Abstract
Organisms exposed to reactive oxygen species, generated endogenously during respiration or by environmental conditions, undergo oxidative stress. Stress response can either repair the damage or activate one of the programmed cell death (PCD) mechanisms, for example apoptosis, and finally end in cell death. One striking characteristic, which accompanies apoptosis in both vertebrates and yeast, is a fragmentation of cellular DNA and mammalian apoptosis is often associated with degradation of different RNAs. We show that in yeast exposed to stimuli known to induce apoptosis, such as hydrogen peroxide, acetic acid, hyperosmotic stress and ageing, two large subunit ribosomal RNAs, 25S and 5.8S, became extensively degraded with accumulation of specific intermediates that differ slightly depending on cell death conditions. This process is most likely endonucleolytic, is correlated with stress response, and depends on the mitochondrial respiratory status: rRNA is less susceptible to degradation in respiring cells with functional defence against oxidative stress. In addition, RNA fragmentation is independent of two yeast apoptotic factors, metacaspase Yca1 and apoptosis-inducing factor Aif1, but it relies on the apoptotic chromatin condensation induced by histone H2B modifications. These data describe a novel phenotype for certain stress- and ageing-related PCD pathways in yeast.
INTRODUCTION
Gene expression in all organisms is regulated at multiple levels, including transcription initiation, mRNA stability and turnover, translation and protein degradation. Not surprisingly, rapid changes in cell metabolism and most responses to environmental stimuli involve significant flux of many cellular RNAs, of which mRNA transcriptome profiles have been most extensively studied to date (1,2). However, also stable RNAs, such as ribosomal, transfer, nuclear and nucleolar RNAs (rRNAs, tRNAs, snRNAs and snoRNAs), are likely to undergo specific transformations in altered conditions. It has been demonstrated that certain pathways of cell death are accompanied by the destruction of nucleic acids. For example, in metazoans the programmed cell death (PCD) called apoptosis, in addition to irreversible DNA damage, which is considered an apoptotic hallmark (3), also involves specific cleavage of several RNA species, including 28S rRNA, U1 snRNA or Ro RNP-associated Y RNAs (4). It was proposed that rRNA degradation could contribute to cell autodestruction, whereas degradation of anti-apoptotic factors mRNAs would accelerate apoptosis. In higher eukaryotes, RNA cleavage is probably carried out by RNase L, a 2′5′-oligoadenylate-dependent endoribonuclease, which functions in RNA decay during the interferon-induced response to viral infection, and whose activation in animal cells causes apoptosis (5–7). However, RNase L-independent cleavage of 28S rRNA in virus infected cells has also been reported (8).
The occurrence of apoptosis was assumed to be limited to metazoans, where elimination of single cells does not kill the whole organism. Nevertheless, recent studies revealed the existence of cell death pathway in yeast, Saccharomyces cerevisiae, and other unicellular eukaryotes, with typical hallmarks of apoptosis: DNA fragmentation, externalization of phosphatidyl serine and chromatin condensation. PCD in yeast is triggered by several different stimuli, including ageing, expression of mammalian pro-apoptotic proteins, exposure to low doses of H2O2, acetic acid, hyperosmotic stress and mating-type α-factor pheromone (9–11). Unicellular organisms are believed to undergo PCD for a variety of reasons, including elimination of old, infected and damaged cells in growth-limiting conditions for better survival of the remaining population, and adaptation of the more fit subpopulation to the ever changing and challenging environment (12).
Orthologues of core regulators of mammalian apoptosis, such as the caspase-related protease Yca1, a homologue of mammalian pro-apoptotic mitochondrial serine protease HtrA2 (Nma111), a yeast EndoG nuclease Nuc1, apoptosis inducing factor (Aif1) involved in chromatin condensation, AIF-homologous mitochondrion-associated inducer of death (AMID) Ndi1 and an inhibitor of apoptosis (IAP) Bir1, are conserved in yeast (13–18). In addition, in both human and yeast cells, histone modifications (histone H2B phosphorylation at serine 14 in human and at serine 10 in yeast and H2B deacetylation at lysine 11 in yeast) play an important role in apoptotic chromatin condensation and cell death (19–21). However, several apoptotic factors are missing in yeast, including the Bcl-2/Bax family and the apoptosis protease activator factor Apaf-1. Also, there is no good homologue of RNase L to execute possible RNA degradation. Nevertheless, yeast apoptosis has recently been shown to be activated in mRNA decay mutants (dcp and lsm) (22,23), supporting the notion that RNA metabolism and apoptosis are linked.
PCD occurs via a number of different mechanisms, e.g. caspase-dependent or independent, however, in all eukaryotes it is thought to be correlated with high levels of reactive oxygen species (ROS). ROS can either be generated exogenously through respiration or originate from exogenous sources such as exposure to hydrogen peroxide (H2O2), superoxide anions or hydroxyl radicals. Excessive ROS results in damage of cellular components (DNA, lipids and proteins), cell cycle arrest, ageing and finally cell death (24). Cells have developed a complex network of defence mechanisms, both enzymatic and non-enzymatic, against adverse consequences of oxidative stress (25). Non-enzymatic system comprises a set of small molecules acting as ROS scavengers (e.g. glutathione, thioredoxin, glutaredoxin and ascorbic acid), whereas enzymatic system eliminates oxygen radicals by the action of specialized cytosolic or mitochondrial enzymes (e.g. catalases, superoxide dismutases, glutathione peroxidases and thioredoxin peroxidases) (25). Most genes encoding components of these systems are induced in response to oxidative stress and are under transcriptional control of specific factors, for example Yap1, Msn2/Msn4 and Skn7 in budding yeast (26). However, it appears that there is no general oxidative stress response. In S. cerevisiae, different response pathways are triggered by specific oxidants and different genes are involved in maintaining efficient cellular resistance to various sources of ROS (1,2). Interestingly, recent genomic approaches to identify these genes showed that strains lacking proteins which function in RNA metabolism were oversensitive to oxidative stress (1,2). These included genes encoding rRNA helicases (Dbp3, Dbp7), rRNA processing factors (Nop12, Nsr1), mRNA deadenylases (Ccr4, Pop2) and several mitochondrial RNA splicing components (2,27). This indicates that RNA processing and degradation may have a role in cellular response to ROS.
In this study, we examined the effects of elevated ROS levels generated by oxidative stress, ageing and other apoptotic-inducing treatments on the status of ribosomal RNA in yeast S. cerevisiae and we have shown that mature rRNAs become specifically fragmented as a result of the cell response to these conditions. RNA degradation coincides with fragmentation of chromosomal DNA but occurs considerably earlier and most likely upstream of the activation of major apoptotic regulators, Yca1 and Aif1. The existence of this mechanism underscores the role of gene expression, namely rRNA turnover, in regulating certain pathways of cell death, in this case most likely through destruction of ribosomes and subsequent inhibition of translation in the early stages of apoptosis.
MATERIALS AND METHODS
Strains, plasmids and media
Yeast strains and plasmids used in this work are listed in Supplementary Table S1. The transformation procedure was as described (28). Strains were grown at 30°C either in YPD, YPGal or YPGly medium (1% yeast extract, 2% Bacto-peptone, 2% glucose or 2% galactose or 2% glycerol, respectively) or in synthetic complete medium (SC, 0.67% yeast nitrogen base, 2% glucose or 2% galactose, supplemented with required amount of amino acids and nucleotide bases). Strains W303-1A-Bax, W303-1A-Bcl-xL, W303-rDNA were grown in SC media without leucine, tryptophan or uracil, respectively.
Yeast treatments
Yeast cultures in early logarithmic phase (OD600 ∼ 0.2) were stressed with H2O2 (0.12–180 mM), menadione (0.05–0.6 mM), cumene hydroperoxide (CHP, 0.1–0.2 mM), tert-butyl hydroperoxide (t-BHP, 1–3 mM), paraquat (0.1–10 mM), diamide (0.2–2 mM) and linoleic acid hydroperoxide (LoaOOH, 0.05–0.2 mM) for 200 min (all reagents from Sigma). Treatment with acetic acid stress was performed as described (29,30). Cells were grown in SC media to early exponential stage, shifted to SC media, pH = 3, and treated with 175 mM acetic acid (Sigma) for 200 min. Hyperosmotic shock was achieved by growth of exponential cells in SC complete media containing 60% (wt/wt) glucose (Fluka) or 30% (wt/wt) sorbitol and 2% glucose (Sigma) for 2–8 h (31). Chronological ageing was performed by constant growth of yeast cultures in SC complete medium for 2–16 days (32). For expression of murine Bax or Bcl-xL proteins, cells were grown to early exponential phase in SC-Leu or SC-Trp, respectively. Expression of murine Bax was induced by shifting the cells grown to early exponential phase in SC-Leu medium containing glucose to SC-Leu medium containing galactose for 6 h. Pre-treatment with ascorbic acid (20 mM, Fluka) and respiratory chain inhibitors oligomycin A (7.5 μg/ml, Sigma) and sodium azide (1.5 mM, Sigma) was performed for 30 or 60 min prior to treatment with H2O2. For inhibition of protein synthesis, cells were treated with 50 μg/ml or 200 μg/ml of cycloheximide (Sigma) for 30 min. Cell fixation was achieved by addition of formaldehyde (final concentration 1%) or EtOH (final concentration 70%) to exponentially growing yeasts and incubation in room temperature for 10 min or 60 min, respectively. Formaldehyde was quenched by addition of glycine to the final concentration of 0.4 M for 5 min. Following the removal of fixation agents, cells were exposed to H2O2 for 200 min as described earlier.
Pulsed field gel electrophoresis
Preparation of samples and analysis of chromosomal DNA fragmentation by pulsed field gel electrophoresis (PFGE) was performed exactly as described (30). PFGE was conducted in a CHEF-DRIII Chiller System (Bio-Rad). One percent agarose gels were run in 0.5% Tris borate–EDTA buffer at 14°C with an angle of 120° with a voltage of 6 V/cm and switch times of 60–90 s for 24 h. Gels stained in ethidium bromide were analysed after destaining using Syngene Gene Genius Bioimaging System.
RNA extraction and analysis
RNA extraction, northern hybridization and primer extension were essentially as described (33,34). Low-molecular weight RNAs were separated on 6% acrylamide gels containing 7M urea and transferred to a Hybond N+ membrane by electrotransfer. High-molecular-weight RNAs were analysed on 1.2% agarose gels and transferred by capillary elution. Oligonucletides used for RNA hybridization and primer extension (W235 and W237) are listed in Supplementary Table S2. Quantification of northern blots was performed using a Storm 860 PhosohorImager and ImageQuant software (Molecular Dynamics). Dideoxy-DNA sequencing was performed on PCR-templates prepared from genomic yeast DNA using the same primers as for primer extension (W235 and W237) and a fMolSeq kit (Promega) according to manufacturer's instructions.
The 3′ RACE assay was carried out on total RNA (20 μg) isolated from untreated cells and treated with 1mM H2O2. DNA ‘adaptor’ oligonucleotide (W242) carrying aminolinker at the 3′-end was ligated with the 3′-end of total RNA using 20U of T4 RNA ligase (NEB). Ligation was performed in the presence of 25% PEG1000 (Sigma) at 37°C. RNA was extracted with phenol: chloroform: isoamyl alcohol (v/v 25:24:1), precipitated and used as a template for cDNA synthesis using W243 primer complementary to the anchor sequence and the Enhanced Avian HS RT-PCR Kit (Sigma) according to manufacturer's instructions. cDNA was amplified using primers W241 and W243, the resulting PCR product was gel purified, cloned into pGEM-T Easy vector and sequenced using primer W241.
RNase H cleavage was performed essentially as described (35). Samples of 10 μg of total RNA were annealed with 40 ng of oligonucleotide complementary to the specific regions within rRNA at 68°C for 10 min and digested with 1.5 U RNase H at 30°C for 1 h. For detection, samples were separated on polyacrylamide gels and analysed by northern hybridization using probes located upstream of RHase H cleavage.
RESULTS
Ribosomal RNAs are fragmented in stress and apoptotic conditions
To examine the existence of the RNA degradation pathway in yeast under oxidative stress, we have performed treatments with low doses of oxidative agents generating different ROS. These included the inorganic H2O2 (concentrations 0.4–3 mM), superoxide-generating menadione (concentrations 0.05–0.6 mM), paraquat (concentrations 0.1–10 mM), thiol oxidant diamide (concentrations 0.2–2 mM), organic CHP (concentrations 0.1–0.2 mM), t-BHP (concentrations 1–3 mM) and a LoaOOH (concentrations 0.05–0.2 mM). Such concentrations of oxidants result in 30–90% of cell death (2,36,37).
Total RNA from wild-type W303 or BY4741 cells grown to early exponential phase (OD600 = 0.25) in YPD media and treated with chemical compounds for 200 min was separated on 1.2% denaturing agarose/formaldehyde gels and analysed by northern hybridization using a probe complementary to the 5′-end of mature 25S rRNA (starting at position +40). This RNA species was chosen in the first place, since effects on 28S rRNA have been observed in apoptotic mammalian cells (8,38,39). On treatment with two oxidants, H2O2 and menadione, extensive decay of the mature 25S and accumulation of specific degradation products was observed, whereas little or no degradation occurred for other chemicals tested (Figure 1A, data shown only for W303 strain and treatment with H2O2, menadione, CHP and t-BHP). This indicates that RNA cleavage accompanies some oxidative stress pathways, as it is known that different oxidants elicit specific cellular responses that, though partly overlapping, induce different groups of genes and require individual sets of specialized defence functions to maintain resistance (1,2,24).
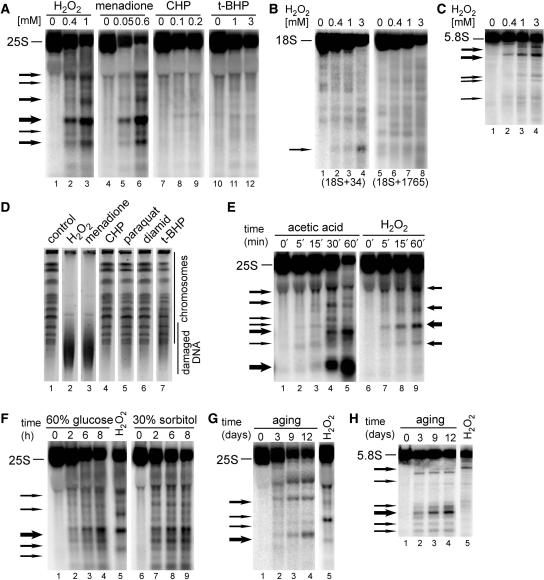
Figure 1.
Ribosomal RNA is fragmented in oxidative stress and apoptotic conditions. Northern hybridizations of total yeast RNA extracted from wild-type W303 cells treated with different stimuli shown above each panel and separated on 1.2% agarose (A, B and E–G) or 6% polyacrylamide (C and H) gels and hybridized with probes against the 25S (007, position +40 relative to the 5′ end), 18S (008, position +34, lanes 1–4 and 029, position +1765, lanes 5–8) or 5.8S rRNA (017, position +30). The position of mature rRNA species is shown on the left of each picture, whereas arrows specify major degradation products. Each experiment was repeated at least 2–3 times, this applies also to Figures 3–7. (A–C) Cells were treated for 200 min with indicated concentrations (mM) of H2O2, menadione, CHP and t-BHP. Hybridizations with probes against 25S (A), 18S (B) and 5.8S (C) are shown. (E) Time courses (min) during exposure to 175 mM acetic acid (lanes 1–5) or 0.6 mM H2O2 (lanes 6–9). (F) Hyperosmotic stress was achieved by addition of 60% glucose (lanes 1–4) or 30% sorbitol (lanes 6–9) for the times indicated (h). RNA from treatment with 0.6 mM H2O2 is shown in lane 5 for comparison. (G–H) Cells were subjected to chronological ageing for times indicated (days, lanes 1–4). RNA from treatment with 0.6 mM H2O2 is shown in lane 5 for comparison. Hybridizations with probes against 25S (G) and 5.8S (H) are shown. (D) Analysis of chromosomal DNA by PFGE. Samples from untreated (lane 1, control) and treated cells for 200 min with 1 mM H2O2 (lane 2), 0.5 mM menadione (lane 3), 0.2 mM CHP (lane 4), 5 mM paraquat (lane 5), 2 mM diamid (lane 6) and 1 mM t-BHP (lane 7) were separated on 1% agarose gels that were stained in ethidium bromide, destained and analysed using Syngene Gene Genius Bioimaging System.
In addition to the 25S rRNA, other rRNA species were also probed for undergoing specific decay. After treatment with H2O2, RNA damage with accumulation of characteristic breakdown products occurred for 5.8S and, to a much lesser extent, for 18S, but not for 5S, (Figure 1B and C; data not shown). In the case of 18S, hardly any degradation intermediates were detected, there was some decay of the mature RNA, however, it was approximately 2.5 to 3-fold weaker than for the mature 25S. Therefore, we conclude that mainly the two components of the large ribosomal subunit, 25S and 5.8S, undergo specific apoptotic degradation.
The oxidants utilized, except for H2O2, had not been tested for apoptotic effects in yeast. One of the most recognized apoptotic markers is fragmentation of chromosomal DNA. Internucleosomal DNA laddering, typical for mammalian apoptosis, has not been detected during PCD in yeast; nevertheless, a higher order chromatin fragmentation to segments of several hundred kilobases also occurs in yeast (30,40). This DNA breakdown can be monitored either by the terminal deoxynucleotidyl transferase dUTP nick-end labelling (TUNEL) assay or by using PFGE of genomic DNA. The latter approach was applied to verify which oxidative agents lead to apoptotic phenotypes. Chromosomal DNA from cells treated with H2O2 (1 mM), menadione (0.5 mM), CHP (0.2 mM), paraquat (5 mM), diamid (2 mM) and t-BHP (1 mM) for 200 min was analysed using PFGE (Figure 1D). Clear DNA degradation was observed only for cells exposed to H2O2 and menadione, other treatments did not result in a visible apoptotic fragmentation. This is in a striking agreement with rRNA degradation that occurred only in H2O2- and menadione-treated cells. This strongly indicates that rRNA decay phenotype can be related to apoptosis.
To confirm this, we have examined other conditions known to provoke apoptosis in yeast, i.e. acetic acid, ageing and hyperosmotic shock (Figure 1E–H). For treatment with acetic acid, cells were grown in SC complete medium (pH 3) to exponential phase and exposed to 175 mM acetic acid for up to 200 min (29,30). Hyperosmotic shock was achieved by growth of exponential cells in SC complete media supplemented with 60% (wt/wt) glucose or 30% (wt/wt) sorbitol (31). And finally, chronological ageing was performed by constant growth of yeast cultures in SC complete medium for 2–16 days (32). This analysis revealed that all apoptotic stimuli tested resulted in the 25S and 5.8 rRNA fragmentation with the degradation pattern specific for each condition (shown in Figure 1E–G for 25S in all apoptotic conditions and in Figure 1H for 5.8S during ageing). The accumulating intermediates generated by some factors were comparable (see for example, cleavages mediated by 60% glucose and 30% sorbitol, acetic acid and H2O2, Figure 1E and F); however, the general outcome of each treatment indicated differences in the course of events during each response.
The occurrence of RNA degradation triggered by H2O2 was monitored during a time course between 5 and 60 min and over a broad range of concentrations (0.12–180mM for 200 min) (Figure 1E, lanes 6–9 and Figure 2A). Degradation was initiated relatively fast, since it was apparent at 5 min for 0.6 mM H2O2 (Figure 1E, lanes 6–9), 15–30 min for 175 mM acetic acid (Figure 1E, lanes 1–5), 2 h for 60% glucose and 30% sorbitol (Figure 1F) and 3 days for ageing (Figure 1G) following the treatment. This onset of rRNA degradation distinctly precedes the timing of DNA damage characterized in apoptotic yeast exposed to the same stimuli (30), indicating that RNA decay process is activated early during the response. Also, in the case of H2O2, low doses of the oxidant, starting with 0.12 mM and optimal at 0.4–3 mM, were sufficient to initiate rRNA degradation with the appearance of specific bands. When high concentration of H2O2 (180 mM), believed to result in cell necrosis, was used, these specific degradation products were absent; however, the level of mature 25S and 18S rRNAs was also significantly reduced (Figure 2A; data not shown).
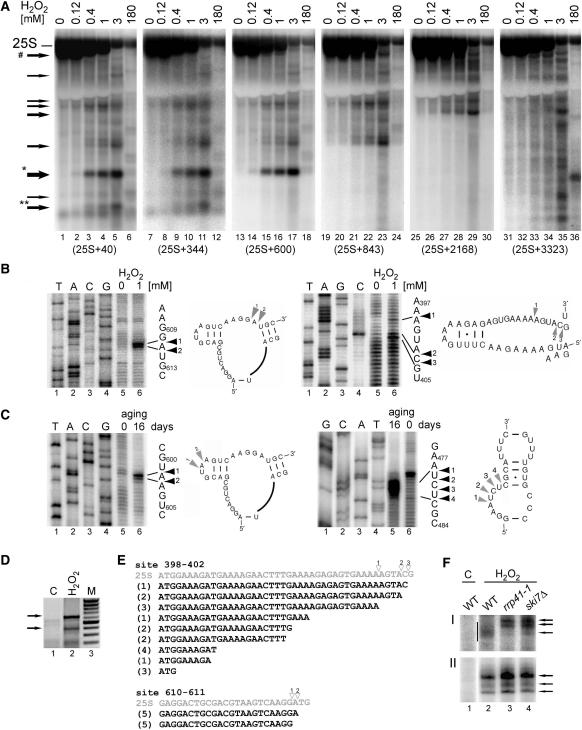
Figure 2.
Main cleavage sites in the 25S rRNA are located in loop regions. (A) Northern hybridizations of total yeast RNA extracted from wild-type W303 cells treated with different concentrations of H2O2. Hybridizations with probes against 25S (probe 007, position +40, lanes 1–6; probe W234, position +344, lanes 7–12; probe W236, position +600, lanes 13–18; probe W238, position +843, lanes 19–24; probe W239, position +2168, lanes 25–30 and probe W240, position +3323, lanes 31–36). Asterisks above the arrows indicate the products that were further analysed. Arrow marked with a hatch shows a band matching the potential 3′ product of the major cleavage, 5′ product is marked with one asterisk. (B–C) Primer extension analysis for two main cleavage sites in the 25S rRNA in W303 cells treated with 1 mM H2O2 (A) and in 16-day old chronologically aged rho0 W303 cells (B). Primer extensions were performed using primers W235 for sites around positions +400 and +470 and W237 for sites around position +600 relative to the 5′ end of the mature 25S. DNA sequencing on a PCR product encompassing the 5′ end of the 25S from +40 to +701, using the same primers was run in parallel on 6% sequencing polyacrylamide gels (lanes 1–4). The sequences with primer extension stops are shown on the right. Secondary structures of the regions in the vicinity of the cleavages, indicated by arrowheads and shown beside corresponding primer extension reactions, were adapted from the website http://rna.icmb.utexas.edu/. (D–E) 3′ ends of cleaved-off products for the major cleavage at positions +610–611 were mapped by 3′ RACE. (D) PCR reactions on cDNA prepared using total RNA from untreated control (lane 1, C) and cells treated with 1 mM H2O2 (lane 2). To generate cDNA, total RNA that had been ligated to an ‘anchor’ oligonucleotide (W242) with T4 RNA ligase, was reverse transcribed using a primer specific for the anchor (W243). This was followed by PCR reaction using the same 3′ primer and the 5′ primer starting at position +50 in the 25S rRNA (W241). Arrows indicate products corresponding to fragments cleaved at +398–404 (lower) and +610–611 (upper). PCR fragments were cloned into pGEM-Teasy and sequenced. (E) Sequences obtained by the 3′ RACE analysis for fragments cleaved at site +398–403 (19 independent clones) and site +610–611 (10 independent clones). The corresponding regions of the 25S with cleavage sites mapped by primer extension and indicated with empty arrowheads are shown above in grey. Figures in parentheses show the number of identical clones. (F) Mapping 3′ ends of two major cleavages sites using RNase H cleavage on total RNA extracted from wild-type, rrp41-1 and ski7Δ cells treated with 1mM H2O2 (lanes, 2–4) and from wild-type untreated control (lane 1, C). RNase H treatment was performed on RNA samples annealed to DNA oligonucleotides W244 and W263 complementary to positions +271 and +510, respectively. Samples were separated on a 8% acrylamide gel and hybridized with probe W234 (F-I) and probe W264 (F-II) to detect 3′ ends of fragments cleaved at +398–403 (F-I) and at +610–611 (F-II), respectively. Arrows show more defined 3′ ends of products cleaved at +610–611 for all strains and at +398–403 in the mutants; vertical bar in F-I indicates heterogenous 3′ ends of products cleaved at +398–403 in wild-type cells.
These data show that different ROS-generating treatments that lead to yeast apoptosis, namely H2O2 and acetic acid, ageing and hyperosmotic shock, induce RNA fragmentation that most likely precedes the DNA damage and, as in higher eukaryotes, can be considered a hallmark of the induction of PCD in yeast.
Cleavages in the 25S rRNA are endonucleolytic and require cellular machinery
Specific cleavages within the 25S rRNA generated in the presence of hydrogen peroxide were monitored by northern hybridization with probes located along the molecule to narrow down the regions to be further analysed (Figure 2A). This analysis showed the accumulation of diverse degradation products, some of which extended from the 5′-end of the molecule (Figure 2A, lanes 1–6), whereas others were also truncated at their 5′ ends (Figure 2A, lanes 25–30). The striking decrease in the level of the mature 25S rRNA at higher doses of the oxidant (1–3 mM) indicates that following specific cleavages the majority of rRNA becomes degraded, possibly by the exosome complex of 3′→5′ exonucleases that participates in the decay of rRNA precursors and excised transcribed spacers (41). The emergence of the characteristic cut-off in the signal at the fragment size corresponding to the position of the probe indicated that major cleavage sites are located around positions +400, +600 and +900 with respect to the 5′-end of the molecule. Two of these cleavages, at positions +610–611 and +398–403, were mapped for treatment with 1 mM H2O2 by primer extension using primers W237 and W235 situated downstream of the expected cleavage sites (Figure 2B). Similarly, major cleavage sites were analysed in 16-day old chronologically aged cells using the same primers (Figure 2C) and mapped at positions +601–602 and +478–501. According to the secondary structure of the 25S rRNA taken from (42), the regions where mapped cleavages occur (shown in Figure 2B and C besides corresponding primer extension reactions) are located at unpaired nucleotides in loops or bulges. This points to the action of single-stranded RNA nucleases.
To establish the nature of the observed RNA fragmentation, 3′-ends of the products generated by the H2O2-mediated cleavage in the 25S at positions +610–611 and +398–403 were determined by the 3′ RACE. To this end, DNA ‘anchor’ oligonucleotide (W242) was ligated with T4 RNA ligase to total RNA from untreated and treated W303 cells to prepare cDNA using a primer specific for the anchor (W243). This served as a template to amplify products containing required fragments using the same 3′ primer and a 5′ primer that covers the 5′-end of 25S RNA starting at position +50 (W241). The ensuing PCR fragments (Figure 2D) were cloned into pGEM-Teasy and sequenced. The results of 10 sequenced clones for the cleavage at +610–611 and 19 clones for the cleavage +398–403 are shown in Figure 2E. In the case of the major site (cuts at positions +610–611), this analysis confirms that the 5′ and 3′ ends of this degradation product overlap (Figure 2E, lower panel), which is consistent with the endonucleolytic mechanism of the cleavage. Mapping the 5′ and 3′ ends at site +398–403 by primer extension and 3′ RACE produced a different pattern: these ends do not match ideally but the products get progressively shorter pointing at the action of 3′→5′ exonucleases (Figure 2E, upper panel). The most likely candidate is the exosome, a large complex with a 3′→5′ exonucleolytic activity involved in the processing and degradation of mRNA, rRNA and other RNA substrates (43). Mutants in the exosome core component Rrp41, the nuclear subunit Rrp6 and the cytoplasmic cofactor Ski7, were used to assess the status of the product 3′ ends by a specific RNase H cleavage. This cleavage, directed by a DNA–RNA hybrid between oligonucleotide W244 and a complementary region in the 25S starting at residue +271, allows higher resolution of analysed RNAs. This analysis shows that products generated at site +398–403 in the exosome mutants rrp41-1 and ski7Δ, but not in rrp6Δ, are 3′ extended and less heterogenous than corresponding fragments in the wild-type strain (Figure 2F–I; data not shown). In contrast, positions of cleavages at site +610–611 are not affected by mutations in the exosome (Fig. 2F–II). This suggests that the cytoplasmic exosome may contribute to rRNA decay by digesting 3′ ends of at least some cleavage products.
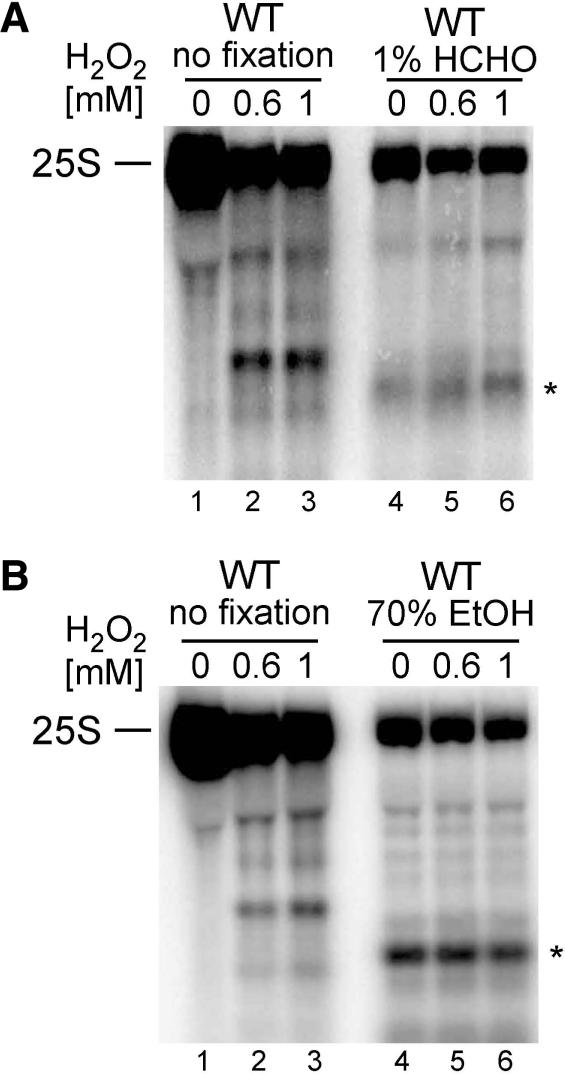
To ascertain that RNA degradation process is enzymatic and not chemically induced by various reactive compounds, yeast cells were fixed with 1% formaldehyde for 10 min or with 70% ethanol for 30 min prior to exposure to increasing concentrations of hydrogen peroxide (Figure 3A and B). Both fixation procedures preserve cellular structures, however, it is known that most fixatives have harmful consequences, e.g. cause some loss of cellular components, including ribosomes. Nevertheless, a similar approach had been used to demonstrate that DNA damage in apoptotic yeast cells was an enzymatic process (30). Also, in the case of RNA, the appearance of specific H2O2-induced degradation products was prevented by fixation, although the overall level of rRNA was reduced. Some faster migrating RNA species were detected in formaldehyde or ethanol fixed cells, however, these were generated also in the absence of the oxidant and did not intensify after treatment (Figure 3A and B, lane 4).
Figure 3.
H2O2-mediated degradation of the 25S rRNA requires cellular response. Northern hybridization of total RNA from untreated wild-type W303 cells (A and B, lanes 1–3), fixed with 1% formaldehyde (HCHO) (A, lanes 4–6) or with 70% EtOH (B, lanes 4–6) prior to exposure to indicated concentrations of H2O2. Hybridization was performed with probe 007 starting at position +40 of the 25S. Asterisks designate unspecific degradation product, detectable only in RNA isolated from fixed cells (A and B lanes 4–6).
Finally, to check whether rRNA destruction during apoptotic response is not due to cessation of translation in dying cells, cells were treated for 200 min with the translation elongation inhibitor cycloheximide (200 μg/μl) and this did not lead to an apparent rRNA degradation (Supplementary Figure S1A). This is also supported by our earlier observations that exposure of yeast cells to many oxidative agents that cause cell death does not result in rRNA decay (Figure 1A).
Together, this strongly suggests that rRNA degradation observed in apoptotic and oxidative stress conditions is not simply a result of cell death but is produced in the process that requires enzymatic activity and functional cellular machinery. rRNA is most likely cleaved endonucleolytically and in some cases, dictated probably by the RNA structure, this is followed by exonucleolytic digestion by the exosome.
rRNA degradation is strongly correlated with ROS levels and is connected with oxidative stress response and apoptosis pathways
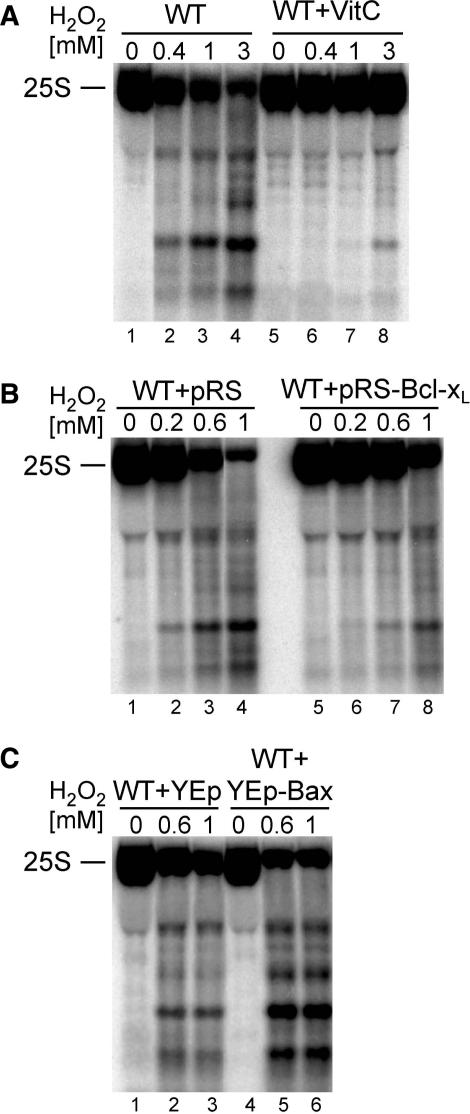
Treatment with oxidative agents and apoptotic stimuli generate elevated levels of ROS in the cell. To test whether there is a direct link between RNA fragmentation and ROS, a potent ROS scavenger, l-ascorbic acid (vitamin C), was used (44). The presence of 10mM ascorbic acid prior to treatment with standard doses of H2O2 almost totally abrogated degradation of the 25S rRNA (Figure 4A). In addition, the ectopic expression of murine Bcl-xL protein of the anti-apoptotic mammalian Bcl-2 family, known to have a protective effect against ROS in yeast (45,46), also strongly safeguarded the 25S rRNA from rapid degradation by H2O2 (Figure 4B). In contrast, expression of the mammalian pro-apoptotic Bax protein that increases ROS level (36,47), additionally enhanced the degradation phenotype (Figure 4C). This confirms the direct correlation between the production of ROS and the fate of cellular nucleic acids, leading not only to DNA but also rRNA damage and destruction of ribosomes.
Figure 4.
Intensity of 25S rRNA degradation is correlated with ROS levels. Northern hybridizations of total RNA from W303 yeast cells exposed to different concentrations of H2O2. The level of ROS was altered by pre-treatment with a potent anti-oxidant ascorbic acid (VitC; A, lanes 5–8) and by the expression of the murine anti-apoptotic protein Bcl-xL (B, lanes 5–8) or the pro-apoptotic murine protein Bax (C, lanes 4–6). Hybridization was performed with probe 007 starting at position +40 of the 25S. Cells in the absence of vitamin C (A, lanes 1–4) and expressing vectors alone (pRS314, B, lanes 1–4 and YEp51, C, lanes 1–3) were subjected to the same amounts of H2O2 by way of control reactions.
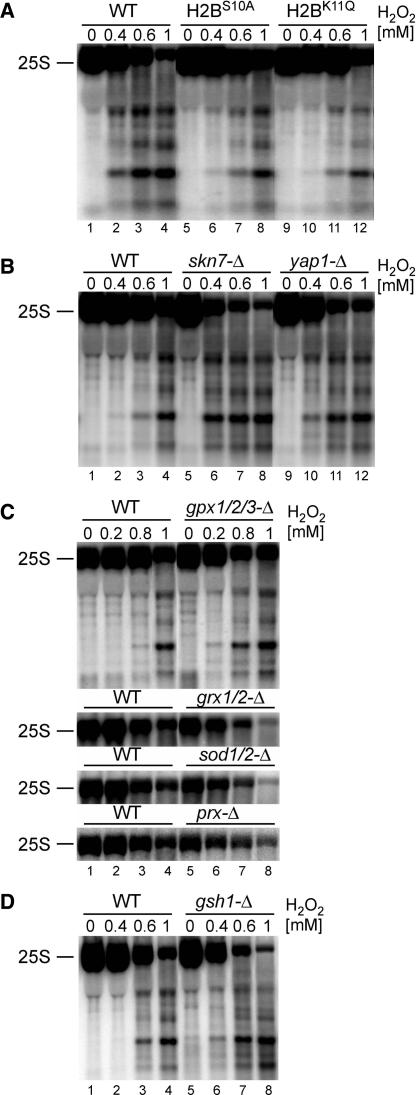
From the data presented so far, it appears that the observed rRNA fragmentation may possibly represent a part of the cellular oxidative stress and apoptotic responses. This was assessed by testing the extent of the 25S rRNA degradation in different mutants defective in these pathways. In the first place, yca1Δ strain, lacking the only identified apoptotic metacaspase Yca1 in yeast, and aif1Δ cells not expressing the yeast apoptosis inducing factor Aif1 (13,15), were assayed for H2O2-induced RNA decay, however, no significant differences were observed (Supplementary Figure S1B and C). Similarly, addition of a broad-range caspase inhibitor z-VAD-FMK (20 μM) that prevents Yca1-dependent cell death in yeast (13,48) had no effect on rRNA fragmentation (data not shown). This indicates that rRNA degradation detected in all apoptotic conditions tested is independent of the two major apoptosis mediators, Yca1 and Aif1, and of other potential yeast caspases. Likewise, treatment with translation inhibitor cycloheximide, that has been shown to prevent apoptotic cell death induced by H2O2 and acetic acid (29,36), had little or no effect on H2O2-mediated rRNA degradation (Supplementary Figure S1A). However, it appears that events in the course of apoptosis that require protein synthesis are rather late, for example DNA fragmentation and chromatin condensation, whereas rRNA decay is initiated relatively fast. In contrast, different outcome was observed for mutants inhibiting chromatin condensation during H2O2-induced apoptosis. Phosphorylation of Serine 10 and deacetylation of Lysine 11, both in histone H2B, were reported to have an essential role for the progress of cell death in yeast (20,21). In agreement, S10A or K11Q mutations in histone H2B that prevent these modifications and abrogate apoptosis resulted in the significant reduction of the 25S rRNA degradation, both the decay of the mature rRNA and the amount of degradation products (Figure 5A). Together, these data show that the destruction of ribosomal RNA in cells treated with H2O2, and possibly with other apoptotic stimuli, is a part of a yeast cell death pathway that involves histone modification and not the caspase-dependent pathway. Remarkably, Nuc1-mediated apoptosis resulting from over-expression of yeast EndoG homologue Nuc1, a major mitochondrial nuclease, was also reported to be Yca1- and Aif1-independent and related to histone modifications (18). Alternatively, it can be envisaged that damage of ribosomes triggered by apoptotic stimuli is an early event during the response, does not require protein synthesis, precedes caspase activation and acts as an upstream signal in the apoptotic pathway. This scenario is consistent with most observations so far.
Figure 5.
The level of 25S rRNA degradation is linked with chromatin condensation and anti-oxidative systems. Northern hybridizations, using the probe 007 starting at position +40 of the 25S, of total RNA from different mutants and their isogenic wild-types (lanes 1–4) treated with indicated concentrations of H2O2. (A) Histone H2B mutants, H2BS10A (lanes 5–8) and H2BK11Q (lanes 9–12). (B) Strains lacking transcriptional factors involved in oxidative stress response, Skn7 (lanes 5–8) and Yap1 (lanes 9–12). (C) Mutants lacking components of anti-oxidant pathways, glutathione peroxidases gpx1/2/3-Δ, glutaredoxins grx1/2-Δ, superoxide dismutases sod1/2-Δ and peroxiredoxins prx-Δ lacking Tsa1, Tsa2, Prx1, Dot5 and Ahp1 enzymes. For the last three mutants, only the level of the mature 25S rRNA is shown. (D) Strain gsh1Δ deficient in glutathione synthesis (lanes 5–8).
As the oxidative stress in yeast proceeds through multiple pathways that involve different response mechanisms (1,2), we tested several known enzymes and factors that regulate these responses. These included two major transcription factors, Yap1 and Skn7, that control expression of several genes induced by oxidative stress and in this way participate in ROS sensing (1,26,49–51), as well as components of antioxidant pathways, e.g. superoxide dismutases Sod1-2, glutathione peroxidases Gpx1-3, glutaredoxins Grx1-2, peroxiredoxins Tsa1-2, Prx1, Dot5 and Ahp1, thioredoxins Trx1-2 and thioredoxin reductases Trr1-2 (24,52). These enzymes are required for protection against ROS either by catalysing the breakdown of oxidative compounds or by restoring natural intracellular redox equilibrium. In addition, as glutathion protects cells against ROS, we also used a gsh1Δ strain lacking a γ-glutamylcysteine synthetase, which, when grown on glutathion-free synthetic medium, leads to glutathion depletion and cell death (36,53). Strains lacking these proteins are more sensitive to several oxidants than their isogenic wild-types (2), and following treatment with H2O2 they showed a marked increase in the 25S rRNA degradation, however, to different degrees depending on the mutant. In Figure 5B–D, yap1Δ, skn7Δ, gpx1/2/3Δ, sod1/2Δ, grx1/2-Δ, prxΔ (tsa1/2Δ/prx1Δ/ahp1Δ/dot5Δ) and gsh1Δ strains are shown, which gave the most evident effects in comparison with their respective isogenic wild-types, particularly when considering the decay rate of the mature 25S rRNA. These data indicate that properly functioning oxidative stress response also protects cellular components such as nucleic acids from the attack by ROS and that defects at any step of this defence result in a more severe and faster breakdown. It is noteworthy that multiple anti-oxidant mutants lacking all components of each enzymatic pathway exhibit a stronger effect on the 25S degradation than single mutants, pointing to the additive protection actions of these systems. Striking effects on rRNA stability in strains lacking stress response transcription factors Yap1 and Skn7 indicate that the synthesis of new anti-oxidant proteins that are induced by oxidative stress might be required for protection of ribosomes. Consistently, blocking protein synthesis by pre-treatment with cycloheximide resulted in somehow stronger rRNA degradation (Supplementary Figure S1A, lanes 1–5 and 11–15).
Taken together, this indicates that targeting rRNA degradation during oxidative stress may directly contribute to cell death.
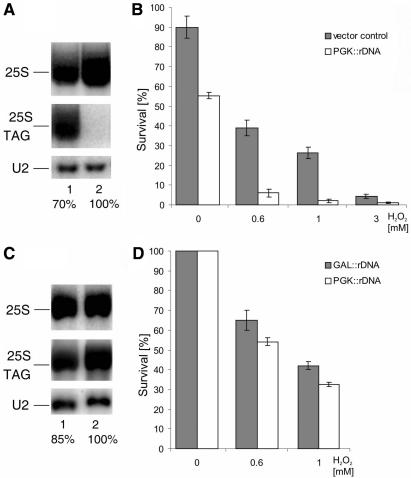
To test whether the level of rRNA, which reflects the amount of cellular ribosomes, may be somehow linked with cell survival under oxidative stress, we attempted to create a situation where the steady-state level of mature rRNAs will be increased or decreased. However, additional copies of rDNA present on a multicopy pNOY102 plasmid under control of the inducible GAL7 promoter (54) did not affect the amount of any mature rRNA species, possibly due to mechanisms that regulate ribosome abundance (data not shown). In contrast, the level of total genomic and plasmid-derived 18S and 25S rRNAs was, to our surprise, reduced to 70% in a strain transformed with the multicopy pJV12 plasmid expressing a tagged rDNA gene under the control of the constitutive PGK1 promoter (55), when compared to a strain transformed with vector alone (Figure 6A). The basis of this effect is unclear, particularly that it was seen even though the tagged rRNA versions were expressed as confirmed by northern blots using probes specific for plasmid borne 25S rRNA (Figure 6A). Nevertheless, the strain carrying pJV12 showed a decreased viability already in the absence of oxidant (1.6-fold) and even more strikingly reduced following treatment with different concentrations of H2O2 (4.6-fold for 0.6 mM and 6.24 for for 1 mM, respectively) relative to the strain with vector alone (Figure 6B). In another approach, we used the NOY504 strain, which carries a temperature sensitive (ts) RNA polymerase I (56). At 37°C, this strain ceases to grow but it sustains slow growth at 30°C due to reduced levels of mature rRNA. Expression of additional copies of rDNA from pNOY102 or pJV12 plasmids improves growth at all temperatures and rescues the ts-lethal phenotype (55,56). Growth of NOY504 expressing additional rDNA under the control of GAL7 (pNOY102) or PGK1 (pJV12) promoters resulted in total rRNA levels lower by 15% in the latter case (Figure 6C). This relatively modest difference in rRNA abundance led to a 10% decrease in survival of cells exposed to oxidative stress (Figure 6D).
Figure 6.
Cell viability and survival is related to the level of rRNA. (A and C) Northern hybridizations of RNA isolated from cells transformed with plasmids carrying additional rDNA repeats and containing a specific tag. rDNA expressed from a constitutive PGK1 promoter (PGK::rDNA, lane 1) compared with vector alone (lane 2), cells grown on glucose (A); rDNA expressed from PGK1 promoter (PGK::rDNA, lane 1) or from a GAL7 (GAL::rDNA lane 2), cells grown on galactose (C). The same membrane was probed with probe 007 starting at position +40 of the 25S rRNA which detects both 25S forms and with probe 046 specific for the tag within the plasmid-encoded 25S. The numbers below each lane, expressed as percentage, show the level of total, i.e. endogenous and plasmid-borne, mature 25S rRNA standardized to a U2 snRNA loading control. Quantification of the northern blot was performed using a Storm 860 PhosphorImager and ImageQuant software (Molecular Dynamics). (B and D) Survival of cells after treatment with increasing concentrations of H2O2 for 200 min. Cells expressed vector alone (grey bars) and PGK::rDNA (white bars) (B) or GAL::rDNA (grey bars) and PGK::rDNA (white bars) (D). Cell viability was measured by the plating assay and the data represent mean ± SEM of 3–4 independent experiments.
This indicates that there may exist a correlation between the quantity of ribosomal subunits and the capacity of the cell to elicit functional defence mechanisms and prevent cell death. It is possible that there is a feedback mechanism that controls this relationship: a healthy cell that contains an adequate number of ribosomes is able to respond more efficiently to stress stimuli to protect cell components from damage, including ribosomes themselves. Therefore, provided that the level of ribosomal RNA monitors cell fitness, its sudden reduction may act as one of the signals to initiate cell death mechanisms, including apoptosis.
rRNA degradation depends on the mitochondrial activity in the cell
Mitochondria are the major source of endogenous ROS generated by oxidative phosphorylation. The extent to which mitochondria are involved in mammalian or yeast apoptosis is still questionable, although it appears that mitochondrial ROS could be important in some signalling pathways (57,58) and have a central role in some apoptotic pathways and less crucial in others (59). For example, apoptotic cell death in yeast induced by acetic acid, pheromone and Bax expression was shown to be mediated by mitochondria (31,60).
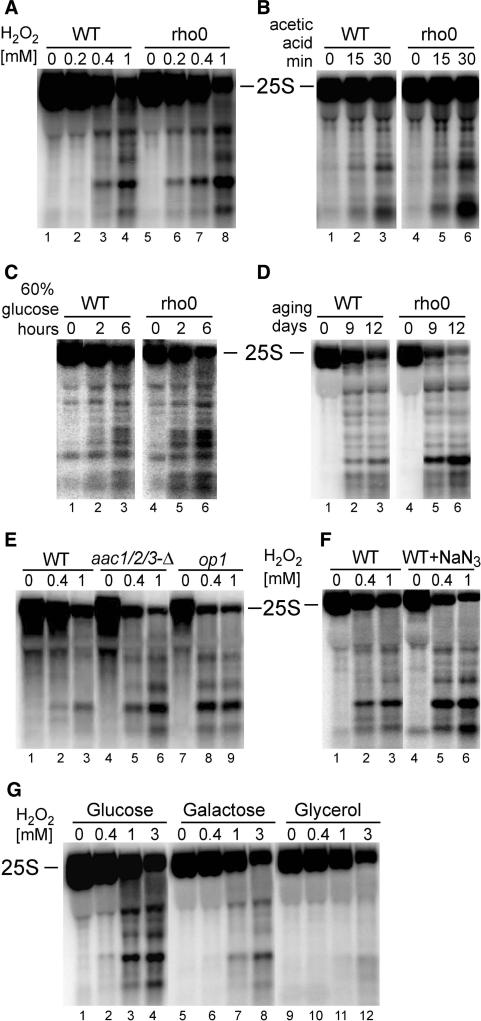
The correlation between mitochondria and rRNA stability was assessed, in the first place, by checking rRNA level in respiratory-deficient rho0 cells lacking mtDNA in conditions inducing apoptosis, i.e. exposed to H2O2 (Figure 7A), 175 mM acetic acid (Figure 7B), hyperosmotic stress (60% glucose, Figure 7C) and during chronological ageing (Figure 7D). All treatments resulted in a remarkably robust degradation of the 25S and 5.8S rRNAs in rho0 strains when compared to the parental W303 and BY4741 strains (Figure 7, Supplementary Figure S1E; and data not shown). This points to the importance of the functioning mitochondria in the stress-induced rRNA degradation. To test the contribution of the oxidative phosphorylation, two mutants in these pathways were used, op1 with a point mutation in a major ADP/ATP carrier AAC2 (Arg96→His96), and a triple aac1/2/3Δ deletion mutant (61) (Figure 7E). Both mutants behaved in a similar manner as rho0 cells and exhibited more pronounced rRNA degradation in the presence of H2O2 than the isogenic wild-type; however, the phenotype was stronger for op1 than upon deletion of the three carrier proteins, possibly due to a dominant negative effect in the point mutant. In addition, treatment with F0-F1 ATPase proton-pump inhibitors, oligomycin A (7.5 μg/ml) and sodium azide (NaN3, 1.5 mM) that block electron transfer and the synthesis of mitochondrial ATP (62–64), resulted in a moderate increase in the rRNA cleavage (Figure 7F and Supplementary Figure S1D). All these experiments indicate that the process of respiration, though generating the endogenous ROS, is also vital for counteracting its adverse effects. This was further supported by the degree of rRNA protection against oxidative damage caused by H2O2 observed for yeast cells grown on different carbon sources, which are known to affect the level of respiration (65). The most extensive RNA decay was observed in glucose, where a process called glucose repression discourages respiration. It was less pronounced in galactose and least of all in the non-fermentable source, glycerol, where mitochondrial respiration is forced (Figure 7G). These experiments directly correlate functional mitochondria and the process of respiration with the defence against oxidative stress triggered by H2O2, acetic acid, hyperosmosis and ageing that, among others, prevents destruction of cellular components, including rRNA.
Figure 7.
Degradation of the 25S rRNA in apoptotic condition is related to mitochondrial respiration activity. Northern analysis, using probe 007 starting at position +40 of the 25S, of RNA from mutants or conditions where mitochondrial respiration is inhibited. (A–D) W303 wild-type or rho0 strains (lacking mtDNA) in different apoptotic conditions: treatment with varying amounts of H2O2 (A, WT lanes 1–4, rho0 lanes 5–8); treatment with 175 mM acetic acid for times indicated (B, WT lanes 1–3, rho0 lanes 4–6); exposure to hyperosmotic stress (60% glucose) for times indicated (C, WT lanes 1–3, rho0 lanes 4–6); chronological ageing for indicated numbers of days (D, WT lanes 1–3, rho0 lanes 4–6). (E) Mitochondrial ATP/ADP carrier mutants, aaa1/2/3-Δ (lanes 4–6) and op1 (lanes 7–9) and the isogenic wild-type (lanes 1–3) treated with increasing concentrations of H2O2. (F) Wild-type W303 strain, untreated (lanes 1–3) and treated with the respiration inhibitor sodium azide (NaN3, lanes 4–6), exposed to indicated concentrations of H2O2. (G) H2O2 treatment of wild-type W303 cells grown on different carbon sources: glucose (lanes 1–4), galactose (lanes 5–8) and glycerol (lanes 9–12).
DISCUSSION
Several cellular responses are regulated at the translational level, particularly by selective translation of specific mRNAs or by inhibition of the ribosome and protein synthesis, as these processes consume a large amount of energy. Such inhibition can follow various stimuli, including endoplasmic reticulum stress and unfolded protein response (UPR), transition into quiescence and different stress-related and cell death-related signals (66). The most straightforward and fastest way to achieve translation inhibition is to target ribosomal RNA. Interestingly, it has been proposed that repression of protein synthesis during UPR in human cells is due to the cleavage of 28S rRNA by hIRE1β, a second homologue of IRE1 (67). Also, during apoptosis in some mammalian cells degradation of 28S rRNA by RNaseL, but also of other RNAs such as Y RNA or some mRNAs, has been suggested to block protein synthesis that contributes to, but could even initiate, cell death. Furthermore, damage to the 28S rRNA may act as a ribotoxic stress and induce an early death-committing signal through activation of SAP and MAP kinases (68,69). These possibilities were not examined in yeast PCD pathways. We have analysed the behaviour of ribosomal RNAs during oxidative stress and in apoptotic pathways that are induced by different stimuli. We have observed that cells exposed to all apoptotic conditions tested, such as H2O2, acetic acid, hyperosmotic stress (60% glucose) and ageing, reveal a significant degradation of the 25S and some of 5.8S rRNAs, with a much lesser effect on the 18S rRNA. The decay of mature rRNAs was accompanied by the accumulation of treatment-specific, yet partly overlapping degradation intermediates.
Although there is no evidence so far that rRNA damage during apoptotic conditions in yeast can directly initiate cell death by activating signalling pathways, it is tempting to speculate that this might be the case. Such signalling could be conveyed either by a critical decrease of the mature 25S rRNA or, alternatively, by the accumulation of degradation intermediates/products, which can function as signal molecules triggering a specific PCD pathway. In most cases, when ribosomal RNAs are depleted (e.g. in pre-rRNA processing mutants), rRNA degradation is conducted rapidly with no or little rRNA fragments detectable; however, lack of Lsm proteins has been reported to result in degradation of ribosomal RNAs with accumulation of unusual intermediates (70). It is noteworthy that one apoptotic pathway that is linked with RNA metabolism is triggered by the defect in mRNA turnover caused by mutations in enzymes involved in 5′ decapping, including components of Dcp1-2 and Lsm1-7 complexes (22).
rRNA fragmentation is an enzymatic process and depends on ROS generation
Each of the applied apoptosis-inducing conditions resulted in a clear-cut pattern of the 25S degradation intermediates or products. This points to the endonucleolytic nature of the reactions, though exonucleolytic destruction, with certain RNA fragments temporarily protected by compact structures or tight interactions with proteins, cannot be excluded. However, mapping 5′ and 3′ boundaries of the major H2O2-induced cleavage product by primer extension and 3′ RACE, respectively, confirmed that both ends strictly overlap and thus result from the endonucleolytic cut.
It has been shown that apoptotic stimuli in yeast generate ROS that are closely correlated with the onset or progression of PCD (71). We saw that, also the degree of rRNA decay corresponded to the cellular level of ROS, which was modified by using ROS scavenger (ascorbic acid) or ectopic expression of pro- or anti-apoptotic proteins (Bax and Bcl-xL, respectively) known to affect ROS generation. Also, rRNA degradation was more robust in oxidative stress defence mutants, both enzymatic and non-enzymatic, where intracellular ROS is not properly neutralized. All these observations argue that there is a direct link between ROS production and rRNA fragmentation. It can be envisaged that various reactive species themselves are able to produce endonucleolytic nicks in RNA molecules that will lead to breakdown, particularly as all mapped cleavages occur in single-stranded regions that constitute loops and bulges and are more accessible to chemical compounds in the solvent. However, this is not the case, given that specific RNA fragmentation did not occur in cells fixed with formaldehyde or ethanol prior to treatment with H2O2. In addition, oxidative agents used in this work generate different forms of ROS such as H2O2, hydroperoxide (LoaOOH, CHP and t-BHP), superoxide anion (menadione and paraquat) and hydroxyl free radical (produced from H2O2 or superoxide anion). Although all were applied at toxic doses, only two of them, H2O2 and menadione, mediated rRNA degradation, supporting the notion that oxidative compounds as such do not provoke cuts in RNA molecules within the cell. Taken together, this strongly suggests that active cellular machinery, such as signalling factors and RNA degrading enzyme(s), is required for this process. Nevertheless, these enzymatic activities are still to be identified. The closest yeast homologue of mammalian RNase L, Ire1, a sensor of the unfolded protein response, functions in the unconventional splicing of Hac1 pre-mRNA and contains protein kinase and endoribonuclease domains similar to those in RNase L (72). Although our unpublished data show that deletion of Ire1 has no effect on the H2O2-induced rRNA degradation (M.S. and J.K.), it does not exclude its participation in other apoptotic pathways, for example those related to endoplasmic reticulum stress and unfolded protein response. We are currently testing several known yeast endo- an exo-nucleases for participation in apoptosis-related rRNA degradation. Preliminary observations suggest that, contrary to expectations, this function may involve not one but a number of unspecific nucleases, including mitochondrial Nuc1p, that act in a redundant fashion (M.S. and J.K., unpublished data).
The reason why treatment with some oxidative agents but not others produce fragmented RNA is not entirely clear, however, it is known that different chemicals induce specific responses leading to expression of distinct sets of genes (1,2), and possibly only a few activate pathways that involve RNA destruction. Our results suggest that rRNA degradation phenotype most likely accompanies apoptosis, and only apoptosis-inducing oxidants result in rRNA decay, whereas cell death caused by others probably occurs via a different mechanism. Another question is why each apoptotic stimuli resulted in slightly different set of cleavage products. Those which were mapped for treatment with H2O2 and in aging cells illustrate that cleavages often occur in loops and bulges in closely located regions within the 25S rRNA, or rather within the accessible RNA elements in the compact RNP structures. The difference in cleavage patterns could be due to stress-induced subtle or severe alterations in ribosome particles that change the local accessibility of the rRNA components presented for cleavage. Alternatively, if rRNA decay is indeed carried out by more than one nuclease, these differences may reflect varying enzyme specificities in each death-inducing condition. It is also possible that somehow different set of nucleases is activated or recruited to rRNA substrates during apoptosis triggered by oxidative stress, acetic acid treatment and ageing.
Mitochondrial respiration protects rRNA against deleterious consequences of ROS
Certain aspects of apoptosis, such as the change in mitochondrial membrane potential, fragmentation of mitochondria and the requirement of cyt c and AIF release to the cytoplasm, are strongly conserved among different organisms and point to the pivotal role of mitochondria. In yeast, PCD pathways triggered by acetic acid, Bax expression and pheromone, are strictly correlated with these events and do not proceed in cells devoid of mtDNA (rho0) (31,60). During chronological ageing and hyperosmotic shock, rho0 strains were reported to have somehow higher survival, which can be attributed to their long doubling time (1.5 to 2-fold), but they still die apoptotically (23,73). However, mitochondrial function is required for resistance to oxidative stress by way of detoxification or repair of the oxidative damage and, consequently, rho0 cells are more sensitive to several oxidants, have higher level of endogenous ROS and undergo apoptosis caused by H2O2 or amino-acid starvation (61,71,74–77). Also, mammalian rho0 cell lines undergo apoptosis in response to some but not all cell death-activating stimuli. It has been postulated that these differences may arise from a distinct mechanism by which rho0 cells maintain membrane potential by way of ATP consumption (78).
Our results show that, in all conditions tested, rRNA degradation is tightly connected with mitochondrial function and the active process of respiration. Rho0 cells that are less resistant to oxidative stress suffer severe rRNA degradation in the presence of H2O2, acetic acid and 60% glucose and during chronological ageing. Also, impediment of oxidative phosphorylation by proton-pump inhibitors or by mutations in ATP/ADP carriers gives a similar outcome. In contrast, rRNA is markedly more stable when mitochondrial respiration is enhanced (e.g. by growth on glycerol versus glucose). This is consistent with the protective role of active mitochondria against the damaging effects of ROS on cellular components, rRNA destruction included. To begin with, several anti-oxidant enzymes localize to mitochondria and are more abundant in respiring cells. In addition, it has been proposed that some anti-oxidant activities may require energy (75).
As some apoptotic pathways depend on the release of mitochondrial factors to the cytoplasm and are not induced in rho0 cells, this poses an important question regarding the link between rRNA degradation and apoptosis.
rRNA degradation—apoptotic or not?
Although several apoptotic mediators (Yca1, Aif1, Nma111, Bir1 and Ndi1) have been identified in yeast, their networking in regulation of apoptosis is not yet fully understood. They may interact and function in a similar fashion as their mammalian counterparts, however, in contrast to mitochondrial mammalian proteins, Nma111 and Bir1 are located in the nucleus, so in yeast there might be some deviations from the mammalian model (13,14,71). Nevertheless, apoptotic cell-death pathways in yeast induced by H2O2, acetic acid, hyperosmotic shock, ageing and increased mRNA stability were reported to require metacaspase Yca1, nominating them as caspase-dependent pathways (13,23,31). Still, deletion of Yca1 in the mRNA turnover mutant lsm1Δ does not attenuate mRNA decay, placing Yca1 action downstream of the signal rising from mRNA level (23). In contrast, histone phosphorylation at Ser10, which requires prior deacetylation at Lys11, has been shown to mediate H2O2-induced apoptosis independently of Yca1 (20,21). Also, the activity of the major mitochondrial nuclease, Nuc1, in the cell-death pathway does not require either apoptotic mediators, Yca1 or Aif1, but is affected by H2B modifications (18). Moreover, two PCD pathways, namely induced by defects in protein N-glycosylation and triggered by ammonia in multicellular yeast colonies, do not rely on Yca1 but on as yet unknown caspase-like activity (79,80).
Degradation of the 25S/5.8S rRNAs is observed in all conditions inducing apoptosis, however, this process is not dependent on Yca1 and Aif1. On the other hand, mutations in histone H2B that inhibit phosphorylation at Ser10 and block the progress of H2O2-induced apoptosis also severely affect rRNA degradation. More importantly, rRNA degradation coincides with apoptotic DNA fragmentation; from several compounds that lead to oxidative stress and cell death, only those that provoked DNA destruction also triggered rRNA decay. Furthermore, at least some apoptotic pathways strictly require the involvement of mitochondria and do not occur in yeast lacking mtDNA, whereas rRNA degradation in all stress conditions tested is more powerful in rho0 cells. To sum up: rRNA decay induced by apoptotic stimuli occurs during cell death pathway that involves ROS generation, mitochondrial activity, fragmentation of chromosomes and histone modification but not apoptotic regulators, Yca1 and Aif1. This could be due to the existence of numerous different but partly overlapping PCD mechanisms, whether caspase- and mitochondria-dependent or independent. Ever increasing numbers of such pathways has been described in the literature in recent years. However, we favour a different model, where all these elements function in concert at different steps of the whole scenario. Stress stimuli induce signals, possibly via ROS, affecting different levels of gene expression, namely chromatin modifications, transcription and translation, which as a result activate defence response. At this level, mitochondrial respiration helps to protect cellular components via adaptive mechanisms with anti-oxidant functions. Even so, when the attack is not successfully pacified, generated ROS molecules initiate the destruction of cellular machineries, targeting in the first place crucial elements such as protein synthesis (i.e. ribosomes). Now, the progression of the response comes to the crossroads—if the conditions are appropriate, the cascade of events leading to apoptosis is triggered (e.g. release of CytC and other mitochondrial factors to the cytoplasm) resulting in cell death with characteristic apoptotic markers. Alternatively, when apoptotic prerequisites are not met, cells do not enter this pathway. The more fit cells escape death, whereas others die anyway, maybe less rapidly and through a different pathway. In this scenario, certain events, including generation of ROS, histone modifications and possibly also RNA fragmentation, occur upstream of subsequent steps, such as activation of caspases and other apoptotic regulators with resulting apoptotic phenotypes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank M. Nomura (University of California, Irvine) for strain NOY504 and plasmid pNOY102; H. Raué and J.C. Vos (University of Vrije, Amsterdam) for plasmid pJV12; J. Kolarov (Comenius University, Bratislava) for plasmids pRS-Bcl-xL and YEp51-Bax and strains aac1/2/3-Δ and op1; Y. Inoue (Kyoto University) for strains YPH250, grx1/2-Δ and gpx1/2/3-Δ; E.B. Gralla (University of California, Los Angeles) for strains EG103 and sod1/2-Δ; C.D. Allis (The Rockefeller University, New York) for strains JHY205, SAY2 and SAY186; R. Wysocki (University of Wroclaw) for plasmid YEp351-YAP1. This work was funded by The Wellcome Trust (067504/Z/02/Z). Funding to pay the Open Access publication charges for this article was provided by The Wellcome Trust.
Conflict of interest statement. None declared.
REFERENCES
- 1.Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thorpe GW, Fong CS, Alic N, Higgins VJ, Dawes IW. Cells have distinct mechanisms to maintain protection against different reactive oxygen species: oxidative-stress-response genes. Proc. Natl Acad. Sci. USA. 2004;101:6564–6569. doi: 10.1073/pnas.0305888101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Samejima K, Earnshaw WC. Trashing the genome: the role of nucleases during apoptosis. Nat. Rev. Mol. Cell Biol. 2005;6:677–688. doi: 10.1038/nrm1715. [DOI] [PubMed] [Google Scholar]
- 4.Degen WG, Pruijn GJ, Raats JM, van Venrooij WJ. Caspase-dependent cleavage of nucleic acids. Cell Death Differ. 2000;7:616–627. doi: 10.1038/sj.cdd.4400672. [DOI] [PubMed] [Google Scholar]
- 5.Kerr IM, Brown RE. pppA2′p5′A2′p5′A: an inhibitor of protein synthesis synthesized with an enzyme fraction from interferon-treated cells. Proc. Natl Acad. Sci. USA. 1978;75:256–260. doi: 10.1073/pnas.75.1.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diaz-Guerra M, Rivas C, Esteban M. Activation of the IFN-inducible enzyme RNase L causes apoptosis of animal cells. Virology. 1997;236:354–363. doi: 10.1006/viro.1997.8719. [DOI] [PubMed] [Google Scholar]
- 7.Castelli JC, Hassel BA, Maran A, Paranjape J, Hewitt JA, Li XL, Hsu YT, Silverman RH, Youle RJ. The role of 2′-5′ oligoadenylate-activated ribonuclease L in apoptosis. Cell Death Differ. 1998;5:313–320. doi: 10.1038/sj.cdd.4400352. [DOI] [PubMed] [Google Scholar]
- 8.Banerjee S, An S, Zhou A, Silverman RH, Makino S. RNase L-independent specific 28S rRNA cleavage in murine coronavirus-infected cells. J. Virol. 2000;74:8793–8802. doi: 10.1128/jvi.74.19.8793-8802.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Skulachev VP. Programmed death in yeast as adaptation? FEBS Lett. 2002;528:23–26. doi: 10.1016/s0014-5793(02)03319-7. [DOI] [PubMed] [Google Scholar]
- 10.Madeo F, Herker E, Wissing S, Jungwirth H, Eisenberg T, Frohlich KU. Apoptosis in yeast. Curr. Opin. Microbiol. 2004;7:655–660. doi: 10.1016/j.mib.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 11.Gourlay CW, Du W, Ayscough KR. Apoptosis in yeast - mechanisms and benefits to a unicellular organism. Mol. Microbiol. 2006;62:1515–1521. doi: 10.1111/j.1365-2958.2006.05486.x. [DOI] [PubMed] [Google Scholar]
- 12.Büttner S, Eisenberg T, Herker E, Carmona-Gutierrez D, Kroemer G, Madeo F. Why yeast cells can undergo apoptosis: death in times of peace, love, and war. J. Cell Biol. 2006;175:521–525. doi: 10.1083/jcb.200608098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Madeo F, Herker E, Maldener C, Wissing S, Lachelt S, Herlan M, Fehr M, Lauber K, Sigrist SJ, et al. A caspase-related protease regulates apoptosis in yeast. Mol. Cell. 2002;9:911–917. doi: 10.1016/s1097-2765(02)00501-4. [DOI] [PubMed] [Google Scholar]
- 14.Fahrenkrog B, Sauder U, Aebi U. The S. cerevisiae HtrA-like protein Nma111p is a nuclear serine protease that mediates yeast apoptosis. J. Cell Sci. 2004;117:115–126. doi: 10.1242/jcs.00848. [DOI] [PubMed] [Google Scholar]
- 15.Wissing S, Ludovico P, Herker E, Buttner S, Engelhardt SM, Decker T, Link A, Proksch A, Rodrigues F, et al. An AIF orthologue regulates apoptosis in yeast. J. Cell Biol. 2004;166:969–974. doi: 10.1083/jcb.200404138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li W, Sun L, Liang Q, Wang J, Mo W, Zhou B. Yeast AMID homologue Ndi1p displays respiration-restricted apoptotic activity and is involved in chronological aging. Mol. Biol. Cell. 2006;17:1802–1811. doi: 10.1091/mbc.E05-04-0333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walter D, Wissing S, Madeo F, Fahrenkrog B. The inhibitor-of-apoptosis protein Bir1p protects against apoptosis in S. cerevisiae and is a substrate for the yeast homologue of Omi/HtrA2. J. Cell Sci. 2006;119:1843–1851. doi: 10.1242/jcs.02902. [DOI] [PubMed] [Google Scholar]
- 18.Büttner S, Eisenberg T, Carmona-Gutierrez D, Ruli D, Knauer H, Ruckenstuhl C, Sigrist C, Wissing S, Kollroser M, et al. Endonuclease G regulates budding yeast life and death. Mol. Cell. 2007;25:233–246. doi: 10.1016/j.molcel.2006.12.021. [DOI] [PubMed] [Google Scholar]
- 19.Cheung WL, Ajiro K, Samejima K, Kloc M, Cheung P, Mizzen CA, Beeser A, Etkin LD, Chernoff J, et al. Apoptotic phosphorylation of histone H2B is mediated by mammalian sterile twenty kinase. Cell. 2003;113:507–517. doi: 10.1016/s0092-8674(03)00355-6. [DOI] [PubMed] [Google Scholar]
- 20.Ahn SH, Cheung WL, Hsu JY, Diaz RL, Smith MM, Allis CD. Sterile 20 kinase phosphorylates histone H2B at serine 10 during hydrogen peroxide-induced apoptosis in S. cerevisiae. Cell. 2005;120:25–36. doi: 10.1016/j.cell.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 21.Ahn SH, Diaz RL, Grunstein M, Allis CD. Histone H2B deacetylation at lysine 11 is required for yeast apoptosis induced by phosphorylation of H2B at serine 10. Mol. Cell. 2006;24:211–220. doi: 10.1016/j.molcel.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 22.Mazzoni C, Mancini P, Verdone L, Madeo F, Serafini A, Herker E, Falcone C. A truncated form of KlLsm4p and the absence of factors involved in mRNA decapping trigger apoptosis in yeast. Mol. Biol. Cell. 2003;14:721–729. doi: 10.1091/mbc.E02-05-0258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mazzoni C, Herker E, Palermo V, Jungwirth H, Eisenberg T, Madeo F, Falcone C. Yeast caspase 1 links messenger RNA stability to apoptosis in yeast. EMBO Rep. 2005;6:1076–1081. doi: 10.1038/sj.embor.7400514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Temple MD, Perrone GG, Dawes IW. Complex cellular responses to reactive oxygen species. Trends Cell Biol. 2005;15:319–326. doi: 10.1016/j.tcb.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 25.Jamieson DJ. Oxidative stress responses of the yeast Saccharomyces cerevisiae. Yeast. 1998;14:1511–1527. doi: 10.1002/(SICI)1097-0061(199812)14:16<1511::AID-YEA356>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 26.Moye-Rowley WS. Transcription factors regulating the response to oxidative stress in yeast. Antioxid. Redox Signal. 2002;4:123–140. doi: 10.1089/152308602753625915. [DOI] [PubMed] [Google Scholar]
- 27.Higgins VJ, Alic N, Thorpe GW, Breitenbach M, Larsson V, Dawes IW. Phenotypic analysis of gene deletant strains for sensitivity to oxidative stress. Yeast. 2002;19:203–214. doi: 10.1002/yea.811. [DOI] [PubMed] [Google Scholar]
- 28.Gietz D, St Jean A, Woods RA, Schiestl RH. Improved method for high efficient transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ludovico P, Sousa MJ, Silva MT, Leao C, Corte-Real M. Saccharomyces cerevisiae commits to a programmed cell death process in response to acetic acid. Microbiology. 2001;147:2409–2415. doi: 10.1099/00221287-147-9-2409. [DOI] [PubMed] [Google Scholar]
- 30.Ribeiro GF, Corte-Real M, Johansson B. Characterization of DNA damage in yeast apoptosis induced by hydrogen peroxide, acetic acid, and hyperosmotic shock. Mol. Biol. Cell. 2006;17:4584–4591. doi: 10.1091/mbc.E06-05-0475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Silva RD, Sotoca R, Johansson B, Ludovico P, Sansonetty F, Silva MT, Peinado JM, Corte-Real M. Hyperosmotic stress induces metacaspase- and mitochondria-dependent apoptosis in Saccharomyces cerevisiae. Mol. Microbiol. 2005;58:824–834. doi: 10.1111/j.1365-2958.2005.04868.x. [DOI] [PubMed] [Google Scholar]
- 32.Herker E, Jungwirth H, Lehmann KA, Maldener C, Frohlich KU, Wissing S, Buttner S, Fehr M, Sigrist S, et al. Chronological aging leads to apoptosis in yeast. J. Cell Biol. 2004;164:501–507. doi: 10.1083/jcb.200310014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tollervey D, Mattaj IW. Fungal small nuclear ribonucleoproteins share properties with plant and vertebrate U-snRNPs. EMBO J. 1987;6:469–476. doi: 10.1002/j.1460-2075.1987.tb04777.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beltrame M, Tollervey D. Identification and functional analysis of two U3 binding sites on yeast pre-ribosomal RNA. EMBO J. 1992;11:1531–1542. doi: 10.1002/j.1460-2075.1992.tb05198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kufel J, Bousquet-Antonelli C, Beggs JD, Tollervey D. Nuclear pre-mRNA decapping and 5′ degradation in yeast require the Lsm2-8p complex. Mol. Cell Biol. 2004;24:9646–9657. doi: 10.1128/MCB.24.21.9646-9657.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Madeo F, Frohlich E, Ligr M, Grey M, Sigrist SJ, Wolf DH, Frohlich KU. Oxygen stress: a regulator of apoptosis in yeast. J. Cell Biol. 1999;145:757–767. doi: 10.1083/jcb.145.4.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Inoue Y, Matsuda T, Sugiyama K, Izawa S, Kimura A. Genetic analysis of glutathione peroxidase in oxidative stress response of Saccharomyces cerevisiae. J. Biol. Chem. 1999;274:27002–27009. doi: 10.1074/jbc.274.38.27002. [DOI] [PubMed] [Google Scholar]
- 38.Houge G, Robaye B, Eikhom TS, Golstein J, Mellgren G, Gjertsen BT, Lanotte M, Doskeland SO. Fine mapping of 28S rRNA sites specifically cleaved in cells undergoing apoptosis. Mol. Cell Biol. 1995;15:2051–2062. doi: 10.1128/mcb.15.4.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.King KL, Jewell CM, Bortner CD, Cidlowski JA. 28S ribosome degradation in lymphoid cell apoptosis: evidence for caspase and Bcl-2-dependent and -independent pathways. Cell Death Differ. 2000;7:994–1001. doi: 10.1038/sj.cdd.4400731. [DOI] [PubMed] [Google Scholar]
- 40.Madeo F, Frohlich E, Frohlich KU. A yeast mutant showing diagnostic markers of early and late apoptosis. J. Cell Biol. 1997;139:729–734. doi: 10.1083/jcb.139.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Allmang C, Mitchell P, Petfalski E, Tollervey D. Degradation of ribosomal RNA precursors by the exosome. Nucleic Acids Res. 2000;28:1684–1691. doi: 10.1093/nar/28.8.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cannone JJ, Subramanian S, Schnare MN, Collett JR, D'Souza LM, Du Y, Feng B, Lin N, Madabusi LV, et al. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002;3:2. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Houseley J, LaCava J, Tollervey D. RNA-quality control by the exosome. Nat. Rev. Mol. Cell Biol. 2006;7:529–539. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- 44.Padh H. Cellular functions of ascorbic acid. Biochem. Cell Biol. 1990;68:1166–1173. doi: 10.1139/o90-173. [DOI] [PubMed] [Google Scholar]
- 45.Manon S, Chaudhuri B, Guerin M. Release of cytochrome c and decrease of cytochrome c oxidase in Bax-expressing yeast cells, and prevention of these effects by coexpression of Bcl-xL. FEBS Lett. 1997;415:29–32. doi: 10.1016/s0014-5793(97)01087-9. [DOI] [PubMed] [Google Scholar]
- 46.Tao W, Kurschner C, Morgan JI. Modulation of cell death in yeast by the Bcl-2 family of proteins. J. Biol. Chem. 1997;272:15547–15552. doi: 10.1074/jbc.272.24.15547. [DOI] [PubMed] [Google Scholar]
- 47.Ligr M, Madeo F, Frohlich E, Hilt W, Frohlich KU, Wolf DH. Mammalian Bax triggers apoptotic changes in yeast. FEBS Lett. 1998;438:61–65. doi: 10.1016/s0014-5793(98)01227-7. [DOI] [PubMed] [Google Scholar]
- 48.Wysocki R, Kron SJ. Yeast cell death during DNA damage arrest is independent of caspase or reactive oxygen species. J. Cell Biol. 2004;166:311–316. doi: 10.1083/jcb.200405016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee J, Godon C, Lagniel G, Spector D, Garin J, Labarre J, Toledano MB. Yap1 and Skn7 control two specialized oxidative stress response regulons in yeast. J. Biol. Chem. 1999;274:16040–10046. doi: 10.1074/jbc.274.23.16040. [DOI] [PubMed] [Google Scholar]
- 50.Cohen BA, Pilpel Y, Mitra RD, Church GM. Discrimination between paralogs using microarray analysis: application to the Yap1p and Yap2p transcriptional networks. Mol. Biol. Cell. 2002;13:1608–1614. doi: 10.1091/mbc.01-10-0472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ikner A, Shiozaki K. Yeast signaling pathways in the oxidative stress response. Mutat. Res. 2005;569:13–27. doi: 10.1016/j.mrfmmm.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 52.Winyard PG, Moody CJ, Jacob C. Oxidative activation of antioxidant defence. Trends Biochem. Sci. 2005;30:453–461. doi: 10.1016/j.tibs.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 53.Brendel M, Grey M, Maris AF, Hietkamp J, Fesus Z, Pich CT, Dafre AL, Schmidt M, Eckardt-Schupp F, Henriques JA. Low glutathione pools in the original pso3 mutant of Saccharomyces cerevisiae are responsible for its pleiotropic sensitivity phenotype. Curr. Genet. 1998;33:4–9. doi: 10.1007/s002940050301. [DOI] [PubMed] [Google Scholar]
- 54.Nogi Y, Yano R, Nomura M. Synthesis of large rRNAs by RNA polymerase II in mutants defective in RNA polymerase I. Proc. Natl Acad. Sci. USA. 1991;88:3962–3966. doi: 10.1073/pnas.88.9.3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Venema J, Dirks-Mulder A, Faber AW, Raué HA. Development and application of an in vivo system to study yeast ribosomal RNA biogenesis and function. Yeast. 1995;10:145–156. doi: 10.1002/yea.320110206. [DOI] [PubMed] [Google Scholar]
- 56.Nogi Y, Yano R, Dodd J, Carles C, Nomura M. Gene RRN4 in Saccharomyces cerevisiae encodes the A12.2 subunit of RNA polymerase I and is essential only at high temperatures. Mol. Cell Biol. 1993;13:114–122. doi: 10.1128/mcb.13.1.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fleury C, Mignotte B, Vayssiere JL. Mitochondrial reactive oxygen species in cell death signaling. Biochimie. 2002;84:131–141. doi: 10.1016/s0300-9084(02)01369-x. [DOI] [PubMed] [Google Scholar]
- 58.Balaban RS, Nemoto S, Finkel T. Mitochondria, oxidants, and aging. Cell. 2005;120:483–495. doi: 10.1016/j.cell.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 59.Danial NN, Korsmeyer SJ. Cell death: critical control points. Cell. 2004;116:205–219. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- 60.Ludovico P, Rodrigues F, Almeida A, Silva MT, Barrientos A, Corte-Real M. Cytochrome c release and mitochondria involvement in programmed cell death induced by acetic acid in Saccharomyces cerevisiae. Mol. Biol. Cell. 2002;13:2598–2606. doi: 10.1091/mbc.E01-12-0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Trancikova A, Weisova P, Kissova I, Zeman I, Kolarov J. Production of reactive oxygen species and loss of viability in yeast mitochondrial mutants: protective effect of Bcl-xL. FEMS Yeast Res. 2004;5:149–156. doi: 10.1016/j.femsyr.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 62.Slater EC. Application of inhibitors and uncouplers for a study of oxidative phosphorylation. Methods Enzymol. 1967;10:48–57. [Google Scholar]
- 63.Weber J, Senior AE. Effects of the inhibitors azide, dicyclohexylcarbodiimide, and aurovertin on nucleotide binding to the three F1-ATPase catalytic sites measured using specific tryptophan probes. J. Biol. Chem. 1998;273:33210–33215. doi: 10.1074/jbc.273.50.33210. [DOI] [PubMed] [Google Scholar]
- 64.Matsuyama S, Xu Q, Velours J, Reed JC. The mitochondrial F0F1-ATPase proton pump is required for function of the proapoptotic protein Bax in yeast and mammalian cells. Mol. Cell. 1998;1:327–336. doi: 10.1016/s1097-2765(00)80033-7. [DOI] [PubMed] [Google Scholar]
- 65.Gancedo JM. Yeast carbon catabolite repression. Microbiol. Mol. Biol. Rev. 1998;62:334–361. doi: 10.1128/mmbr.62.2.334-361.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Holcik M, Sonenberg N. Translational control in stress and apoptosis. Nat. Rev. Mol. Cell Biol. 2005;6:318–327. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- 67.Iwawaki T, Hosoda A, Okuda T, Kamigori Y, Nomura-Furuwatari C, Kimata Y, Tsuru A, Kohno K. Translational control by the ER transmembrane kinase/ribonuclease IRE1 under ER stress. Nat. Cell Biol. 2001;3:158–164. doi: 10.1038/35055065. [DOI] [PubMed] [Google Scholar]
- 68.Narayanan S, Surendranath K, Bora N, Surolia A, Karande AA. Ribosome inactivating proteins and apoptosis. FEBS Lett. 2005;579:1324–1331. doi: 10.1016/j.febslet.2005.01.038. [DOI] [PubMed] [Google Scholar]
- 69.Biggiogera M, Bottone MG, Scovassi AI, Soldani C, Vecchio L, Pellicciari C. Rearrangement of nuclear ribonucleoprotein (RNP)-containing structures during apoptosis and transcriptional arrest. Biol. Cell. 2004;96:603–615. doi: 10.1016/j.biolcel.2004.04.013. [DOI] [PubMed] [Google Scholar]
- 70.Kufel J, Allmang C, Petfalski E, Beggs J, Tollervey D. Lsm proteins are required for normal processing and stability of ribosomal RNAs. J. Biol. Chem. 2003;278:2147–2156. doi: 10.1074/jbc.M208856200. [DOI] [PubMed] [Google Scholar]
- 71.Ludovico P, Madeo F, Silva M. Yeast programmed cell death: an intricate puzzle. IUBMB Life. 2005;57:129–135. doi: 10.1080/15216540500090553. [DOI] [PubMed] [Google Scholar]
- 72.Sidrauski C, Walter P. The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell. 1997;90:1031–1039. doi: 10.1016/s0092-8674(00)80369-4. [DOI] [PubMed] [Google Scholar]
- 73.Pozniakovsky AI, Knorre DA, Markova OV, Hyman AA, Skulachev VP, Severin FF. Role of mitochondria in the pheromone- and amiodarone-induced programmed death of yeast. J. Cell Biol. 2005;168:257–269. doi: 10.1083/jcb.200408145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jamieson DJ. Saccharomyces cerevisiae has distinct adaptive responses to both hydrogen peroxide and menadione. J. Bacteriol. 1992;174:6678–6681. doi: 10.1128/jb.174.20.6678-6681.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Grant CM, MacIver FH, Dawes IW. Mitochondrial function is required for resistance to oxidative stress in the yeast Saccharomyces cerevisiae. FEBS Lett. 1997;410:219–222. doi: 10.1016/s0014-5793(97)00592-9. [DOI] [PubMed] [Google Scholar]
- 76.Heeren G, Jarolim S, Laun P, Rinnerthaler M, Stolze K, Perrone GG, Kohlwein SD, Nohl H, Dawes IW, et al. The role of respiration, reactive oxygen species and oxidative stress in mother cell-specific ageing of yeast strains defective in the RAS signalling pathway. FEMS Yeast Res. 2004;5:157–167. doi: 10.1016/j.femsyr.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 77.Eisler H, Frohlich KU, Heidenreich E. Starvation for an essential amino acid induces apoptosis and oxidative stress in yeast. Exp. Cell Res. 2004;300:345–353. doi: 10.1016/j.yexcr.2004.07.025. [DOI] [PubMed] [Google Scholar]
- 78.Chandel NS, Schumacker PT. Cells depleted of mitochondrial DNA (rho0) yield insight into physiological mechanisms. FEBS Lett. 1999;454:173–176. doi: 10.1016/s0014-5793(99)00783-8. [DOI] [PubMed] [Google Scholar]
- 79.Vachova L, Palkova Z. Physiological regulation of yeast cell death in multicellular colonies is triggered by ammonia. J. Cell Biol. 2005;169:711–717. doi: 10.1083/jcb.200410064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hauptmann P, Riel C, Kunz-Schughart LA, Frohlich KU, Madeo F, Lehle L. Defects in N-glycosylation induce apoptosis in yeast. Mol. Microbiol. 2006;59:765–778. doi: 10.1111/j.1365-2958.2005.04981.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.