Abstract
DNA polymerase μ is a member of the mammalian pol X family and reduces deletion during chromosome break repair by nonhomologous end joining (NHEJ). This biological role is linked to pol μ's ability to promote NHEJ of ends with noncomplementary 3′ overhangs, but questions remain regarding how it performs this role. We show here that synthesis by pol μ in this context is often rapid and, despite the absence of primer/template base-pairing, instructed by template. However, pol μ is both much less active and more prone to possible template independence in some contexts, including ends with overhangs longer than two nucleotides. Reduced activity on longer overhangs implies pol μ is less able to synthesize across longer gaps, arguing pol μ must bridge both sides of gaps between noncomplementary ends to be effective in NHEJ. Consistent with this argument, a pol μ mutant defective specifically on gapped substrates is also less active during NHEJ of noncomplementary ends both in vitro and in cells. Taken together, pol μ activity during NHEJ of noncomplementary ends can thus be primarily linked to pol μ's ability to work together with core NHEJ factors to bridge DNA ends and perform a template-dependent gap fill-in reaction.
INTRODUCTION
Nonhomologous end joining (NHEJ) is a key pathway for repair of chromosome breaks in mammals. A number of factors are required for efficient rejoining of broken chromosome ends regardless of the structure of the broken end (core factors). These include a ligase complex of XRCC4 and DNA ligase IV (XL), a DNA end binding heterodimer, Ku70 and Ku80 (Ku), the DNA-dependent protein kinase (DNA-PKcs) and the recently described XL accessory factor XLF/Cernunnos. With rare exception, deficiency in any of these core factors results in immunodeficiency, general sensitivity to double-strand break inducing agents and premature cellular senescence (1,2).
When the structure of a broken end is not immediately amenable for efficient ligation, NHEJ also employs a variety of factors to process the ends, including DNA polymerases. Terminal deoxynucleotidyl transferase (TdT), DNA polymerase μ (pol μ) and pol λ are members of the mammalian pol X family of DNA polymerases and all three polymerases form a specific complex with the NHEJ core factors Ku and XL (3–7), implicating them as end-processing proteins in NHEJ (8).
The contribution of TdT to NHEJ is clear. It is expressed only in lymphocytes active in assembly of antigen-specific receptors by V(D)J recombination. It promotes diversity in these receptors through its ability to extend 3′ overhanging ends in a template independent manner during repair of intermediates by NHEJ. On the other hand, the contributions of pol μ and pol λ to NHEJ are less clear (8). These polymerases are distinguishable from TdT in that they are both more widely expressed (9,10) and are both capable of template-dependent synthesis in vitro (9,10). However, pol μ deficient mice, but not pol λ deficient mice, have fewer B cells due to inaccurate NHEJ of light-chain recombination intermediates (11). This cannot be attributed to insufficient levels of pol λ as this polymerase is expressed at roughly equivalent levels after birth (including those cells active in IgL chain recombination) (12). Moreover, even when overexpressed, pol λ is unable to impact the accuracy of Igκ recombination to the same degree as pol μ (5). Maintenance of accurate NHEJ during repair of Igκ recombination intermediates can thus be linked to some unique activity of pol μ.
We previously showed this unique biological role can be linked to pol μ's ability to promote NHEJ when aligned ends possess no complementary sequence: a mutant of pol μ specifically defective in this function in vitro (pol μ Δloop) is unable to improve the accuracy of Igκ recombination (5). We further argued that pol μ was active in this context because it has the unprecedented ability to prime synthesis from a 3′ overhang from one DNA end and use a 3′ noncomplementary overhang from a second end as template. Such an activity would define pol μ as clearly distinct both from the canonical DNA polymerases (including pol λ and pol β) that require some degree of pairing between the primer and template, but also TdT, an obligately template-independent polymerase. We therefore suggested pol μ occupies an intermediate position in a ‘gradient’ of template dependence in pol X activities. This argument is further supported by analysis of the relative contributions of these different polymerases to NHEJ in a yeast model (13).
Alternate models explaining pol μ's ability to promote NHEJ of noncomplementary ends have also been proposed (14,15). Like TdT, pol μ is capable of template-independent extension of 3′ overhangs. Even a single such template independent addition could subsequently promote NHEJ when it is fortuitously complementary to sequence in the second end, in part due to ligase IV's surprising ability to sustain end joining activity in the presence of mismatches and gaps in aligned ends (7,14,16,17).
We therefore address here how pol μ synthesis activity varies with differences in end sequence and structure during NHEJ in vitro. We show pol μ activity within NHEJ is efficient only when adding nucleotides that are complementary to a template end (‘template dependent’), consistent with pol μ's higher intrinsic template-dependent activity (10). However, systematic analysis of substrates also reveals exceptions where pol μ activity in NHEJ is both much lower and more prone to noncomplementary additions, including overhangs in excess of 2 nt. Study of a targeted mutation in pol μ indicates this length dependency is a function of pol μ's need to interact with both sides of a short gap, in order for it to efficiently participate in NHEJ. Taken together, our results indicate pol μ's ability to act during NHEJ can be closely correlated with its ability to interact productively with a template end through end bridging interactions.
MATERIALS AND METHODS
Expression constructs and purified proteins
Human recombinant Ku, XL and polymerases were expressed and purified as previously described (4,5,18,19). The R175A (amino acid numbering as in full-length native pol μ) mutation was introduced independently in a human pol μ bacterial expression construct as well as a mouse pol μ retroviral construct by the Quickchange (Stratagene, Cedar Creek, TX, USA) protocol, and the open reading frames for these constructs verified by sequencing as correct.
In vitro assays
Substrates for end-joining and synthesis assays were generated by amplification of a 275 bp mouse genomic fragment including the Jκ1 coding region. Varied end structures were generated by using primers with different restriction enzymes sites appended 5′ of a common sequence complementary to the upstream end of the 275 bp κ-locus fragment (GTGGACGTTCGGTGGAGGC; referred to as ‘U’ below) and 5′ of a sequence complementary to the downstream end of this fragment (GGCTACCCTGCTTCTTTGAGC; referred to as ‘D’ below). We list in order (i) the structure of both fragment ends generated after digestion, (ii) the restriction enzyme used to generate this structure and (iii) the sequences appended to the common 3′ tails needed to generate the enzyme site: Blunt, Pvu II, CTGCCGCAGCTGTC-U and CGGACCAGCTG-D; 3′T, Ahd I, CTGCCGGACATTCAGTC-U and CGGCAGACCGTCAGTC-D; 3′TT, Bts CI, CTGCCGAACATCC-U and CGGCAGAACATCC-D; 3′TTT, Bgl I, TCATGGCCTAAACGGCT-U and CGGCAGCCTAAAT-D; 3′TTTT, Bst XI, CGGCACCATAAAACT-U and TCATGCCATAAAACTGGA-D; 3′GT, Bts CI, CTGCCGCACATCCG-U and CGGCAGCACATCC-D; 3′GTT, Bgl I, TCATGGCCTCAACGGCT-U and CGGCAGCCTCAAT-D; 3′GTTT, Bst XI, TCATGCCATCAAACTGGA-U and CGGCACCATCAAACT; 3′GG, Bts CI, CTGCCGCCCATCC-U and CGGCAGCCCATCC-D; 3′AA, Bts CI, CTGCCGTTCATCC-U and CGGCAGTTCATCC-D; 3′CC, Bts CI, CTGCCGGGCATCC–U and CGGCAGGGCATCC-D; 3′A, Ahd I, CTGCCGGACATACAGTC-U and CGGCAGACCGACAGTC-D; 3′C, Ahd I, CTGCCGGACATCCAGTC-U and CGGCAGACCGCCAGTC-D; 3′G, Ahd I, CTGCCGGACATGCAGTC-U and CGGCAGACCGGCAGTC-D.
PCR products of these primer pairs were introduced into the TOPO-TA 2.1 vector (Invitrogen, Carlsbad, CA, USA), and sequenced to verify the accuracy of the insert. Plasmid DNA for each insert was then amplified with the 5′ biotinylated vector-specific primers DAR470 (AGTGTGCTGGAATTCGCCCTT) and DAR471 (GTGATGGATATCTGCAGAATTCGCCCT) in the presence of [α-32P]dCTP. After digestion with the appropriate enzyme, the fragment was purified by depletion of uncut or partially extended products with magnetic streptavidin beads (Roche, Basel, Switzerland), as well as a Qiaquick PCR purification step (Qiagen, Valencia, CA, USA) with a 35% guanadine hydrochloride wash included.
End-joining assays were performed by preincubating 25 nM Ku, 50 nM XL and 25 nM polymerase with 5 nM DNA substrate for 10 min in our standard reaction buffer [25 nM Tris (pH 7.5), 1 mM DTT, 150 mM KCl, 40 μg/ml bovine serum albumin (BSA), 4% glycerol, 0.1 mM EDTA and 10% (wt/vol) polyethylene glycol (MW >8000 kDa) (PEG)]. Ligation was initiated by addition of dNTPs or individual ddNTPs to 25 μM (or 25 μM each), MgCl2 to 5 mM and 200 ng supercoiled plasmid DNA (Litmus38; New England Biolabs, Ipswich, MA, USA). Reactions were incubated at 37°C for the noted times, stopped and deproteinized. Relative to conditions used in prior work (5), use of ∼5-fold higher concentrations of protein and substrate increases the rate of joining significantly (Supplementary Figure 1; new conditions in lane 1, and conditions similar to prior work in lanes 2 and 3). Reactions performed with dNTPs (GE Biosciences, Piscataway, NJ, USA) were analyzed by electrophoresis on a 5% native polyacrylamide gel (PAGE). Reactions performed with ddNTPs (Roche) were first digested with HinfI prior to analysis on a denaturing 8% PAGE. All analysis of ddNTP reactions focuses on the substrate end fragment that is sufficiently small (55 nt) to resolve single nucleotide additions to the substrate band. Head-to-tail junctions of in vitro reactions were sequenced for Table 1 by cloning products of 25 cycles of amplification of 1/1 × 103 of a reaction with the primers 5′GCTGGGAATAGGCTAGACATG and 5′GCCACAGACATAGACAACGG.
Oligonucleotide substrates for pol μ assays (Figure 4 and Table 2) were generated by annealing the primer Cy3-5′GCTTGAAGACTGGTGAAGACTTGAG (SNM34) to the template 5′CCATGAATCGACCTGTACCTCAAGTCTTCACCAGTCTTCA (SNM36), generating PE-1. A single nucleotide gapped substrate (‘Gap’) was made by adding to annealing reactions the downstream strand 5′TACAGGTCGATTCATGGAGT (SNM35). This downstream strand was typically 5′ phosphorylated by T4 polynucleotide kinase (New England Biolabs) to make the ‘Gap P+’ substrate, but this kinase step was omitted to make the ‘Gap P−‘substrate also referred to in Table 2. A substrate to measure extension from a 3′ overhang (TdT-like primer extension; PE-2) was made by annealing Cy3-5′GTAGGGCTCATGTTAGATCTATCGAGCAAGTGCATCTGCAGTACTCATATGGAATTCCCAGCTGAG (DAR167) to 5′CAGCTGGGAATTCCATATGAGTACTGCAGATGCACTTGCTCGATAGATCT AACATGAGCC (DAR166).
Figure 4.
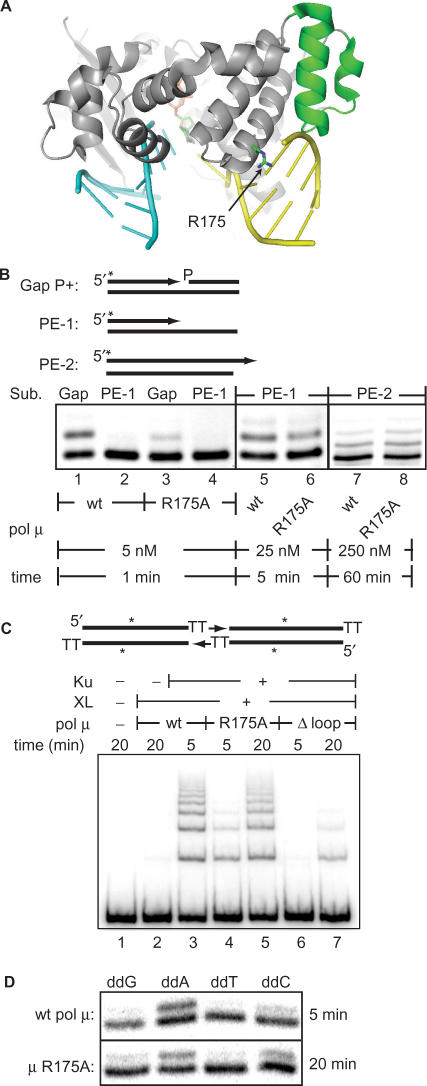
Activity of pol μ R175A in vitro. (A) An arrow locates R175 (in stick representation) in a structure of pol μ bound to a gapped DNA substrate with an incoming ddTTP (also in stick representation) (PDB code, 2IHM) (20). The helix–hairpin–helix motif is in green. Upstream DNA duplex, including the primer, is in cyan, while downstream DNA duplex (‘template end’) is in yellow. Two nucleotides opposite the primer terminus (T6 and T7 in 2IHM) were omitted. (B) Gap and primer extension substrates (PE-1, template dependent; PE-2, template-independent) are shown, with 5′ end labels noted by an asterisk. Assays were done with 5 nM substrate and indicated amounts of wt pol μ or pol μ R175A and incubated for the lengths of time noted. Ku and XL were not included. (C) Reactions performed as in Figure 1B contained Ku, XL and wt pol μ, pol μ R175A or pol μ Δloop as indicated. (D) Reactions contained Ku, XL, wt pol μ or pol μ R175A and 25 μM of the indicated ddNTP as in Figure 1C for 5 or 20 min.
Gap filling and primer extension reactions were incubated at 37°C with 25 mM Tris pH 7.5, 0.1 mM EDTA, 1.1 mM DTT, 25 mM KCl, 125 mM NaCl, 1% glycerol, 10 μg/ml BSA and 5 mM MgCl2 in the presence of 0.1 mM each dNTP and 5 nM DNA substrate. The reactions were then stopped by addition of formamide loading dye and analyzed by denaturing 10% PAGE. Extension from a 3′ overhang was analyzed by incubation at 37°C in a similar reaction buffer, except using 31.25 mM KCl, 118.75 mM NaCl and 10% PEG. Reactions were stopped by addition of formamide loading dye and analyzed by denaturing 8% PAGE.
Analysis of Igκ recombination in pol μ R175A overexpressing cells
Pol μ R175A overexpressing cells were made by infection of SP-9 with a pBABE-puro retroviral construct containing the pol μ R175A mutant cDNA as previously described for SP-9 cells overexpressing wild-type pol μ and pol μ Δloop (5). Western analysis was performed on whole-cell extracts using an anti-myc antibody (#2272; Cell Signaling, Beverly, MA, USA) and an anti-actin antibody (A2066; Sigma, St Louis, MO, USA) followed by visualization with an anti-rabbit-horseradish peroxidase secondary antibody and ECL+ chemiluminescence (GE Biosciences). VJκ1 recombination junctions were generated and analyzed as previously described (5).
RESULTS
Pol μ typically adds complementary nucleotides more efficiently than noncomplementary nucleotides
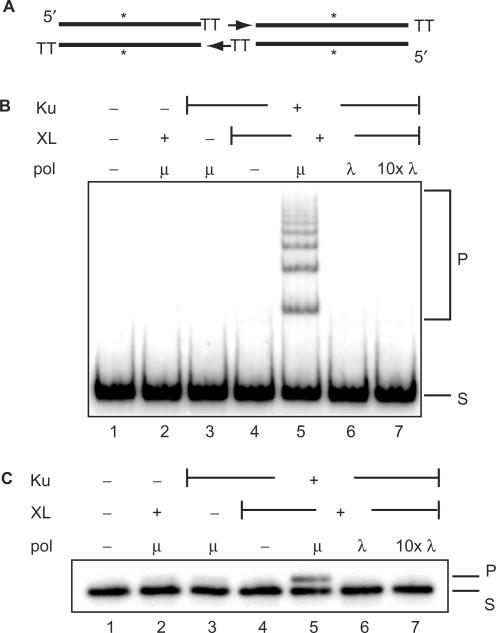
We determined how well two pol X family polymerases are able to help NHEJ join noncomplementary ends by using an in vitro assay. A 32P labeled linear DNA fragment (∼300 bp) with 3′TT overhangs (Figure 1A) was incubated with Ku, XL, polymerases and dNTPs. This fragment's overhangs are not complementary, thus levels of ligation in the presence of the purified NHEJ core factors Ku and XL alone are negligible (Figure 1B, lane 4). As previously described (5), the further addition of pol μ to these reactions allowed for efficient generation of concatamer ligation products (Figure 1B, lane 5). Pol λ, another pol X member that also interacts with Ku and XL, was unable to substitute for pol μ even when in 10-fold excess of pol μ (Figure 1B, lanes 6 and 7).
Figure 1.
Factors required for joining of noncomplementary ends. (A) A diagram of the standard 280 bp substrate labeled internally with 32P (asterisk), and possessing 3′ TT overhangs. Arrows indicate direction of synthesis by pol μ after alignment of ends by core NHEJ factors Ku and XL. (B) All reactions used 5 nM DNA substrate as illustrated in (A), and products analyzed after 5 min reactions. 25 nM Ku and 50 nM XL were added as indicated (+). Polymerase μ or λ was added at 25 nM or 250 nM (10×). S, substrate; P, concatamer ligation products. (C) Reactions performed as in B except ddNTPs substituted for dNTPs, and synthesis at one end analyzed by denaturing PAGE as described in methods. S, substrate; P, +1 synthesis product.
XLF and DNA-PKcs have also been implicated as NHEJ core factors in cells. Under the conditions used here, further addition of purified XLF and DNA-PKcs to our in vitro reactions does not stimulate joining. However, we note that addition of XLF (but not DNA-PKcs) allows for significant joining of noncomplementary ends independent of the addition of pol μ, consistent with recent reports (16,17). Nevertheless, this pol μ-independent, XLF-dependent joining of this substrate was still much less (∼6-fold) than that observed in the presence of pol μ (our unpublished data). Moreover, neither XLF nor DNA-PKcs allowed pol λ to perform synthesis-dependent joining. Ku and pol μ are thus both necessary and sufficient for efficient ligation of ends by XL under these conditions.
We next addressed if pol μ's synthesis activity is dependent on both XL and Ku. To separate synthesis from ligation, we repeated experiments as described earlier, but substituted ddNTPs for standard dNTPs. This assessed synthesis directly, rather than assessing only synthesis that gave rise to an end structure that permitted ligation. Interestingly, synthesis by pol μ was dependent on the presence of both Ku and XL (Figure 1C, compare lanes 2 and 3 to lane 5).
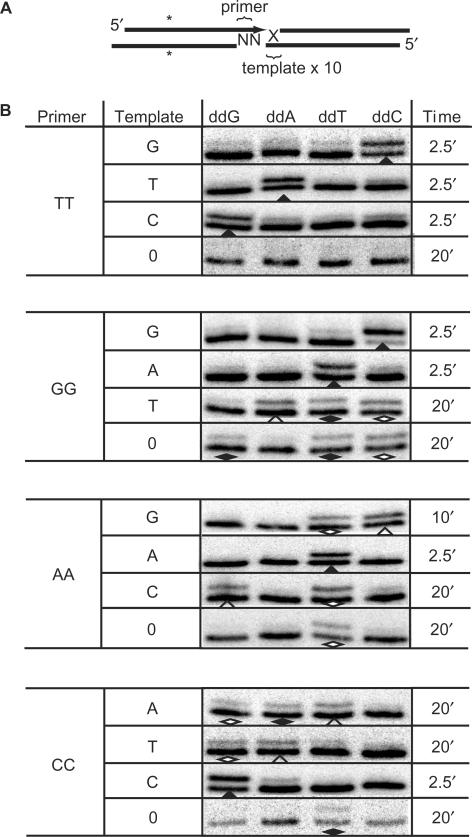
How does pol μ promote NHEJ in this context? We previously proposed pol μ within a bridged end complex is uniquely able to synthesize from a primer on one DNA end, but use as template the overhang sequence of a second DNA end (5). However, only a limited number of substrate contexts have been explored, leaving open several alternate explanations. We therefore devised the following assay to definitively resolve this issue (Figure 2A) (20). As shown previously (Figure 1C; compare lanes 3 and 5), XL is essential for significant pol μ synthesis activity under our conditions, thus we continued to use ddNTP substrates to permit analysis of synthesis products in the presence of XL independently of whether synthesis can contribute to a joined product. Additionally, synthesis could conceivably only appear template-dependent under prior assay conditions if primer sequence affects which nucleotides can be added by pol μ. The ability to vary template end sequence independent of the primer end was achieved by mixing a small amount of radiolabeled (hot) fragment with an excess of un-labeled (cold) DNA fragment. Ends of the hot fragment will thus most frequently interact with ends of the cold fragment, allowing us to define the hot ends as the primer, and the overhangs of the cold ends as the primary template. The relative efficiency of synthesis for each of the four ddNTPs from different priming end sequences can then be evaluated, while independently varying the sequence of the template end. Importantly, this setup has limitations when the intended template is used inefficiently, as is apparent below.
Figure 2.
Effect of overhang sequence on pol μ activity in NHEJ. (A) Diagram of the 3′ overhang substrates, emphasizing how primer ends are distinguished from template ends by radiolabeling of the former, and inclusion of a 10-fold excess of the latter. (B) Column 1 lists the different primer overhang sequences and Column 2 the different template overhang sequences. The ‘0’ in column 2 identifies reactions where a blunt-ended fragment was substituted for the 1 nt 3′ overhanging template ends used elsewhere. Synthesis with each mixture of primer and template end was assayed in the presence of pol μ, Ku and XL as in Figure 1C, except each of the four ddNTPs were included individually at 25 μM. Reaction times were varied as noted in the final column to allow for significant accumulation of product. Filled triangles mark combinations of primer, template and ddNTP that generated significant product within 2.5 min. For combinations of primer, template and dNTP that required 10–20 min for significant product, open triangles identify additions complementary to the intended template, open diamonds identify additions complementary to the primer, while filled diamonds identify additions that are definitively template independent (not-complementary to either primer or intended template).
We assessed activity using the previously described 3′TT overhang substrate as a primer, but varied the identity of potential template ends. For all three potential noncomplementary template ends (G, T and C), high activity (30–50% of substrate is converted to n + 1 product) is observed only when the ddNTP that is complementary to the template end sequence is supplied (Figure 2B, top panel, first three rows). In contrast, levels of activity with noncomplementary ddNTPs are typically over 10-fold lower. Results were similar when standard dNTPs were used in one experiment (template T; our unpublished data), indicating inefficient use of noncomplementary ddNTPs is not a function of the missing 3′ hydroxyl. Interestingly, when using dATP (complementary to template), the unligated products of extension from an end were detectable, but were less abundant (approximately one-third) than the levels of the synthesis-dependent ligation products involving the same end. Ligation is thus well-coupled to synthesis. We next addressed what the consequences would be of the absence of template under the same conditions. We substituted a blunt ended fragment (template ‘0’) for the standard 3′ overhang containing ‘template-end’ fragments and failed to detect activity with any of the ddNTPs (Figure 2B, top panel, bottom row), even after extended incubation (20 min). Pol μ activity using the TT overhanging primer is thus primarily both template-directed and template dependent: template directed because it is most efficient using the ddNTP complementary to the sequence of a template end overhang, and template dependent because activity is much lower when pol μ-containing NHEJ complexes interact with ends that lack a potential template (blunt ends).
We then repeated this analysis for the remaining nine different mismatching primer and template-end combinations. For four of these combinations (7/12 total), pol μ remained both efficient and accurate: over 20% of the primer is extended within 2.5 min when using a ddNTP complementary to template sequence (reactions noted by filled triangles in Figure 2B), while activity using noncomplementary ddNTPs was typically not detectable in the same time frame. We did not detect significant addition for any of the 4 ddNTPs after 2.5 min using GG, AA and CC primers when a blunt-ended fragment was substituted for template (our unpublished data). Thus, as with the TT primer, we conclude pol μ is most active in the presence of a potential template (template dependent).
However, pol μ was not equally active for the remaining five combinations. For these combinations, activity was barely detectable over the same time frame regardless of the identity of the ddNTP (unpublished observations), and much longer reactions (10–20 min) were needed to observe significant levels of activity. In these extended incubations, addition of at least one of the presumed noncomplementary ddNTPs (diamonds in Figure 2B) can also approach that seen with the complementary ddNTP (open triangles in Figure 2B). Significant activity can also be seen in the absence of added template (template ‘0’). It is important to note that when nucleotides added are not complementary to the intended template (or are observed using template ‘0’), they are often complementary to the primer (open diamonds in Figure 2B), consistent with use of a second primer molecule as template. This is probably the appropriate explanation for examples where ddC is added, since activity using this nucleotide is significant only in the presence of the GG primer or when G is the intended template. However, there remain several examples (most often additions of ddT) where the nucleotide added is not complementary to sequence in either the intended template or the primer overhang (filled diamonds in Figure 2B). These represent definitive template-independent activity.
Thus, while high levels of pol μ activity are template dependent and template instructed, specific combinations of primer/template mismatches lead to much lower template-instructed activity. Possible explanations for pol μ's reduced activity in these circumstances, as well as their potential impact on cellular NHEJ, will be addressed in the Discussion section.
End-bridging interactions help pol μ fill short gaps between two ends
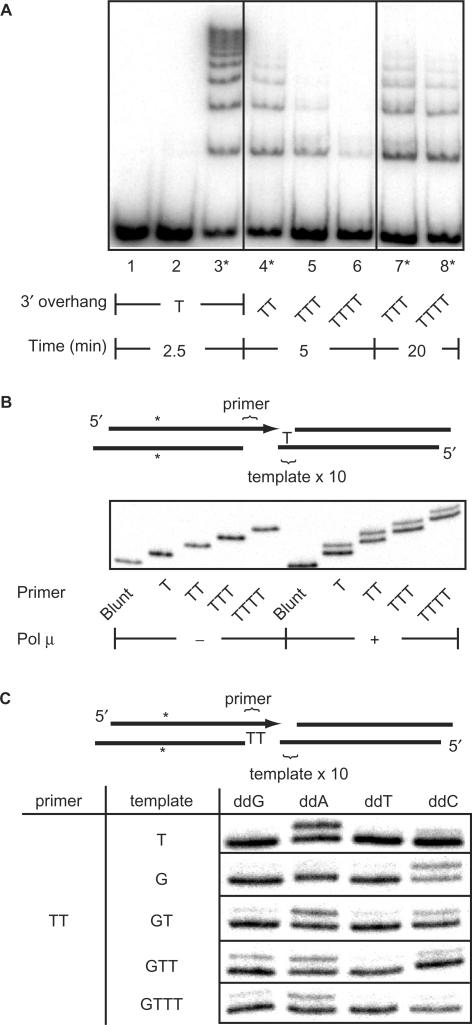
Pol μ activity in NHEJ is thus affected by the sequence of the overhangs. We next determined if there were limitations on the length of the overhang. Comparison of joining activity on substrates with 1, 2, 3 and 4 nt 3′ overhangs indicates pol μ activity declines sharply as overhang length increases (Figure 3A, lanes 3–6) (Table 1). For example, a single nucleotide overhang substrate was efficiently joined within 2.5 min, while a 4 nt overhang substrate required 20 min for comparable levels of joining (Figure 3A; compare lanes 3, 6 and 8). Additionally, sequences of the junctions from these reactions indicate that on the longer overhangs (3–4 nt), one of the overhangs is entirely present, but 1–2 nt of the second overhang were typically lost (Table 1). This is best explained if pol μ is unable to efficiently fill in gaps longer than 2 nt.
Figure 3.
Effect of overhang length on pol μ activity in NHEJ. (A) Substrates with the noted 3′ overhang sequences were incubated with dNTPs, Ku, XL and pol μ as in Figure 1B, except control lanes 1 (no protein) and 2 (no pol μ), for the indicated time periods. Asterisks identify reactions where the products were amplified, sequenced and reported in Table 1. (B) A diagram of the substrates used in this panel. Primer molecules of varying length, as noted, were incubated as in Figure 2B, with Ku, XL, pol μ, 3′ T overhang template and 25 μM each ddNTP. (C) A diagram of the substrates used in this panel. Assays were carried out as in Figure 2B with a 3′ overhang TT primer, varying the template overhang sequence and ddNTP added as noted. Images of reactions with T and G templates are reproduced from Figure 2B to aid in comparison.
Table 1.
Sequences of junctions from reactions noted with asterisks in Figure 3A
| Substratea | Activityb (%) | Synthesis in junctionsc | |
|---|---|---|---|
| T | 100 | A (17) | Accurate fill-in |
| TT | 16 | AA (13) | Accurate fill-in |
| C (1) | Mis-insertion | ||
| TTT | 5 | AA (8) | 1 nt deletion |
| AAA (4) | Accurate fill-in | ||
| GTAAA (1) | N-addition+fill-in | ||
| TTTT | 1 | AA (9) | 2 nt deletion |
| AAA (4) | 1 nt deletion |
a300 bp substrates with the noted 3’ overhangs. bJoining activity, relative to the most active substrate (single T overhang; see Figure 3A). cNucleotides synthesized by pol μ in junctions. In parentheses, the number of examples of each junction.
We further explored this question by assaying synthesis activity, while independently varying the length of the primer and template overhangs (Figure 3B and C), using the experimental setup described earlier (Figures 1C and 2B). Using a template 3′ overhang of fixed length (1 nt), we determined that while pol μ is relatively inactive using a blunt-end primer, 3′ overhang primers from 1 to 4 nt in length can all be used with only a slight reduction in pol μ activity with increasing overhang length (Figure 3B). We next addressed the impact of different length 3′ overhang templates, using a 2 nt 3′ overhanging primer. At the same time, we addressed two possible explanations why an increased length of template overhang might generate junctions with deletions (Table 1 and Figure 3C). Pol μ might initiate synthesis correctly using an overhang terminus as template, but because the overhangs were homopolymeric in these experiments, the primer terminus might have slipped and reannealed with an internal site in the template overhang. Alternatively, pol μ might be unable to use the terminal nucleotide effectively as a template on the longer (3–4 nt) overhangs: in this case, pol μ presumably initiates synthesis using an internal nucleotide as template instead. To distinguish these two possibilities, we used a series of substrates where the identity of the terminal template nucleotide (G) was different from internal template nucleotide(s) (T). As single nucleotide overhangs, both template nucleotides can be used efficiently with the 3′TT primer (Figure 3C, top two rows; reproduced from Figure 2B for comparison purposes). In contrast, on longer overhangs pol μ is progressively less able to add the terminal nucleotide (ddC), while activity with ddA is less affected (Figure 3C, lower three rows). The loss of overhang nucleotides in the junctions described in Table 1 can thus be best explained if pol μ cannot efficiently use the terminal nucleotide as a template when the template overhang is over 2 nt long. Instead, pol μ uses internally located template nucleotides, presumably because these nucleotides are closer to where the template end is double stranded.
Pol μ activity thus appears limited by the spacing between the 3′ terminus of the primer end and the 5′ terminus of the strand ‘downstream’ of synthesis activity on the template end, implying pol μ must interact with both strand termini for optimum activity. The ability to interact with both sides of a gap is a unique and distinguishing feature of the pol X family (21). Importantly, during NHEJ of noncomplementary ends, interaction with both sides of the gap will ‘bridge’ the two ends: end bridging interactions should have a key role in pol μ's activity in this context only if pol μ acts by our proposed template-dependent mechanism. In contrast, end-bridging interactions should be dispensable if pol μ promotes NHEJ of noncomplementary ends by a TdT-like, template independent, activity.
We therefore sought to further address the relative contribution of template-dependent and template-independent activity by altering pol μ such that it is less able to interact with the downstream strand. The structure of pol μ bound to a gapped DNA substrate has recently been solved (20). Like most pol X members, pol μ interacts with the strand downstream of the primer terminus in a gapped substrate through its ‘8 kD’ domain. This domain contacts the downstream strand both through a helix–hairpin–helix motif, as well as a positively charged pocket that envelops the 5′ terminus of the downstream strand. Notably, the structure highlights a residue in this positively charged pocket (R175, Figure 4A) that could make a hydrogen bond with the 5′ phosphate of the downstream strand, and thus might be expected to play an important role in gap bridging interactions (20). Consistent with this structure's predictions, mutation of R175 to alanine results in a specific defect in gap-filling. R175A is 4-fold less active than wild-type pol μ in primer extension in the presence of a 5′ phosphorylated downstream strand (Figure 4B, gapped substrate, compare lanes 1 and 3), but possesses activity more similar to wild-type pol μ in primer extension when the downstream strand is missing (Figure 4B, compare lanes 5 and 6; Table 2) or even when only the downstream strand's 5′ phosphate is missing (Table 2).
Table 2.
Polymerase activity on oligonucleotide substrates
| Substratea |
Polymerase activityb |
|
|---|---|---|
| Wt | R175A | |
| Gap filling P+ (GF P+) | 1 | 0.26 |
| Gap filling P− | 0.28 | 0.17 (0.6) |
| Primer extension (PE-1) | 3.8×10−2 | 2.3 × 10−2 (0.6) |
| TdT-like (PE-2) | 2.3×10−4 | 3.3 × 10−4 (1.4) |
aGF P+, PE-1 and PE-2 refer to substrates described in Figure 4B. The single nucleotide gap substrate was also tested without a 5′ phosphate on the downstream strand (P−). PE-2 assays required the presence of 10% polyethylene glycol to recover detectable activity. bSpecific activities are expressed relative to wt pol µ on a 1 nt gap with a 5′ phosphate on the downstream strand (GF P+). The same substrate was also tested without this 5′ phophate (P−); other substrates as in Figure 4B. For R175A, we also list in parenthesis the specific activity of pol µ R175A relative to wt pol µ on the same substrate. TdT-like activity is the average activity from reactions with each dNTP tested individually.
Much greater concentrations of enzyme and longer incubations are required to see significant levels of TdT-like extension under these conditions (Figure 4B; Table 2). Importantly, R175A is as active as wild-type pol μ in this assay (Figure 4B, compare lanes 7 and 8; Table 2). The R175A mutant thus has significantly reduced activity only on gapped substrates. However, we note that while R175A's defect in gap filling is significant in comparison to the wild-type protein, R175A is still much more active (10-fold) on gapped substrates than on those where the downstream strand is absent (Figure 4B, compare lanes 3 and 4; Table 2). Pol μ is thus clearly able to interact in a functionally significant way with the downstream strand even when R175 is mutated (e.g. through the helix–hairpin–helix motif), making this mutation a partial loss of function only.
We next tested R175A's ability to promote NHEJ of noncomplementary ends. Strikingly, R175A is 4-fold less active than wild-type pol μ in this context (Figure 4C, compare lanes 3 and 4). Since this mutant has near wild-type levels of activity on substrates without a downstream strand as well as wild-type levels of TdT-like activity, we conclude gap recognition is an important component to pol μ's ability to efficiently promote NHEJ of noncomplementary ends in vitro. However, we note R175A is still much more active than pol μ Δloop (Δ369–385) (Figure 4C, compare lanes 5 and 7), a mutant previously defined as specifically defective in promoting NHEJ of noncomplementary ends (5). Moreover, synthesis by R175A remains most efficient with ddATP, the nucleotide complementary to template (Figure 4D). R175A thus possesses high residual template-dependent activity in the NHEJ assay, consistent with it retaining interactions with the downstream strand that are independent of R175 (see above).
Does mutation of R175 have a similar impact on NHEJ in cells? We previously used a pre-B-cell line to assess relative activities of variant pol μ constructs in cellular NHEJ (5). In mice, pol μ is essential for accurate resolution of intermediates in V(D)J recombination at Igκ loci (12). A pre-B-cell line transformed with a temperature sensitive variant of the Abelson murine leukemia virus (ts-abl) can be induced to undergo high levels of Igκ recombination in culture (22), but levels of deletion in Igκ recombination junctions approach that seen in pre-B cells from pol μ deficient mice. Critically, stable overexpression of wild-type pol μ, but not catalytically defective pol μ, is sufficient to correct this apparent defect (5).
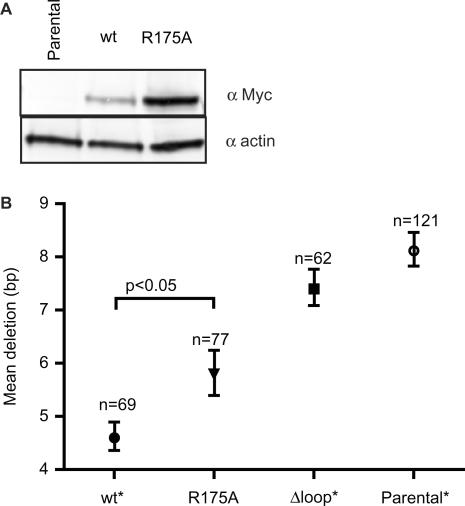
We therefore generated a variant clone of this pre-B-cell line that overexpresses R175A (Figure 5A), and compared the accuracy of Igκ recombination junctions to that seen in cells overexpressing wild-type pol μ (Figure 5B). Igκ recombination in cells overexpressing R175A is less accurate than when wild-type pol μ is overexpressed (P < 0.05; Mann–Whitney test), but more accurate than in the parental line, or in a line overexpressing pol μ Δloop. Thus, the phenotype of the R175A mutant in cellular NHEJ is consistent with in vitro NHEJ results, where R175A's activity is similarly intermediate between wild-type pol μ and the severely defective pol μ Δloop mutant (Figure 4C). Given that R175A is only partly defective in recognition of gapped substrates (Figure 4B; Table 2), we conclude pol μ's ability to bridge gaps between ends is an important component of its ability to promote NHEJ both in vitro and in cells.
Figure 5.
Activity of pol μ R175A in cells. (A) Western blot of whole-cell extracts from the parental ts-abl pre-B-cell line (SP-9), a line stably over-expressing a myc-epitope tagged wild-type pol μ (wt), and a line stably over-expressing the myc epitope tagged R175A mutant pol μ (R175A). Extracts were probed with an anti-myc antibody or, to verify similar quantities of extract loaded, an antibody to actin. (B) Average number of deletions from recombined Igκ loci in the pre-B-cell line for wt pol μ, pol μ R175A, pol μ Δloop and the parental cell line. Error bars mark the SEM, and the number of Igκ junctions sequenced for each cell line is noted. Asterisks identify data from ref. (5).
DISCUSSION
We previously argued that pol μ is active in joining noncomplementary ends because it is uniquely able to synthesize across a broken template, by taking advantage of the ability of a pol μ/Ku/XL complex to bridge two DNA ends (5). However, pol μ could also promote NHEJ of noncomplementary ends by extending 3′ overhangs in a TdT-like, template-independent manner. In this alternate model, cycles of random, TdT-like addition could promote joining as soon as one of these additions is complementary to the ‘acceptor’ end. Is pol μ template dependent or template independent during NHEJ?
We show here that pol μ, when efficient, is primarily template dependent: synthesis from one end is typically over 10-fold more active with nucleotides complementary to the sequence in a second end, relative to synthesis with noncomplementary nucleotides (Figure 2). We further identified three contexts where wild-type pol μ was relatively inactive: (i) when the primer end is aligned with a blunt end; (ii) on template overhangs >2 nt, and (iii) on specific primer/template sequence combinations. In at least the first two examples (and likely the third as well; see the following paragraph), pol μ's reduced activity correlates with a reduced ability to ‘see’ its template. In the first context (interaction with a blunt end), there is no template. In the second context (long overhangs), we argue pol μ's relative inability to effectively interact with both sides of gaps between ends that are >2 nt in length (‘end bridging’) indirectly reduces its ability to see a template. Consistent with this latter argument, a pol μ mutant designed to be specifically defective in end-bridging interactions (and which consequently retains wild-type levels of template-independent activity) has reduced activity in NHEJ both in vitro and in cells. Thus, with regard to whether pol μ is template dependent or template independent during NHEJ, we note these two mechanisms are not mutually exclusive, and both presumably make some contribution. However, our results indicate that (i) participation of pol μ in NHEJ is mostly dependent on activity within a bridged-end complex, and (ii) when pol μ is active in this complex, it adds nucleotides complementary to template significantly more efficiently than noncomplementary nucleotides (template-instructed).
Systematic analysis of substrates identified a surprisingly striking effect of specific combinations of overhang sequence on activity. Pol μ is neither efficient nor accurate for 5 of the 12 different combinations of noncomplementary primer/template end sequence: G/T, A/C, C/A, A/G and C/T. Notably, three of the four pyrimidine/purine mismatches (G/T, A/C and C/A) are represented in this set. Recent work indicates the NHEJ core factors Ku and XL are able to align and join ends with pyrimidine/purine mismatches directly, without synthesis, particularly in the presence of XLF (16). Therefore, reduced pol μ activity in these three contexts could simply reflect the frequent absence of a suitable substrate (a gap) when Ku and XL align such ends. This is also consistent with evidence that ligase IV suppresses needless processing of already ligatable ends, independent of whether ligase IV joins the end (23). Possible explanations for reduced pol μ activity on the primer/template combinations A/G and C/T are less obvious. However, it should be noted that pol μ will retain high activity when the identities of the ends acting as primer and template in these latter two combinations are switched (i.e. primer/template ends with overhanging G/A and T/C; Figure 2B). Thus, for example, while synthesis might be inefficient from an end terminating with A when using G as template, these ends can still be accurately joined on one strand by synthesis primed from the G overhang, and using the A as template. The remaining single strand gap can then be resolved independently of NHEJ.
Finally, we must emphasize that the alternate ‘template-independent’ model could also contribute significantly in these latter substrate contexts, where template-instructed activity is low. In particular, a hybrid model is suggested by the observations that the most frequent template-independent addition is T [Figure 2B, and (14)], and pol μ is most consistently active and template-instructed when using a primer with a terminal T (Figure 2B). Therefore, addition of a template-independent T may be a useful means of converting an inefficiently used substrate context to one where pol μ is now both efficient and template-instructed.
Pol μ possesses an activity intermediate in a ‘gradient’ of template dependency observed with mammalian members of the pol X family (pol β > pol λ > pol μ > TdT). Prior studies focused on elements that helped explain why both pol μ and TdT might be less dependent on template than the other pol X members (pol β and pol λ) (5,15,20). Here, we identify an element in pol μ that helps distinguish pol μ's activity in NHEJ from TdT. R175, as well as the 8 kD domain in general, promotes pol μ's activity in NHEJ through its ability to help bridge ends in the presence of Ku and XL. R175 is part of a positively charged pocket in the 8 kD domain of pol μ that grasps the 5′ phosphate of the downstream strand (20,21). This pocket has a higher density of positive charges in pol λ and pol β relative to pol μ, but even pol μ has a significantly higher density of positive charges in the same region relative to TdT (21). In particular, R175 is conserved among pol μ orthologues and is functionally analogous to conserved positively charged residues in pol β and pol λ [K35 and R275 respectively; see Figure 6 of (21)]. In contrast, a positively charged residue is not observed in this region in TdT. Thus, with respect to the gradient in template dependency observed among these four pol X members, it is interesting that the increasing strength of interactions with the terminus of the downstream strand correlates with an increase in template dependency. This has parallels to our prior correlation of progressive lengthening of a different element (‘loop1’) in pol X members and decreasing template dependence (5,15).
A recent structure of the polymerase domain of ligD from Mycobacterium tuberculosis determined it too interacts with the terminus of the strand downstream of its primer (24). This polymerase is a member of the archaeo-eukaryotic primase superfamily and, while not related to the pol X family, is also implicated in NHEJ. Moreover, as shown here for pol μ, the ability of the ligD pol domain to recognize the downstream strand was shown to be critical for its ability to act in a NHEJ-relevant context. We suggest end-bridging through gap recognition may be a feature common to NHEJ-associated polymerases, regardless of origin. Future work is needed to confirm if end bridging is also important for the function of pol λ in NHEJ (and possibly pol β as well).
We show here end-bridging interactions are an important feature to pol μ's ability to promote NHEJ of ends with noncomplementary 3′ overhangs. End-bridging interactions are intrinsic, through pol μ's ability to bind the primer terminus and the downstream strand, but probably also extrinsic, through interaction with NHEJ core factors. Synthesis activity within such a context is primarily template dependent, and consequently is sufficient to make ends at least partially complementary. This is uniquely useful for NHEJ, as it allows NHEJ to ‘match’ ends with fully noncomplementary overhangs for ligation without resorting to nucleolytic removal of flanking sequence. We suggest this has particular significance during V(D)J recombination, as diversity in terminal sequence will often generate overhanging ends with no complementary sequence. By comparison, double strand breaks arising from damage are typically staggered strand breaks (25–27), and overhangs are more likely to possess at least partially complementary sequence. More typical DNA polymerases (those that are dependent on template primer pairing; e.g. pol λ) are thus probably sufficient for NHEJ in contexts other than V(D)J recombination, and may even be ‘safer’ in these other contexts. For example, budding yeast's Pol4 promotes the aberrant fusion of chromosomes at telomeres in a RAP1 deficient background, an activity argued to be dependent on an ability to promiscuously match ends similar to that shown by pol μ (28). In general, DNA repair in mammals might thus employ pol μ as a backup to other pol X members that are less tolerant of noncomplementary sequence, and consequently less prone to inappropriate joining of ends.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank the Ramsden lab for helpful discussion and S. Roberts, Dr T Kunkel and Dr S. Nick McElhinny, and Dr M. Burkhalter for critical reading of the article. This work was supported by US Public Health Service Grant CA-97096 to D.A.R., a Leukemia and Lymphoma Society Scholar. Open Access publication charges for this article was provided by National Institute of Health research grant CA-97096 to D.A.R.
Conflict of interest statement. None declared.
REFERENCES
- 1.Mills KD, Ferguson DO, Alt FW. The role of DNA breaks in genomic instability and tumorigenesis. Immunol. Rev. 2003;194:77–95. doi: 10.1034/j.1600-065x.2003.00060.x. [DOI] [PubMed] [Google Scholar]
- 2.Sekiguchi JM, Ferguson DO. DNA double-strand break repair: a relentless hunt uncovers new prey. Cell. 2006;124:260–262. doi: 10.1016/j.cell.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 3.Fan W, Wu X. DNA polymerase lambda can elongate on DNA substrates mimicking non-homologous end joining and interact with XRCC4-ligase IV complex. Biochem. Biophys. Res. Commun. 2004;323:1328–1333. doi: 10.1016/j.bbrc.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 4.Mahajan KN, Nick McElhinny SA, Mitchell BS, Ramsden DA. Association of DNA polymerase mu (pol mu) with Ku and ligase IV: role for pol mu in end-joining double-strand break repair. Mol. Cell. Biol. 2002;22:5194–5202. doi: 10.1128/MCB.22.14.5194-5202.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nick McElhinny SA, Havener JM, Garcia-Diaz M, Juarez R, Bebenek K, Kee BL, Blanco L, Kunkel TA, Ramsden DA. A gradient of template dependence defines distinct biological roles for family X polymerases in nonhomologous end joining. Mol. Cell. 2005;19:357–366. doi: 10.1016/j.molcel.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 6.Nick McElhinny SA, Ramsden DA. Polymerase mu is a DNA-directed DNA/RNA polymerase. Mol. Cell. Biol. 2003;23:2309–2315. doi: 10.1128/MCB.23.7.2309-2315.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma Y, Lu H, Tippin B, Goodman MF, Shimazaki N, Koiwai O, Hsieh CL, Schwarz K, Lieber MR. A biochemically defined system for mammalian nonhomologous DNA end joining. Mol. Cell. 2004;16:701–713. doi: 10.1016/j.molcel.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 8.Nick McElhinny SA, Ramsden DA. Sibling rivalry: competition between Pol X family members in V(D)J recombination and general double strand break repair. Immunol. Rev. 2004;200:156–164. doi: 10.1111/j.0105-2896.2004.00160.x. [DOI] [PubMed] [Google Scholar]
- 9.Aoufouchi S, Flatter E, Dahan A, Faili A, Bertocci B, Storck S, Delbos F, Cocea L, Gupta N, Weill JC, et al. Two novel human and mouse DNA polymerases of the polX family. Nucleic Acids Res. 2000;28:3684–3693. doi: 10.1093/nar/28.18.3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dominguez O, Ruiz JF, Lain de Lera T, Garcia-Diaz M, Gonzalez MA, Kirchhoff T, Martinez AC, Bernad A, Blanco L. DNA polymerase mu (Pol mu), homologous to TdT, could act as a DNA mutator in eukaryotic cells. EMBO J. 2000;19:1731–1742. doi: 10.1093/emboj/19.7.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bertocci B, De Smet A, Berek C, Weill JC, Reynaud CA. Immunoglobulin kappa light chain gene rearrangement is impaired in mice deficient for DNA polymerase mu. Immunity. 2003;19:203–211. doi: 10.1016/s1074-7613(03)00203-6. [DOI] [PubMed] [Google Scholar]
- 12.Bertocci B, De Smet A, Weill JC, Reynaud CA. Nonoverlapping functions of DNA polymerases mu, lambda, and terminal deoxynucleotidyltransferase during immunoglobulin V(D)J recombination in vivo. Immunity. 2006;25:31–41. doi: 10.1016/j.immuni.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 13.Daley JM, Laan RL, Suresh A, Wilson TE. DNA joint dependence of pol X family polymerase action in nonhomologous end joining. J. Biol. Chem. 2005;280:29030–29037. doi: 10.1074/jbc.M505277200. [DOI] [PubMed] [Google Scholar]
- 14.Gu J, Lu H, Tippin B, Shimazaki N, Goodman MF, Lieber MR. XRCC4:DNA ligase IV can ligate incompatible DNA ends and can ligate across gaps. EMBO J. 2007;26:1010–1023. doi: 10.1038/sj.emboj.7601559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Juarez R, Ruiz JF, McElhinny SA, Ramsden D, Blanco L. A specific loop in human DNA polymerase mu allows switching between creative and DNA-instructed synthesis. Nucleic Acids Res. 2006;34:4572–4582. doi: 10.1093/nar/gkl457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gu J, Lu H, Tsai AG, Schwarz K, Lieber MR. Single-stranded DNA ligation and XLF-stimulated incompatible DNA end ligation by the XRCC4-DNA ligase IV complex: influence of terminal DNA sequence. Nucleic Acids Res. 2007;35:5755–5762. doi: 10.1093/nar/gkm579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsai CJ, Kim SA, Chu G. Cernunnos/XLF promotes the ligation of mismatched and noncohesive DNA ends. Proc. Natl Acad. Sci. USA. 2007;104:7851–7856. doi: 10.1073/pnas.0702620104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia-Diaz M, Bebenek K, Sabariegos R, Dominguez O, Rodriguez J, Kirchhoff T, Garcia-Palomero E, Picher AJ, Juarez R, Ruiz JF, et al. DNA polymerase lambda, a novel DNA repair enzyme in human cells. J. Biol. Chem. 2002;277:13184–13191. doi: 10.1074/jbc.M111601200. [DOI] [PubMed] [Google Scholar]
- 19.Nick McElhinny SA, Snowden CM, McCarville J, Ramsden DA. Ku recruits the XRCC4-ligase IV complex to DNA ends. Mol. Cell. Biol. 2000;20:2996–3003. doi: 10.1128/mcb.20.9.2996-3003.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moon AF, Garcia-Diaz M, Bebenek K, Davis BJ, Zhong X, Ramsden DA, Kunkel TA, Pedersen LC. Structural insight into the substrate specificity of DNA Polymerase mu. Nat. Struct. Mol. Biol. 2007;14:45–53. doi: 10.1038/nsmb1180. [DOI] [PubMed] [Google Scholar]
- 21.Moon AF, Garcia-Diaz M, Batra VK, Beard WA, Bebenek K, Kunkel TA, Wilson SH, Pedersen LC. The X family portrait: structural insights into biological functions of X family polymerases. DNA Repair. 2007;6:1709–1725. doi: 10.1016/j.dnarep.2007.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen YY, Wang LC, Huang MS, Rosenberg N. An active v-abl protein tyrosine kinase blocks immunoglobulin light-chain gene rearrangement. Genes Dev. 1994;8:688–697. doi: 10.1101/gad.8.6.688. [DOI] [PubMed] [Google Scholar]
- 23.Budman J, Kim SA, Chu G. Processing of DNA for nonhomologous end-joining is controlled by kinase activity and XRCC4/ligase IV. J. Biol. Chem. 2007;282:11950–11959. doi: 10.1074/jbc.M610058200. [DOI] [PubMed] [Google Scholar]
- 24.Brissett NC, Pitcher RS, Juarez R, Picher AJ, Green AJ, Dafforn TR, Fox GC, Blanco L, Doherty AJ. Structure of a NHEJ polymerase-mediated DNA synaptic complex. Science. 2007;318:456–459. doi: 10.1126/science.1145112. [DOI] [PubMed] [Google Scholar]
- 25.Gulston M, de Lara C, Jenner T, Davis E, O’Neill P. Processing of clustered DNA damage generates additional double-strand breaks in mammalian cells post-irradiation. Nucleic Acids Res. 2004;32:1602–1609. doi: 10.1093/nar/gkh306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang N, Galick H, Wallace SS. Attempted base excision repair of ionizing radiation damage in human lymphoblastoid cells produces lethal and mutagenic double strand breaks. DNA Repair. 2004;3:1323–1334. doi: 10.1016/j.dnarep.2004.04.014. [DOI] [PubMed] [Google Scholar]
- 27.Ward JF. The complexity of DNA damage: relevance to biological consequences. Int. J. Radiat. Biol. 1994;66:427–432. doi: 10.1080/09553009414551401. [DOI] [PubMed] [Google Scholar]
- 28.Pardo B, Ma E, Marcand S. Mismatch tolerance by DNA polymerase Pol4 in the course of nonhomologous end joining in Saccharomyces cerevisiae. Genetics. 2006;172:2689–2694. doi: 10.1534/genetics.105.053512. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.