Abstract
Gene transfer has been used to correct inherited immunodeficiencies, but in several patients integration of therapeutic retroviral vectors activated proto-oncogenes and caused leukemia. Here, we describe improved methods for characterizing integration site populations from gene transfer studies using DNA bar coding and pyrosequencing. We characterized 160 232 integration site sequences in 28 tissue samples from eight mice, where Rag1 or Artemis deficiencies were corrected by introducing the missing gene with gamma-retroviral or lentiviral vectors. The integration sites were characterized for their genomic distributions, including proximity to proto-oncogenes. Several mice harbored abnormal lymphoproliferations following therapy—in these cases, comparison of the location and frequency of isolation of integration sites across multiple tissues helped clarify the contribution of specific proviruses to the adverse events. We also took advantage of the large number of pyrosequencing reads to show that recovery of integration sites can be highly biased by the use of restriction enzyme cleavage of genomic DNA, which is a limitation in all widely used methods, but describe improved approaches that take advantage of the power of pyrosequencing to overcome this problem. The methods described here should allow integration site populations from human gene therapy to be deeply characterized with spatial and temporal resolution.
INTRODUCTION
Successful correction of human genetic diseases has been achieved for X-linked severe combined immunodeficiency (SCID) (1), adenosine deaminase deficiency (ADA-SCID) (2) and chronic granulomatous disease (CGD) (3). However, several serious adverse events occurred during therapeutic gene transfer for X-SCID, in which the gamma-retrovirus-based vectors used for gene correction integrated near cellular proto-oncogenes and contributed to transformation (4,5). For this reason there is intense interest in improving methods for analyzing integration site placement during gene therapy, both to allow monitoring of possible incipient adverse events and to guide the development of safer integrating vectors. Here, we describe the use of DNA bar coding and pyrosequencing to allow efficient determination of integration site sequences from many samples, using murine models of therapeutic gene transfer.
Human severe combined immunodeficiencies have been identified that involve mutations in the genes for the Artemis and Rag1 proteins, prompting interest in devising gene correction strategies (6–8). For the Artemis gene, two groups have devised lentiviral vectors capable of gene correction (9,10). For Rag1, an MLV-based vector transducing Rag1 is available (11). However, questions of genotoxicity have arisen in both models, which we have investigated in this study.
For the Artemis model, the mice are tumor prone due to the loss of Artemis itself, particularly in combination with conditioning prior to cell transfer with irradiation, as was used in pilot studies (12). Thus elevated transformation rates were expected after gene transduction, and it was observed that some of the transformed cells harbored integrated lentiviral vectors. Similarly, lymphoproliferations were seen in Rag1 gene corrected mice, and analysis of transformed cells showed that they contained integrated gamma-retroviral vectors (11). It was thus of interest to investigate whether transformation might involve insertional activation of proto-oncogenes.
Here, we describe the use of DNA bar coding and pyrosequencing to monitor genotoxicity during gene correction. We used the massively parallel pyrosequencing method commercialized by 454 Life Sciences (13), which at the time this work was carried out was capable of generating up to 200 000 sequence reads of about 100 bp each per run (since then, the numbers of reads available per run and the read lengths have increased). The DNA bar coding method (14–16) allowed amplicons from different samples to be pooled for sequencing, then separated afterwards using information contained in the DNA bar codes. In the study reported here, we demonstrate the utility of these methods by analyzing the spatial distribution of integration sites in gene-corrected mice with lymphoproliferations or healthy controls. We analyzed 28 tissue samples from eight mice, each sample with a unique DNA bar code. After quality control, 160 232 sequence reads could be mapped to unique sites on the murine genome. Because of the very large number of sequence reads available, integration sites could be annotated with their frequency of recovery, providing information relevant to evaluating possible insertional activation of proto-oncogenes and the distribution of transduced cell clones. We also show that the typical recovery methods used for integration sites isolation are highly biased, but the use of pyrosequencing has the potential to overpower the problem. Together these data illustrate a collection of methods that substantially improves our ability to monitor populations of integration sites generated during therapeutic gene transfer.
MATERIALS AND METHODS
Vector transduction and preparation of tissue samples
The construction and preparation of lentiviral and gamma-retroviral vectors, and transduction of hematopoietic stem cells are described in (10,11). To identify integration site sequences, mice were sacrificed and genomic DNA was isolated from tissues indicated in Table 1, as described (10,11).
Table 1.
Tissues analyzed by integration site sequencing and frequency of integration near Cancergenes
| Mouse | Disease state | Tissue | Integration sites | Sites within 50 kb cancergenes 5′ end | Percentage of Cancergenes (tissue) | Percentage of Cancergenes (mouse) |
|---|---|---|---|---|---|---|
| Total reads (unique sites) | Total reads (unique sites) | Total reads (unique sites) | Total reads (unique sites) | |||
| Gamma-retroviral vector in rag-deficient mice | ||||||
| 1 | Healthy control | LN | 2360 (274) | 269 (45) | 11.4 (16.4) | 10.1 (15.3) |
| marrow | 2701 (101) | 137 (17) | 5.1 (16.8) | |||
| thymus | 3262 (103) | 436 (11) | 13.4 (10.7) | |||
| 215 | Lymphoproliferation | LN | 6950 (60) | 262 (6) | 3.8 (10.0) | 3.3 (13.6) |
| marrow | 1893 (33) | 39 (3) | 2.1 (9.1) | |||
| spleen | 4734 (48) | 173 (7) | 3.7 (14.6) | |||
| thymus | 4044 (43) | 104 (9) | 2.6 (20.9) | |||
| X | Lymphoproliferation | liver | 4049 (40) | 1344 (10) | 33.2 (25.0) | 33.3 (23.5) |
| marrow | 4160 (53) | 1433 (11) | 34.4 (20.8) | |||
| spleen | 5287 (43) | 1717 (11) | 32.5 (25.6) | |||
| Lentiviral (EF1α) vector in artemis-deficient mice | ||||||
| 22 | Healthy control | LN | 7789 (325) | 268 (21) | 3.4 (6.5) | 5.1 (6.0) |
| marrow | 3387 (89) | 322 (4) | 9.5 (4.5) | |||
| spleen | 7028 (252) | 482 (16) | 6.9 (6.3) | |||
| thymus | 3325 (50) | 25 (2) | 0.8 (4.0) | |||
| 31 | Healthy cells pretransplantation | Sca1+ | 9574 (337) | 735 (25) | 7.7 (7.4) | 7.7 (7.4) |
| Lymphoproliferation | LN | 3687 (215) | 89 (20) | 2.4 (9.3) | 1.7 (9.0) | |
| spleen | 1669 (6) | 0 (0) | 0 (0) | |||
| 8 | Healthy cells pretransplantation | Sca1+ | 10648 (265) | 597 (18) | 5.6 (6.8) | 5.6 (6.8) |
| Lymphoproliferation | LN | 737 (46) | 3 (3) | 0.4 (6.5) | 0.04 (3.9) | |
| liver | 14041 (11) | 0 (0) | 0 (0) | |||
| pleural fluid | 7183 (55) | 13 (3) | 0.2 (5.5) | |||
| spleen | 7179 (18) | 0 (0) | 0 (0) | |||
| thymus | 12290 (23) | 0 (0) | 0 (0) | |||
| Lentiviral (PGK) vector in artemis-deficient mice | ||||||
| 401 | Healthy control | LN | 1647 (65) | 117 (4) | 7.1 (6.2) | 4.3 (9.5) |
| marrow | 379 (41) | 41 (4) | 10.8 (9.8) | |||
| spleen | 1265 (40) | 40 (4) | 3.2 (10.0) | |||
| thymus | 2487 (53) | 50 (7) | 2.0 (13.2) | |||
| 613 | Healthy cells pretransplantation | Sca1+ | 26477 (37) | 0 (0) | 0 (0) | 0 (0) |
| Lymphoproliferationa | LN | 42 (5) | 0 (0) | 0 (0) | 1.0 (5.9) | |
| marrow | 31 (6) | 1 (1) | 3.2 (16.7) | |||
| thymus | 26 (6) | 0 (0) | 0 (0) | |||
| MLV-based vector in MEF | 7420 (4828) | 1003 (632) | 13.5 (13.1) | |||
| HIV-based vector in MEF | 3929 (2441) | 329 (209) | 8.4 (8.6) | |||
aThese samples were sequenced using the Sanger method only.
LN, lymph nodes.
As controls, murine embryonic fibroblasts (MEFs) were transduced with either a lentiviral vector or a gamma-retroviral vector, followed by integration site analysis. MEFs were cultured in DMEM supplemented with 10% heat-inactivated FCS, 2 mM glutamine, 0.1 mM β-mercaptoethanol, nonessential amino acids, 1 mM sodium pyruvate and 50 μg/ml gentamycin. The lentiviral vector was prepared as described previously (17,18). The gamma-retroviral vector was prepared by transfection of the MLV vector segment (pMLV LTR-GFP), the packaging construct pCGP producing the MLV Gag and Pol polyproteins (pCGP), and the vesicular stomatitis virus G-producing plasmid (pMD.G). Viral supernatant was harvested 48 h after transfection, filtered through 0.45 μm filters, concentrated, treated with DNaseI, and stored frozen at −80°C. To isolate integration sites, MEFs (3 × 106 in a 10 cm dish) were inoculated with 1 μg of p24 capsid antigen for 7–15 h in the presence of 10 μg/ml DEAE-dextran, washed, and cultured for additional 48 h in culture medium. Cells were harvested, genomic DNA was isolated and isolation of the integration sites was performed as described subsequently.
Isolation of integration site sequences
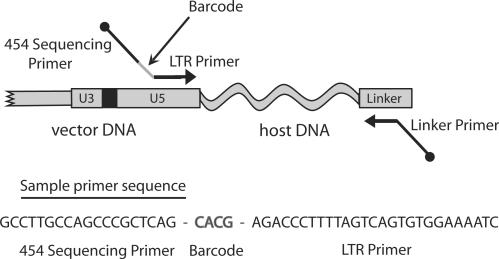
To determine vector integration sites on the mouse genome, DNA fragments from host–vector junction were prepared using ligation-mediated PCR (6,17–20). Briefly, each DNA sample (1–1.5 μg) was digested with MseI. The digested samples were ligated to linkers, and then amplified by nested PCR. In order to sequence all the samples in a single sequencing experiment, primers that contain unique 4-bp barcodes were used in the second PCR step (Figure 1, Supplementary Table 1). The PCR products were gel purified, pooled, and then subjected to pyrosequencing as implemented by 454 Life Sciences (13). To minimize bar code ‘crossover’, the mouse samples were separated into four quadrants on a single picotiter sequencing plate. Inspection of the integration sites revealed 23 instances (<0.01% of total sequence reads) in which an identical integration site was observed in different mice in the same quadrant with barcodes of edit distance 1 (i.e. ‘barcode collisions’). These barcode collisions were resolved by removing all collision sites from the sample with the lower number of clones (which was usually one).
Figure 1.
The DNA bar coding strategy. Each LTR primer used in ligation-mediated PCR contained a unique DNA barcode that specified the mouse and tissue of origin. Each barcode consists of a unique 4-bp nucleotide sequence, inserted between the sequencing primer binding site and the LTR specific primer segment. Thus all sequencing reads begin with the 4-bp barcode identifiers. A sample primer with a bar code is shown at the bottom of the diagram.
Integration sites were judged to be authentic if the sequences began within 3 bp of vector LTR ends, had either a >98% sequence match or no more than one base mismatch if the read length was <50 bp, and had a unique best hit when aligned to the draft mouse genome (mm8) using BLAT. The sequence data from this study have been submitted to GenBank under accession nos. ET648700-ET656552.
The lymphoproliferation samples from mouse 8, 31 and 613 were additionally analyzed by LAM-PCR (21). Briefly, 5′ Biotinylated primers LTRI (5′-GAGCTCTCTGGCTAACTAGG-3′) and LTRII (5′-GAACCCACTGCTTAAGCCTCA-3′) were used for pre-amplification of the vector-host junctions. After magnetic capture, hexanucleotide priming, and restriction digestion with Tsp509I, a linker cassette was ligated to the 3′ end of the genomic sequence. First exponential amplification of the vector-host junction was performed using linker cassette primer LCI and vector LTR specific primer LTRIII (5′-AGCTTGCCTTGAGTGCTTCA-3′), followed by second exponential PCR using primer LCII and LTRIV (5′-AGTAGTGTGTGCCCGTCTGT-3′). The final PCR product was separated on a spreadex high- resolution gel (Elchrom, Cham, Switzerland). Isolated specific DNA bands or LAM-PCR amplicons were purified, shotgun cloned into TOPO TA vector (Invitrogen, Carlsbad, CA, USA) and sequenced (GATC, Konstanz, Germany) using the Sanger method. Integration site sequences were aligned to the mouse genome as described before.
Cancer gene database (‘Cancergenes’)
To determine integration frequency near proto-oncogenes and tumor suppressors from tissue samples, a ‘Cancergene’ database was compiled from seven sources: (i) http://atlasgeneticsoncology.org (22); (ii) http://rtcgd.abcc.ncifcrf.gov (23); (iii) http://www.sanger.ac.uk/genetics/CGP/Census/(24); (iv) http://cc.ucsf.edu/people/waldman/GENES/completechroms.html; (v) Sjoblom et al. (25); (vi) a custom list of lymphoid-specific proto-oncogenes and (vii) a custom list of proto-oncogenes and tumor suppressors compiled from literature sources. A composite of these seven lists, totaling 1650 genes, was used to annotate the integration sites. This ‘Cancergenes’ list (allOnco) can be found at http://microb230.med.upenn.edu/protocols/cancergenes.html.
Bioinformatic and statistical analysis
Detailed bioinformatic methods can be found in (26) and (18), and Supplementary Data 1–4. All analysis was carried out using the mm8 genome draft.
RESULTS
The murine models studied
In each model studied, Sca1+ bone marrow cells were harvested and transduced ex vivo. Recipient mice were irradiated to depopulate bone marrow precursors and stem cells, and to increase the selective advantage of the injected transduced cells. Two vectors were used for gene correction of an Artemis knockout strain. Both were lentiviral vectors derived from the TRIPΔU3-vector (27). In one, the EF1α promoter drove expression of the Artemis gene; in the second, the PGK promoter drove expression (10). In the Rag1 correction model, the Rag1 gene was delivered using a gamma-retroviral vector Lagresle-Peyrou et al. (11). In both disease models, abnormal lymphoproliferative events were detected in some of the mice (Table 1; detailed pathology data is presented in Supplementary Tables 2 and 3).
Six months following Rag1 gene transfer, one mouse (mouse X) was euthanized because of a sudden onset of slow-moving behavior. Hepatosplenomegaly was found due to the presence of monomorphic undifferentiated lymphocytes containing a high transgene copy number (20 copies/cell). Studies to analyze the T-cell receptor diversity (‘Immunoscope’ method) demonstrated that the acute leukemic proliferation was due to the expansion of cells containing a unique T-cell receptor gene rearrangement (Vβ6Vα6). In an effort to detect additional adverse events, 12 mice were subjected to secondary transplantation from primary gene-corrected mice, and one of these (mouse 215) developed an enlarged spleen infiltrated with polymorphic and polyclonal B lymphocytes (10 transgene copies/cell) at 6 months following secondary transplantation. Secondary transplantation is known to be associated with elevated rates of abnormal lymphoproliferation (28), leaving the possible involvement of insertional activation during gene correction uncertain.
For the Artemis study, 9 months following Artemis gene transfer, one mouse (mouse 8) died, and hepatosplenomegaly was observed. Immunohistochemical studies revealed that spleen, liver, lung and thymus were infiltrated with small cells (Supplementary Table 3). A second mouse (mouse 31) also developed lymphoproliferation 4 months after gene transfer. Tissues were harvested from these mice and integration site distributions analyzed. Control mice without lymphoproliferations were also included for comparison.
Several sets of control integration sites were analyzed for comparison. For several of the mice, samples of the initially transduced cell population prior to engraftment were available and could be analyzed (labeled ‘Sca1+’ in Table 1). As additional controls, two large data sets were generated by ex vivo transduction of MEFs with either a lentiviral vector or a gamma-retroviral vector, followed by harvesting of integration sites 55–63 h after transduction (Table 1). These data sets are from a different cell type but provide large numbers of integration sites in the murine genome for comparison.
DNA bar coding and pyrosequencing
To prepare samples for pyrosequencing, DNA fragments from host–virus junctions were amplified using ligation-mediated PCR (3,17–20,29–34). Each DNA sample was first cleaved with MseI, which recognizes a four-base site. The digested cellular DNA was ligated to DNA linkers, and the junctions between the viral and cellular DNA were amplified using nested PCR.
In order to allow multiplex sequencing of many DNA samples in a single experiment, we took advantage of DNA bar coding (14–16). For the second PCR step, we used primers that contain unique 4-bp barcodes to identify each sequence (Figure 1). The PCR amplicons from each sample were gel purified and pooled together, then subjected to pyrosequencing using the method commercialized by 454 Life Sciences (13). We obtained a total of 274 575 raw sequence reads, averaging ∼100 bp in length. Each sequence was then assigned to a sample according to its barcode. Sequences from all samples were recovered in good yield (Table 1). After quality control, 160 232 total integration site sequence reads were available for analysis, which correspond to 2726 unique integration sites on the murine genome. The redundancy in sequence reads per integration site likely arose both from cell division in vivo and PCR amplification during DNA isolation. Integration sites generated by acute infection of MEFs were isolated in a similar fashion and yielded a total of 11 349 reads, which correspond to 7269 unique integration sites.
For mouse 8 and 31, an additional 104 sites were cloned from the tissues with lymphoproliferation by cleaving genomic DNA with the enzyme Tsp509I, linker ligation, PCR amplification and cloning in bacteria, followed by sequencing using the Sanger method. Subsequently, we compare the distributions of these sites with those analyzed using MseI cleavage and pyrosequencing.
Overview of the distribution of vector integration sites in mice
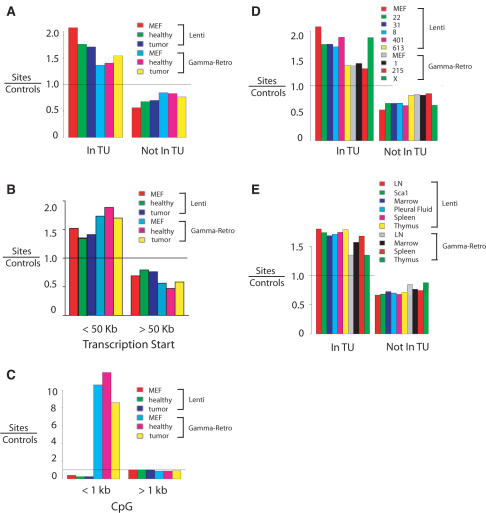
An analysis of integration site distributions in mice and cell culture controls are shown in Figure 2. More detailed catalogs of integration site distributions are presented in Supplementary Data 1–3. To maximize the sensitivity of statistical tests, the integration site data were pooled in several ways, and integration frequency assessed near recognizable genomic features. In one approach, the integration site data was pooled to allow comparison of lymphoproliferation samples versus healthy tissue, and subdivided by lentiviral vector and gamma-retroviral vector (Supplementary Data 1). As in previous studies (6,17,19,20,30–33,35,36), integration by the lentiviral vectors was strongly favored within transcription units (Figure 2A). The gamma-retroviral vectors showed a weaker though still significant preference, also paralleling previous work (19,20,22,23,35–37). Integration site populations recovered from mice were not greatly different from sites in MEFs, indicating that evolution of transduced cells in vivo did not strongly affect the site distributions relative to transcription units.
Figure 2.
Lentiviral vector and gamma-retroviral vector integration site distributions in the murine genome. Vector integration sites for pooled samples were compared to their matched random control sites. In the matching procedure (20), each unique integration site was matched with 10 control sites in the genome randomly selected in silico that were constrained to lie the same distance from an MseI recognition site as the experimentally determined integration site. Comparison of experimentally determined integration sites to the matched random controls thus ‘washed out’ any possible biases introduced by the use of MseI cleavage. A value above 1 indicates favored integration relative to random control sites; a value below 1 indicates disfavored integration. (A) Frequency of integration in transcription units. MEF: control integration sites from cultured murine embryonic fibroblasts. Healthy: control healthy mice. Tumor: mice with lymphoproliferation. (B) Frequency of integration near transcription start site. (C) Frequency of integration near CpG islands. (D) Frequency of integration in transcription units. In this analysis the integration site data sets are pooled for all tissues from each mouse. (E) Frequency of integration in transcription units for integration sites pooled by tissue of origin (samples from liver are not included in this analysis due to the low number of integration sites). Comparisons between the lentiviral and gamma-retroviral vectors in each of the panels achieved P <0.0001 (Fisher's exact test). Comprehensive analysis of integration frequency relative to many genomic features can be found in Supplementary Data 1–3.
The gamma-retrovirus-based vectors showed preferential integration near transcription start sites compared to random controls (Figure 2B). CpG islands are probable regulatory regions associated with gene 5′ ends, and the gamma-retroviral vectors showed particularly strongly favored integration near these features (Figure 2C), as anticipated from previous studies (19,20). Integration by the lentiviral vectors was disfavored within 1 kb of CpG islands.
Integration near transcription units was examined for each mouse individually, and integration was found to be favored near transcription units in each (Figure 2D and Supplementary Data 2). For some of the samples the numbers of available sites were modest, diminishing the resolution of the data. In general, the samples from animals transduced with the lentiviral vector showed more frequent integration within transcription units than the gamma-retroviral vector, though mouse X was an outlier (we return to the distinctive features in mouse X subsequently).
We also investigated whether there were systematic differences in integration site distributions by the organ of origin of the tissue sample. Comparison over a variety of genomic features showed no notable trends associated with specific organs (Figure 2E and Supplementary Data 3).
Analysis of integration frequency near proto-oncogenes and tumor suppressors
To identify integration sites near proto-oncogenes and tumor supressors that might contribute to insertional activation, we assembled a database of reported proto-oncogenes and tumor suppressors (summarized in http://microb230.med.upenn.edu/protocols/cancergenes.html) and used them to annotate the integration sites isolated from the murine samples. To assemble as comprehensive a database as possible, proto-oncogenes and tumor suppressors identified in diverse vertebrates were analyzed to identify their murine orthologs, and these genes added to the database (termed ‘Cancergenes’ subsequently). The Cancergenes list was then used in queries against integration sites. The large number of pyrosequencing reads allowed us to quantify the percent of integration sites within 50 kb of a Cancergene 5′ end as a fraction of total number of sites (Table 1). Separate percentages are shown for each mouse and tissue combination analyzed individually, and for pooled integration sites within each mouse. In each case, the frequencies of integration near Cancergenes were evaluated both based on the number of raw sequences reads, and unique integration sites generated after de-replicating duplicate reads.
Starting with the gamma-retroviral vector samples, we could compare one control mouse (mouse 1) to two mice harboring lymphoproliferative events (mice 215 and X). As an estimate of the initial frequency of integration near Cancergenes, we also analyzed data from MEF cells transduced in culture with the gamma-retroviral vector. Control mouse 1 showed 10.1% of total integration site sequence reads (15.3% of unique integration sites) within 50 kb of a Cancergene 5′ end, and for the MEF control, 13.5% of total integration site sequence reads (13.1% unique integration sites) were within 50 kb of a Cancergene 5′ end. Similar frequencies of integration near Cancergene 5′ ends are seen for gamma-retroviral vectors in human cells (38), suggesting that the frequency may not be strongly cell-type specific.
Pooled sites from mouse X showed a significantly higher proportion of integration near Cancergene 5′ ends compared to the MEF control (P < 0.0001; chi-square) based on total integration site sequence reads. Such a trend was not seen for both mouse 1 and mouse 215, and in fact the proportion of sites near Cancergenes were lower compared to the MEF control (P < 0.0001; chi-square). In addition, the proportion of sites near Cancergenes in mouse X was higher than the proportion in control mouse 1 (P < 0.0001; chi-square) and also mouse 215 (P < 0.0001; chi-square). This suggests that cells harboring integration sites near Cancergenes in mouse X became more abundant during growth in vivo. Analysis of the lymphoproliferations in these mice revealed that the expanded cell population in mouse X was of monoclonal origin, consistent with insertional activation [(11) and Supplementary Table 2]. In contrast, the lymphoproliferative event in mouse 215 was polyclonal and integration near Cancergene 5′ ends was not significantly enriched.
For the study using lentiviral vectors to correct the Artemis defect, the combination of the Artemis knockout and conditioning by irradiation is known to result in a high level of transformation. Thus, although transformed cells did harbor integrated lentiviral vectors, it was uncertain whether the observed lymphoproliferative events were a result of insertional activation or background transformation in the model. Five gene-corrected mice were studied, two controls (mice 22 and 401) and three with abnormal lymphoproliferation (mice 31, 8 and 613; Table 1). For all three lentiviral vector-corrected mice with abnormal lymphoproliferations, control samples of transduced Sca1+ cells from before transplantation were also available for comparison. Because the numbers of integration sites were low for some of the mouse and organ combinations, integration site reads from all organs were pooled for initial analysis. The proportions of integration near Cancergene 5′ ends for the mice ranged from 0% to 7.7% based on total sequence reads (0% to 9.5% based on unique integration sites). In the lentiviral vector-transduced MEF control, 8.4% of total integration site sequence reads (8.6% of unique integration sites) were near the 5′ ends of Cancergenes. None of the values for lentivirus-transduced lymphoproliferation samples was significantly greater than the value from the lentiviral integration in the MEF control (in fact, all the values based on total sequence reads were lower). Thus these data show no enrichment of integration near Cancergenes in the lymphoproliferation samples and so fail to strengthen the idea that insertional activation by the lentiviral vector contributed to transformation.
Insertional activation and cell proliferation assessed using integration site sequence counts
The large numbers of pyrosequencing reads allowed us to carry out another form of analysis, taking advantage of not just the locations of sequenced integration sites, but also the number of times each sequence was recovered. In this analysis, we assumed that the frequency of integration sites in the initial pool of cells is related to the number of sequence reads recovered, though as is discussed subsequently there are probable distortions due to differential isolation.
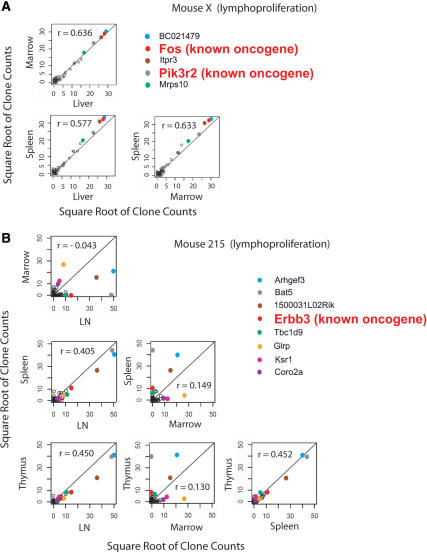
One useful analysis involved the comparison of the location and numbers of integration sites among tissues within individual mice. Infiltration of tissues by transformed cells is expected to be associated with advanced disease. Thus the identification of identical integration sites near Cancergenes that are abundant in multiple different tissues indicates probable insertional activation and spread after transformation. For mouse X, which harbored the apparent insertional activation event, a small number of integration sites were extremely abundant, and clones that were highly abundant in any one of the three tissues analyzed were also abundant in the other two (Figure 3; a detailed analysis of all mice studied is presented in Supplementary Data 4). This tendency is reflected in the higher Spearman correlations of clone counts in pairs of tissues and in scatterplots of those counts (Figure 3A). Analysis of the most abundant integration site sequences in mouse X showed that two of the four most frequently recovered sites were near the proto-oncogenes c-Fos and Pik3r2. Both integration sites are about 29 000 bp upstream of the respective proto-oncogene transcription start sites, consistent with the idea that integration activated transcription, leading to dissemination of the transformed cells.
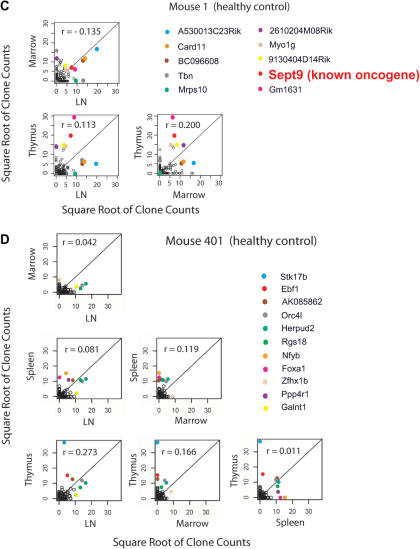
Figure 3.
Correlations of clone counts between tissues in mice with abnormal lymphoproliferation (A and B) or healthy controls (C and D). The mouse studied is indicated on the figure, along with the genes nearest the integration site. Each point represents an individual integration site; the values on the x- and y-axes indicate the number of sequences for each clone. The square root of clone counts for each tissue is plotted to allow very high counts to be displayed conveniently. Integration sites at known proto-oncogenes are indicated by the larger red lettering. The r-values indicate the Spearman correlations for counts between tissues. For detailed analysis of all mice see Supplementary Data 4. We note that it is probable that not all potential cancer-related genes have been identified. Also, for any given insertion, additional studies are required to establish whether the integration event up-regulates Cancergene transcription.
Mouse 215 (Figure 3B) is an intermediate case. The Spearman correlations for clone counts between tissues are lower than for mouse X, and only the fourth most abundant integration site is near a proto-oncogene (Erbb3). The data are consistent with development of early stage insertional activation following the secondary bone marrow transplantation, but from these data alone there is no strong support for transformation by insertional activation.
As controls, results are shown for two healthy mice in Figure 3C and 3D. In these cases the Spearman correlations are lower, and the most abundant proto-oncogene insertion is only the ninth most abundant (Sept9 in mouse 1). Thus frequency information from pyrosequencing reads could be used to distinguish an example of probable insertional activation and spread to multiple tissues from polyclonal repopulation.
Differential recovery of integration sites associated with the restriction enzymes used to cleave genomic DNA
For some of the samples, integration site sequences were recovered after using either of two methods: (i) cleavage of the murine genome with Tsp509I followed by cloning in bacteria and Sanger sequencing or (ii) cleavage by MseI followed by pyrosequencing. The tissue samples are expected to have many copies of each provirus due to division of the transduced cells, allowing us to ask how often identical sites were recovered from the same DNA sample after recovery using the two methods.
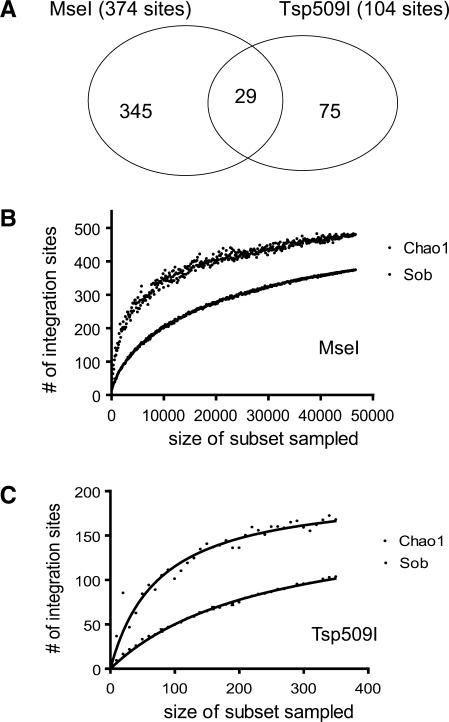
A total of 374 unique integration sites were recovered using MseI and 104 recovered using Tsp509I. A comparison of the data sets for MseI and Tsp509I showed that global trends in integration targeting were not significantly different between the two (data not shown). However, only 29 integration sites were common between the two data sets (Figure 4A).
Figure 4.
Severe biases in recovery of integration sites arising due to use of different restriction enzymes. (A) Venn diagram indicating the overlap between integration sites isolated by cleaving genomic DNA with MseI versus Tsp509I. (B) Collector's curve (rarefaction) analysis of integration sites recovered after MseI cleavage. Repeated samples of integration site subsets were used to evaluate whether further sampling would likely yield additional integration sites (rarefaction analysis), as indicated by whether the curve has reached a plateau value. The y-axis indicates the number of integration site sequences detected, the x-axis the number of integration sites in the subset analyzed. ‘Sob’ (Species-observed) indicates rarefaction on the original data. The Chao1 estimator was used to estimate the number of undetected integration sites from frequency of isolation information. ‘Chao1’ indicates collector's curve analysis on Chao1 estimates for sequence subsets. (C) Collector's curve analysis for integration sites recovered after cleavage of genomic DNA with Tsp509I. Markings as in (B).
The relatively modest overlap between the two data sets could arise for either of two reasons. It could simply be due to sparse sampling, so that drawing two samples of a few hundred integration sites from a much larger pool by chance yielded little overlap. The other possibility is that sampling was near saturation for both the MseI and Tsp509I data sets, but that the isolation methods are so biased that only a few integration sites are in common.
Consistent with the bias model, the average number of sequence reads per unique site recovered was high—208 in the MseI set and 3.5 in the Tsp509I set. However, it could still be that a few hyper-abundant sites were recovered many times, accounting for the high average number of duplicates. To test this, a collector's curve (rarefaction) analysis was carried out, which is a technique from quantitative ecology for comparing species richness. In this method, the set of sequence reads is subdivided into many subsets of different sizes, and the number of unique sites plotted (Figure 4B and C). If a plateau number of unique sites is reached at numbers of sequence reads well below the total sampled, then one can conclude that further sampling will not yield many new integration sites. As can be seen in Figure 4B and C (curves labeled ‘Sob’ for ‘Species-observed’), the curves are approaching, though not at, a plateau value, indicating that many of the integration sites recoverable with each method have been isolated.
The total number of integration site sequences in the samples could be estimated using Chao1, which uses frequency of isolation information to estimate the unseen number of species (integration sites) in a sample. Collector's curve analysis was also applied to the Chao1 estimates. As can be seen from Figure 4B and C (‘Chao1’), the estimated values are higher than the experimental values, indicating that the estimated total number of integration sites is greater than the observed number. The Chao1 estimates also approach a plateau at the highest values. The Chao1 estimates for the total number of recoverable integration sites in each sample was 480 for MseI and 167 for Tsp509I.
The data in Figure 4B and C indicated that more than half of the estimated number of integration sites was recovered for both MseI and Tsp509I methods, yet the shared integration sites were only 7.8% of the MseI data set and 28% of the Tsp509I data set. We conclude that cleavage with either of these restriction enzymes allows recovery of only a fraction of the integration sites in the sample, and that the recovery methods are biased by the restriction enzymes chosen.
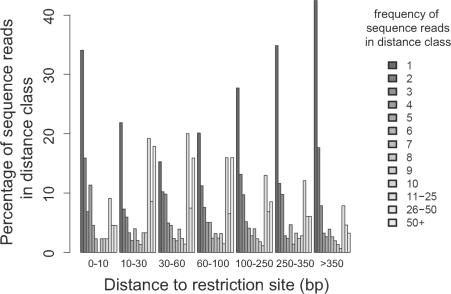
The origin of the recovery bias has not been fully clarified. Possible factors could include proximity to a restriction enzyme recognition site used to cleave genomic DNA, and possible factors affecting PCR efficiency such as G/C content or inverted repeat sequences within the amplicon. To begin to investigate the origin of the recovery bias, we asked whether proximity to MseI sites influenced isolation frequency in the MseI data set. Figure 5 plots frequency of isolation as a function of this distance. The counts of identical sequence reads are highest when the MseI site is 30–60 bases away from the integration site. Lower frequencies were seen for integration sites that were very close or very far from the restriction sites. Thus, proviruses integrated too near or too far from recognition sites for the restriction enzyme chosen will be recovered only inefficiently, potentially leading to missing data. This bias due to restriction enzymes was also seen in other integration site studies where multiple pyrosequencing data sets were compared (our unpublished data).
Figure 5.
Bias in recovery of integration sites due to the distance between integration sites and the closest MseI sites. The distribution of counts of identical sequence reads is shown as a function of distance to the nearest restriction site. The y-axis represents the percentage of integration sites with the indicated frequencies of isolation, and the x-axis is the range of distances between integration sites and their nearby MseI restriction sites used for binning.
DISCUSSION
Here we present examples of the use of DNA bar coding and pyrosequencing to monitor possible insertional activation of proto-oncogenes during therapeutic gene transfer. As models, we tested lymphoproliferations that arose during gene correction in mice lacking Artemis or Rag1. To illustrate the potential of this technology, we carried out a study of the spatial distribution of integration sites in mice with abnormal lymphoproliferations or healthy controls. Because such a large number of sequence reads (160 232) were available for analysis, additional types of analysis became possible, taking advantage of not just information on the positions of integration sites, but their frequency of isolation.
A comparison among the tissue distributions of integration sites in different mice revealed a pattern consistent in one mouse with transformation and accumulation of transformed cells in diverse tissues. Mouse X had a monoclonal lymphoproliferation based on analysis of TCR rearrangements. In this mouse, we found that a relatively small number of integration sites were recovered from each tissue, but that the most abundant sites were mostly the same between tissues. Two proto-oncogenes were located near the most abundant integration sites. This pattern is as expected for insertional activation of proto-oncogenes, followed by spread of the transformed cells to multiple tissues.
One of the goals of this study and others like it is to identify patterns in integration site data associated with adverse events, so that similar trends may be monitored in samples taken early after transplantation from patients undergoing therapeutic gene transfer. For mouse X, the high correlation coefficient for integration site identity and abundance among tissues provides one such read out. In cases where integration site data are available from multiple tissues from human gene transfer patients, such a trend can be assayed for information on possible progression toward adverse events. Understanding of the molecular basis of genotoxicity may also allow improved vector design for future gene therapy trials.
Here, we demonstrate the use of DNA bar coding and pyrosequencing to analyze the spatial distribution of transduced clones, but the method also can be applied to efficient analysis of longitudinal samples. Several groups have now reported the use of large numbers of DNA bar codes in single pyrosequencing experiments [this work and (14–16)]. This allows samples over many time-points to be analyzed in parallel at reasonable expense.
Another reason for carrying out this study was to evaluate possible genotoxicity by the lentiviral and gamma-retroviral vectors used. The numbers of mice studied here were too small for statistical analysis, but the integration site data does allow an assessment of whether insertional activation of proto-oncogenes was likely to be involved for the individual mice studied. In both the Artemis and Rag1 correction models, there is expected to be background transformation rates due to the mutations and irradiation used during conditioning, so the finding of integrated vectors in tumor tissues is not itself evidence for insertional activation of proto-oncogenes or inactivation of tumor suppressors. For the case of Artemis correction using lentiviral vectors, no increase in the frequency of integration near Cancergenes 5′ ends was seen in tissues from abnormal lymphoproliferation, nor was integration near Cancergenes more frequent than in control Sca1 + cells (transduced ex vivo) or fibroblasts transduced in cell culture. This observation does not strengthen the idea that lentiviral vector integration was involved in causing transformation—instead, it seems more likely that the effects of irradiation and the Artemis genetic defect were responsible. For mouse X, the integrated gamma-retroviral vectors do seem likely to have contributed to lymphoproliferation by insertional activation.
We note that there are two possible mechanisms for the increase in frequency of cells harboring integration near proto-oncogenes or tumor suppressors during growth in vivo. First, integration sites near proto-oncogenes may become more abundant as a fraction of the total during cell growth, as a result of selective growth advantage conferred by integration near a gene promoting cellular proliferation. Second, only a small subset of integration sites near proto-oncogenes will cause full transformation. These events probably need to be accompanied by secondary genomic alterations such as chromosomal rearrangements that contribute additional ‘hits’ toward transformation.
Lastly, we note that the problem of restriction bias in integration site recovery can be severe. Sampling from the genomic DNA tested was near saturation for each of the two enzymes studied in Figures 4 and 5, but only a small proportion of the integration sites were common to the two (7.8% of the MseI data set and 28% of the Tsp509I data set). We have also applied this method to studies of integration sites in human cells from adverse events during X-SCID gene therapy, and found examples of sites involved in adverse events that were difficult or impossible to isolate after use of certain restriction enzymes to cleave genomic DNA (39). Most previously reported methods use restriction enzyme cleavage of genomic DNA to recover integration sites and so likely suffer from this bias (40). However, using the DNA bar coding and pyrosequencing protocol described here, it is possible to examine very large numbers of sequence reads in a single experiment. Thus it is feasible to use several different restriction enzymes for cleavage and linker ligation for each sample of interest, thereby maximizing the diversity of sequences recovered. A combination of statistical modeling and empirical testing should allow estimation of the number of different enzymes and pyrosequence reads needed for complete cataloging of all the integrated vectors present in a genomic DNA sample.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank members of the Bushman laboratory for materials and helpful discussions. The MLV GagPol plasmid is a gift from Paul Bates and Bridget Puffer. Murine embryonic fibroblast is a kind gift from Thomas Jenuwein. This work was supported by NIH grants AI52845, and AI66290 to F.D.B, and by grants from INSERM, Agence nationale de la recherche (ANR) 05-MRAR-004 and programme hospitalier de recherche clinique (PHRC) AOM 04052 UA6461. F.B. was supported by a fellowship from the Association Française contre les Myopathies (AFM). A.C. was supported in part by a fellowship from the Swiss National Science Foundation. G.P.W. was supported by NIH NIAID T32 AI07634 (Training Grant in Infectious Diseases) and University of Pennsylvania School of Medicine Department of Medicine Measey Basic Science Fellowship Award. Funding to pay the Open Access publication charges for this article was provided by National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1.Cavazzana-Calvo M, Hacein-Bey S, de Saint Basile G, Gross F, Yvon E, Nusbaum P, Selz F, Hue C, Certain S, Casanova JL, et al. Gene therapy of human severe combined immunodeficiency (SCID)-X1 disease. Science. 2000;288:669–672. doi: 10.1126/science.288.5466.669. [DOI] [PubMed] [Google Scholar]
- 2.Aiuti A, Slavin S, Aker M, Ficara F, Deola S, Mortellaro A, Morecki S, Andolfi G, Tabucchi A, Carlucci F, et al. Correction of ADA-SCID by stem cell gene therapy combined with nonmyeloablative conditioning. Science. 2002;296:2410–2413. doi: 10.1126/science.1070104. [DOI] [PubMed] [Google Scholar]
- 3.Ott MG, Schmidt M, Schwarzwaelder K, Stein S, Siler U, Koehl U, Glimm H, Kuhlcke K, Schilz A, Kunkel H, et al. Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1. Nat. Med. 2006;12:401–409. doi: 10.1038/nm1393. [DOI] [PubMed] [Google Scholar]
- 4.Hacein-Bey-Abina S, von Kalle C, Schmidt M, Le Deist F, Wulffraat N, McIntyre E, Radford I, Villeval JL, Fraser CC, Cavazzana-Calvo M, et al. A serious adverse event after successful gene therapy for X-linked severe combined immunodeficiency. N. Engl. J. Med. 2003;348:255–256. doi: 10.1056/NEJM200301163480314. [DOI] [PubMed] [Google Scholar]
- 5.Hacein-Bey-Abina S, von Kalle C, Schmidt M, McCormack MP, Wulffraat N, Leboulch P, Lim A, Osborne CS, Pawliuk R, Morillon E, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:400–401. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 6.Schwarz K, Gauss GH, Ludwig L, Pannicke U, Li Z, Lindner D, Friedrich W, Seger RA, Hansen-Hagge TE, Desiderio S, et al. RAG mutations in human B cell-negative SCID. Science. 1996;274:97–99. doi: 10.1126/science.274.5284.97. [DOI] [PubMed] [Google Scholar]
- 7.Corneo B, Moshous D, Gungor T, Wulffraat N, Philippet P, Le Deist FL, Fischer A, de Villartay JP. Identical mutations in RAG1 or RAG2 genes leading to defective V(D)J recombinase activity can cause either T-B-severe combined immune deficiency or omenn syndrome. Blood. 2001;97:2772–2776. doi: 10.1182/blood.v97.9.2772. [DOI] [PubMed] [Google Scholar]
- 8.Moshous D, Callebaut I, de Chasseval R, Corneo B, Cavazzana-Calvo M, Le Deist F, Tezcan I, Sanal O, Bertrand Y, Philippe N, et al. Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell. 2001;105:177–186. doi: 10.1016/s0092-8674(01)00309-9. [DOI] [PubMed] [Google Scholar]
- 9.Mostoslavsky G, Fabian AJ, Rooney S, Alt FW, Mulligan RC. Complete correction of murine artemis immunodeficiency by lentiviral vector-mediated gene transfer. Proc. Natl Acad. Sci. USA. 2006;103:16406–16411. doi: 10.1073/pnas.0608130103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Benjelloun F, Garrigue A, Demerens C, Malassis-Séris M, Stockholm D, Blondeau J, Rivière J, Lim A, David SP, Dutrillaux R, et al. Mol. Ther. Stable and functional lymphoid reconstitution in artemis-deficient mice following ex-vivo gene transfer with lentiviral vector. in press. [Google Scholar]
- 11.Lagresle-Peyrou C, Yates F, Malassis-Seris M, Hue C, Morillon E, Garrigue A, Liu A, Hajdari P, Stockholm D, Danos O, et al. Long-term immune reconstitution in RAG-1-deficient mice treated by retroviral gene therapy: a balance between efficiency and toxicity. Blood. 2006;107:63–72. doi: 10.1182/blood-2005-05-2032. [DOI] [PubMed] [Google Scholar]
- 12.Rooney S, Sekiguchi J, Zhu C, Cheng HL, Manis J, Whitlow S, DeVido J, Foy D, Chaudhuri J, Lombard D, et al. Leaky scid phenotype associated with defective V(D)J coding end processing in artemis-deficient mice. Mol. Cell. 2002;10:1379–1390. doi: 10.1016/s1097-2765(02)00755-4. [DOI] [PubMed] [Google Scholar]
- 13.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoffmann C, Minkah N, Leipzig J, Wang G, Arens MQ, Tebas P, Bushman FD. DNA bar coding and pyrosequencing to identify rare HIV drug resistance mutations. Nucleic Acids Res. 2007;35:e91. doi: 10.1093/nar/gkm435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Binladen J, Gilbert MT, Bollback JP, Panitz F, Bendixen C, Nielsen R, Willerslev E. The use of coded PCR primers enables high-throughput sequencing of multiple homolog amplification products by 454 parallel sequencing. PLoS ONE. 2007;2:e197. doi: 10.1371/journal.pone.0000197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shoemaker DD, Lashkari DA, Morris D, Mittmann M, Davis RW. Quantitative phenotypic analysis of yeast deletion mutants using a highly parallel molecular bar-coding strategy. Nat. Genet. 1996;14:450–456. doi: 10.1038/ng1296-450. [DOI] [PubMed] [Google Scholar]
- 17.Ciuffi A, Llano M, Poeschla E, Hoffmann C, Leipzig J, Shinn P, Ecker JR, Bushman FD. A role for LEDGF/p75 in targeting HIV DNA integration. Nat. Med. 2005;11:1287–1289. doi: 10.1038/nm1329. [DOI] [PubMed] [Google Scholar]
- 18.Wang GP, Ciuffi A, Leipzig J, Berry CC, Bushman FD. HIV integration site selection: analysis by massively parallel pyrosequencing reveals association with epigenetic modifications. Genome Res. 2007;17:1186–1194. doi: 10.1101/gr.6286907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitchell R, Beitzel B, Schroder A, Shinn P, Chen H, Berry C, Ecker JR, Bushman FD. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2004;2:E234. doi: 10.1371/journal.pbio.0020234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu X, Li Y, Crise B, Burgess SM. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003;300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- 21.Schmidt M, Zickler P, Hoffmann G, Haas S, Wissler M, Muessig A, Tisdale JF, Kuramoto K, Andrews RG, Wu T, et al. Polyclonal long-term repopulating stem cell clones in a primate model. Blood. 2002;100:2737–2743. doi: 10.1182/blood-2002-02-0407. [DOI] [PubMed] [Google Scholar]
- 22.Huret JL, Minor SL, Dorkeld F, Dessen P, Bernheim A. Atlas of genetics and cytogenetics in oncology and haematology, an interactive database. Nucleic Acids Res. 2000;28:349–351. doi: 10.1093/nar/28.1.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Akagi K, Suzuki T, Stephens RM, Jenkins NA, Copeland NG. RTCGD: retroviral tagged cancer gene database. Nucleic Acids Res. 2004;32:D523–527. doi: 10.1093/nar/gkh013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, Rahman N, Stratton MR. A census of human cancer genes. Nat. Rev. Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 26.Berry CC, Hannenhalli S, Leipzig J, Bushman FD. Selection of target sites for mobile DNA integration in the human genome. 2, 2006:e157. doi: 10.1371/journal.pcbi.0020157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sirven A, Ravet E, Charneau P, Zennou V, Coulombel L, Guetard D, Pflumio F, Dubart-Kupperschmitt A. Enhanced transgene expression in cord blood CD34(+)-derived hematopoietic cells, including developing T cells and NOD/SCID mouse repopulating cells, following transduction with modified trip lentiviral vectors. Mol. Ther. 2001;3:438–448. doi: 10.1006/mthe.2001.0282. [DOI] [PubMed] [Google Scholar]
- 28.Holyoake TL, Freshney MG, Samuel K, Ansell J, Watson GE, Wright EG, Graham GJ, Pragnell IB. In vivo expansion of the endogenous B-cell compartment stimulated by radiation and serial bone marrow transplantation induces B-cell leukaemia in mice. Br. J. Haematol. 2001;114:49–56. doi: 10.1046/j.1365-2141.2001.02903.x. [DOI] [PubMed] [Google Scholar]
- 29.Schroder A, Shinn P, Chen H, Berry C, Ecker JR, Bushman FD. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- 30.Barr SD, Ciuffi A, Leipzig J, Shinn P, Ecker JR, Bushman FD. HIV integration site selection: Targeting in macrophages and the effects of different routes of viral entry. Mol. Ther. 2006;14:218–225. doi: 10.1016/j.ymthe.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 31.Barr SD, Leipzig J, Shinn P, Ecker JR, Bushman FD. Integration targeting by avian sarcoma-leukosis virus and human immunodeficiency virus in the chicken genome. J. Virol. 2005;79:12035–12044. doi: 10.1128/JVI.79.18.12035-12044.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ciuffi A, Mitchell RS, Hoffman C, Leipzig J, Shinn P, Ecker JR, Bushman FD. Integration site selection by HIV-based vectors: targeting in dividing and nondividing IMR-90 lung fibroblasts. Mol. Ther. 2006;13:366–373. doi: 10.1016/j.ymthe.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 33.Lewinski M, Bisgrove D, Shinn P, Chen H, Verdin E, Berry CC, Ecker JR, Bushman FD. Genome-wide analysis of chromosomal features repressing HIV transcription. J. Virol. 2005;79:6610–6619. doi: 10.1128/JVI.79.11.6610-6619.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aiuti A, Cassani B, Andolfi G, Mirolo M, Biasco L, Recchia A, Urbinati F, Valacca C, Scaramuzza S, Aker M, et al. Multilineage hematopoietic reconstitution without clonal selection in ADA-SCID patients treated with stem cell gene therapy. J. Clin. Invest. 2007;117:2233–2240. doi: 10.1172/JCI31666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lewinski M, Yamashita M, Emerman M, Ciuffi A, Marshall H, Crawford G, Collins F, Shinn P, Leipzig J, Hannenhalli S, et al. Retroviral DNA integration: viral and cellular determinants of target site selection. PLOS Pathogens. 2006;2:e60. doi: 10.1371/journal.ppat.0020060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Palma M, Montini E, Santoni de Sio FR, Benedicenti F, Gentile A, Medico E, Naldini L. Promoter trapping reveals significant differences in integration site selection between MLV and HIV vectors in primary hematopoetic cells. Blood. 2005;105:2307–2315. doi: 10.1182/blood-2004-03-0798. [DOI] [PubMed] [Google Scholar]
- 37.Turlure F, Maertens G, Rahman S, Cherepanov P, Engelman A. A tripartite DNA-binding element, comprised of the nuclear localization signal and two AT-hook motifs, mediates the association of LEDGF/p75 with chromatin in vivo. Nucleic Acids Res. 2006;34:1653–1675. doi: 10.1093/nar/gkl052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cattoglio C, Facchini G, Sartori D, Antonelli A, Miccio A, Cassani B, Schmidt M, von Kalle C, Howe S, Thrasher AJ, et al. Hot spots of retroviral integration in human CD34+ hematopoietic cells. Blood. 2007;110:1770–1778. doi: 10.1182/blood-2007-01-068759. [DOI] [PubMed] [Google Scholar]
- 39.Hacein-Bey-Abina S, Garrigue A, Wang GP, Soulier J, Lim A, Morillon E, Clappier E, Caccavelli L, Delabesse E, et al. 2008. Oncogenesis by insertional activation in four patients afters retrovirus-mediated gene therapy of SCID-X1, submitted for publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmidt M, Hoffmann G, Wissler M, Lemke N, Mubig A, Glimm H, Williams DA, Ragg S, Hesemann C.-., von Kalle C. Detection and direct genomic sequencing of multiple rare unknown flanking DNA in highly complex samples. Hum. Gene Ther. 2001;12:743–749. doi: 10.1089/104303401750148649. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.