Abstract
Hepatitis C virus (HCV) is a human RNA virus encoding 10 proteins in a single open reading frame. In the +1 frame, an ‘alternate reading frame’ (ARF) overlaps with the core protein-encoding sequence and encodes the ARF protein (ARFP). Here, we investigated the molecular regulatory mechanisms of ARFP expression in HCV target cells. Chimeric HCV-luciferase reporter constructs derived from the infectious HCV prototype isolate H77 were transfected into hepatocyte-derived cell lines. Translation initiation was most efficient at the internal AUG codon at position 86/88, resulting in the synthesis of a truncated ARFP named 86/88ARFP. Interestingly, 86/88ARFP synthesis was markedly enhanced in constructs containing an inactivated core protein reading frame. This enhancement was reversed by co-expression of core protein in trans, demonstrating suppression of ARFP synthesis by HCV core protein. In conclusion, our results indicate that translation of ARFP occurs mainly by alternative internal initiation at position 86/88 and is regulated by HCV core protein expression. The suppression of ARFP translation by HCV core protein suggests that ARFP expression is inversely linked to the level of viral replication. These findings define key mechanisms regulating ARFP expression and set the stage for further studies addressing the function of ARFP within the viral life cycle.
INTRODUCTION
More than 170 million people worldwide are currently infected with the hepatitis C virus (HCV). HCV infection represents a major cause of chronic liver disease and hepatocellular carcinoma (1). HCV is an enveloped virus belonging to the Flaviviridae family and is the unique member of the hepacivirus genus. The HCV genome is a single-stranded positive RNA of about 9.6 kb. The 5′ untranslated region (UTR) forms an internal ribosome entry site (IRES) that directs the translation of a precursor polyprotein of about 3000 amino acids. This polyprotein is cleaved co- and post-translationally by cellular and viral proteases, resulting in the synthesis of both structural and non-structural (NS) proteins. The structural proteins core, E1 and E2 are processed by host signal peptidases. The core protein presumably forms the viral nucleocapsid (2), whereas E1 and E2 are glycoproteins anchored in the viral envelope (3). The NS proteins NS2, NS3, NS4A, NS4B, NS5A and NS5B are essential for replication of the viral genome (4). The 3′ UTR is a highly conserved and structured region playing a key role in the initiation and regulation of viral replication.
Besides the polyprotein open reading frame, sequence analyses have shown the existence of an alternate reading frame (ARF) overlapping with the core protein encoding sequence in the +1 frame (5). Xu et al. (6) have suggested that this overlapping reading frame encodes a 17 kDa protein. This protein is named ‘F’ (for frameshift) or ‘ARFP’ [for Alternate Reading Frame Protein (7)]. Previous studies have suggested that ARFP's synthesis results from a +1 ribosomal frameshifting event near codon 11. This event occurs after initiation at the AUG initiator codon of the polyprotein (6). ARFP's existence is supported by a high conservation of its open reading frame (ORF) in all genotypes. Furthermore, peptides corresponding to this +1 reading frame react with antiviral antibodies present in the sera from HCV-infected patients. These findings suggest that ARFP is synthesized in vivo and its synthesis results in the induction of anti-ARFP immune responses (8,9). The function of the ARFP is currently unknown. Following translation, the protein is localized to the endoplasmic reticulum (10,11).
Programmed ribosomal frameshifting is relatively frequent in viral translation (12,13), and is used in particular by viruses to express a maximum of information through a limited genome. Several studies [reviewed by Giedroc et al. (14)], have mapped the optimal context required to direct a −1 ribosomal frameshifting event. These include a sequence formed by redundant runs of bases, coupled with a stem-loop often folded in a pseudoknot conformation. In this context, the presence of strong RNA structures most likely results in the induction of a ribosomal pause during translation and induces a ribosomal shift from its initial reading frame to the overlapping frame.
In contrast, mechanisms responsible for +1 ribosomal frameshifting can depend on the stability of the ribosome–RNA interaction and on specific RNA structures (15,16). The bacterial RF2 gene (17), the yeast TY1 and TY3 retrotransposones (18,19) and the eukaryotic antizyme (20) are well-characterized examples of the proteins synthesized by +1 ribosomal frameshifting.
The occurrence of a ribosomal frameshifting event during HCV translation, first described by Xu et al. (6), is controversial, since the sequence used to demonstrate the +1 ribosomal frameshift contains a unique 10 adenine stretch at the beginning of the core protein sequence. As suggested by Xu et al. (6), this adenine stretch could account for ribosomal pausing. On the other hand, sequences composed of consecutive adenines have been shown to support transcriptional slippage by inducing RNA polymerase stuttering. Stuttering may lead to modification of the final protein amino acid sequence through the insertion of non-template nucleotides during RNA synthesis (21,22). Moreover, alternative translation mechanisms of the overlapping reading frame have been proposed. These include initiation of translation at different sites resulting in the synthesis of shorter forms of the ARFP. Baril and Brakier-Gingras (23) have proposed that codon 26 is recognized by ribosomes as a site of non-AUG translation initiation in HCV genotype 1a isolates. On the other hand, Vassilaki and Mavromara (24) have shown that translation of the +1 frame can initiate on an AUG codon at position 86 or 88 of the polyprotein. The relevance of these mechanisms in translation of functional infectious HCV isolates in target cells of HCV infection is unknown. The ARFP products, presumably resulting from these translation mechanisms, have been named fARFP, 26ARFP and, 86/88ARFP for ribosomal frameshifting, initiation at codon 26, and initiation at codon 86/88, respectively. Furthermore, mechanisms of regulation of ARFP expression are not understood.
To define the molecular mechanisms of ARFP expression, we transfected chimeric HCV-H77-luciferase constructs into Huh-7 cells. We demonstrate that initiation of ARFP translation occurs mainly at position 86/88 and is regulated by HCV core protein expression. Furthermore, we show that ARFP translation is down-regulated by HCV core protein. Thus, ARFP expression is inversely linked to the level of viral replication which may explain the low-level expression of ARFP during viral infection.
MATERIALS AND METHODS
Cell culture and transfection
The human hepatoma cell line Huh-7 was kindly provided by R. Bartenschlager (University of Heidelberg, Germany). Embryonic kidney cells (HEK 293) were obtained from American Type Culture Collection (ATCC). The cells were grown and transfected as previously described (25). Medium, fetal calf serum and cell culture additives were purchased from Gibco-BRL.
HCV H77 luciferase reporter constructs
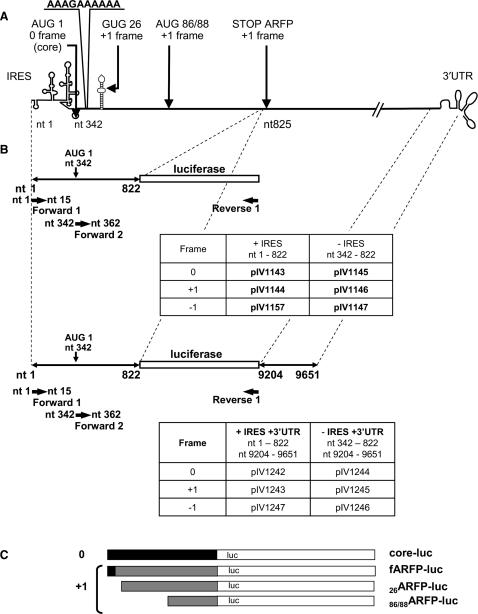
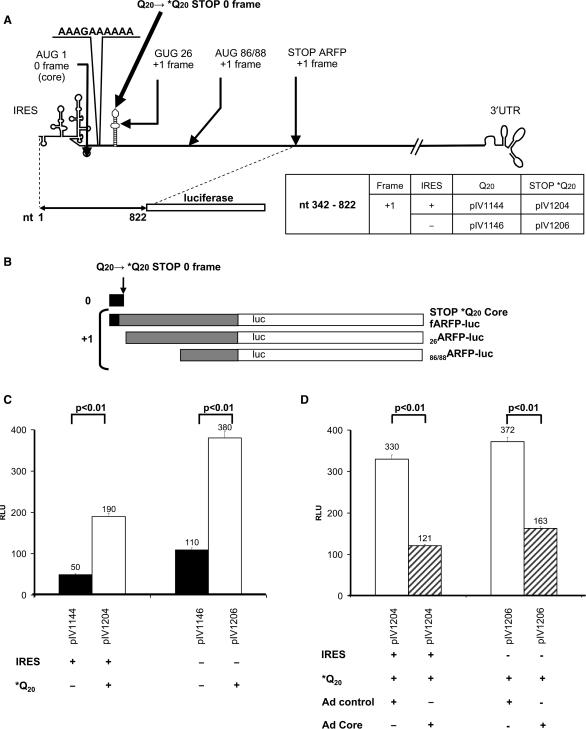
A schematic outline of the plasmids is shown in Figure 1. Nucleotide numbering refers to the HCV H77 genome sequence (genotype 1a) encoded by p90/HCV FL-long pU (26). This plasmid was used as a template for DNA amplifications. Plasmid pIV1066 was obtained by inserting the luciferase gene of pBI-L (Clontech) into the EcoRI-XbaI sites of a pCIneo plasmid vector (Promega). The first nine nucleotides (including the ATG) of the luciferase gene were deleted by PCR, and the resulting fragment was cloned into the EcoRI-XbaI sites of pCIneo. The resulting plasmid was named pIV1115. For plasmids pIV1143 to 1147, and pIV1157, fragments of the HCV core protein encoding sequence (with or without 5′ UTR) were amplified by PCR using p90/HCV FL-long pU as a template. The amplified fragments obtained were then ligated into the EcoRI site of pIV1115. Plasmids pIV1143, pIV1144 and pIV1157 include the 5′ UTR and nucleotides (nts) 342–822 of the core protein sequence. This sequence was fused in the 0, +1 or −1 frame with the luciferase gene. pIV1145, pIV1146 and pIV1147 are the corresponding plasmids without 5′ UTR. Plasmids pIV1204 and pIV1206 contain a single nucleotide substitution at nt 399 of the HCV H77 sequence introduced by the QuickChange® Site-directed Mutagenesis Kit (Stratagene) into plasmids pIV1144 and pIV1146. This substitution leads to a stop codon in the core protein sequence (0 frame) (Q20 → TAG) without affecting the ARFP amino acid sequence. Plasmids pIV1203 and pIV1240 contain a single nucleotide substitution at nt 407 of the HCV H77 sequence introduced by the QuickChange® Site-directed Mutagenesis Kit (Stratagene) into plasmids pIV1143 and pIV1144. This substitution leads to a stop codon in the ARFP protein sequence (+1 frame) (S22 → TGA) without affecting the core amino acid sequence. For plasmids pIV1242 to pIV1247, an XbaI - SalI restriction fragment encoding the 3′ UTR of HCV (nts 9204–9651) was amplified by PCR. This PCR product was added to the 3′ end of the luciferase gene sequence in all previous plasmid sequences.
Figure 1.
Design of core-luciferase and ARFP-luciferase expression constructs. (A) Schematic outline of the HCV genome. HCV (core/ARFP) potential translation initiation sites are indicated. Numbers correspond to respective codons. (B) Schematic outline of the plasmid constructs. Plasmid constructs contain the core/ARFP sequence fused with the luciferase sequence in the presence or absence of the IRES and the 3′ UTR. The names, the genomic organization of the plasmids and the nucleotide limits are indicated on the panels. Black arrows indicate the position of the primers (Forward 1 and 2, Reverse 1) used for RT-PCR of HCV-luc transcripts. (C) Potential proteins resulting from each type of plasmid translation in the 0 and +1 frames (core in black, ARFP in grey, and luciferase in white).
RT–PCR analysis of transcripts derived from HCV-luc constructs
Huh-7 cells were transfected with chimeric HCV-luc expression constructs as described earlier. Total RNA was isolated using the RNeasy kit (Qiagen). Reverse transcription was performed using 5 µg of purified total cellular RNA, SuperScript III reverse transcriptase (Invitrogen), and random primers (Invitrogen), following the manufacturer's recommendations. Subsequent PCR was performed on the reverse transcribed cDNA using Taq DNA Polymerase (Invitrogen), a reverse primer corresponding to the 3′ region of the luciferase gene (Reverse 1, 5′CCTCTAGAACTAGTGGATCCCCCC3′) and a forward primer corresponding to the HCV 5′UTR (nts 1–15) for IRES-containing constructs (Forward 1, 5′GGAATTCGCAGCCCCCTGATG3′) or to the 5′ region of the core protein encoding sequence (nts 342–362) for constructs lacking IRES (Forward 2, 5′GGAATTCATGAGCACGAATCCTAAACCT3′). PCR products were then separated on a 1% agarose gel.
Recombinant adenovirus vectors
Recombinant adenoviral genomes were generated as infectious plasmids by homologous recombination in E. coli as previously described (27). Briefly, a DNA fragment encoding the full-length core protein (nts 342–914) was obtained by PCR using p90/HCV FL-long pU as a template, and was inserted into the EcoRI-XbaI sites of the adenoviral shuttle plasmid pTG13337. The resulting vector, pIV1210, was used for homologous recombination with adenoviral sequences of the backbone vector pTG6624 (27), resulting in Ad core. Recombinant adenoviruses were synthesized as recently described (4).
Luciferase assay
At 48 h post-transfection, cells were washed twice in PBS and lysed in 250 µl Reporter Lysis Buffer (Promega). Luciferase expression was determined using a TD-20/20 Luminometer (Turner Design). Briefly, 5 µl of cell lysate were mixed with 25 µl of Luciferase Reagent (Promega), and luciferase activity was assessed as described by the manufacturer. Each transfection or infection was performed in triplicate and repeated at least three times. Each set of experiments was normalized to the luciferase activity in cells transfected by a control plasmid encoding the luciferase gene in the absence of the HCV sequence (pIV1066).
Western blot analysis
Huh-7 cells were lysed in Reporter Lysis Buffer (Promega). The lysates were subjected to low-speed centrifugation (500 × g; 15 s) to remove nuclei and debris. Proteins were then resolved by SDS–PAGE, transblotted onto Hybond-P membranes (GE Healthcare), and probed with anti-luciferase Mab (Sigma, dilution 1/500 in PBS, 0.1% Tween; 1% skimmed milk) for 1 h at room temperature. After extensive washing, an anti-mouse IgG conjugated to horseradish peroxidase (Sigma, diluted to 1/5000 in PBS, 0.1% Tween, 1% skimmed milk) was added to the blots for 1 h at room temperature. Following an additional wash step, the bound antibodies were detected by chemiluminescence (GE Healthcare).
RESULTS
Mapping of HCV genetic elements required for ARFP synthesis
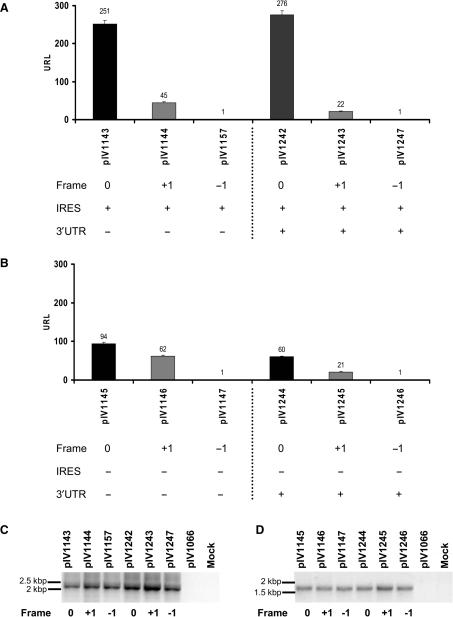
Since ribosomal frameshifting efficiency can be optimized by specific RNA structures, we evaluated the impact of the RNA context on core/ARFP translation efficiency. To address this question, we fused nts 342–822 of the core protein coding sequence to a luciferase reporter gene cDNA. We assessed translation efficiency in the three frames (0, +1, −1) by adding zero, one or two nucleotides, respectively, between the core protein and luciferase sequences. To study the impact of HCV IRES on ARFP translation a second set of similar plasmids lacking the IRES element was constructed (Figure 1). The chimeric HCV-luciferase reporter plasmids were transfected into Huh-7 cells. To exclude translation initiation at the luciferase AUG, the first nine nucleotides, including the ATG, were deleted in all HCV chimeric luciferase constructs. Thus, for each plasmid, luciferase activity represented translation of the respective frame. The normalization of luciferase values, for each experiment and between different experiments, was performed using transfections with pIV1066 control plasmid lacking HCV sequences.
Translation of the core protein-encoding sequence corresponding to the full-length ARF sequence (nts 342–822) was analyzed by transfection of the HCV-Luc chimeric constructs into Huh-7 hepatoma cells. Compared to translation in the 0 frame (core-luc), a significant level of translation in the +1 frame (ARFP-luc) was observed as shown in Figure 2. The translation ratio ARFP-luc/core-luc was higher when the IRES was absent (compare 45/251 RLU in Figure 2A and 62/94 RLU in Figure 2B). These results clearly show that ARFP translation is not dependent on the presence of the IRES. Luciferase activity was absent in cells transfected with the −1 fusion constructs. These data confirm that translation in the +1 frame, although occurring at low levels, was present (Figure 2). Addition of the proteasome inhibitor MG132 did not modify translation efficiency (data not shown), suggesting that differences in protein expression levels were not due to differences in proteaosomal degradation of HCV-luc fusion proteins.
Figure 2.
Translation of chimeric HCV core-luciferase sequences in frames 0, +1 and −1 in hepatoma cells. Plasmids containing nts 342–822 of the core coding sequence fused to a luciferase gene reporter cDNA were transfected into Huh-7 cells. The translation of HCV 5′ sequences was studied in the three frames [0 (core frame) in black, +1 (ARFP frame) in gray and −1 frame in white] and in the presence (A) or absence (B) of the IRES sequence, using luciferase activity quantification. The integrity of the RNA transcripts was investigated in Huh-7 cells transfected with HCV-luc plasmids containing the IRES (C) or lacking the IRES (D). RT-PCR of HCV-luc transcripts. Total RNA was purified from Huh-7 cells transfected with the indicated plasmids and reverse transcribed as described earlier. The resulting cDNAs were amplified by PCR using HCV (Forward 1 and 2) and Luc-specific primers (Reverse 1, see Figure 1A for primer position). Plasmid pIV1066 encoding luciferase and Huh-7 cells transfected with an irrelevant plasmid were used as controls.
We next investigated the impact of the HCV 3′ UTR on ARFP translation. Viral 3′ regions have been shown to enhance translation and replication. Furthermore, this region has been shown to play a potential role in HCV genome stabilization and/or encapsidation (28). The HCV 3′ UTR consists of three regions: a variable sequence, an internal poly (U/UC) stretch and the conserved X region. The poly (U/UC) tail and the highly structured and conserved X region are essential for replication in the chimpanzee (29,30). Furthermore, the requirement of the HCV 3′ UTR for core protein translation has been recently demonstrated (31,32). To study whether the 3′ UTR has an impact on core/ARFP translation, we added the HCV 3′ UTR sequence downstream of the luciferase gene coding sequence in plasmids containing the full-length ARF sequence (nts 342–822) (Figure 1). Prior to transfection into Huh-7 cells, plasmids were linearized following the 3′ UTR sequence to generate blunt-ended extremities similar to the authentic HCV RNA. As shown in Figure 2, the addition of the 3′ UTR had no effect on IRES-mediated core-luc synthesis (Figure 2A, compare pIV1143, 251 RLU and pIV1243, 276 RLU). Taken together, these results show that the 3′ UTR has no effect on IRES-mediated core-luc translation as previously shown (25). The addition of the HCV 3′ UTR induced only a minor decrease in the +1 frame translation, suggesting that this HCV genetic element is not required for ARFP translation (Figure 2B).
Finally, we investigated the integrity of the RNA transcripts by performing a RT-PCR of transcripts derived from the luciferase reporter constructs in Huh-7 transfected cells. The reverse transcriptase product was amplified using a forward primer corresponding to the HCV 5′ UTR for IRES-containing plasmids (Forward 1, Figure 1A) or to the 5′ region of the core protein encoding gene for plasmids without IRES (Forward 2, Figure 1A), and a reverse primer corresponding to the 3′ part of the luciferase gene (Reverse 1, Figure 1A). The expected size of the corresponding RT-PCR products was 2.2 kb in constructs containing the HCV IRES and 1.8 kb in constructs without IRES. As shown in Figure 2C and D, analysis of the corresponding amplified RT-PCR products demonstrates the perfect integrity of all transcript templates. No signal was detected in cells transfected with a control plasmid (pIV1066) or mock transfected cells. These data clearly rule out the possibility of cryptic promoters, splicing or other events.
Taken together, these data demonstrate that ARFP translation is not dependent on the presence of HCV IRES and 3′UTR.
Translation of ARFP in the +1 frame occurs by alternative initiation
In our model, translation in the +1 frame can account for different fusion proteins: 26ARFP-luc and 86/88ARFP-luc as well as fARFP-luc. Indeed, since the translation machinery can recognize the core AUG (0 frame) even in the absence of the IRES sequence (compare pIV1143 Figure 2A with pIV1145 Figure 2B), the possibility of a ribosomal frameshifting or a transcriptional slippage event cannot be excluded.
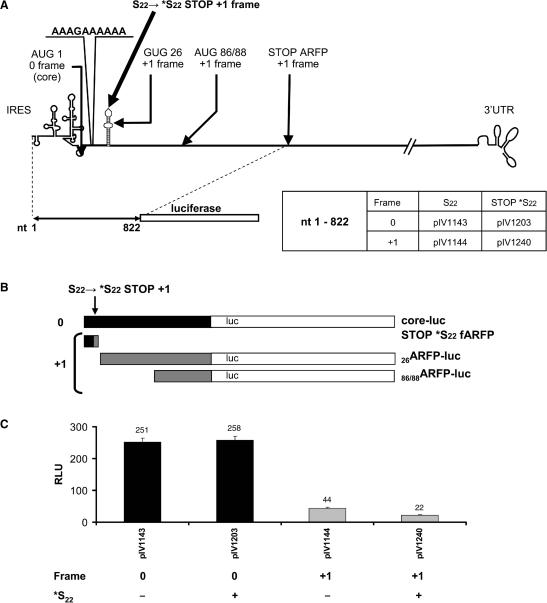
To distinguish ribosomal frameshifting or transcriptional slippage from alternative initiation of translation, we suppressed fARFP synthesis. Using site-directed mutagenesis, a stop codon in the ARF-coding sequence at position 22 (*S22) was introduced (Figure 3A). The mutation of the ARFP codon 22 into *S22 stop codon does not modify the core protein amino acid sequence but terminates fARFP-luc protein synthesis at codon 22 (Figure 3B). To demonstrate that the *S22 (ARFP) stop mutation had no impact at the core-luc protein translation level, we measured the core-luc (0 frame) translation rates of transfected *S22 ‘0’ frame fusion plasmids and compared those rates to those of regular 0 frame transfected plasmids. As expected, core-luc protein translation (frame 0) was not affected by the *S22 stop mutation (Figure 3C).
Figure 3.
Alternative initiation of ARFP translation in the +1 frame. (A) Plasmid constructs containing the *S22 stop codon mutation in the +1 frame resulting in knock-out of fARFP expression. Plasmid numbers are indicated. (B) Potential proteins resulting from translation in the 0 and +1 frames. (C) Huh-7 cells were transfected with the constructs shown in (A) and luciferase activity was assessed as described above. Black and grey bars correspond to HCV translation in the 0 and +1 frame, respectively.
To determine the fraction of +1 frame translation that results from alternative initiation of translation, we transfected *S22 +1 frame fusion plasmids into Huh-7 cells (Figure 3A). Using these reporter constructs, we detected significant translation in the +1 frame (Figure 3C). The luciferase expression measured in these constructs, accounts for alternative initiation of translation on codon 26 (26ARFP) and/or codon 86/88 (86/88ARFP) in the +1 frame, but not for fARFP. The alternative initiation of translation measured corresponded to 50% of the regular non mutated +1 frame translation. Therefore, our experiments demonstrate that translation occurring in the +1 frame is mediated at least in a large part by alternative initiation of translation occurring downstream of the *S22 stop codon introduced in the ARFP coding-sequence.
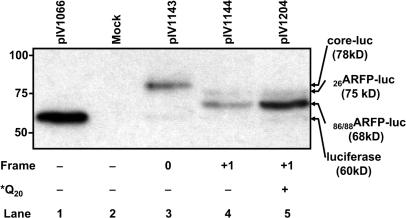
To further address the mechanism of ARFP translation in prototype isolate HCV H77 (frameshifting or transcriptional slippage versus alternative internal initiation), we characterized translation products synthesized in the +1 frame in Huh-7 cells transfected with recombinant plasmids containing the HCV H77 ARFP coding sequence (nts 342–822). Since monoclonal anti-ARFP antibodies directed against epitopes of the ARFP N-terminal domain are unavailable, ARFP-luc fusion proteins were analyzed using an anti-luc monoclonal antibody. Expected fusion protein sizes were 60 kDa for luciferase, 78 kDa for the core/fARFP-luc fusion proteins and 75 and 68 kDa for fusion proteins synthesized by alternative initiation at positions 26 (26ARFP-luc) and 86/88 (86/88ARFP-luc), respectively. Background translation of luciferase in the fusion constructs was minimal, compared to expression of the fusion proteins (Figure 4, lanes 4 and 5). The two forms detected for ARFP synthesis corresponded to products derived from alternative initiation sites: the major band corresponded to 86/88ARFP, whereas 26ARFP was synthesized at a lower level (Figure 4). In our experimental system, fARFP was not detected. Since the addition of proteasome inhibitor MG-132 did not modify protein expression (data not shown), it is likely that the absence of fARFP detection was not due to a problem with protein stability.
Figure 4.
Identification of luciferase fusion proteins. Lysates of Huh-7 cells transfected with the luciferase encoding plasmid (pIV1066, lane 1), the empty pCI-neo vector (mock, lane 2), plasmids containing sequences encoding core-luc (pIV1143, lane 3), ARF-luc (pIV1144, lane 4) or ARF-luc containing the *Q20 core mutation (pIV1204, lane 5) were analyzed by Western blotting using an anti-luciferase monoclonal antibody as described in ‘Material and methods’ section. Identified translation products are indicated on the right of the panel.
Thus, we conclude that the observed +1 frame (ARFP-luc) translation occurs from alternative initiation of translation. The decrease in ARFP-luc translation observed in the *S22-mutated constructs (Figure 3C) is likely the result of a decrease in alternative initiation of translation at position 26, due to the *S22 mutation itself, rather than a suppression of a putative ribosomal frameshifting event.
Regulation of ARFP alternative initiation by HCV core protein expression
Core protein is known to play a role in HCV polyprotein translation. It has been shown to decrease the translation activity of the IRES through interactions involving 5′ regions (33). Whether this inhibition depends on RNA–RNA (34,35) or core-RNA interactions (36,37) remains unclear. Thus, we aimed to study whether core protein plays a role in the regulation of ARFP translation.
First, we investigated the impact of core protein on the +1 frame translation in cis. To address this question, we constructed plasmids containing a stop codon (*Q20) in the core-encoding sequence fused in the +1 frame to the luc gene (Figure 5A). In these plasmids, core protein is not synthesized, whereas ARFP translation is maintained. Interestingly, in the absence of the core protein, we observed a marked, 4-fold enhancement of ARFP-luc fusion protein expression in the +1 frame (Figure 5C, compare black and white bars). Thus, the premature stop of core protein translation at codon 20 in the 0 frame resulted in a marked increase of ARFP synthesis. This observation suggested a negative regulation of core protein on ARFP synthesis. Moreover, the core protein inhibitory effect was corroborated by Western blot analysis of the expressed proteins. As shown in Figure 4, the expression of 86/88ARFP-luc fusion protein (pIV1204, lane 5) was markedly increased in Huh-7 cells transfected with plasmids containing the *Q20 core stop mutation, compared to plasmids allowing core protein expression in cis (pIV1144, lane 4). Thus, when core protein translation was absent (pIV1204, lane 5), 86/88ARFP represented the major ARFP translation product.
Figure 5.
Regulation of ARFP translation by HCV core protein. (A) Plasmids containing a stop codon in the 0 frame (*Q20). Names of the plasmids are indicated on the panel. (B) Proteins resulting from plasmid expression in the 0 and +1 frames of constructs lacking the core ORF. (C) To study the effect of core in cis, we transfected cells with +1 frame constructs with or without the *Q20 mutation. Luciferase activity corresponds to the translation in the +1 frame without mutation (black bars) and with the *Q20 stop mutation (white bars). (D) To study the effect of core in trans, cells were transfected with +1 frame constructs containing the *Q20 mutation and transduced with a control adenovirus (Ad control, white bars) or a HCV core-encoding adenovirus (Ad core, hatched bars). Luciferase activity was analyzed in the presence or absence of the HCV IRES as described earlier.
To confirm the negative regulation of ARFP translation by core protein, we studied the impact of core protein expressed in trans on ARFP synthesis. To ensure that core protein was expressed in the entire transfected cell population, we constructed a recombinant adenovirus encoding the full-length core protein (Ad core). As a control, we used an adenovirus encoding the green fluorescent protein (Ad control).
To test the hypothesis of a direct effect of core protein expression on ARFP translation, we infected Huh-7 cells transfected with the plasmids containing the *Q20 stop mutation with the core-encoding recombinant adenovirus (Ad core). The impact of the adenovirus on translation of these plasmids was first evaluated by using the GFP-encoding adenovirus as a control. The control adenovirus induced a global enhancement of translation (Figure 5C and D). However, expression of core in trans by the recombinant adenovirus (Ad core) decreased the +1 frame (ARFP-luc) translation to levels similar to those obtained with non-mutated core-encoding plasmids (Figure 5C and D). Since translation in the +1 frame was enhanced in the absence of core protein and was markedly reduced when core protein is added in trans, we conclude that core protein expression has an inhibitory effect on ARFP synthesis.
DISCUSSION
It has been suggested that the core protein encoding sequence directs the synthesis of three different products: (i) the core protein in the 0 frame, (ii) the ARFP in the +1 frame and (iii) a 13 amino-acid peptide in the −1 frame (5). The ARFP itself can result from a +1 ribosomal frameshifting event (6) (fARFP) and/or alternative initiation in the +1 frame at codon 26 (23) (26ARFP) and 86/88 (24) (86/88ARFP). In addition, a transcriptional slippage event cannot be excluded. To compare the translation levels of these proteins, we designed plasmids containing the HCV core/ARFP sequence fused in the 0, +1 or −1 frame to a luciferase reporter gene cDNA. Using hepatoma cells transfected with chimeric HCV-luciferase reporter constructs as a model system, we demonstrate that the predominant form of ARFP is a 70 amino acid protein initiated at codon 86/88. Finally, we show that core protein regulates ARFP synthesis by inhibiting 86/88ARFP translation.
Evidence for a +1 ribosomal frameshifting event was demonstrated in vitro by Xu et al. (6) using a unique sequence of genotype 1a containing a 10-adenine stretch in the first 50 nt of the core protein coding sequence (6). However, the incidence of this A stretch appears to be very rare since only a single HCV sequence of this type is found in the European HCV database (http://euhcvdb.ibcp.fr/) (38). Thus, conclusions derived from translation studies using this unique sequence may not generally apply for other, more prevalent HCV strains. To study mechanisms of ARFP translation in well characterized infectious and functional HCV strains, we used prototype infectious clone HCV H77 (39). Analysis of the translation products of HCV H77 luc plasmids (Figure 4) clearly demonstrates that ARFP synthesis in the HCV H77 strain occurs by alternative internal initiation of translation.
The IRES-independence of the ARFP translation could be related to the nature of codon GUG at position 26 in the HCV genotype 1a sequence that we used. Indeed, in a recent report, Baril and Brakier-Gingras (23) showed that the ARFP translation is IRES independent when a GUG codon is present at position 26, which is the case in the 1a genotype (23). Besides IRES-independence, the mechanism responsible for initiation at codon 26 and/or 86/88 remains to be determined. Although our results cannot exclude a linear scanning event, such a mechanism seems to be highly unlikely in the presence of the IRES because of the stability of its structure. At least two other alternative mechanisms could explain the recruitment of ribosomal subunits far upstream of the initiation codon: first, internal initiation could occur through an internal ribosome entry site present in the viral sequence downstream of the HCV IRES; second, specific ribosomal shunting or ribosomal tethering and clustering (40) could be involved in initiation of ARFP translation. The latter model is highly interesting because it explains how translation can initiate on AUG codons located either downstream or upstream of an IRES. According to this hypothesis, ARFP translation would be dependent on an IRES located downstream of codon 86/88.
The addition of the HCV 3′ UTR induced only a minimal decrease in translation in the 0 or +1 frame, suggesting that this HCV genetic element is not required for ARFP translation. Nevertheless, the HCV-luc model system is limited to the ARFP and core protein coding sequences. Thus, we cannot exclude that results might be different using a full-length infectious clone as a model system. However, the difficulty studying translation of the short +1 frame protein in the full-length clone currently precludes the use of this system to address this question.
Most importantly, we demonstrate that HCV core protein plays a key role in the regulation of 86/88ARFP translation in HCV target cells. Using a stop mutation preventing core protein expression, we demonstrate that core protein inhibits 86/88ARFP synthesis. Furthermore, inhibition of ARFP synthesis can be restored by expression of core protein in trans. Since the two proteins partially share the same RNA sequence, a switch between ARFP and core protein translation can be imagined during the viral cycle. Core protein is the capsid protein. Its level may vary during the viral life cycle (i.e. during encapsidation), allowing a modification of ARFP translation efficiency. ARFP could also be synthesized when core protein is expressed at a lower rate, i.e. during the very early or the late phase of chronic infection, when levels of viral replication can be low. Suppression of ARFP translation by HCV core protein suggests that ARFP expression is inversely linked to viral replication. Furthermore, these findings may explain the apparent low-level expression of ARFP during viral infection, precluding its easy detection during HCV infection in vivo or in vitro.
Core-mediated regulation of ARFP translation may have important implications for the role of ARFP in the viral cycle. The interaction of core protein with its RNA coding sequence has been described (33). Thus, ARFP translation may be silenced in the presence of core protein bound to RNA, and this inhibition is reversed when core protein is expressed at low levels or is channeled into nucleocapsid assembly.
It is conceivable that the observed inhibition of ARFP expression by core protein may also play an important role in suppressing the putative ARFP function. Such a mechanism could result in the establishment of a stoichiometric balance between ARFP and core protein, reminiscent of translation regulation in HIV, where Env protein expression is high and related to a weak Vpu protein synthesis (41).
The impact of the ARFP role in the viral cycle is still unknown. Since the existence of an additional protein in an alternate reading frame has not yet been described for other members of the Flaviviridae family, we looked for the presence of similar proteins in other virus families. Similar to HCV, the translation of picornaviruses is IRES-dependent, resulting in the synthesis of a polyprotein cleaved into structural and non-structural proteins. For encephalomyocarditis virus (EMCV), foot and mouth disease virus (FMDV), hepatitis A virus and Theiler's murine encephalomyelitis virus (TMEV), additional initiation sites exist in the same frame and have been documented in vitro (EMCV, hepatitis A and FMDV) (42–44) and in vivo (FMDV) (44). Initiation sites for L* protein in the +1 frame both in vitro and in vivo have been identified for TMEV (45). The TMEV L* out-of-frame protein was first thought to be an artifact of in vitro translation. However, more recent studies indicate that this protein indeed has an anti-apoptotic activity and is involved in viral pathogenesis and persistence (46). Thus, it is conceivable that ARFP may also play a functional role in the HCV viral life cycle. Although a recent study has demonstrated that ARFP may not be required for viral replication (47), the constructs used in the latter study (47) did not prevent expression of 86/88ARFP. Thus, a functional role for 86/88ARFP in viral replication is still conceivable. Interestingly, HCV-infected patients mount an easily detectable, humoral immune response against ARFP, suggesting that this protein is expressed in vivo. The short protein half-life of about 10 min (10,11) could point to a punctual action during the viral cycle.
In conclusion, suppression of ARFP expression by core protein suggests that ARFP expression is inversely linked to viral replication. The identification of negative regulation by HCV core protein expression may set the stage for further studies addressing the function of 86/88ARFP within the viral life cycle and the pathogenesis of HCV infection.
ACKNOWLEDGEMENTS
We gratefully acknowledge Charles M. Rice (The Rockefeller University, NY, USA) for providing the p90/HCV FL-Long pU clone, TRANSGENE S. A. (Strasbourg, France) for generously supplying shuttle plasmids pTG13387 and pTG6624, Glaucia Paranhos-Baccala (BioMérieux, Lyon, France) for the kind gift of monoclonal anti-ARFP antibodies, and Marie Parnot for excellent technical assistance with sequencing. We thank Joseph Puglisi (Stanford University School of Medicine, CA, USA) and Pascale Romby (Institut de Biologie Moléculaire et Cellulaire, Strasbourg, France) for helpful discussions and critical reading of the manuscript. M.W. was supported in part by the French Ministry of Research grant (MRT) and a grant from the Association pour la Recherche sur le Cancer. The study was funded by the foundation FRM-BNP Paribas (C.S.), the chair of excellence program of the Agence Nationale de la Recherche (ANR-05-CEXC-008; T.F.B.), France, the Agence Nationale de la Recherche sur le SIDA et les Hépatites Virales (ANRS Grant #2005/002 C.S.; ANRS Grant #06221; T.F.B.), the 6th framework program of the European Union (LSHM-CT-2004-503359, NoE ‘VIRGIL’, T.F.B.), Inserm, and the Louis Pasteur University, Strasbourg France. Funding to pay the Open Access publication charges for this article was provided by FRM-BNP Paribas.
Conflict of interest statement. None declared.
REFERENCES
- 1.Wasley A, Alter MJ. Epidemiology of hepatitis C: geographic differences and temporal trends. Semin Liver Dis. 2000;20:1–16. doi: 10.1055/s-2000-9506. [DOI] [PubMed] [Google Scholar]
- 2.Santolini E, Migliaccio G, La Monica N. Biosynthesis and biochemical properties of the hepatitis C virus core protein. J. Virol. 1994;68:3631–3641. doi: 10.1128/jvi.68.6.3631-3641.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matsuura Y, Harada S, Suzuki R, Watanabe Y, Inoue Y, Saito I, Miyamura T. Expression of processed envelope protein of hepatitis C virus in mammalian and insect cells. J. Virol. 1992;66:1425–1431. doi: 10.1128/jvi.66.3.1425-1431.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dimitrova M, Imbert I, Kieny MP, Schuster C. Protein-protein interactions between hepatitis C virus nonstructural proteins. J. Virol. 2003;77:5401–5414. doi: 10.1128/JVI.77.9.5401-5414.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walewski JL, Keller TR, Stump DD, Branch AD. Evidence for a new hepatitis C virus antigen encoded in an overlapping reading frame. RNA. 2001;7:710–721. doi: 10.1017/s1355838201010111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu Z, Choi J, Yen TS, Lu W, Strohecker A, Govindarajan S, Chien D, Selby MJ, Ou J. Synthesis of a novel hepatitis C virus protein by ribosomal frameshift. EMBO J. 2001;20:3840–3848. doi: 10.1093/emboj/20.14.3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Varaklioti A, Vassilaki N, Georgopoulou U, Mavromara P. Alternate translation occurs within the core coding region of the hepatitis C viral genome. J. Biol. Chem. 2002;277:17713–17721. doi: 10.1074/jbc.M201722200. [DOI] [PubMed] [Google Scholar]
- 8.Bain C, Parroche P, Lavergne JP, Duverger B, Vieux C, Dubois V, Komurian-Pradel F, Trepo C, Gebuhrer L, Paranhos-Baccala G, et al. Memory T-cell-mediated immune responses specific to an alternative core protein in hepatitis C virus infection. J. Virol. 2004;78:10460–10469. doi: 10.1128/JVI.78.19.10460-10469.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Komurian-Pradel F, Rajoharison A, Berland JL, Khouri V, Perret M, Van Roosmalen M, Pol S, Negro F, Paranhos-Baccala G. Antigenic relevance of F protein in chronic hepatitis C virus infection. Hepatology. 2004;40:900–909. doi: 10.1002/hep.20406. [DOI] [PubMed] [Google Scholar]
- 10.Xu Z, Choi J, Lu W, Ou JH. Hepatitis C virus F protein is a short-lived protein associated with the endoplasmic reticulum. J. Virol. 2003;77:1578–1583. doi: 10.1128/JVI.77.2.1578-1583.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roussel J, Pillez A, Montpellier C, Duverlie G, Cahour A, Dubuisson J, Wychowski C. Characterization of the expression of the hepatitis C virus F protein. J. Gen. Virol. 2003;84:1751–1759. doi: 10.1099/vir.0.19065-0. [DOI] [PubMed] [Google Scholar]
- 12.Farabaugh PJ, Bjork GR. How translational accuracy influences reading frame maintenance. EMBO J. 1999;18:1427–1434. doi: 10.1093/emboj/18.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baranov PV, Gurvich OL, Hammer AW, Gesteland RF, Atkins JF. Recode 2003. Nucleic Acids Res. 2003;31:87–89. doi: 10.1093/nar/gkg024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giedroc DP, Theimer CA, Nixon PL. Structure, stability and function of RNA pseudoknots involved in stimulating ribosomal frameshifting. J. Mol. Biol. 2000;298:167–185. doi: 10.1006/jmbi.2000.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baranov PV, Gesteland RF, Atkins JF. Recoding: translational bifurcations in gene expression. Gene. 2002;286:187–201. doi: 10.1016/s0378-1119(02)00423-7. [DOI] [PubMed] [Google Scholar]
- 16.Namy O, Rousset J, Napthine S, Brierley I. Reprogrammed genetic decoding in cellular gene expression. Cell. 2004;13:157–168. doi: 10.1016/s1097-2765(04)00031-0. [DOI] [PubMed] [Google Scholar]
- 17.Craigen WJ, Cook RG, Tate WP, Caskey CT. Bacterial peptide chain release factors: conserved primary structure and possible frameshift regulation of release factor 2. Proc. Natl Acad. Sci. USA. 1985;82:3616–3620. doi: 10.1073/pnas.82.11.3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Belcourt MF, Farabaugh PJ. Ribosomal frameshifting in the yeast retrotransposon Ty: tRNAs induce slippage on a 7 nucleotide minimal site. Cell. 1990;62:339–352. doi: 10.1016/0092-8674(90)90371-K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Farabaugh PJ, Zhao H, Vimaladithan A. A novel programmed frameshift expresses the POL3 gene of retrotransposon Ty3 of yeast: frameshifting without tRNA slippage. Cell. 1993;74:93–103. doi: 10.1016/0092-8674(93)90297-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ivanov IP, Anderson CB, Gesteland RF, Atkins JF. Identification of a new antizyme mRNA +1 frameshifting stimulatory pseudoknot in a subset of diverse invertebrates and its apparent absence in intermediate species. J. Mol. Biol. 2004;339:495–504. doi: 10.1016/j.jmb.2004.03.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moon S, Byun Y, Han K. FSDB: a frameshift signal database. Comput. Biol. Chem. 2007;31:298–302. doi: 10.1016/j.compbiolchem.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Penno C, Hachani A, Biskri L, Sansonetti P, Allaoui A, Parsot C. Transcriptional slippage controls production of type III secretion apparatus components in Shigella flexneri. Mol. Microbiol. 2006;62:1460–1468. doi: 10.1111/j.1365-2958.2006.05456.x. [DOI] [PubMed] [Google Scholar]
- 23.Baril M, Brakier-Gingras L. Translation of the F protein of hepatitis C virus is initiated at a non-AUG codon in a +1 reading frame relative to the polyprotein. Nucleic Acids Res. 2005;33:1474–1486. doi: 10.1093/nar/gki292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vassilaki N, Mavromara P. Two alternative translation mechanisms are responsible for the expression of the HCV ARFP/F/core+1 coding open reading frame. J. Biol. Chem. 2003;278:40503–40513. doi: 10.1074/jbc.M305504200. [DOI] [PubMed] [Google Scholar]
- 25.Imbert I, Dimitrova M, Kien F, Kieny MP, Schuster C. Hepatitis C virus IRES efficiency is unaffected by the genomic RNA 3′NTR even in the presence of viral structural or non-structural proteins. J. Gen. Virol. 2003;84:1549–1557. doi: 10.1099/vir.0.18907-0. [DOI] [PubMed] [Google Scholar]
- 26.Kolykhalov AA, Agapov EV, Blight KJ, Mihalik K, Feinstone SM, Rice CM. Transmission of hepatitis C by intrahepatic inoculation with transcribed RNA. Science. 1997;277:570–574. doi: 10.1126/science.277.5325.570. [DOI] [PubMed] [Google Scholar]
- 27.Chartier C, Degryse E, Gantzer M, Dieterle A, Pavirani A, Mehtali M. Efficient generation of recombinant adenovirus vectors by homologous recombination in Escherichia coli. J. Virol. 1996;70:4805–4810. doi: 10.1128/jvi.70.7.4805-4810.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dreher TW. Functions of the 3′-untranslated regions of positive strand RNA viral genomes. Annu. Rev. Phytopathol. 1999;37:151–174. doi: 10.1146/annurev.phyto.37.1.151. [DOI] [PubMed] [Google Scholar]
- 29.Yanagi M, St Claire M, Emerson SU, Purcell RH, Bukh J. In vivo analysis of the 3′ untranslated region of the hepatitis C virus after in vitro mutagenesis of an infectious cDNA clone. Proc. Natl Acad. Sci. USA. 1999;96:2291–2295. doi: 10.1073/pnas.96.5.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kolykhalov AA, Mihalik K, Feinstone SM, Rice CM. Hepatitis C virus-encoded enzymatic activities and conserved RNA elements in the 3′ nontranslated region are essential for virus replication in vivo. J. Virol. 2000;74:2046–2051. doi: 10.1128/jvi.74.4.2046-2051.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song Y, Friebe P, Tzima E, Junemann C, Bartenschlager R, Niepmann M. The Hepatitis C Virus RNA 3′-untranslated region strongly enhances translation directed by the Internal Ribosome Entry Site. J. Virol. 2006;80:11579–11588. doi: 10.1128/JVI.00675-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bradrick SS, Walters RW, Gromeier M. The hepatitis C virus 3′-untranslated region or a poly(A) tract promote efficient translation subsequent to the initiation phase. Nucleic Acids Res. 2006;34:1293–1303. doi: 10.1093/nar/gkl019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shimoike T, Mimori S, Tani H, Matsuura Y, Miyamura T. Interaction of hepatitis C virus core protein with viral sense RNA and suppression of its translation. J. Virol. 1999;73:9718–9725. doi: 10.1128/jvi.73.12.9718-9725.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang TH, Rijnbrand RC, Lemon SM. Core protein-coding sequence, but not core protein, modulates the efficiency of cap-independent translation directed by the internal ribosome entry site of hepatitis C virus. J. Virol. 2000;74:11347–11358. doi: 10.1128/jvi.74.23.11347-11358.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim YK, Lee SH, Kim CS, Seol SK, Jang SK. Long-range RNA-RNA interaction between the 5′ nontranslated region and the core-coding sequences of hepatitis C virus modulates the IRES-dependent translation. RNA. 2003;9:599–606. doi: 10.1261/rna.2185603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang J, Yamada O, Yoshida H, Iwai T, Araki H. Autogenous translational inhibition of core protein: implication for switch from translation to RNA replication in hepatitis C virus. Virology. 2002;293:141–150. doi: 10.1006/viro.2001.1270. [DOI] [PubMed] [Google Scholar]
- 37.Li D, Takyar ST, Lott WB, Gowans EJ. Amino acids 1-20 of the hepatitis C virus (HCV) core protein specifically inhibit HCV IRES-dependent translation in HepG2 cells, and inhibit both HCV IRES- and cap-dependent translation in HuH7 and CV-1 cells. J. Gen. Virol. 2003;84:815–825. doi: 10.1099/vir.0.18697-0. [DOI] [PubMed] [Google Scholar]
- 38.Combet C, Penin F, Geourjon C, Deleage G. HCVDB: hepatitis C virus sequences database. Appl. Bioinformatics. 2004;3:237–240. doi: 10.2165/00822942-200403040-00005. [DOI] [PubMed] [Google Scholar]
- 39.Yanagi M, Purcell RH, Emerson SU, Bukh J. Transcripts from a single full-length cDNA clone of hepatitis C virus are infectious when directly transfected into the liver of a chimpanzee. Proc. Natl Acad. Sci. USA. 1997;94:8738–8743. doi: 10.1073/pnas.94.16.8738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chappell SA, Edelman GM, Mauro VP. Ribosomal tethering and clustering as mechanisms for translation initiation. Proc. Natl Acad. Sci. USA. 2006;103:18077–18082. doi: 10.1073/pnas.0608212103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schwartz S, Felber BK, Fenyo EM, Pavlakis GN. Env and Vpu proteins of human immunodeficiency virus type 1 are produced from multiple bicistronic mRNAs. J. Virol. 1990;64:5448–5456. doi: 10.1128/jvi.64.11.5448-5456.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaminski A, Howell MT, Jackson RJ. Initiation of encephalomyocarditis virus RNA translation: the authentic initiation site is not selected by a scanning mechanism. EMBO J. 1990;9:3753–3759. doi: 10.1002/j.1460-2075.1990.tb07588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tesar M, Harmon SA, Summers DF, Ehrenfeld E. Hepatitis A virus polyprotein synthesis initiates from two alternative AUG codons. Virology. 1992;186:609–618. doi: 10.1016/0042-6822(92)90027-m. [DOI] [PubMed] [Google Scholar]
- 44.Clarke BE, Sangar DV, Burroughs JN, Newton SE, Carroll AR, Rowlands DJ. Two initiation sites for foot-and-mouth disease virus polyprotein in vivo. J. Gen. Virol. 1985;66:2615–2626. doi: 10.1099/0022-1317-66-12-2615. [DOI] [PubMed] [Google Scholar]
- 45.Kong WP, Roos RP. Alternative translation initiation site in the DA strain of Theiler's murine encephalomyelitis virus. J. Virol. 1991;65:3395–3399. doi: 10.1128/jvi.65.6.3395-3399.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Eyll O, Michiels T. Non-AUG-initiated internal translation of the L* protein of Theiler's virus and importance of this protein for viral persistence. J. Virol. 2002;76:10665–10673. doi: 10.1128/JVI.76.21.10665-10673.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McMullan LK, Grakoui A, Evans MJ, Mihalik K, Puig M, Branch AD, Feinstone SM, Rice CM. Evidence for a functional RNA element in the hepatitis C virus core gene. Proc. Natl Acad. Sci. USA. 2007;104:2879–2884. doi: 10.1073/pnas.0611267104. [DOI] [PMC free article] [PubMed] [Google Scholar]