99)00135-0. [DOI] [PubMed] [Google Scholar]
- 21.El Rifai W, Moskaluk CA, Abdrabbo MK, Harper J, Yoshida C, Riggins GJ, Frierson HF, Powell SM. Gastric cancers overexpress S100A calcium-binding proteins. Cancer Res. 2002;62:6823–6826. [PubMed] [Google Scholar]
- 22.Hough CD, Cho KR, Zonderman AB, Schwartz DR, Morin PJ. Coordinately up-regulated genes in ovarian cancer. Cancer Res. 2001;61:3869–3876. [PubMed] [Google Scholar]
- 23.Hsieh HL, Schafer BW, Sasaki N, Heizmann CW. Expression analysis of S100 proteins and RAGE in human tumors using tissue microarrays. Biochem. Biophys. Res. Commun. 2003;307:375–381. doi: 10.1016/s0006-291x(03)01190-2. [DOI] [PubMed] [Google Scholar]
- 24.Lauriola L, Michetti F, Maggiano N, Galli J, Cadoni G, Schafer BW, Heizmann CW, Ranelletti FO. Prognostic significance of the Ca(2+) binding protein S100A2 in laryngeal squamous-cell carcinoma. Int. J. Cancer. 2000;89:345–349. doi: 10.1002/1097-0215(20000720)89:4<345::aid-ijc5>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 25.Nagy N, Brenner C, Markadieu N, Chaboteaux C, Camby I, Schafer BW, Pochet R, Heizmann CW, Salmon I, Kiss R, et al. S100A2, a putative tumor suppressor gene, regulates in vitro squamous cell carcinoma migration. Lab. Invest. 2001;81:599–612. doi: 10.1038/labinvest.3780269. [DOI] [PubMed] [Google Scholar]
- 26.Wang H, Zhang Z, Li R, Ang KK, Zhang H, Caraway NP, Katz RL, Jiang F. Overexpression of S100A2 protein as a prognostic marker for patients with stage I non small cell lung cancer. Int. J. Cancer. 2005;116:285–290. doi: 10.1002/ijc.21035. [DOI] [PubMed] [Google Scholar]
- 27.Mueller A, Schafer BW, Ferrari S, Weibel M, Makek M, Hochli M, Heizmann CW. The calcium-binding protein S100A2 interacts with p53 and modulates its transcriptional activity. J. Biol. Chem. 2005;280:29186–29193. doi: 10.1074/jbc.M505000200. [DOI] [PubMed] [Google Scholar]
- 28.Tsai WC, Tsai ST, Jin YT, Wu LW. Cyclooxygenase-2 is involved in S100A2-mediated tumor suppression in squamous cell carcinoma. Mol. Cancer Res. 2006;4:539–547. doi: 10.1158/1541-7786.MCR-05-0266. [DOI] [PubMed] [Google Scholar]
- 29.Murray-Zmijewski F, Lane DP, Bourdon JC. p53/p63/p73 isoforms: an orchestra of isoforms to harmonise cell differentiation and response to stress. Cell Death. Differ. 2006;13:962–972. doi: 10.1038/sj.cdd.4401914. [DOI] [PubMed] [Google Scholar]
- 30.Schmale H, Bamberger C. A novel protein with strong homology to the tumor suppressor p53. Oncogene. 1997;15:1363–1367. doi: 10.1038/sj.onc.1201500. [DOI] [PubMed] [Google Scholar]
- 31.Senoo M, Seki N, Ohira M, Sugano S, Watanabe M, Tachibana M, Tanaka T, Shinkai Y, Kato H. A second p53-related protein, p73L, with high homology to p73. Biochem. Biophys. Res. Commun. 1998;248:603–607. doi: 10.1006/bbrc.1998.9013. [DOI] [PubMed] [Google Scholar]
- 32.Trink B, Okami K, Wu L, Sriuranpong V, Jen J, Sidransky D. A new human p53 homologue. Nat. Med. 1998;4:747–748. doi: 10.1038/nm0798-747. [DOI] [PubMed] [Google Scholar]
- 33.Yang A, Kaghad M, Wang Y, Gillett E, Fleming MD, Dotsch V, Andrews NC, Caput D, McKeon F. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol. Cell. 1998;2:305–16. doi: 10.1016/s1097-2765(00)80275-0. [DOI] [PubMed] [Google Scholar]
- 34.Flores ER, Tsai KY, Crowley D, Sengupta S, Yang A, McKeon F, Jacks T. p63 and p73 are required for p53-dependent apoptosis in response to DNA damage. Nature. 2002;416:560–564. doi: 10.1038/416560a. [DOI] [PubMed] [Google Scholar]
- 35.Brunner HG, Hamel BC, van Bokhoven H. The p63 gene in EEC and other syndromes. J. Med. Genet. 2002;39:377–381. doi: 10.1136/jmg.39.6.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398:708–713. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- 37.Rinne T, Hamel B, van Bokhoven H, Brunner HG. Pattern of p63 mutations and their phenotypes–update. Am. J. Med. Genet. A. 2006;140:1396–1406. doi: 10.1002/ajmg.a.31271. [DOI] [PubMed] [Google Scholar]
- 38.van Bokhoven H, Hamel BC, Bamshad M, Sangiorgi E, Gurrieri F, Duijf PH, Vanmolkot KR, van Beusekom E, van Beersum SE, Celli J, et al. p63 Gene mutations in eec syndrome, limb-mammary syndrome, and isolated split hand-split foot malformation suggest a genotype-phenotype correlation. Am. J. Hum. Genet. 2001;69:481–492. doi: 10.1086/323123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Bokhoven H, McKeon F. Mutations in the p53 homolog p63: allele-specific developmental syndromes in humans. Trends Mol. Med. 2002;8:133–139. doi: 10.1016/s1471-4914(01)02260-2. [DOI] [PubMed] [Google Scholar]
- 40.Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT, Tabin C, Sharpe A, Caput D, Crum C, et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–718. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- 41.Yang A, McKeon F. P63 and P73: P53 mimics, menaces and more. Nat. Rev. Mol. Cell Biol. 2000;1:199–207. doi: 10.1038/35043127. [DOI] [PubMed] [Google Scholar]
- 42.Ortt K, Sinha S. Derivation of the consensus DNA-binding sequence for p63 reveals unique requirements that are distinct from p53. FEBS Letters. 2006;580:4544–4550. doi: 10.1016/j.febslet.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 43.Osada M, Park HL, Nagakawa Y, Yamashita K, Fomenkov A, Kim MS, Wu GJ, Nomoto S, Trink B, Sidransky D. Differential recognition of response elements determines target gene specificity for p53 and p63. Mol. Cell. Biol. 2005;25:6077–6089. doi: 10.1128/MCB.25.14.6077-6089.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dietz S, Rother K, Bamberger C, Schmale H, Mössner J, Engeland K. Differential regulation of transcription and induction of programmed cell death by human p53-family members p63 and p73. FEBS Lett. 2002;525:93–99. doi: 10.1016/s0014-5793(02)03093-4. [DOI] [PubMed] [Google Scholar]
- 45.Yu J, Zhang L, Hwang PM, Rago C, Kinzler KW, Vogelstein B. Identification and classification of p53-regulated genes. Proc. Natl Acad. Sci. USA. 1999;96:14517–14522. doi: 10.1073/pnas.96.25.14517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hagiwara K, McMenamin MG, Miura K, Harris CC. Mutational analysis of the p63/p73L/p51/p40/CUSP/KET gene in human cancer cell lines using intronic primers. Cancer Res. 1999;59:4165–4169. [PubMed] [Google Scholar]
- 47.Yoshikawa H, Nagashima M, Khan MA, McMenamin MG, Hagiwara K, Harris CC. Mutational analysis of p73 and p53 in human cancer cell lines. Oncogene. 1999;18:3415–3421. doi: 10.1038/sj.onc.1202677. [DOI] [PubMed] [Google Scholar]
- 48.Haugwitz U, Wasner M, Wiedmann M, Spiesbach K, Rother K, Mössner J, Engeland K. A single cell cycle genes homology region (CHR) controls cell cycle-dependent transcription of the cdc25C phosphatase gene and is able to cooperate with E2F or Sp1/3 sites. Nucleic Acids Res. 2002;30:1967–1976. doi: 10.1093/nar/30.9.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bressac B, Galvin KM, Liang TJ, Isselbacher KJ, Wands JR, Ozturk M. Abnormal structure and expression of p53 gene in human hepatocellular carcinoma. Proc. Natl Acad. Sci. USA. 1990;87:1973–1977. doi: 10.1073/pnas.87.5.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krause K, Wasner M, Reinhard W, Haugwitz U, Lange-zu Dohna C, Mössner J, Engeland K. The tumour suppressor protein p53 can repress transcription of cyclin B. Nucleic Acids Res. 2000;28:4410–4418. doi: 10.1093/nar/28.22.4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wasner M, Haugwitz U, Reinhard W, Tschöp K, Spiesbach K, Lorenz J, Mössner J, Engeland K. Three CCAAT-boxes and a single cell cycle genes homology region (CHR) are the major regulating sites for transcription from the human cyclin B2 promoter. Gene. 2003;312:225–237. doi: 10.1016/s0378-1119(03)00618-8. [DOI] [PubMed] [Google Scholar]
- 52.Wasner M, Tschöp K, Spiesbach K, Haugwitz U, Johne C, Mössner J, Mantovani R, Engeland K. Cyclin B1 transcription is enhanced by the p300 coactivator and regulated during the cell cycle by a CHR-dependent repression mechanism. FEBS Lett. 2003;536:66–70. doi: 10.1016/s0014-5793(03)00028-0. [DOI] [PubMed] [Google Scholar]
- 53.Contente A, Dittmer A, Koch MC, Roth J, Dobbelstein M. A polymorphic microsatellite that mediates induction of PIG3 by p53. Nat. Genet. 2002;30:315–320. doi: 10.1038/ng836. [DOI] [PubMed] [Google Scholar]
- 54.Boyd KE, Wells J, Gutman J, Bartley SM, Farnham PJ. c-Myc target gene specificity is determined by a post-DNA binding mechanism. Proc. Natl Acad. Sci. USA. 1998;95:13887–13892. doi: 10.1073/pnas.95.23.13887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Spiesbach K, Tannapfel A, Mössner J, Engeland K. TAp63gamma can substitute for p53 in inducing expression of the maspin tumor suppressor. Int. J. Cancer. 2005;114:555–562. doi: 10.1002/ijc.20766. [DOI] [PubMed] [Google Scholar]
- 56.el-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B. Definition of a consensus binding site for p53. Nat. Genet. 1992;1:45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- 57.Hupp TR, Meek DW, Midgley CA, Lane DP. Regulation of the specific DNA-binding function of P53. Cell. 1992;71:875–886. doi: 10.1016/0092-8674(92)90562-q. [DOI] [PubMed] [Google Scholar]
- 58.van Bokhoven H, Brunner HG. Splitting p63. Am. J. Hum. Genet. 2002;71:1–13. doi: 10.1086/341450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Petitjean A, Cavard C, Shi H, Tribollet V, Hainaut P, Caron dF. The expression of TA and DeltaNp63 are regulated by different mechanisms in liver cells. Oncogene. 2005;24:512–519. doi: 10.1038/sj.onc.1208215. [DOI] [PubMed] [Google Scholar]
- 60.Mills AA. p63: oncogene or tumor suppressor? Curr. Opin. Genet. Dev. 2006;16:38–44. doi: 10.1016/j.gde.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 61.Yang A, Zhu Z, Kapranov P, McKeon F, Church GM, Gingeras TR, Struhl K. Relationships between p63 binding, DNA sequence, transcription activity, and biological function in human cells. Mol. Cell. 2006;24:593–602. doi: 10.1016/j.molcel.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 62.Ianakiev P, Kilpatrick MW, Toudjarska I, Basel D, Beighton P, Tsipouras P. Split-hand/split-foot malformation is caused by mutations in the p63 gene on 3q27. Am. J. Hum. Genet. 2000;67:59–66. doi: 10.1086/302972. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.