Abstract
Microarray analyses of mRNAs over-expressed in strains lacking the nuclear exosome component Rrp6 identified the transcript encoding the ARE-binding protein Cth2, which functions in cytoplasmic mRNA stability. Subsequent northern analyses revealed that exosome mutants accumulate a 3′-extended transcript at the expense of the mature CTH2 mRNA. The 3′ ends of the CTH2 mRNA were mapped to a [GU3–5]5 repeat, unlike any previously characterized polyadenylation site. CTH2 mRNA accumulation was not inhibited by mutations in 3′-cleavage and polyadenylation factors, Rna14, Rna15 and Pap1, which block accumulation of other mRNAs. The 3′-extended CTH2 pre-mRNA strongly accumulated in strains with mutations in the TRAMP4 polyadenylation complex or the Nrd1/Nab3/Sen1 complex, and contains multiple Nrd1 and Nab3 binding sites. CTH2 carries a consensus ARE element and levels of the pre-mRNA and mRNA were elevated by mutation of the ARE or inactivation of the nuclear 5′-exonuclease Rat1. We propose that CTH2 mRNA is processed from a 3′-extended primary transcript by the exosome, TRAMP and Nrd1/Nab3/Sen1 complexes. This unusual pathway may allow time for nuclear, ARE-mediated regulation of CTH2 levels involving Rat1.
INTRODUCTION
The exosome is a complex with 3′-5′ exonuclease activity that has been implicated in the processing and degradation of many different RNA species (1). The complex contains 10 essential components, often termed the core exosome that are common to the nuclear and cytoplasmic forms of the exosome. This can associate with the 3′-exonuclease Rrp6 in the nucleus and with the GTPase Ski7 in the cytoplasm. Known substrates include mRNA precursors that have failed to undergo normal co-transcriptional cleavage and polyadenylation due to mutations in the Rna14 and Rna15 components of the cleavage and polyadenylation machinery (2). Previous analyses by transcription run on had shown that under these circumstances, RNA polymerase II (Pol II) generates long 3′-extended transcripts (3); however, these were essentially undetectable by northern hybridization in the single mutant strains, presumably due to their rapid degradation. The long 3′-extended species were greatly stabilized by the combination of rna14-1 and rna15-2 mutations with depletion of components of the core exosome or the putative RNA helicase Mtr4. The Trf4/5, Air1/2, Mtr4 polyadenylation (TRAMP) complexes function together with the exosome in the degradation of many different transcripts (1). These observations therefore suggested that the aberrant 3′-extended pre-mRNAs were recognized and very rapidly degraded by the TRAMP/exosome system.
An unexpected feature of these analyses was the finding that synthesis of functional mRNAs in rna14-1 strains could largely be restored by loss of the nuclear-specific exosome component Rrp6 (2). This indicated that functional mRNAs could be generated by exonuclease digestion by the exosome, followed by polyadenylation that was uncoupled from co-transcriptional cleavage by the cannonical mRNA cleavage and polyadenylation system. This suggested the existence of a novel 3′-processing pathway for mRNA synthesis, but no mRNAs were known that followed this pathway.
There has been extensive analysis of the regulation of mRNA levels by cytoplasmic mRNA turnover. However, we previously speculated that a class of mRNAs might be regulated by exosome-mediated RNA degradation within the nucleus. With the aim of identifying such mRNAs we conducted microarray analyses on strains lacking the nuclear-specific exosome component Rrp6, or carrying conditional mutations in the core exosome component Rrp41/Ski6 or the nuclear exosome co-factor Nrd1 (4). Amongst the up-regulated transcripts identified was the CTH2 mRNA, which we selected for further analysis.
Cth2 and Cth1 are yeast homologues of the human zinc finger protein tristetraprolin (TTP)/Tis11, which is known to be involved in the activated mRNA degradation pathway, ARE-mediated decay (5). This pathway targets mRNAs containing an A+U rich sequence element (ARE), generally localized in the 3′ UTR, which is the binding site for TTP and several other human proteins. Human ARE-containing mRNAs encode many proteins that function as growth factors, expression of which must be very tightly controlled. Under normal growth conditions, TTP binding promotes the rapid degradation of ARE-containing mRNAs. However, under stress conditions the stability and translation of ARE-containing mRNAs can be specifically enhanced (6), showing these to be versatile regulatory elements. Characterized ARE-binding proteins associate with pre-mRNAs in the nucleus and are exported to the cytoplasm in complex with the mRNA. It has been generally assumed that ARE-mediated decay is a cytoplasmic process, but this has not been assessed for most substrates. Insertion of an ARE into the U1 small nuclear RNA resulted in its destabilization (7), but the localization of degradation was not tested.
In yeast, the identified targets of Cth2 are mRNAs encoding proteins involved in iron (Fe) metabolism (8). Under conditions of Fe deficiency, Cth2 binds to and stimulates the degradation of many mRNAs that encode proteins involved in pathways that require Fe availability. Human TTP is auto-regulated via a consensus ARE in the 3′ UTR of its own mRNA. This has not been assessed for yeast CTH2, but inspection of the sequence of the 3′-UTR reveals the presence of a perfect consensus ARE sequence (UUAUUUAUU), and deletion of the gene encoding Cth1 up-regulates CTH2 mRNA levels (9).
Here, we report that CTH2 mRNA has a novel biosynthetic pathway and appears to be subject to ARE-mediated regulation in the nucleus.
MATERIAL AND METHODS
Strains, culture, media and yeast genetics
Strains were grown in YPD medium containing 2% peptone, 1% yeast extract and 2% glucose or YNB medium containing 0.67% yeast nitrogen base, 0.5% ammonium sulphate, 2% glucose and supplemented with the required amino acids. Strains used in this study are listed in Supplementary Table S1. Transformation was performed as described (10). Half-lives of the (mini-PGK1/CTH2-3′-UTR) reporter constructs were determined by adding doxycycline to a final concentration of 2 µg ml–1 to inhibit transcription before harvesting samples at different time points.
RNA extraction and northern hybridization
RNA extractions were made as previously described (11). Twenty micrograms of total RNA for each sample were analysed on glyoxal–agarose gels (12). Analyses of reporter construct expression were made by resolving 7 µg of total RNA on a 6% polyacrylamide/8.3 M urea gel. Random primed probes of the coding sequence of CTH2 mRNA, made with Ambion Strip-EZ DNA Kit, were used to detect both mature and pre-CTH2 mRNA species. Other oligonucleotides used are listed in Supplementary Table S2. Hybridization was performed using Ambion ULTRAhyb or ULTRAhyb–Oligo according to the manufacturer's conditions.
Plasmid constructions, cloning and mutagenesis
The mini-PGK1 construct used (13) was cloned under the control of a tetoff promoter in the pCM189 plasmid after removal of the CYC1 transcriptional terminator. The mini-PGK1 3′-UTR was removed and replaced by the 2000-nt downstream of the stop codon of CTH2 [see Figure 4C for structure of the (Mini-PGK1/CTH2-3′-UTR) plasmid]. The ARE sequence AUUUA located 51–55 after the translation stop codon was mutated to AGGUA by directed mutagenesis with oligonucleotides listed in Supplementary Table S2.
Figure 4.
Mutation of Nrd1, Nab3 or Sen1 induces accumulation of 3′ extended CTH2 mRNA. (A) Accumulation of 3′ extended transcript of Cth2 in mutants of Nrd1/Nab3 complex. Northern-blot analysis of RNA extracts from wild-type, nrd1-102 and nab3-11 mutants grown at permissive temperature (23°C lanes) and then shifted for 1 h at non-permissive temperature (37°C lanes). The 3′-extended transcript is labelled pre-CTH2 and the asterisk indicates a putative processing intermediate. The asterisk marks a species 150–200 nt longer than the mature mRNA. (B) Accumulation of 3′-extended CTH2 in strain carrying a mutation in SEN1. Northern blot analysis of RNA extracts from isogenic wild-type and sen1-1 mutant grown at permissive temperature (23°C lanes) and then shifted for 1 h at non-permissive temperature (37°C lanes). (C) Representation of the mini-PGK1-CTH2 reporter. The reporter is under the control of a tetoff promoter (tetO7 Pr), followed by the mini-PGK1 ORF sequence (Mini-PGK1), a poly(G) cassette (G18) followed by the full-length CTH2 3′-UTR (2000-nt downstream of the CTH2 ORF). (D) Accumulation of a mini-PGK1-CTH2 extended transcript in strains carrying mutation in Nrd1 and Nab3. Northern blot analysis of the Mini-PGK1-CTH2 reporter expression in from wild-type, nrd1-102 and nab3-11 mutants grown at permissive temperature (23°C lanes) and then shifted for 1 h at non-permissive temperature (37°C lanes).
RNase H treatment
Deadenylation was performed essentially as described (14). Samples (20 µg) of RNA were annealed with a gene-specific primer located 161–180 nt 3′ to the stop codon (Supplementary Table S2), with or without 400 ng oligo(dT) for 10 min at 68°C and digested with 1.5 U RNase H at 30°C for 1 h.
3′-end rapid amplification of cDNA ends (RACE) and PCR
To map the CTH2 poly(A) site, 5 µg of wild-type mRNA were reverse transcribed with superscript II RT for 50 min at 44°C in presence of the oligo V(T)25-Adaptor. cDNA was then amplified with a reverse adaptor oligonucleotide and a forward oligonucleotide at the end of the coding sequence (Cth2-CDS-F-750). A second round of PCR amplification was performed with a forward oligonucleotide located 30-nt downstream (Cth2-CDS-F-780). After gel purification, PCR products were cloned into pGEM-T and 19 clones were sequenced. The same procedure was used to characterize the end of the 3′-extended transcript, using tfr4Δ mRNA, V(T)25-Adaptor oligonucleotide for the reverse transcription and forward primers Cth2-3′-UTR-F-1300 and Cth2-3′-UTR-F-1350, with the reverse adaptor oligonucleotide for the nested-PCR. To characterize the presence of an extended transcript (Figure 2), we reverse transcribed 5 µg of wild-type mRNA or tfr4Δ mRNA in presence of random hexanucleotides. Primers Cth2-CDS-F-750 and Cth2-3′-UTR-R-1340, located on either side of the predicted poly(A) site were used for the PCR reaction. Oligonucleotide sequences are listed in Supplementary Table S2.
Figure 2.
Extended Cth2 transcripts are detected in wild type and trf4 strains. (A) Locations of PCR primers for detection of 3′ extended CTH2 transcripts. (B) Some CTH2 mRNA is not cleaved co-transcriptionally. RT–PCR reactions were performed on RNA samples from wild-type, trf4Δ and cth2Δ strains with PCR primers located on either side of the CTH2 poly(A) site, and separated on an agarose gel.
RESULTS
Synthesis of CTH2 mRNA does not require the activity of the normal cleavage and polyadenylation machinery
The 3′ ends of almost all characterized yeast mRNAs are generated by co-transcriptional cleavage and coupled polyadenylation. To test whether this is also the case for CTH2, we tested temperature-sensitive lethal (ts-lethal) mutations in the Rna14 and Rna15 components of cleavage factor 1 (CF1) and the poly(A) polymerase Pap1 (Figure 1A). Transfer of rna14-1, rna15-2, pap1-2 or pap1-5 strains to the non-permissive temperature (37°C) for 1 h led to the complete loss of RPL30 mRNA, as expected. Strikingly, accumulation of the CTH2 mRNA was not abolished in the mutant strains at 37°C and, indeed, the levels were elevated relative to 23°C.
Figure 1.
Accumulation of CTH2 mRNA does not require the normal mRNA 3′ cleavage and polyadenylation machinery and the mRNA has an unusual site of polyadenylation. (A) Effect of ts-lethal mutations of the cleavage and polyadenylation machinery on CTH2 mRNA accumulation. RNA samples from indicated strains grown at 23°C and then shifted at 37°C for 1 h were analysed for CTH2, RPL30 mRNA expression and SCR1 as a loading control by northern blot. (B) CTH2 species present in rna15-2, rna14-1 and pap1-5 mutants at non-permissive temperature are deadenylated. Total RNA extracts of rna15-2, rna14-1 and pap1-5 mutants, grown at non-permissive temperature (37°C) for 1 h, were digested by RNase H in presence of an oligonucleotide located 50 nt before the estimated poly(A) site and with or without an oligo dT. Poly(A)+ and poly(A)– species are indicated on the right of the panel. (C) The poly(A) tail is added on the CTH2 mRNA at a non-conventional poly(A) site. 3′-end RACE was performed on total RNA extracted from the wild-type strain. The poly(A) sites identified were located in the [GU3–5]5 region and are indicated by arrows.
In the rna14-1, pap1-2 and pap1-5 strains the migration of CTH2 mRNA appeared faster than in the wild-type at 37°C, suggesting that deadenylated mRNA was accumulated (Figure 1A). To confirm this conclusion, we performed oligonucleotide directed RNase H cleavage (Figure 1B) on RNA extracted from the rna14-1, rna15-2 and pap1-5 strains 1 h after transfer to 37°C. All samples were cleaved with an oligo positioned 50-nt 5′ to the estimated 3′ end of the mature CTH2 mRNA, to release a smaller 3′ fragment of the mRNA, and in the presence or absence of oligo(dT) to remove the poly(A) tail. This experiment confirmed that a deadenylated form of CTH2 mRNA accumulated in rna14-1 and pap1-5 strains at non-permissive temperature, whereas CTH2 mRNA was polyadenylated in the rna15-2 strain.
These results indicated that 3′ end formation on CTH2 mRNA does not require the normal activity of Rna14 or Rna15, which are necessary for the stable accumulation of many different mRNA previously tested. Formation of the poly(A) tail requires the activities of Rna14 and Pap1 but, unusually, polyadenylation is also not required for CTH2 mRNA accumulation. In the mRNA 3′ processing mutants, the CTH2 mRNA level was substantially elevated at 37°C relative to 23°C. Under these conditions, almost all other newly synthesized pre-mRNAs are being degraded by the RNA surveillance system, and this competition may reduce turnover of CTH2 mRNA.
These analyses indicated that 3′-end formation on CTH2 was different from other characterized mRNAs. We therefore identified the polyadenylation site of the mature CTH2 mRNA transcript by 3′-end RACE to determine whether this also showed unusual features (Figure 1C). PCR products were cloned in pGEM-T and 19 clones were sequenced; the 17 relevant clones each identified poly(A) sites located within a [GU3–5]5 repeat sequence, with most transcripts terminating between the third and the fourth repeats.
To assess whether the 3′ ends of the CTH2 mRNA were generated by transcription termination, the [GU3–5]5 sequence and 40 nt of flanking sequence were inserted into the 3′ flanking region of a mini-PGK1 reporter (see below). Discrete 3′ ends were not detected in the vicinity of the [GU3–5]5 element (data not shown), indicating that it does not function as a transcription terminator.
The [GU3–5]5 repeat is unrelated to the consensus site for cleavage and poly(A) or any other characterized site of mRNA polyadenylation in yeast. Moreover, the flanking region also lacks sequence features expected at yeast poly(A) sites. These generally include a positioning element (PE) comprised of AAUAAA and related sequences, and an efficiency element that acts as a binding site for Nab4 (also known as Hrp1). In CTH2 a potential binding site for Nab4 (15) is located 40-nt 5′ to the [GU3–5]5 element but we could not detect any evident PE.
CTH2 mRNA can be generated from a 3′-extended precursor
Our data indicated that the 3′ ends of the mature CTH2 transcripts are not generated by conventional co-transcriptional cleavage by the polyadenylation machinery, which suggested that longer 3′-extended transcripts would be detectable in wild-type strains. This was assessed by RT–PCR using primers on either side of the 3′ end of the mature mRNA. The forward primer is located just before the stop codon within the ORF, whereas the reverse primer is located 1300-nt 3′ to the stop codon (Figure 2A). The RT–PCR product was detected in the wild-type strain, showing that a non-cleaved CTH2 transcript extends at least 1.3 kb beyond the 3′ end of the mRNA (Figure 2B). The PCR product is specific because it was not detected in a RT minus sample or in RNA extracted from a CTH2Δ strain. Consistent with data presented below, the signal was elevated in a strain lacking the Trf4 component of the TRAMP4 complex.
Our previous microarray analysis (4) indicated that the abundance of the CTH2 mRNA was elevated in strains lacking the nuclear-specific exosome component Rrp6 compared to the wild-type. This result suggested that CTH2 mRNA expression might be regulated by nuclear degradation (4). To test this hypothesis, we examined CTH2 mRNA expression by northern hybridization in strains with defects in the activity of the nuclear exosome.
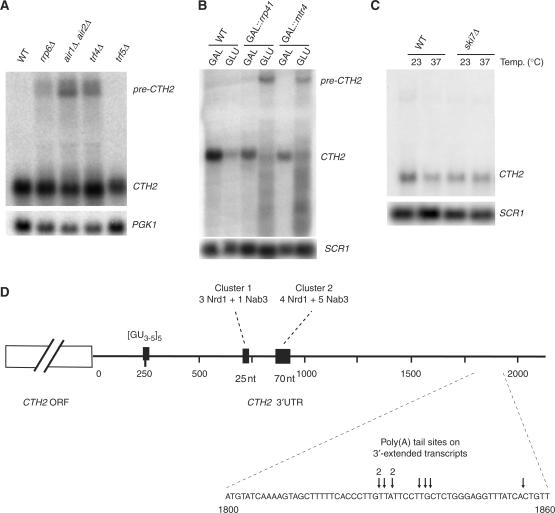
The rrp6Δ strain showed levels of mature CTH2 mRNA that were similar to wild-type, but accumulated an extended transcript of about 2.8 kb in length plus a smear of intermediate-sized RNAs (Figure 3A). The extended RNA species were detectable at low levels in the wild-type, consistent with the RT–PCR data. The TRAMP complexes function together with the nuclear exosome in the degradation of many RNA substrates, and we therefore also analysed strains lacking TRAMP components. The 3′-extended, 2.8-kb CTH2 RNA accumulated in strains with a double deletion for the functionally redundant AIR1 and AIR2 genes and in a trf4Δ strain lacking the poly(A) polymerase from the TRAMP4 complex (Figure 3A). In contrast, no accumulation of the extended transcripts was detected in a trf5Δ strain lacking the poly(A) polymerase from the TRAMP5 complex (Figure 3A). The DEVH-box, putative RNA helicase Mtr4 is common to both the TRAMP4 and TRAMP5 complexes, and is essential for viability. A conditional GAL::mtr4 strain was therefore depleted of Mtr4 by growth on glucose medium (Figure 3B). The extended CTH2 transcript was accumulated, also accompanied by substantial loss of the mature mRNA (Figure 3B). In addition, strong accumulation was seen for CTH2 RNA fragments, mostly shorter than the mRNA. A very similar phenotype was seen for a conditional GAL::rrp41 strain depleted of the core exosome component Rrp41. Strains with ts-lethal mutations in core exosome components, mtr3-1, rrp4-1 and rrp44-1 also showed accumulation of 3′ extended CTH2 mRNA (data not shown). In contrast, no effects on CTH2 were seen in a ski7Δ strain that lacks a cytoplasmic exosome component (Figure 3C).
Figure 3.
Inactivation of the exosome or TRAMP complex induces accumulation of 3′ extended CTH2 mRNA. (A) Northern-blot analysis for CTH2 mRNA expression of total RNA samples extracted from wild-type, rrp6Δ, air1Δ+air2Δ, trf4Δ and trf5Δ strains grown at 23°C. The 3′-extended transcript is labelled pre-CTH2. (B) Northern-blot analysis of RNA samples extracted from wild-type strain grown in glucose, GAL::rrp41 and GAL::mtr4 strains pre-grown in galactose (GAL) and then shifted to glucose medium for 20 h (GLU). (C) Northern-blot analysis of RNA extracts from isogenic wild-type and ski7Δ mutant grown at permissive temperature (23°C lanes) and then shifted for 1 h at non-permissive temperature (37°C lanes). (D) Characterization of the 3′-extended transcript. 3′-end RACE was performed on total RNA extracted from the trf4Δ strain. The poly(A) tails in nine clones sequenced were located around 1850 nt after the stop codon (illustrated with arrows on the figure), while the remaining clone was located 120 nt closer to the CTH2 ORF.
Levels of mature CTH2 mRNA were much more strongly affected in the PGAL-MTR4 strain than trf4Δ. This suggests that Trf5, and therefore the TRAMP5 complex, may participate in degradation of pre-CTH2, at least in the absence of Trf4. However, processing of several characterized exosome substrates, notably the precursors to the 5.8S rRNA, strictly require the activity of Mtr4, but do not clearly involve the TRAMP complexes. Mtr4 is present in excess over the other TRAMP components and can apparently independently activate the exosome on some substrates.
Oligo(dT) selection demonstrated that the 3′-extended CTH2 transcripts are polyadenylated, even in the trf4Δ strain (data not shown). We therefore determined the end points of the extended transcripts using 3′-end RACE on total RNA extracted from a trf4Δ strain. PCR products from the 3′ end RACE were cloned in pGEM-T and 10 clones were sequenced. Nine clones identified 3′ ends of the extended transcripts within a region of 25 nt centred around 1850-nt 3′ to the translation stop codon (Figure 3D). The sequence flanking these sites also lacked clear resemblance to consensus mRNA polyadenylation signals. The remaining clone terminated 120 nt closer to the CTH2 ORF (position +1722 relative to the translation stop codon). The positions of the majority of the stops would be consistent with the gel migration of the 3′-extended CTH2 transcripts, representing the CTH2 ORF (855 nt), together with a 3′ UTR of 1.85 kb. Inspection of the northerns also suggests that there is some length heterogeneity in the extended CTH2 transcripts, but the positions around +1850 appear to represent the major 3′ ends.
These data indicate that CTH2 mRNA has an unusual mode of synthesis. The major mRNA species has a poly(A) tail added at a sequence unrelated to any characterized polyadenylation site, and appears to be generated from a long 3′ extended species, which is processed by the nuclear exosome together with Mtr4 and the TRAMP4 complex.
3′ extended CTH2 pre-mRNA is a target for Nrd1, Nab3 and Sen1
Co-transcriptional binding of the Nrd1/Nab3 heterodimer of RNA-binding proteins to nascent transcripts leads to transcription termination on genes encoding snRNAs, snoRNAs and cryptic unstable intergenic transcripts (CUTs) (16–20). In addition, Nrd1/Nab3 are involved in exosome recruitment onto RNA (21) and a high-throughput screen identified CTH2 as an mRNA down-regulated by depletion of Nab3 (22). This suggested that Nrd1/Nab3 might be involved in recruiting the exosome to the 3′-extended CTH2 pre-mRNA. Inspection of the pre-mRNA sequence revealed multiple consensus binding sequences for Nrd1 and Nab3 (10 predicted Nrd1 binding sites and 24 Nab3 binding sites) along the 1.85 kb of the 3′-UTR, with two major clusters centred around positions +720 and +930 relative to the translation termination codon (Figure 3D).
Expression of the CTH2 mRNA was examined in strains with ts-lethal mutations in the Nrd1/Nab3 complex, nrd1-102 and nab3-11. In the nrd1-102 strain, the pre-CTH2 was mildly accumulated (1.9-fold) following transfer to 37°C, although the mature transcript was still detected (Figure 4A). In the nab3-11 strain, the phenotype was more pronounced (5-fold accumulation), with almost complete disappearance of the mature mRNA (Figure 4A). Nrd1 and Nab3 frequently function together with the RNA helicase Sen1 (16–18,21,23), and a ts-lethal sen1-1 mutant also showed accumulation of the 3′-extended pre-CTH2 at non-permissive temperature (Figure 4B). In addition to the species corresponding to the 3′ ends at +1850, a shorter extended transcript was observed in the nrd1, nab3 and sen1 mutants (asterisk in Figure 4A and B). This species is extended by 150–200 nt and we speculate that it reflects pausing during 3′ exonuclease digestion.
To assess processing of the CTH2 3′ UTR in the absence of the remainder of the gene, a 2-kb region was inserted into a reporter plasmid expressing a well-characterized mini-PGK1 construct (24,25) under the control of a doxycyclin-repressible tet07 promoter (Figure 4C). The reporter also contains a poly(G) cassette, which results in around 30% inhibition of degradation in the cytoplasm (26), but it is unclear whether it has any effects in the nucleus. The reporter generated a prominent transcript with the gel mobility expected for a species extended to +1850 (Figure 4D). This was strongly elevated in nrd1-102 and nab3-11 strains, consistent with the model that the extended CTH2 RNA is a target for Nrd1/Nab3. The level of the 3′-extended PGK1-CTH2 RNA relative to the ‘mature’ RNA was higher in the reporter than for endogenous CTH2, suggesting that additional features of the CTH2 sequence and/or genomic context contribute to the normally very rapid maturation.
ARE-mediated regulation of CTH2 mRNA levels
In human cells, Tis11/TTP is subject to auto-regulation via an ARE sequence within its 3′ UTR (27,28). Inspection of the 3′ UTR of yeast CTH2 revealed a perfect consensus ARE sequence (UUAUUUAUU) located 50-nt 3′ to the ORF (Figure 1C), suggesting that it too might be subject to ARE-mediated regulation. To assess this, we tested the effects of double-point mutations (UUAUUUAUU -> UUAGGUAUU) in the mini-PGK1 reporter plasmid. Transcription of the reporter was inhibited and the decay of the mRNA and 3′-extended pre-mRNA was followed over time. In the strain expressing the reporter with the double-point mutation (ΔARE), the levels of both the mRNA and extended forms of the RNA were elevated under induced conditions (Figure 5A, 0-min lanes). Following doxycyclin addition the mRNA carrying the ΔARE mutation decayed more slowly (Figure 5A and quantified for the mRNA in 5B). We conclude that CTH2 is subject to ARE-mediated decay.
Figure 5.
CTH2 mRNA expression is regulated by ARE-dependent degradation and by the nuclear-specific 5′-exonuclease Rat1. (A) Half-life analysis of the mini-PGK1-CTH2 reporter plasmid with or without mutation of the ARE element. Wild-type strains were transfected with Mini-PGK1-CTH2 plasmid (Figure 4C) containing either the wild-type ARE element (WT) or a double-point mutated ARE element (ΔARE). Half-lives of reporters (WT and ΔARE) have been determined by northern blot analysis of total RNA extracted from cultures harvested at different time points after addition of doxycycline (2 µg/ml) to inhibit transcription. (B) Quantification of half-live measurements. The graph shows mean values ± standard deviations for the mini-PGK1-CTH2 transcript, obtained from quantification of three independent experiments and normalized to the SCR1 loading control. (C) Regulation of CTH2 mRNA expression by Rat1. Northern blot analysis of total RNA extracted from isogenic wild-type and rat1-2 strains at permissive temperature (23°C) and 30 min, 1 h and 2 h after transfer to non-permissive temperature (37°C). (D) Quantification of the previous experiment. The graph shows mean values ± standard deviations for the endogenous CTH2 transcript, obtained from quantification of three independent experiments and normalized to the SCR1 loading control.
Yeast contains two related proteins Cth1 and Cth2. To assess whether Cth1 or Cth2 was responsible for targeting the CTH2 mRNA, the stability of the reporter was assessed in the cth1Δ and cth2Δ single mutants and the cth1Δ cth2Δ double mutant. Over multiple experiments the reproducibility of the data was poor, presumably due to subtle differences in growth conditions and the complexity of CTH2 regulation. Strains lacking Cth1 showed clearly increased levels of the reporter in some experiments, but over multiple experiments the data could not be demonstrated to be statistically significant.
In the cytoplasm, ARE-mediated decay can stimulate both 3′ and 5′ degradation. In at least some cases, the largest effect is on 5′ degradation, which is performed by the cytoplasmic 5′ to 3′ exonuclease Xrn1 (29). We speculated that the ARE-mediated decay of CTH2 mRNA might be a nuclear event and therefore tested the effects of mutation of the exclusively nuclear 5′ to 3′ exonuclease Rat1. For this we made use of the ts-lethal tap1-1 allele (30) (hereafter referred to as rat1-2). Transfer of the wild-type strain to 37°C led to a ∼2- fold reduction in CTH2 levels. In contrast, transfer of the rat1-2 strain to non-permissive temperature led to a marked increase in the level of both the CTH2 mRNA and 3-extended pre-mRNA (Figure 5C and D). The increase was most marked 30 min after transfer and subsequently declined. This decline may reflect the rapid growth inhibition seen in the rat1-2 strain at non-permissive temperature. Increased levels of pre-CTH2 and CTH2 RNAs were also seen in strains carrying MET::rat1 following addition of methionine to the medium (Supplementary Figure S1).
These data indicate that CTH2 is a target for both ARE-mediated decay and nuclear degradation by Rat1.
DISCUSSION
We have presented several lines of data showing that the synthesis of CTH2 follows an unusual pathway (Figure 6). Accumulation of the mRNA is resistant to transfer of the rna14-1, rna15-2 and pap1 mutant strains to non-permissive temperature. Indeed, CTH2 mRNA levels were strikingly increased in rna14-1, rna15-2 and pap1-5 strains at 37°C. In these strains, massive nuclear pre-mRNA degradation is presumably occurring, as the surveillance machinery attempts to degrade the aberrant 3′ extended forms of the products of other protein coding genes (2). The increased level of CTH2 mRNA may be a consequence of competition for nuclear surveillance and degradation systems. This would imply a considerable level of nuclear degradation of CTH2 mRNA in wild-type cells. The major site of the poly(A) tails present in the mature CTH2 mRNA comprised a [GU3–5]5 repeat element that differs from all previously characterized yeast polyadenylation sites. Inspection of the yeast genome revealed the presence of related sequences located 3′ to several other ORFs (data not shown), but the functional significance of these elements has not yet been assessed. Together, these results indicated that the mature CTH2 mRNA is generated by the processing of a 3′-extended pre-mRNA transcript, followed by polyadenylation that is not coupled to pre-mRNA cleavage. It seems likely that related pathways will be used for other genes and in other eukaryotes.
Figure 6.
Mature Cth2 mRNA processing model. Data presented here suggest that CTH2 mRNA transcription goes far beyond the poly(A) signal and that co-transcriptional cleavage at the poly(A) site does not occur. Binding of the Nrd1/Nab3 complex onto the transcript, in association with the action of Sen1, might terminate transcription and/or recruit TRAMP and the exosome complexes onto the transcript. With the help of the TRAMP complex, the exosome is then able to degrade the primary transcript but pauses at the [GUn]5 site, where the transcript is subsequently polyadenylated by Pap1.
Processing of the CTH2 pre-mRNA is inhibited by inhibition of the core exosome or loss of the nuclear-specific exosome component Rrp6. Efficient maturation also requires the TRAMP4 polyadenylation complex and the Nrd1/Nab3/Sen1 complex, both of which have previously been identified as co-factors for the nuclear exosome (19–21,31–34). Nrd1 and Nab3 are believed to bind target RNAs co-transcriptionally, since they can promote transcription termination on other transcripts (16,17,19,20). The extended 3′-UTR of the CTH2 pre-mRNA contains numerous potential binding sites for Nrd1 and Nab3, including two conspicuous clusters. We predict that the Nrd1/Nab3/Sen1 complex binds the nascent CTH2 transcript, and recruits the TRAMP4 complex and the nuclear exosome. The exosome then rapidly degrades the pre-mRNA, soon after transcription termination generates a free 3′ end (Figure 6).
The 3′ UTR of CTH2 includes a sequence of 12 adenosine residues (labelled A12 in Figure 1C). We initially speculated that proteins (e.g. Pab1) bound to the A12 sequence might halt the exosome allowing poly(A) addition. However, all sequenced CTH2 cDNAs terminated further 3′ than the A12 element. Previous analysis of the NAB2 gene indicated that the auto-regulation of mRNA levels involves an encoded A26 sequence (35). Whether the A12 sequence in CTH2 plays a role in regulation remains to be determined. We also considered that the Nab4 protein might contribute to halting exonuclease progression, since a putative binding site (UAUAUA) is located 40-nt 5′ to the [GU3–5]5 element. Strains carrying nab4-1, nab4-4 and nab4-7 mutations did not clearly reduce CTH2 mRNA formation (data not shown), arguing against this model. However, a caveat is that different alleles of NAB4 show distinctly different phenotypes (36), so it is conceivable that other alleles would have stronger effects.
A potential explanation is that at least some of the components of the normal cleavage and polyadenylation machinery associate with sequences around the [GU3–5]5 element and block progression of the exosome, as previously proposed for 3′ extended pre-mRNAs in rna14 mutants (2). In the rna14-1 mutant strain, the CTH2 mRNA lacks a poly(A) tail. This shows that Rna14 is involved in polyadenylation, implying that it is present, even though it is not required for 3′-end formation. The poly(A) tail is also absent in pap1 mutants, indicating that the 3′-processed CTH2 mRNA is polyadenylated by Pap1.
In yeast, intergenic regions are generally short, and the extended CTH2 transcripts run through much of the length of the SLX4 (YLR135W) gene, which is encoded on the opposite strand. Both polyadenylation site of the extended transcript and the cluster of Nrd1/Nab3 binding sites lie within the ORF of SLX4. Comparison of the locations of the Nrd1/Nab3 binding sites in the CTH2 transcript to the protein-coding capacity of SLX4 yielded no clear pattern. Some predicted Nrd1/Nab3 binding sites correspond to evolutionarily conserved amino acids, while others do not. An obvious difficulty in interpretation is that it is currently unclear whether all of the predicted Nrd1/Nab3 binding sites actually function in targeting the transcript for processing. No significant alterations in SLX4 expression were detected in the microarray analyses of exosome mutants in which increased CTH2 levels were observed (4).
Regulation of CTH2 levels
Cth2 is involved in ARE-dependent regulation of mRNA stability, particularly in the context of Fe deprivation, allowing coordinated inhibition of the expression of several genes. The mammalian homologue TTP auto-regulates its own mRNA via ARE sequences present in its 3′-UTR (27,28). The 3′-UTR of CTH2 mRNA contains a perfect consensus ARE nonamer (UUAUUUAUU) in a U-rich environment, located 50 nt after the translation stop codon. This sequence is functionally important, since a double-point mutation (UUAGGUAUU) significantly stabilized both the 3′ extended and mature forms of a reporter construct containing the entire 3′ flanking region of CTH2. Cytoplasmic ARE-mediated degradation is reported to promote mRNA decapping and 5′ degradation by the 5′ exonuclease Xrn1 (29). Since all our data indicated that CTH2 is subject to nuclear processing, we tested the effects of mutation or depletion of the nuclear 5′ exonuclease Rat1. The levels of both pre-CTH2 and CTH2 were rapidly increased by inhibition of Rat1. This suggests that the ARE stimulates 5′-degradation of pre-CTH2 and CTH2 by Rat1. It was previously reported that deletion of the gene encoding Cth1, a homologue of Cth2, up-regulates CTH2 mRNA levels (9), strongly suggesting that Cth1 participates in the ARE-mediated regulation of CTH2 mRNA. Consistent with this, we saw increased levels of the mini-PGK1-CTH2 reporter in strains lacking Cth1, but the data could not be shown to be statistically significant.
We postulate that CTH2 is subject to ARE-mediated degradation, possibly mediated by Cth1, and that this involves the nuclear 5′ exonuclease activity of Rat1. The unusual 3′ processing pathway deduced for CTH2 mRNA, may allow time for nuclear surveillance and degradation to occur on the pre-mRNA, prior to export of the mRNA to the cytoplasm. The nuclear localization of ARE-mediated degradation of CTH2 may be important for its physical and functional separation from the cytoplasmic ARE-mediated degradation of target RNAs.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Aziz El Hage for providing the MET::rat1, xrn1Δ strain, Claudia Schneider and Jon Houseley for critical reading of the article. This work was funded by the Wellcome Trust and a FEBS long-term fellowship to M.T.B. Funding to pay the Open Access publication charges for this article was provided by The Wellcome Trust.
Conflict of interest statement. None declared.
REFERENCES
- 1.Houseley J, LaCava J, Tollervey D. RNA-quality control by the exosome. Nat. Rev. Mol. Cell. Biol. 2006;7:529–539. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- 2.Torchet C, Bousquet-Antonelli C, Milligan L, Thompson E, Kufel J, Tollervey D. Processing of 3′ extended read-through transcripts by the exosome can generate functional mRNAs. Mol. Cell. 2002;9:1285–1296. doi: 10.1016/s1097-2765(02)00544-0. [DOI] [PubMed] [Google Scholar]
- 3.Birse CE, Minvielle-Sebastia L, Lee BA, Keller W, Proudfoot NJ. Coupling termination of transcription to messenger RNA maturation in yeast. Science. 1998;280:298–301. doi: 10.1126/science.280.5361.298. [DOI] [PubMed] [Google Scholar]
- 4.Houalla R, Devaux F, Fatica A, Kufel J, Barrass D, Torchet C, Tollervey D. Microarray detection of novel nuclear RNA substrates for the exosome. Yeast. 2006;23:439–454. doi: 10.1002/yea.1369. [DOI] [PubMed] [Google Scholar]
- 5.Ciais D, Cherradi N, Bailly S, Grenier E, Berra E, Pouyssegur J, Lamarre J, Feige JJ. Destabilization of vascular endothelial growth factor mRNA by the zinc-finger protein TIS11b. Oncogene. 2004;23:8673–8680. doi: 10.1038/sj.onc.1207939. [DOI] [PubMed] [Google Scholar]
- 6.Vasudevan S, Steitz JA. AU-rich-element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell. 2007;128:1105–1118. doi: 10.1016/j.cell.2007.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fan XC, Myer VE, Steitz JA. AU-rich elements target small nuclear RNAs as well as mRNAs for rapid degradation. Genes Dev. 1997;11:2557–2568. doi: 10.1101/gad.11.19.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Puig S, Askeland E, Thiele DJ. Coordinated remodeling of cellular metabolism during iron deficiency through targeted mRNA degradation. Cell. 2005;120:99–110. doi: 10.1016/j.cell.2004.11.032. [DOI] [PubMed] [Google Scholar]
- 9.Thompson MJ, Lai WS, Taylor GA, Blackshear PJ. Cloning and characterization of two yeast genes encoding members of the CCCH class of zinc finger proteins: zinc finger-mediated impairment of cell growth. Gene. 1996;174:225–233. doi: 10.1016/0378-1119(96)00084-4. [DOI] [PubMed] [Google Scholar]
- 10.Gietz RD, Schiestl RH, Willems AR, Woods RA. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- 11.Tollervey D. A yeast small nuclear RNA is required for normal processing of pre-ribosomal RNA. EMBO J. 1987;6:4169–4175. doi: 10.1002/j.1460-2075.1987.tb02763.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 13.Mitchell P, Tollervey D. An NMD pathway in yeast involving accelerated deadenylation and exosome-mediated 3′–>5′ degradation. Mol. Cell. 2003;11:1405–1413. doi: 10.1016/s1097-2765(03)00190-4. [DOI] [PubMed] [Google Scholar]
- 14.Muhlrad D, Parker R. Mutations affecting stability and deadenylation of the yeast MFA2 transcript. Genes Dev. 1992;6:2100–2111. doi: 10.1101/gad.6.11.2100. [DOI] [PubMed] [Google Scholar]
- 15.Zhao J, Hyman L, Moore C. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 1999;63:405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steinmetz EJ, Conrad NK, Brow DA, Corden JL. RNA-binding protein Nrd1 directs poly(A)-independent 3′-end formation of RNA polymerase II transcripts. Nature. 2001;413:327–331. doi: 10.1038/35095090. [DOI] [PubMed] [Google Scholar]
- 17.Kim M, Vasiljeva L, Rando OJ, Zhelkovsky A, Moore C, Buratowski S. Distinct pathways for snoRNA and mRNA termination. Mol. Cell. 2006;24:723–734. doi: 10.1016/j.molcel.2006.11.011. [DOI] [PubMed] [Google Scholar]
- 18.Morlando M, Ballarino M, Greco P, Caffarelli E, Dichtl B, Bozzoni I. Coupling between snoRNP assembly and 3′ processing controls box C/D snoRNA biosynthesis in yeast. EMBO J. 2004;23:2392–2401. doi: 10.1038/sj.emboj.7600254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thiebaut M, Kisseleva-Romanova E, Rougemaille M, Boulay J, Libri D. Transcription termination and nuclear degradation of cryptic unstable transcripts: a role for the nrd1-nab3 pathway in genome surveillance. Mol. Cell. 2006;23:853–864. doi: 10.1016/j.molcel.2006.07.029. [DOI] [PubMed] [Google Scholar]
- 20.Arigo JT, Eyler DE, Carroll KL, Corden JL. Termination of cryptic unstable transcripts is directed by yeast RNA-binding proteins Nrd1 and Nab3. Mol. Cell. 2006;23:841–851. doi: 10.1016/j.molcel.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 21.Vasiljeva L, Buratowski S. Nrd1 Interacts with the Nuclear Exosome for 3′ Processing of RNA Polymerase II Transcripts. Mol. Cell. 2006;21:239–248. doi: 10.1016/j.molcel.2005.11.028. [DOI] [PubMed] [Google Scholar]
- 22.Mnaimneh S, Davierwala AP, Haynes J, Moffat J, Peng WT, Zhang W, Yang X, Pootoolal J, Chua G, Lopez A, et al. Exploration of essential gene functions via titratable promoter alleles. Cell. 2004;118:31–44. doi: 10.1016/j.cell.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 23.Ursic D, Himmel KL, Gurley KA, Webb F, Culbertson MR. The yeast SEN1 gene is required for the processing of diverse RNA classes. Nucleic Acids Res. 1997;25:4778–4785. doi: 10.1093/nar/25.23.4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hagan KW, Ruiz-Echevarria MJ, Quan Y, Peltz SW. Characterization of cis-acting sequences and decay intermediates involved in nonsense-mediated mRNA turnover. Mol. Cell. Biol. 1995;15:809–823. doi: 10.1128/mcb.15.2.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang S, Ruiz-Echevarria MJ, Quan Y, Peltz SW. Identification and characterization of a sequence motif involved in nonsense-mediated mRNA decay. Mol. Cell. Biol. 1995;15:2231–2244. doi: 10.1128/mcb.15.4.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cao D, Parker R. Computational modeling and experimental analysis of nonsense-mediated decay in yeast. Cell. 2003;113:533–545. doi: 10.1016/s0092-8674(03)00353-2. [DOI] [PubMed] [Google Scholar]
- 27.Brooks SA, Connolly JE, Rigby W.FC. The role of mRNA turnover in the regulation of tristetraprolin expression: evidence for an extracellular signal-regulated kinase-specific, AU-rich element-dependent, autoregulatory pathway. J. Immunol. 2004;172:7263–7271. doi: 10.4049/jimmunol.172.12.7263. [DOI] [PubMed] [Google Scholar]
- 28.Tchen CR, Brook M, Saklatvala J, Clark AR. The stability of tristetraprolin mRNA is regulated by mitogen-activated protein kinase p38 and by tristetraprolin itself. J. Biol. Chem. 2004;279:32393–32400. doi: 10.1074/jbc.M402059200. [DOI] [PubMed] [Google Scholar]
- 29.Stoecklin G, Mayo T, Anderson P. ARE-mRNA degradation requires the 5′-3′ decay pathway. EMBO Rep. 2006;7:72–77. doi: 10.1038/sj.embor.7400572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Di Segni G, McConaughy BL, Shapiro RA, Aldrich TL, Hall BD. TAP1, a yeast gene that activates the expression of a tRNA gene with a defective internal promoter. Mol. Cell. Biol. 1993;13:3424–3433. doi: 10.1128/mcb.13.6.3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vanacova S, Wolf J, Martin G, Blank D, Dettwiler S, Friedlein A, Langen H, Keith G, Keller W. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 2005;3:e189. doi: 10.1371/journal.pbio.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kadaba S, Krueger A, Trice T, Krecic AM, Hinnebusch AG, Anderson J. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev. 2004;18:1227–1240. doi: 10.1101/gad.1183804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wyers F, Rougemaille M, Badis G, Rousselle J.-C, Dufour M.-E, Boulay J, Régnault B, Devaux F, Namane A, Séraphin B, et al. Cryptic Pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 34.LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;21:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 35.Roth KM, Wolf MK, Rossi M, Butler JS. The nuclear exosome contributes to autogenous control of NAB2 mRNA levels. Mol. Cell. Biol. 2005;25:1577–1585. doi: 10.1128/MCB.25.5.1577-1585.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kessler MM, Henry MF, Shen E, Zhao J, Gross S, Silver PA, Moore CL. Hrp1, a sequence-specific RNA-binding protein that shuttles between the nucleus and the cytoplasm, is required for mRNA 3′-end formation in yeast. Genes Dev. 1997;11:2545–2556. doi: 10.1101/gad.11.19.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.