Abstract
In Saccharomyces cerevisiae, multiple approaches have arrived at a consensus TATA box sequence of TATA(T/A)A(A/T)(A/G). TATA-binding protein (TBP) affinity alone does not determine TATA box function. To discover how a minimal set of factors required for basal and activated transcription contributed to the sequence requirements for a functional TATA box, we performed transcription reactions using highly purified proteins and CYC1 promoter TATA box mutants. The TATA box consensus sequence is a good predictor of promoter activity. However, several nonconsensus sequences are almost fully functional, indicating that mechanistic requirements are not the only selective pressure on the TATA box. We also found that the effect of a mutation at a certain position is often dependent on other bases within a particular TATA box. Although activators and coactivators strongly influence TBP recruitment and stability at promoters, neither Mediator, the activator Gal4-V16, nor TFIID specifically compensate for the low transcription levels of the weak TATA boxes. The addition of Mediator to purified transcription reactions did, however, increase the functional selectivity for certain consensus TATA sequences. Transcription in whole-cell extracts or in vivo with these TATA box mutants indicated that factors, other than those in our purified system, may help initiate transcription from weak TATA boxes.
INTRODUCTION
Efficient and accurate transcription initiation of eukaryotic genes requires specific sequences in the core promoter. The core promoter of a gene includes the transcription start site as well as the region immediately surrounding this site. Various functional DNA motifs, known as core promoter elements, assist in the recruitment, assembly and initiation of the RNA polymerase II (RNA Pol II) transcription machinery (1). Originally assumed to be largely invariant in strength and sequence, core promoters and their elements have been revealed to be highly variable (2). Hence, the core promoter can be a point of regulation for gene-specific transcription. Little is known, however, about what mechanisms and transcription factors are required to read and execute the information encoded by core promoter elements (1). Although core promoter elements are currently better defined in metazoans, the core promoter elements in the yeast Saccharomyces cerevisiae appear to share many of the same features (2). The TATA box is perhaps the best-known core promoter element, but the exact requirement for, and role of, TATA-like sequences in a core promoter is still not well understood.
Early studies led to the view that most mRNA synthesis requires a TATA box in order to recruit the TATA-binding protein (TBP) to a promoter. Over the past decade, however, many studies have demonstrated large classes of promoters that do not follow this rule (3). Among the most striking were studies that revealed the TATA box is not a general component of all Pol II core promoters, despite the TBP requirement for all Pol II transcription. Detailed analyses have shown that only ∼20% of yeast core promoters (4) and ∼10% of mammalian core promoters (2) have a readily identifiable TATA box. Why the TATA box is required for the transcription of some genes, but not others, and whether the mechanism of initiation differs according to the presence of a TATA box are open questions.
Biochemical, genetic and bioinformatic approaches have been taken to determine the sequence requirements for the TATA box. Initial genetic studies on the TR TATA element of the S. cerevisiae HIS3 promoter indicated very stringent sequence requirements for the TR element in vivo (5,6) and in an extract-based transcription system in vitro (7). A similar, but not identical, pattern was observed for the HIS3 TR TATA box in a reconstituted system using a partially fractionated basal transcription system (8). Although replacing single bases within the TR element led to severe impairment of HIS3 transcription in vivo (5), the fact that the replacement of the entire TR element by a variety of DNA sequences could lead to high levels of HIS3 transcription (9) suggested a greater variability in functional TATA boxes.
Initial efforts to identify functional TATA boxes and define the TATA box consensus sequence focused on single-gene analyses. A recent study (4), however, reevaluated the S. cerevisiae TATA box consensus sequence using a genome-wide bioinformatic approach. This statistical method, based on the combination of genome-wide sequence conservation and experimental data, identified the extent of TATA box usage and the most highly utilized TATA box sequences. The consensus sequence TATA(T/A)A(T/A)(A/G) determined in this study revealed a previously unidentified preference for (A/G) at the eighth position. It is currently unknown what dictates the selection of this consensus sequence. Indirect factors, such as the counter-selection of sequences bound by other specific DNA-bound proteins may influence the consensus sequence. Direct factors, on the other hand, shape the consensus based on their direct mechanistic requirements for transcription.
TBP binding to the TATA box (10–12) is a direct determinant of the consensus sequence. TBP binds to the minor groove of the DNA helix and has many contact points on the TATA box. Even though TBP affinity to TATA boxes plays a role in dictating the consensus (13), experiments have failed to show any direct correlation between TBP affinity and transcriptional activity (14,15). TATA box DNA bound by TBP exhibits a dramatic 80° bend (10,11,16). Although the structure of the TBP–DNA complex and the bending angle of the DNA provide important clues to how the sequence of the TATA box influences transcription, it is still unclear exactly how differences in transcriptional activity correspond to differences in the TBP–DNA structure (15,17–20). Although TBP affinity for TATA boxes and the DNA bending induced by TBP play an important role in dictating the consensus TATA box, TBP may not be the only factor influencing the sequence.
Activators, coactivators and general transcription factors (GTFs) other than TBP may also dictate the bases of the TATA box. Activators interact physically with both components of the basal transcription machinery (21) and with coactivator complexes (22–24). The activator fusion protein Gal4-VP16 can stabilize TBP on TATA boxes, and this effect is more pronounced on weak TATA boxes (25). It has also been shown that TATA box utilization in a partially defined basal transcription system differs from activated transcription in vivo (8). The role played by activators in TATA box utilization also may include coactivators, such as Mediator and TFIID.
Mediator enhances basal transcription, and is required for activated transcription in vivo and in vitro (26). It has physical and functional interactions with both activators and the core transcription machinery. Biochemical and structural studies have shown that Mediator is likely to interact with many components of the core RNA Pol II transcription machinery (27), including TBP (28,29). TBP recruitment to certain promoters in vivo is impaired in Mediator mutants (30–33), and the recruitment of GTFs to a preinitiation complex assembled from extracts is impaired by Mediator mutants (34). The functional interplay between TBP and Mediator suggests Mediator may influence TATA box utilization.
Another coactivator complex that is likely to be important for TATA box usage is TFIID. TFIID consists of TBP and TBP-associated factors (TAFs) and is important for the expression of, and TBP recruitment to, a subset of genes in vivo (35–37). Genome-wide studies have revealed both a physical and a functional interaction between TFIID and TATA-less promoters in vivo (4,38). Other studies have shown that mutations, which weaken the TATA box of the yeast CYC1 promoter, convert this promoter from a TFIID independent to a TFIID-dependent promoter (39).
To address the origin of the TATA box consensus sequence and determine what components of the transcription machinery influence its usage, we have functionally evaluated the yeast TATA box consensus sequence using a biochemical approach. This is the first systematic analysis of transcriptional activities of different TATA box sequences in vitro using a highly purified S. cerevisiae transcription system capable of activated transcription. In addition, we have compared the transcription levels in the purified system to levels seen in a whole-cell extract-based system, and to the transcription levels of the same TATA box variants in a reporter assay in vivo. Our studies show that the ability of the purified apparatus to initiate transcription is a strong, but not exclusive, determinant of a consensus TATA box. There are likely mechanistic subtleties to TATA box utilization that involve both factors tested in our current study as well as factors yet to be characterized.
MATERIALS AND METHODS
Transcription templates
All transcription templates that were used in this study are derivatives of the DNA plasmid pJJ470 (40). This template contains a truncated CYC1 core promoter (−139 to −36, relative to the translational start site) that retains only one of the two primary functional TATA boxes identified in studies of the native CYC1 promoter (41). Mutations in the wild-type CYC1 TATA box in our template (TATATAAA) were introduced by amplifying an XhoI–AflIII fragment from pJJ470 using PCR primer pairs consisting of the primer pUCAflR2 (5′-GCTGGCCTTTTGCTCACATG-3′) and one of the primers listed in Supplementary Table S1, depending on the desired mutation(s). The PCR products were digested with XhoI and AflIII and ligated into pJJ470 that had been digested with the same enzymes, creating pJJ470(M) plasmids. Two minor TATA-like sequences have also been identified in the CYC1 core promoter sequence (41). Mutations in these TATA-like sequences were introduced by amplifying the pJJ470(M) plasmids using the primers pJJ470F1 (5′-CGGCAGGTCCTTTGTAGCATAAATTACTA-3′) and pJJ470R1 (5′-CTACAAAGGACCTAACGTAGAAGGAAAGAATCTTTAGAGAAAAG-3′). The product was digested with PpuMI and re-ligated, creating pJJ470(MA) plasmids. All constructs were confirmed by DNA sequencing.
Protein purification and extract preparation
TFIID was purified from a yeast strain SHY761 (a generous gift from Dr Steven Hahn) in which both Taf1p and Taf7p are Flag-tagged (24). The cells were grown and lyzed in a blender as previously described (42). The extract (2 g of protein) was adjusted to 175 mM KOAc via dialysis and fractionated on a 400-ml BioRex70 column as described (43) with the following modifications: the loading buffer was 50 mM Hepes–KOH (pH 7.6), 1 mM EDTA, 10% glycerol, 175 mM KOAc, 5 mM β-mercaptoethanol and protease inhibitors. The column was washed once with the loading buffer and twice in the same buffer with 300 mM KOAc. The elution buffer was 20 mM HEPES–KOH (pH 7.6), 10% glycerol, 1 M KOAc, 0.5 mM DTT and protease inhibitors. Peak fractions were pooled and dialyzed against dialysis buffer [20 mM HEPES–KOH (pH 7.6), 0.01% NP-40, 10% glycerol, 150 mM KOAc, 0.5 mM DTT, protease inhibitors] to 300 mM KOAc. FLAG purification was performed as described (44) except that the elution buffer had 0.001% NP-40. The peak fractions from four 200 μl FLAG columns were pooled and fractionated on a Mono S 5/5 column as described (43). The TFIID fraction was concentrated by dialysis in a membrane with a molecular weight cutoff of 12–14 000 kDa against 20 mM HEPES–KOH pH 7.6, 10% glycerol, 0.001% NP-40, 150 mM KOAc, 30% (w/v) PEG 20 000, 1 mM DTT and protease inhibitors.
The transcription factors RNA Pol II, rTBP, TFIIA, rTFIIB, rTFIIE, TFIIF, TFIIH, Mediator and Gal4-VP16 were purified as previously described (45). Transcription competent whole-cell extracts were prepared as previously described (46).
G-less cassette transcription assays
Transcription activities of CYC1 TATA box mutants with purified factors were measured in a G-less cassette assay as previously described (47) with the following modifications. The final salt concentration of the reactions was 142 mM KOAc (unless otherwise noted) when measuring basal transcription, and 165 mM KOAc when measuring enhanced basal and activated transcription. The template and the protein factors were preincubated for 10 min without the nucleotides, followed by addition of nucleotides and an incubation time of 30 min for basal transcription and 60 min for enhanced basal and activated transcription. Fifty nanograms of the DNA template were used in each reaction. Reactions containing TFIID and TFIIA were run as previously described (48).
Transcription activities of TATA box mutants in the presence of a whole-cell extract were measured using the G-less cassette assay as described (46) with the following modifications. The final salt concentration in the reactions was 121.7 mM KOAc. One hundred nanograms of the template were used in each reaction. The template and the extract were preincubated for 10 min without the nucleotides, followed by an incubation time of 20 min after the addition of nucleotides.
β-Galactosidase assay
The CYC1 TATATAAA box in the lacZ reporter plasmid pLGSD5 (49) was replaced with mutated TATA boxes in the following manner: An XhoI–BamHI fragment was amplified from pJJ470(MA) plasmids with primers JJ470XhoI (5′-CGCCCTCGAGGCATGTGCTCTGTA-3′) and JJ470lacZ (5′-CCGGGGATCCGGTCATTATTAATTTAGTGTGTGTATTTGTGTTTGCGTGTCTATAGAAGTATAGTAATTTATGCTAC-3′). After digestion with XhoI and BamHI this fragment was ligated into pLGSD5 which had been cut with the same restriction enzymes. Yeast strain DY1880 (Mat a ade2 can1 leu2 trp1 ura3) (50) was transformed with a low-copy number plasmid expressing the Gal4-VP16 (F456A) fusion protein under the control of the ADH1 promoter and the pLGSD5 reporter plasmids. β-Galactosidase activities in reporter strains were measured as previously described (47).
RESULTS
Functional evaluation of the yeast consensus TATA sequence TATA(T/A)A(T/A)(A/G) using a purified basal transcription system
To functionally evaluate the TATA(T/A)A(T/A)(A/G) consensus sequence identified by Pugh and colleagues (4), we tested transcriptional activities of 23 variations (Figure 1B) of this sequence using a G-less cassette assay with the minimal set of purified factors capable of basal transcription (51,52). The G-less cassette assay template pJJ470 (40) has a Gal4-binding site upstream of a CYC1 core promoter fragment, which has the consensus TATA box TATATAAA (41). Downstream of the transcriptional start site is a G-less cassette (Figure 1A). We also introduced three other mutations in TATA-like sequences that had been identified (41) downstream of the TATA box (Figure 1A). This was done to prevent these sequences from being used as TATA boxes in the transcription reactions. These mutations, however, did not have an effect on transcription levels of either a wild-type template or a mutant template (data not shown) suggesting these sequences were not functional core promoter elements in our purified system.
Figure 1.
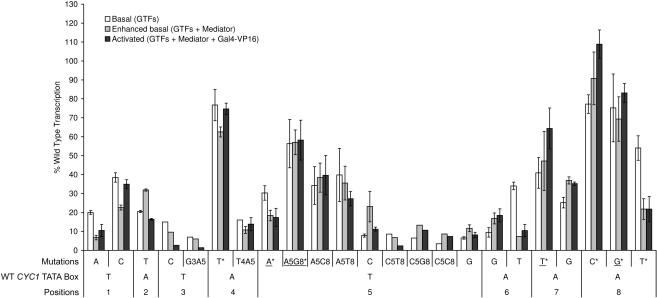
The CYC1 promoter of the transcription templates used in this work. (A) Above is shown the double-stranded sequence of the CYC1 promoter fragment fused to the G-less cassette in the original pJJ470 plasmid (40). Potential transcriptional start sites (41,65,66) located in the G-less cassette are indicated by dots. Below is the double-stranded sequence of the modified CYC1 promoter from the pJJ470(MA) plasmid. Mutations in the TATA-like sequences are shown in bold. (B) List of the wild-type and all 23 TATA box mutants that were used in this study. Whether the sequence was a consensus TATA box and/or TBP sensitive were based on a previous study (4,53). TBP-sensitive promoters show decreased transcription in yeast strains with mutations in the DNA-binding surface of TBP (53).
Basal transcription activities of the CYC1 TATA box mutants in vitro were measured in the presence of the general transcription factors TBP, TFIIB, TFIIE, TFIIF, TFIIH and RNA Pol II. In our assay, most consensus TATA boxes showed high transcription activities, whereas most nonconsensus TATA boxes showed low transcription activities (Figure 2). A second TATA-like, but nonconsensus, sequence (TATAAAAC) in the CYC1 promoter is offset from the wild-type TATA box by +2 bp. This sequence makes a small, if any, contribution to CYC1 transcription. The mutation of T to A in the first position of the wild-type sequence, which does not affect the alternate sequence, almost eliminates all transcription in our purified reactions (Figure 2). As observed in previous studies (8), the difference in transcription levels cannot be attributed only to differences in TBP affinity since the reactions were performed in the presence of excess TBP. This conclusion is also supported by our data showing that increasing the amount of TBP did not have a differential effect on the transcription of a wild-type template as compared to one of the weaker mutant TATA boxes (Supplementary Figure S1).
Figure 2.
Transcription activities of CYC1 TATA box mutants in a purified system in vitro. Mutations that represent another consensus base (4) at that position are underlined. Mutants that are sensitive to mutations in the DNA-binding surface of TBP (4) are marked with an asterisk. Transcription levels were measured in a G-less cassette assay. Basal transcription was measured in the presence of the highly purified general transcription factors RNA Pol II, TBP, TFIIB, TFIIE, TFIIF and TFIIH. Enhanced transcription was measured in the presence of purified GTFs and Mediator. Activated transcription was measured in the presence of purified GTFs, Mediator and Gal4-VP16. Shown is the quantification of transcription signals, expressed as a percentage of the signal from a wild-type TATA box template. Representative primary data are shown in Figures 3–5. The CYC1 core promoter/G-less cassette has two major start sites for transcription. We chose only the upstream start site (longer transcript) for quantification. Most columns represent the mean of at least three independent experiments. Reactions with signals that were almost undetectable were only measured twice. Error bars indicate the SEM. RNA recovery controls were run for most reactions and showed a consistent recovery over all samples checked.
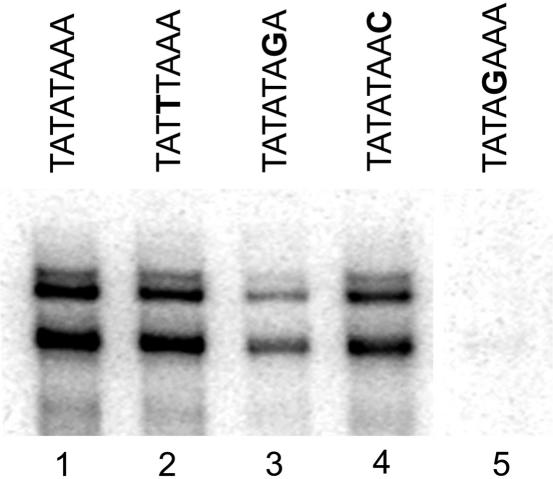
All TATA consensus and nonconsensus sequences tested had transcription signals lower than the wild-type, indicating that the TATATAAA in the CYC1 promoter is optimized for its transcriptional activity. Mutation to a nonconsensus base in the central positions (T3–A6) of the TATA box generally had the most detrimental effects on transcription (Figure 2). Very little selectivity was observed for consensus bases in the eighth position (Figure 2) in reactions reconstituted from the purified basal transcription machinery. A nonconsensus C or T at the eighth position gave a transcription signal similar to a consensus G and wild-type A. The only other nonconsensus single base mutation that gave >70% of the signal observed with the wild type was the mutation of the fourth position to a T (Figure 3). One of the criteria used to define consensus TATA boxes via the statistical approach (4) was TBP sensitivity. Promoters classified as ‘TBP sensitive’ showed decreased transcription in vivo in yeast strains with mutations in the DNA-binding surface of TBP (53). The three nonconsensus TATA boxes that exhibited high levels of transcription in our assay (TATTTAAA, TATATAAC and TATATAAT) (Figures 2 and 3), as well as the consensus sequences, all were scored as being TBP sensitive (4). Among the criteria used in the study, TBP sensitivity may be the most accurate in predicting a TATA box that will function with the basal transcription machinery. Generally, however, the yeast consensus TATA sequence [TATA(T/A)A(T/A)(A/G)] defined through bioinformatics approaches (4), is a good predictor of transcriptional activity of the CYC1 TATA box in vitro. One aspect of TATA box mechanism apparent in our studies that was not clearly apparent from the bioinformatic approach was the functional codependency of changes in different positions in the TATA box.
Figure 3.
Certain nonconsensus TATA boxes facilitate high rates of transcription. Shown are basal transcription activities of wild-type and mutant CYC1 TATA boxes in a purified system in vitro. Mutated bases are shown in bold. Lanes 2 and 4 show nonconsensus TATA boxes with high transcription signals. Lanes 3 and 5 serve as controls and show nonconsensus TATA boxes that give low transcription signals. Basal transcription was measured in a G-less cassette assay that was performed in the presence of highly purified RNA Pol II, TBP, TFIIB, TFIIE, TFIIF and TFIIH.
Codependence of bases within the CYC1 TATA box for transcription
Two results in the initial mutation analysis, where single base changes were made to the CYC1 TATA box sequence, helped reveal a functional coupling between bases in the CYC1 TATA box. In basal reactions with purified factors, the consensus TATA box TATAAAAA gave a surprisingly low transcription signal (<40% of wild type) and the nonconsensus TATA box TATTTAAA gave a surprisingly high transcription signal (>75% of wild type) (Figure 2). As described above, the nonconsensus TATA box TATTTAAA is TBP sensitive in vivo, which correlated with its ability to facilitate transcription in vitro. However, if a second change is made in this TATA box such that the T in the fifth position is changed to another consensus base (A), the resulting TATTAAAA sequence drops transcription below 20% of wild type (Figure 2). In accordance with our observations above, TATTAAAA was not classified as TBP sensitive (4). Of the two possible consensus bases in the fifth position, only T allows a TATA box with a T in the fourth position to facilitate basal transcription with purified factors. Changing the consensus T to a consensus A in the fifth position is also coupled to the eighth position.
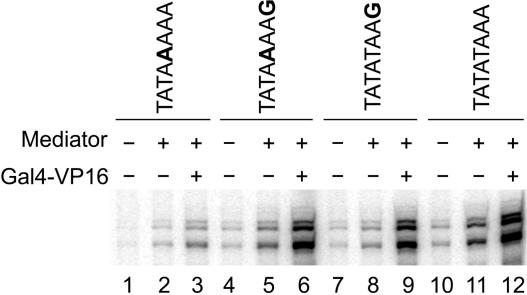
Although many yeast promoters contain and conserve the consensus TATAAAAA TATA box (4), using this sequence in our basal reactions led to low levels of relative transcription (Figures 2 and 4, lanes 1–3 versus 10–12). An A in the fifth position, however, does not exclusively lead to low levels of transcription in vitro. The Adenovirus Major Late (AdML) promoter has an A in the fifth position and has been shown to give strong transcription signals in vitro in both yeast and metazoan transcription systems (54,55). We noticed that the AdML TATA box (TATAAAAG) had a consensus G in the eighth position as opposed to the A in the eighth position of the CYC1 TATA box. We therefore decided to test if substituting the A in the eighth position of TATAAAAA for a G, creating TATAAAAG, would work better in the context of the CYC1 promoter. The double mutant gave significantly higher relative transcription levels than the single mutant (Figures 2 and 4). Thus, our results show that in the context of the CYC1 promoter, the ability to use an A in the fifth position of the TATA box relies on having a G in the eighth position. Although the substitution of a G for an A in the eighth position led to the largest suppression of the in vitro defect of the A in the fifth position, both a nonconsensus C and T in the eighth position also reproducibly led to small increases in transcription. The suppressing effect of G in the eighth position was specific for an A in the fifth position as neither a G nor any other alternative base suppressed the basal transcription defect associated with having a C in the fifth position (Figure 2).
Figure 4.
Substituting a G for the A in the eighth position of the TATA box suppresses the transcription defect in the TATA consensus sequence TATAAAAA. Shown are basal, enhanced basal and activated transcription activities of CYC1 TATA box mutants and wild type in a purified system in vitro. Lanes 10–12 show the transcription with the wild-type TATA box. Lanes 1–3 show the transcription decrease with a T5A mutation in the TATA box. Placing a G in the eighth position compensates for the transcription defect in the TATAAAAA sequence (lanes 4–6). Placing a G in the eighth position of the wild-type TATA box, however, leads to no increase in transcription (lanes 7–9). G-less cassette assays were performed in the presence of highly purified RNA Pol II, TBP, TFIIB, TFIIE, TFIIF and TFIIH, in addition to the factors indicated. Salt concentration was 165 mM KOAc and reaction time was 60 min for all reactions. Mutated bases are shown in bold.
Mediator increases the selectivity for consensus bases at the sixth and eighth position of the TATA box, but neither Mediator nor Gal4-VP16 specifically compensate for low transcription levels of weak TATA boxes
The minimal purified basal transcription system is not capable of responding to DNA-bound transcriptional activators (40). The addition of purified Mediator to the minimal basal transcription system enhances basal transcription and enables this machinery to respond to activators (40 and Figure 5, lanes 1–3). We repeated the transcription assays on the wild-type and mutant CYC1 TATA boxes with Mediator and activators to determine whether the step(s) of transcription affected by these factors functionally interacted with the TATA box sequence.
Figure 5.
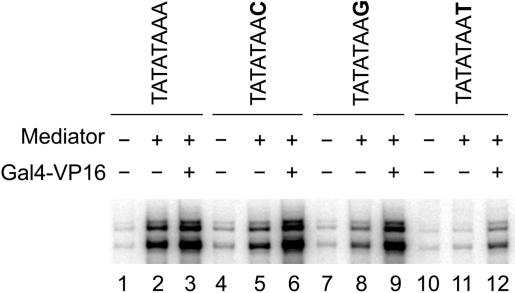
Mediator-enhanced basal transcription is disabled by substitution of a T in the eighth position of the CYC1 TATA box, but is functional with either A, C or G. Shown are basal, enhanced basal and activated transcription activities of CYC1 TATA box mutants and wild type in a purified system in vitro. Substitution of a T in the eighth position of the CYC1 TATA box specifically disables Mediator enhanced basal transcription (lanes 10–12 versus lanes 1–3), while substitution with a C (lanes 4–6) or G (lanes 7–9) has no effect. G-less cassette assays were performed in the presence of highly purified RNA Pol II, TBP, TFIIB, TFIIE, TFIIF and TFIIH, in addition to the factors indicated. Salt concentration was 165 mM KOAc and reaction time was 60 min for all reactions. Mutated bases are shown in bold.
First, we tested how Mediator-enhanced basal transcription was affected by the TATA sequence. The pattern of TATA box-dependent transcription observed using the purified general transcription factors was largely unchanged when Mediator was added to the system (Figure 2). Mediator does not specifically compensate for low transcription levels of weak TATA boxes. Mediator's stabilizing physical and/or functional interactions with TBP and, perhaps, the DNA itself do not specifically facilitate the step of transcription initiation governed by the TATA box sequence. The addition of Mediator to the reactions, however, did enhance the selectivity of the transcription apparatus for consensus bases at the sixth and eighth position. The replacement of a consensus A at either the sixth or eighth position with a nonconsensus T led to a much greater decrease in relative transcription in the presence of Mediator than in its absence (Figures 2 and 5). In both of these cases, the substitution of the T for the A prevented Mediator from enhancing basal transcription (Figures 2 and 5, lanes 1, 2 versus 10, 11). The ability of a nonconsensus base at these positions to prevent the enhancement of basal transcription by Mediator was specific to T. Neither the replacement of A with G at the sixth position nor the replacement of A with C at the eighth position had a similar effect as replacement with T (Figures 2 and 5).
Second, we looked at the effect of the hybrid activator Gal4-VP16 on TATA box utilization. We measured transcriptional activities of the CYC1 TATA box mutants in a system containing Mediator and Gal4-VP16 in addition to the factors used to measure basal transcription. In the pJJ470 vector, Gal4-VP16 binds to a Gal4 site upstream of the CYC1 core promoter and stimulates transcription in the presence of Mediator. The addition of Gal4-VP16 to the Mediator based reactions did not change the pattern observed in the absence of the activator (Figure 2). Interestingly, even though Mediator had little to no enhancement of basal transcription with the TATATAAT and TATATTAA TATA boxes, Mediator was still able to facilitate a similar fold activation of transcription as with the wild-type TATA box (Figures 2 and 5, lanes 2, 3 versus 11,12). The addition of the activator Gal4-VP16 leads to no further TATA box selectivity beyond that generated by Mediator alone.
TFIID does not specifically compensate for low transcription levels of weak TATA boxes
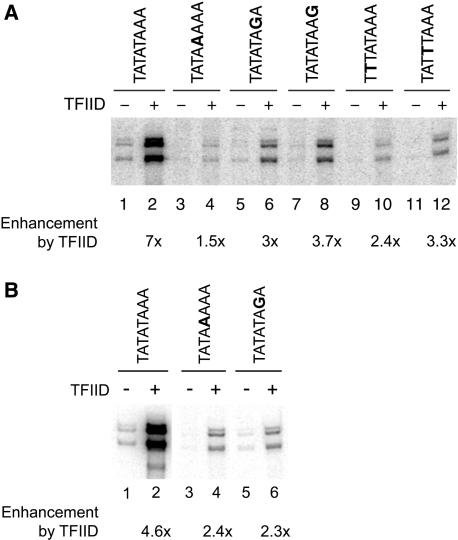
TATA-less promoters (4,38) as well as some TATA containing promoters with a weak mutated TATA box (39) have been shown to be TFIID dependent, while most promoters with strong consensus TATA box are not (4,38). Hence, we decided to test if TFIID can promote high levels of transcription from weak TATA boxes. If TFIID facilitates transcription initiation at promoters without a strong consensus TATA box in vitro, then we would expect to see a larger effect of purified TFIID on weak TATA box mutants than on the wild type or strong mutants. We chose a subset of mutants and tested their transcription activities in a purified system with or without TFIID. TFIID was purified from a strain that had a Flag tag on the Taf1p and Taf7p subunits of TFIID (24). We supplemented all TFIID transcription reactions with enough recombinant TBP to establish an ∼1:1 stoichiometry among TBP and the TAFs as well as to assure identical TBP concentrations in reactions with and without TFIID (48). In addition to TFIID and/or TBP, the transcription reactions were performed in the presence of RNA Pol II, TFIIB, TFIIE, TFIIF, TFIIH, TFIIA and Mediator as described (48). TFIID dependency was tested both in the presence and absence of Gal4-VP16. As shown previously, TFIID stimulated transcription with the consensus CYC1 TATA box (48). TFIID showed an equal fold stimulation of transcription on a variety of mutant TATA boxes of varying strength (Figure 6). These results show that purified TFIID alone does not specifically compensate for low transcription levels of weak TATA boxes.
Figure 6.
TFIID does not lead to greater enhancement of transcription from weak CYC1 TATA boxes than from strong TATA boxes in a purified system. Transcription was measured in a G-less cassette assay that was performed in the presence of highly purified RNA Pol II, TBP, TFIIB, TFIIE, TFIIF, TFIIH, TFIIA and Mediator. The reactions were performed in the absence (A) or presence (B) of the activator Gal4-VP16. Quantitation of the fold-enhancement by TFIID is shown for each TATA box. Mutated bases are shown in bold.
Components of a whole-cell extract are able to enhance transcription from weak TATA boxes
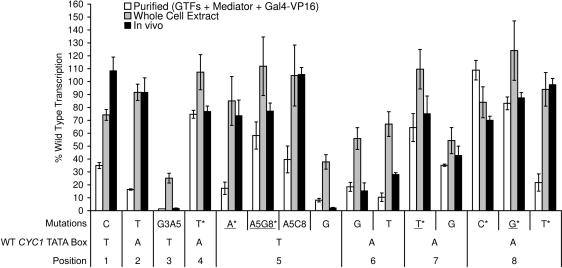
Our results have shown that neither Mediator, Gal4-VP16, nor TFIID compensate specifically for low transcription levels from weak TATA boxes in a purified system. Additional factors, independently or in association with these minimal components of the transcription machinery, may compensate specifically for low relative transcription levels from mutant TATA boxes. In order to test whether weak mutant TATA boxes had the potential to direct high levels of transcription, we performed transcription assays with a subset of our templates using a transcription competent whole-cell extract (46). The whole-cell extract was supplemented with Gal4-VP16 in the transcription reactions, while all other factors were supplied by the extract. The results from the transcription assays performed with the whole-cell extract show that transcription levels normalized to the wild-type TATA box were overall higher than in the purified system (Figure 7). Specifically, we observed an increase in the relative transcription levels of some of the weak TATA box mutants (Figure 7). The overall pattern of TATA box dependence remained similar to the purified system as the very weakest TATA boxes in the purified system (TAGAAAAA and TATAGAAA) also gave the weakest transcription in the extract system (Figure 7). The very strongest mutants in the purified system (relative transcription to wild type >70%) exhibited only a slight increase in the extract transcription system (Figure 7). Only one mutant TATA box (TATATAAG) showed a relative transcription signal slightly >100%. These results show that the wild-type TATA box is still optimal for transcription in the extract system. The ability of the extract system to utilize some nonconsensus TATA boxes more efficiently than the purified system indicates that there likely are additional factors in the whole-cell extract participating in the assembly of active preinitiation complexes on weak TATA box promoters. Also of note is that the codependence between TATA box bases in the fifth and the eighth positions (TATAAAAA versus TATAAAAG) and the discrimination between bases in the eighth position (TATATAAT versus TATATAAC or TATATAAG) observed in the complete purified system (see above) do not occur in the extract-based system (Figure 7).
Figure 7.
Transcription activities of wild-type and mutant CYC1 TATA boxes in an extract-based system in vitro, and in a reporter assay in vivo. Consensus mutants (4) are underlined. Mutants that are sensitive to mutations in the DNA-binding surface of TBP (4) are marked with an asterisk. Activated transcription levels with purified factors are from Figure 2 and are repeated here for ease of comparison. The transcription levels in an extract-based system are results from G-less cassette assays performed in the presence of a whole-cell extract supplemented with the activator Gal4-VP16. The final concentration of TBP was about 10 times lower in the extract-based system than in the purified system. RNA recovery controls were run for the purified and extract-based transcription reactions and showed a consistent recovery over all samples checked. Shown is the quantification of transcription signals, expressed as a percentage of the wild-type signal. Most columns represent the mean of at least three independent experiments. Some reactions whose signals were almost undetectable were only measured twice. Transcription levels in vivo were measured in a β-galactosidase reporter assay in a yeast strain expressing the Gal4-VP16 (F456A) fusion protein. Shown is the quantification of β-galactosidase activity units expressed as a percentage of the wild type. The columns represent the mean of three independent experiments. Error bars indicate the SEM.
Transcription activities of TATA box mutants in vivo correlate with their activities in an extract-based system
To determine whether the results of either the extract transcription system or the purified transcription system accurately reflected the discrimination of TATA boxes in vivo, we performed a reporter assay with the CYC1 promoter TATA box and mutant variants. We introduced our mutated TATA boxes into a lacZ reporter plasmid in which a GAL1, 10 upstream activation sequence (UAS) in front of the CYC1 promoter drives the expression of the reporter gene (49). This system requires an activator to obtain a detectable signal. We induced transcription by expression of Gal4-VP16(F456A). Gal4-VP16(F456A), a point mutant with slightly attenuated activation potential, is used since expression of wild-type Gal4-VP16 is toxic to yeast (49). We found that the results of the extract-based system in vitro are highly representative of signal measured using a β-galactosidase assay, with respect to the relative transcription activities of the TATA box mutants (Figure 7). The only notable difference was that the reporter assay showed a much greater decrease in relative transcription for the very weakest TATA boxes compared to the extract system (Figure 7). Generally, these results indicate that the extract-based system in vitro is a good indicator of how different TATA boxes direct transcription in vivo in the presence of chromatin.
DISCUSSION
We have performed a systematic analysis to functionally evaluate the recently derived yeast TATA box consensus sequence (4) using a highly purified transcription system capable of basal, Mediator-enhanced, TFIID-enhanced and activated transcription. We have compared these results to transcription using an identical promoter in extracts and in vivo. Considerable effort has gone into studying the intricate mechanisms and structural details of TBP binding to the TATA box (56). Comprehensive structural studies have noted the disparity that cocrystal structures of TBP with multiple mutant TATA boxes are largely the same despite 20-fold differences in transcription among these mutants in vivo (15). These studies have concluded that factors beyond the static structure of the complex must be responsible for variations in transcriptional efficiency. Ultimately the critical ‘output’ of a TATA box is the ability to facilitate accurate and efficient transcription initiation. Our studies represent the first effort to evaluate the functional properties of a yeast TATA box using a highly purified transcription system capable of activated transcription. Our study demonstrates that the disparity between structural compatibility and transcription activity extends to a minimal purified system. We have made several important observations.
In general, the functional TATA box sequences best utilized by the purified transcription machinery corresponded closely to the consensus sequence TATA(T/A)A(T/A)(A/G) derived via a genome-wide statistical approach (4). It is likely that selective pressure on TATA boxes in gene expression originates from the minimal set of components required to initiate transcription from a core promoter. These results are complimentary to those obtained for the HIS3 TR TATA box using a partially defined transcription system (8). The high levels of relative transcription for the nonconsensus sequences TATTTAAA and TATATAAC observed in our purified system as well as the genome-wide statistical study ranking these sequences as ‘TBP-sensitive’ demonstrate that these two sequences are mechanistically consensus TATA boxes. Why these two sequences did not score highly in the criteria of conservation and location in the genome-wide statistical study is unclear, although it is almost certainly the result of indirect rather than direct selection.
The inability of the consensus TATA box TATAAAAA to produce high levels of transcription in our purified system is in opposition to the results of the genome-wide statistical approach. This TATA box scored high in both the location analysis and TBP sensitivity criteria used in this study (4). One possibility is that this consensus sequence produces less efficient transcription by design and is used as part of a broad-based regulatory scheme. A second possibility is that this difference may be a result of context dependence. A TATA box that works well in one promoter context might not work well in another context. TBP binding to TATA boxes has recently been shown to be dependent on the flanking sequences (57). It is likely that promoter context, either flanking sequence or the presence of another core promoter element such as an initiator (2), may influence the ability for certain TATA box sequences to function. Our studies have already shown that other bases within the TATA box influence the effect of an A in the fifth position on transcription. The suppression of the transcription defect in the consensus TATAAAAA sequence by the replacement of an A in the eighth position by a consensus G provides initial evidence that the A5 is locally context dependent.
Despite extensive contacts between Mediator and the core transcription machinery, Mediator does not specifically compensate for low levels of weak TATA boxes. Previous studies from our lab have shown that Mediator can enhance basal transcription in purified reactions by compensating for limiting amounts of a general transcription factor, such as TBP (58). Since our data and others (8) show that increasing TBP concentration does not increase relative transcription from a mutant TATA box, it is perhaps not surprising that Mediator does not compensate for weak TATA boxes. Mediator does, however, seem to interact functionally with the sixth and the eighth position of the TATA box in the purified system. Substitution of a T in the sixth or the eighth position of the CYC1 TATA box prevents Mediator from enhancing basal transcription. Structural models of Mediator within a preinitiation complex (27) suggest it makes contacts with several components of the basal transcription apparatus. It is currently unclear how Mediator makes formation of an active preinitiation complex more efficient. The result that a T in the sixth or eighth position inhibits this action, however, suggests that Mediator may allosterically modulate interactions between general transcription factors that occur near the 3′ end of the TATA box, such as the TBP–TFIIB interaction. In contrast to these results, the fold activation by Gal4-VP16, facilitated by Mediator in the purified transcription system, does not vary with the sequence of the TATA box. It is unlikely that the step of transcription initiation stimulated by Gal4-VP16 in the purified reactions is the same step as regulated by the TATA box sequence.
TFIID, in vivo, is specifically required for the transcription of most TATA-less promoters (4,38) as well as the CYC1 promoter with a weak mutated TATA box (39). Generally, TFIID has less widespread effects on TATA-containing promoters (38). Hence, it was somewhat surprising that holo-TFIID did not specifically compensate for weak relative transcription from nonconsensus TATA boxes in the purified system. Several explanations for this result are possible. First, TFIID might require factors in addition to those in the purified system to help initiate transcription from TATA-less promoters. Second, TFIID may have some DNA sequence requirements for its action that are not present in the fragment of the CYC1 promoter fused to the G-less cassette transcription template. Lastly, it is possible that TFIID exerts its influence on TATA-less promoters in vivo through interactions with nucleosomes. TFIID is known to have interactions with acetylated (59) and methylated (60) histones. Although neither Mediator nor TFIID specifically compensated for weak TATA boxes in the CYC1 promoter, some factor in yeast extracts does.
The results from the transcription assays performed with the whole-cell extract show that transcription levels normalized to the wild-type TATA box were overall higher than in the purified system. This pattern was largely replicated using the same promoter in a reporter assay system in vivo. There was, however, a notable difference in the weak TATA boxes TAGAAAAA, TATAGAAA, TATATGAA and TATATTAA, which showed considerably lower relative signals in the reporter assays in vivo compared to the signals in the extract transcription system in vitro. The source of this difference is unknown, but one explanation could be that there is a factor in vivo which has a negative effect on initiation of certain weak TATA boxes. Perhaps this factor is not present in its active form in the extract that was used in our experiments. Given the extensive coupling between mRNA transcription and processing (61), it is also possible that weak TATA boxes could lead to less efficiently processed, and hence less stable, transcripts in vivo. Lastly, chromatin-based mechanisms of repression have been shown to suppress transcription initiation from weak promoter like elements (62,63). These processes would, of course, not be reflected in our nonchromatin transcription system in vitro.
The relative lack of sensitivity to nonconsensus TATA mutants in our extract and transcription assays in vivo is somewhat surprising given the greater sensitivity to TATA box mutations observed in some other yeast studies. Detailed analyses of the HIS3 TR TATA box in extracts (7) and in vivo (5,64) has revealed a much greater sensitivity to nonconsensus mutations in this promoter. However, this could be due to the fact that the native HIS3 TR TATA box (TATAAAGT) already has two nonconsensus bases in the seventh and eighth positions that may sensitize it to further nonconsensus mutations in the first six positions. The ability of the extract to facilitate higher levels of relative transcription than the purified system for nonconsensus TATA boxes suggests that there are additional factors that assist in transcription of weak TATA containing or TATA-less promoters. Fractionating these extracts and adding them back to our purified system is a promising approach for future attempts to identify these factors.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Dr S. Hahn for the Flag-tagged TFIID strain and Dr P.A. Weil for TFIID antibodies. We also thank Benjamin Guidi for assistance in the preparation of purified general transcription factors. Funding for this work was provided by an institutional research grant from the Hitchcock Foundation and by the NIH (GM62483). G.B. was supported in part by a Rosaline Borrison Graduate Fellowship and the Fulbright Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1.Smale ST, Kadonaga JT. The RNA polymerase II core promoter. Annu. Rev. Biochem. 2003;72:449–479. doi: 10.1146/annurev.biochem.72.121801.161520. [DOI] [PubMed] [Google Scholar]
- 2.Yang C, Bolotin E, Jiang T, Sladek FM, Martinez E. Prevalence of the initiator over the TATA box in human and yeast genes and identification of DNA motifs enriched in human TATA-less core promoters. Gene. 2007;389:52–65. doi: 10.1016/j.gene.2006.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Müller F, Demény MA, Tora L. New problems in RNA polymerase II transcription initiation: Matching the diversity of core promoters with a variety of promoter recognition factors. J. Biol. Chem. 2007;282:14685–14689. doi: 10.1074/jbc.R700012200. [DOI] [PubMed] [Google Scholar]
- 4.Basehoar AD, Zanton SJ, Pugh BF. Identification and distinct regulation of yeast TATA box-containing genes. Cell. 2004;116:699–709. doi: 10.1016/s0092-8674(04)00205-3. [DOI] [PubMed] [Google Scholar]
- 5.Chen W, Struhl K. Saturation mutagenesis of a yeast his3 “TATA element”: Genetic evidence for a specific TATA-binding protein. Proc. Natl Acad. Sci. USA. 1988;85:2691–2695. doi: 10.1073/pnas.85.8.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harbury PAB, Struhl K. Functional distinctions between yeast TATA elements. Mol. Cell Biol. 1989;9:5298–5304. doi: 10.1128/mcb.9.12.5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ponticelli AS, Struhl K. Analysis of Saccharomyces cerevisiae his3 transcription in vitro: Biochemical support for multiple mechanisms of transcription. Mol. Cell Biol. 1990;10:2832–2839. doi: 10.1128/mcb.10.6.2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wobbe CR, Struhl K. Yeast and human TATA-binding proteins have nearly identical DNA sequence requirements for transcription in vitro. Mol. Cell Biol. 1990;10:3859–3867. doi: 10.1128/mcb.10.8.3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singer VL, Wobbe CR, Struhl K. A wide variety of DNA sequences can functionally replace a yeast TATA element for transcriptional activation. Genes Dev. 1990;4:636–645. doi: 10.1101/gad.4.4.636. [DOI] [PubMed] [Google Scholar]
- 10.Kim JL, Nikolov DB, Burley SK. Co-crystal structure of TBP recognizing the minor groove of a TATA element. Nature. 1993;365:520–527. doi: 10.1038/365520a0. [DOI] [PubMed] [Google Scholar]
- 11.Kim Y, Geiger JH, Hahn S, Sigler PB. Crystal structure of a yeast TBP/TATA-box complex. Nature. 1993;365:512–520. doi: 10.1038/365512a0. [DOI] [PubMed] [Google Scholar]
- 12.Nikolov DB, Chen H, Halay ED, Usheva AA, Hisatake K, Lee DK, Roeder RG, Burley SK. Crystal structure of a TFIIB-TBP-TATA-element ternary complex. Nature. 1995;377:119–128. doi: 10.1038/377119a0. [DOI] [PubMed] [Google Scholar]
- 13.Wong JM, Bateman E. TBP-DNA interactions in the minor groove discriminate between A:T and T:A base pairs. Nucleic Acids Res. 1994;22:1890–1896. doi: 10.1093/nar/22.10.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hahn S, Buratowski S, Sharp PA, Guarente L. Yeast TATA-binding protein TFIID binds to TATA elements with both consensus and nonconsensus DNA sequences. Proc. Natl Acad. Sci. USA. 1989;86:5718–5722. doi: 10.1073/pnas.86.15.5718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patikoglou GA, Kim JL, Sun L, Yang SH, Kodadek T, Burley SK. TATA element recognition by the TATA box-binding protein has been conserved throughout evolution. Genes Dev. 1999;13:3217–3230. doi: 10.1101/gad.13.24.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nikolov DB, Chen H, Halay ED, Hoffmann A, Roeder RG, Burley SK. Crystal structure of a human TATA box-binding protein/TATA element complex. Proc. Natl Acad. Sci. USA. 1996;93:4862–4867. doi: 10.1073/pnas.93.10.4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoopes BC, LeBlanc JF, Hawley DK. Contributions of the TATA box sequence to rate-limiting steps in transcription initiation by RNA polymerase II. J. Mol. Biol. 1998;277:1015–1031. doi: 10.1006/jmbi.1998.1651. [DOI] [PubMed] [Google Scholar]
- 18.Starr DB, Hoopes BC, Hawley DK. DNA bending is an important component of site-specific recognition by the TATA binding protein. J. Mol. Biol. 1995;250:434–446. doi: 10.1006/jmbi.1995.0388. [DOI] [PubMed] [Google Scholar]
- 19.Wu J, Parkhurst KM, Powell RM, Brenowitz M, Parkhurst LJ. DNA bends in TATA-binding protein-TATA complexes in solution are DNA sequence-dependent. J. Biol. Chem. 2001;276:14614–14622. doi: 10.1074/jbc.M004402200. [DOI] [PubMed] [Google Scholar]
- 20.Strahs D, Barash D, Qian X, Schlick T. Sequence-dependent solution structure and motions of 13 TATA/TBP (TATA-box binding protein) complexes. Biopolymers. 2003;69:216–243. doi: 10.1002/bip.10409. [DOI] [PubMed] [Google Scholar]
- 21.Burley SK, Roeder RG. Biochemistry and structural biology of transcription factor IID (TFIID) Annu. Rev. Biochem. 1996;65:769–799. doi: 10.1146/annurev.bi.65.070196.004005. [DOI] [PubMed] [Google Scholar]
- 22.Hall DB, Struhl K. The VP16 activation domain interacts with multiple transcriptional components as determined by protein-protein cross-linking in vivo. J. Biol. Chem. 2002;277:46043–46050. doi: 10.1074/jbc.M208911200. [DOI] [PubMed] [Google Scholar]
- 23.Bhaumik SR, Raha T, Aiello DP, Green MR. In vivo target of a transcriptional activator revealed by fluorescence resonance energy transfer. Genes Dev. 2004;18:333–343. doi: 10.1101/gad.1148404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fishburn J, Mohibullah N, Hahn S. Function of a eukaryotic transcription activator during the transcription cycle. Mol. Cell. 2005;18:369–378. doi: 10.1016/j.molcel.2005.03.029. [DOI] [PubMed] [Google Scholar]
- 25.Mishra AK, Vanathi P, Bhargava P. The transcriptional activator Gal4-VP16 regulates the intra-molecular interactions of the TATA-binding protein. J. Biosci. 2003;28:423–436. doi: 10.1007/BF02705117. [DOI] [PubMed] [Google Scholar]
- 26.Björklund S, Gustafsson CM. The yeast Mediator complex and its regulation. Trends Biochem. Sci. 2005;30:240–244. doi: 10.1016/j.tibs.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 27.Asturias FJ. RNA polymerase II structure, and organization of the preinitiation complex. Curr. Opin. Struct. Biol. 2004;14:121–129. doi: 10.1016/j.sbi.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 28.Kang JS, Kim SH, Hwang MS, Han SJ, Lee YC, Kim YJ. The structural and functional organization of the yeast Mediator complex. J. Biol. Chem. 2001;276:42003–42010. doi: 10.1074/jbc.M105961200. [DOI] [PubMed] [Google Scholar]
- 29.Larivière L, Geiger S, Hoeppner S, Röther S, Sträßer K, Cramer P. Structure and TBP binding of the Mediator head subcomplex Med8-Med18-Med20. Nat. Struct. Mol. Biol. 2006;13:895–901. doi: 10.1038/nsmb1143. [DOI] [PubMed] [Google Scholar]
- 30.Qiu H, Hu C, Yoon S, Natarajan K, Swanson MJ, Hinnebusch AG. An array of coactivators is required for optimal recruitment of TATA binding protein and RNA polymerase II by promoter-bound Gcn4p. Mol. Cell Biol. 2004;24:4104–4117. doi: 10.1128/MCB.24.10.4104-4117.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang F, Sumibcay L, Hinnebusch AG, Swanson MJ. A triad of subunits from the Gal11/tail domain of Srb Mediator is an in vivo target of transcriptional activator Gcn4p. Mol. Cell Biol. 2004;24:6871–6886. doi: 10.1128/MCB.24.15.6871-6886.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Govind CK, Yoon S, Qiu H, Govind S, Hinnebusch AG. Simultaneous recruitment of coactivators by Gcn4p stimulates multiple steps of transcription in vivo. Mol. Cell Biol. 2005;25:5626–5638. doi: 10.1128/MCB.25.13.5626-5638.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He Q, Battistella L, Morse RH. Mediator requirement downstream of chromatin remodeling during transcriptional activation of CHA1 in yeast. J. Biol. Chem. 2007 doi: 10.1074/jbc.M708266200. 10.1074/jbc.M708266200 [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 34.Ranish JA, Yudkovsky N, Hahn S. Intermediates in formation and activity of the RNA polymerase II preinitiation complex: holoenzyme recruitment and a postrecruitment role for the TATA box and TFIIB. Genes Dev. 1999;13:49–63. doi: 10.1101/gad.13.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee TI, Causton HC, Holstege FCP, Shen WC, Hannett N, Jennings EG, Winston F, Green MR, Young RA. Redundant roles for the TFIID and SAGA complexes in global transcription. Nature. 2000;405:701–704. doi: 10.1038/35015104. [DOI] [PubMed] [Google Scholar]
- 36.Li XY, Bhaumik SR, Zhu X, Li L, Shen WC, Dixit BL, Green MR. Selective recruitment of TAFs by yeast upstream activating sequences: Implications for eukaryotic promoter structure. Curr. Biol. 2002;12:1240–1244. doi: 10.1016/s0960-9822(02)00932-6. [DOI] [PubMed] [Google Scholar]
- 37.Shen WC, Bhaumik SR, Causton HC, Simon I, Zhu X, Jennings EG, Wang TH, Young RA, Green MR. Systematic analysis of essential yeast TAFs in genome-wide transcription and preinitiation complex assembly. EMBO J. 2003;22:3395–3402. doi: 10.1093/emboj/cdg336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huisinga KL, Pugh BF. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol. Cell. 2004;13:573–585. doi: 10.1016/s1097-2765(04)00087-5. [DOI] [PubMed] [Google Scholar]
- 39.Tsukihashi Y, Kawaichi M, Kokubo T. Requirement for yeast TAF145 function in transcriptional activation of the RPS5 promoter that depends on both core promoter structure and upstream activating sequences. J. Biol. Chem. 2001;276:25715–25726. doi: 10.1074/jbc.M102416200. [DOI] [PubMed] [Google Scholar]
- 40.Kim YJ, Björklund S, Li Y, Sayre MH, Kornberg RD. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 41.Li WZ, Sherman F. Two types of TATA elements for the CYC1 gene of the yeast Saccharomyces cerevisiae. Mol. Cell Biol. 1991;11:666–676. doi: 10.1128/mcb.11.2.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takagi Y, Chadick JZ, Davis JA, Asturias FJ. Preponderance of free Mediator in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 2005;280:31200–31207. doi: 10.1074/jbc.C500150200. [DOI] [PubMed] [Google Scholar]
- 43.Sanders SL, Weil PA. Identification of two novel TAF subunits of the yeast Saccharomyces cerevisiae TFIID complex. J. Biol. Chem. 2000;275:13895–13900. doi: 10.1074/jbc.275.18.13895. [DOI] [PubMed] [Google Scholar]
- 44.Baidoobonso SM, Guidi BW, Myers LC. Med19(Rox3) regulates intermodule interactions in the Saccharomyces cerevisiae Mediator complex. J. Biol. Chem. 2007;282:5551–5559. doi: 10.1074/jbc.M609484200. [DOI] [PubMed] [Google Scholar]
- 45.Myers LC, Leuther K, Bushnell DA, Gustafsson CM, Kornberg RD. Yeast RNA polymerase II transcription reconstituted with purified proteins. Methods Companion Methods Enzymol. 1997;12:212–216. doi: 10.1006/meth.1997.0473. [DOI] [PubMed] [Google Scholar]
- 46.Myers LC, Lacomis L, Erdjument-Bromage H, Tempst P. The yeast capping enzyme represses RNA polymerase II transcription. Mol. Cell. 2002;10:883–894. doi: 10.1016/s1097-2765(02)00644-5. [DOI] [PubMed] [Google Scholar]
- 47.Myers LC, Gustafsson CM, Hayashibara KC, Brown PO, Kornberg RD. Mediator protein mutations that selectively abolish activated transcription. Proc. Natl Acad. Sci. USA. 1999;96:67–72. doi: 10.1073/pnas.96.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Auty R, Steen H, Myers LC, Persinger J, Bartholomew B, Gygi SP, Buratowski S. Purification of active TFIID from Saccharomyces cerevisiae. Extensive promoter contacts and co-activator function. J. Biol. Chem. 2004;279:49973–49981. doi: 10.1074/jbc.M409849200. [DOI] [PubMed] [Google Scholar]
- 49.Berger SL, Piña B, Silverman N, Marcus GA, Agapite J, Regier JL, Triezenberg SJ, Guarente L. Genetic isolation of ADA2: A potential transcriptional adaptor required for function of certain acidic activation domains. Cell. 1992;70:251–265. doi: 10.1016/0092-8674(92)90100-q. [DOI] [PubMed] [Google Scholar]
- 50.Jiang YW, Stillman DJ. Epigenetic effects on yeast transcription caused by mutations in an actin-related protein present in the nucleus. Genes Dev. 1996;10:604–619. doi: 10.1101/gad.10.5.604. [DOI] [PubMed] [Google Scholar]
- 51.Sayre MH, Tschochner H, Kornberg RD. Reconstitution of transcription with five purified initiation factors and RNA polymerase II from Saccharomyces cerevisiae. J. Biol. Chem. 1992;267:23376–23382. [PubMed] [Google Scholar]
- 52.Svejstrup JQ, Feaver WJ, LaPointe J, Kornberg RD. RNA polymerase transcription factors IIH holoenzyme from yeast. J. Biol. Chem. 1994;269:28044–28048. [PubMed] [Google Scholar]
- 53.Chitikila C, Huisinga KL, Irvin JD, Basehoar AD, Pugh BF. Interplay of TBP inhibitors in global transcriptional control. Mol. Cell. 2002;10:871–882. doi: 10.1016/s1097-2765(02)00683-4. [DOI] [PubMed] [Google Scholar]
- 54.Sawadogo M, Roeder RG. Factors involved in specific transcription by human RNA polymerase II: analysis by a rapid and quantitative in vitro assay. Proc. Natl Acad. Sci. USA. 1985;82:4394–4398. doi: 10.1073/pnas.82.13.4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lue NF, Flanagan PM, Sugimoto K, Kornberg RD. Initiation by yeast RNA polymerase II at the adenoviral major late promoter in vitro. Science. 1989;246:661–664. doi: 10.1126/science.2510298. [DOI] [PubMed] [Google Scholar]
- 56.Matangkasombut O, Auty R, Buratowski S. Structure and function of the TFIID complex. Adv. Protein Chem. 2004;67:67–92. doi: 10.1016/S0065-3233(04)67003-3. [DOI] [PubMed] [Google Scholar]
- 57.Faiger H, Ivanchenko M, Cohen I, Haran TE. TBP flanking sequences: asymmetry of binding, long-range effects and consensus sequences. Nucleic Acids Res. 2006;34:104–119. doi: 10.1093/nar/gkj414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nair D, Kim Y, Myers LC. Mediator and TFIIH govern carboxyl-terminal domain-dependent transcription in yeast extracts. J. Biol. Chem. 2005;280:33739–33748. doi: 10.1074/jbc.M506067200. [DOI] [PubMed] [Google Scholar]
- 59.Martinez-Campa C, Politis P, Moreau JL, Kent N, Goodall J, Mellor J, Goding CR. Precise nucleosome positioning and the TATA box dictate requirements for the histone H4 tail and the bromodomain factor Bdf1. Mol. Cell. 2004;15:69–81. doi: 10.1016/j.molcel.2004.05.022. [DOI] [PubMed] [Google Scholar]
- 60.Vermeulen M, Mulder KW, Denissov S, Pijnappel WWMP, van Schaik FMA, Varier RA, Baltissen MPA, Stunnenberg HG, Mann M, Timmers HTM. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell. 2007;131:58–69. doi: 10.1016/j.cell.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 61.Maniatis T, Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- 62.Kaplan CD, Laprade L, Winston F. Transcription elongation factors repress transcription initiation from cryptic sites. Science. 2003;301:1096–1099. doi: 10.1126/science.1087374. [DOI] [PubMed] [Google Scholar]
- 63.Struhl K. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat. Struct. Mol. Biol. 2007;14:103–105. doi: 10.1038/nsmb0207-103. [DOI] [PubMed] [Google Scholar]
- 64.Stewart JJ, Fischbeck JA, Chen X, Stargell LA. Non-optimal TATA elements exhibit diverse mechanistic consequences. J. Biol. Chem. 2006;281:22665–22673. doi: 10.1074/jbc.M603237200. [DOI] [PubMed] [Google Scholar]
- 65.Hahn S, Hoar ET, Guarente L. Each of three “TATA elements” specifies a subset of the transcription initiation sites at the CYC-1 promoter of Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 1985;82:8562–8566. doi: 10.1073/pnas.82.24.8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang Z, Dietrich FS. Mapping of transcription start sites in Saccharomyces cerevisiae using 5′SAGE. Nucleic Acids Res. 2005;33:2838–2851. doi: 10.1093/nar/gki583. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.