Abstract
RNA interference (RNAi) is a powerful approach to inhibit human immunodeficiency virus type 1 (HIV-1) replication. However, HIV-1 can escape from RNAi-mediated antiviral therapy by selection of mutations in the targeted sequence. To prevent viral escape, multiple small interfering RNAs (siRNAs) against conserved viral sequences should be combined. Ideally, these RNA inhibitors should be expressed simultaneously from a single transgene transcript. In this study, we tested a multiplex microRNA (miRNA) expression strategy by inserting multiple effective anti-HIV siRNA sequences in the miRNA polycistron mir-17-92. Individual anti-HIV miRNAs that resemble the natural miRNA structures were optimized by varying the siRNA position in the hairpin stem to obtain maximal effectiveness against luciferase reporters and HIV-1. We show that an antiviral miRNA construct can have a greater intrinsic inhibitory activity than a conventional short hairpin (shRNA) construct. When combined in a polycistron setting, the silencing activity of an individual miRNA is strongly boosted. We demonstrate that HIV-1 replication can be efficiently inhibited by simultaneous expression of four antiviral siRNAs from the polycistronic miRNA transcript. These combined results indicate that a multiplex miRNA strategy may be a promising therapeutic approach to attack escape-prone viral pathogens.
INTRODUCTION
RNAi is an evolutionary conserved and sequence-specific gene silencing mechanism in eukaryotes (1,2). RNAi can be induced by double-stranded RNA that is processed by the RNase III-like enzyme Dicer into 21–25 bp siRNAs (3–5). The siRNA is incorporated into the RNA-induced silencing complex (RISC) in the cytoplasm and directs RISC to degrade an mRNA that is perfectly complementary to one (guide) strand of the siRNA (5).
Cellular miRNAs are the natural inducers of RNAi. Most miRNAs are synthesized as part of longer primary RNA transcripts (pri-miRNAs) (6–8). The pri-miRNAs are cleaved by the nuclear Drosha-DGCR8 complex to produce miRNA precursors (pre-miRNAs) of ∼70 nt (9–12). Pre-miRNAs are transported by Exportin-5 to the cytoplasm, where they are cleaved by Dicer to produce the miRNA duplex of ∼22 bp. The single-stranded mature miRNA programs RISC for mRNA cleavage (perfect complementarity) or translational repression (incomplete complementarity) (13,14). Several miRNAs are encoded in genomic clusters that are transcribed as polycistronic pri-miRNAs, allowing the production of multiple miRNAs from a single transcription unit (15,16). RNAi can be induced using RNA polymerase III promoter-driven shRNA expression vectors that direct siRNA expression. Another siRNA expression strategy uses a pre-miRNA backbone that is transcribed by polymerase II or III (17–20). An optimized pre-miRNA design includes the single-stranded flanks of the pri-miRNA (21–23).
Currently, RNAi has been employed to inhibit the replication of a wide range of viruses including HIV-1, hepatitis C virus (HCV), hepatitis B virus (HBV), dengue virus, poliovirus, influenza virus A, coronavirus, herpesvirus and picornavirus (24,25). For HIV-1, potent inhibition has been reported with single shRNA and miRNA expression constructs (17,26–28). However, the therapeutic use of a single inhibitor is limited because of the rapid emergence of HIV-1 escape mutants (27,29,30). Minor sequence changes in the target sequence, sometimes even a single-point mutation, are sufficient to overcome RNAi-mediated inhibition (30), thus demonstrating the exquisite sequence-specificity of RNAi.
Strategies to reduce the chance of viral escape include the simultaneous use of multiple shRNAs in a combinatorial RNAi approach, which increases the genetic barrier for viral escape (31,32). Similar strategies have previously been validated for antisense DNA and ribozymes (33–35). However, expression of these shRNAs necessitates multiple expression cassettes and the construction of rather complex vectors that will not easily provide equimolar shRNA expression levels. Furthermore, when the same promoter is reiterated in a lentiviral vector, recombination occurs with high frequency on the repeated sequences (36). Alternative anti-escape strategies include the use of a second-generation of siRNAs that target-specific escape variants (37), the use of tandem siRNA transcripts (38), long hairpin RNAs (39) or the targeting of cellular co-factors that are critically involved in viral replication (40–44). Another attractive approach is to express multiple antiviral siRNAs from a single polycistronic miRNA transcript, such as a natural genomic miRNA cluster that can be expressed from an RNA polymerase II promoter. This strategy is of particular interest for antiviral purposes because miRNA-like transcripts were shown to be more effective antivirals than regular shRNAs (17,45). Furthermore, using an RNA polymerase II promoter will allow lower and regulated expression, thereby reducing the risk of toxicity due to oversaturation of the RNAi machinery (46).
In this study, we designed a polycistronic transcript based on the mir-17-92 backbone to simultaneously express four anti-HIV siRNAs. To generate this transcript, we first constructed individual anti-HIV miRNAs that resemble the natural pri-miRNA structures. These hairpins were optimized for viral inhibition by varying the siRNA position in the hairpin stem. We show that the expression of individual miRNAs is greatly enhanced in multiplex hairpin transcripts that are properly processed into functional miRNAs. HIV-1 replication can be potently inhibited by simultaneous expression of four antiviral miRNAs. These combined results indicate that the multiplex miRNA strategy is a promising therapeutic approach against escape-prone viral pathogens.
MATERIALS AND METHODS
DNA constructs
The wild-type mir-17-19b polycistron was amplified from genomic DNA of 293T cells with primers OncoF1 and OncoR1 and TA-cloned in TOPO2.1 (Invitrogen). The mir-17-19b polycistron sequence was verified and is identical to the sequence in the NCBI database (NT_009952.14). The TOPO2.1/mir-17-19b construct was used as a template to generate the antiviral miRNA constructs. The construction consists of a four-step fusion PCR as shown in the Supplementary Figure 1. Briefly, the 5′-flank of the pri-miRNA was amplified with a forward primer encoding a BamHI site and a reverse primer encoding the HIV-1 sequence at its 3′-end (step 1). Similarly, the 3′-flank of the pri-miRNA was amplified with a forward primer containing HIV-1 sequences and a reverse primer encoding BglII and XhoI sites (step 2). Two complementary oligonucleotides, creating the stem-loop structure of the antiviral miRNA, were annealed as described previously (step 3) (32). The partial sequence similarity between the fragments generated in steps 1, 2 and 3 allowed their fusion by PCR with the outer forward F1 and reverse R2 primers (step 4), resulting in the antiviral miRNA. Supplementary Table 1 lists all oligonucleotides used in this study.
PCR amplification was performed in a 50 μl reaction containing 1 × PCR amplification buffer (Invitrogen), 1.5–2 mM MgCl2 (optimized for each reaction), 25 pmol of each primer, 0.2 mM dNTPs and 2.5 units of AmpliTaq DNA polymerase (Perkin Elmer Applied Biosystem). The PCR program was as follows: 95°C for 5 min, 35 cycles of 2 min at 94°C, 1.5 min at 60°C, 2.5 min at 72°C and a final extension for 8 min at 72°C. The PCR products were separated on a 1% agarose gel stained with ethidium bromide and compared to a standard DNA size marker (Eurogentec). miRNA PCR products were excised from gel, purified with the QIAquick gel extraction kit (Qiagen), digested with BamHI and XhoI and cloned in the corresponding sites of pcDNA6.2-GW/EmGFP-miR (Invitrogen). All miRNA constructs were sequenced with primers GFPseqF and mirR using the BigDye Terminator v1.1 Cycle Sequencing Kit (Perkin Elmer Applied Biosystem).
Multimerization of the individual pri-miRNA units was performed by digestion of a single miRNA hairpin construct with BamHI and XhoI and religation into the BglII/XhoI sites of pcDNA6-miRNA. By repeating this procedure we obtained constructs expressing different combinations of 1, 2, 3, 4 and 6 pri-miRNAs. The RNA structures formed by the transcripts were predicted with the Mfold program (47) at http://frontend.bioinfo.rpi.edu/applications/mfold/ and found to be similar to the predicted conformation of the wild-type pri-miRNAs.
The firefly luciferase (FL) reporters containing HIV-1 target sequences pol47 (Luc-Apol47), pol1 (Luc-Bpol1), gag5 (Luc-Cgag5), r/t5 (Luc-Dr/t5), ldr9 (Luc-Eldr9) and the anti-HIV shRNAs have been described previously (32). The full-length HIV-1 molecular clone LAI (accession number AF33819.3) (48) was used to produce wild-type virus and to study its inhibition by the antiviral miRNAs and shRNAs.
Cells, DNA transfection and virus infection
Human embryonic kidney (HEK) 293T cells were grown as a monolayer in DMEM (Invitrogen) supplemented with 10% fetal calf serum (FCS) (Hybond), minimal essential medium nonessential amino acids, penicillin (100 U/ml) and streptomycin (100 μg/ml) at 37°C and 5% CO2. Cells were trypsinized one day before transfection, resuspended in DMEM without antibiotics and seeded in 24-well plates at a density of 1.2 × 105 cells per well. Cells were co-transfected with the indicated DNA constructs using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions. One nanogram of pRL plasmid (Promega) expressing renilla luciferase (RL) from the CMV promoter was added as an internal control for cell viability and transfection efficiency. All transfection experiments were controlled for equal DNA input by adding pBluescript SK- (Promega). Luciferase and renilla expression was measured with the dual-luciferase reporter assay system (Promega) according to the manufacturer's instructions. Virus production was determined by measuring the CA-p24 levels in the culture supernatant by ELISA (Abbott) (49). Subsequently, the cells were lysed to measure the renilla luciferase activities using the Renilla Luciferase Assay System (Promega) according to the manufacturer's protocol. Transfection experiments were corrected for between session variations as described previously (50).
The human T-cell line SupT1 was cultured in 25 cm2 flasks in RPMI 1640 medium supplemented with 10% FCS, penicillin (100 U/ml) and streptomycin (100 μg/ml) at 37°C and 5% CO2. 200.000 SupT1 cells were infected with equal amounts virus (0.5 ng CA-p24) produced in 293T cells. When HIV-induced cytopathic effects were observed, cell and supernatant samples were stored at −80°C. Virus spread was followed by measuring the CA-p24 levels in the culture supernatant by ELISA.
siRNA detection by Northern blotting
One day before transfection 7.5 × 105 293T cells were plated in 6-well plates. Cells were transfected with 39–5000 ng of the shRNA construct and 2500 or 5000 ng of the miRNA-expression construct using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions. In all transfection experiments we added pBluescript SK- (Promega) to obtain identical DNA concentrations. Total cellular RNA was extracted 2 days post-transfection with the mirVana miRNA isolation kit (Ambion) according to the manufacturer's protocol. For Northern blot analysis, 15 μg total RNA was heated for 5 min at 95°C before electrophoresis on a 15% denaturing polyacrylamide gel (precast Novex TBU gel, Invitrogen). To check for equal sample loading, the gel was stained with 2 μg/ml ethidium bromide in milliQ water for 20 min. Destaining was performed by rinsing the gel three times with milliQ water for 10 min. The ribosomal RNA (rRNA) bands were visualized under UV light. The RNA samples were electrotransferred to a positively charged nylon membrane (Boehringer Mannheim, GmbH, Mannheim, Germany) and crosslinked to the membrane using UV light at a wavelength of 254 nm (1200 μJ × 100). LNA oligonucleotide probes were 5′-end labeled with the kinaseMax kit (Ambion) in the presence of 1 μl [γ-32P]ATP (0.37 MBq/μl Amersham Biosciences). To remove unincorporated nucleotides, the probes were purified on Sephadex G-25 spin columns (Amersham Biosciences) according to the manufacturer's protocol. Hybridizations were performed at 42°C with labeled LNA oligonucleotides in 10 ml ULTRAhyb hybridization buffer (Ambion) according to the manufacturer's instructions. We used the oligonucleotide probes (LNA-positions underlined): 5′-GTGAAGGGGCAGTAGTAAT-3′ (pol47 probe), 5′-ACAGGAGCAGATGATACAG-3′ (pol1 probe), 5′-GAAGAAATGATGACAGCAT-3′ (gag5 probe), 5′-ATGGCAGGAAGAAGCGGAG-3′ (r/t5 probe) and 5′-AGATGGGTGCGAGAGCGTC-3′ (ldr9 probe). The membranes were washed twice for 5 min at 42°C in 2 × SSC/0.1% SDS and twice for 15 min at 42°C in 0.1 × SSC/0.1% SDS. Signals were detected and quantified using a phosphorimager (Amersham Biosciences).
Lentiviral vector production and transduction
Lentiviral vector plasmids are derived from the construct pLenti6/V5-Dest (Invitrogen), which we renamed pLV. The miRNA cassettes, containing a GFP marker, were inserted into pLV using the Gateway-adapted BLOCK-iT Lentiviral Pol II miR RNAi Expression System (Invitrogen) according to the manufacturer's instructions. The sequences of all miRNA constructs were verified using the primers CMVf and V5r. The miRNA inhibitory potential was determined by co-transfection with appropriate luciferase reporters (results not shown). Lentiviral vector was produced in 293T cells (2.2 × 106) seeded in a 25 cm2 flask. The next day, medium was replaced with 2.2-ml medium without antibiotics. Subsequently, pLV vectors expressing a miRNA (2.4 μg) were co-transfected with packaging plasmids pSYNGP (1.5 μg) (51), RSV-rev (0.6 μg) and pVSVg (0.8 μg) (52) with 16 μl of Lipofectamine 2000 reagent and 1.5 ml Optimem (Gibco BRL). The second day, medium was replaced with fresh medium. On the third and fourth day, medium containing lentiviral vector was harvested and pooled. Cellular debris was removed by filtration through a FP30/0.45 CA-S filter (Schleicher and Schuell MicroScience) and 1-ml aliquots were stored at −80°C. Lentiviral stocks were titrated on SupT1 and 293T cells to determine the vector titer. SupT1 and 293T cells (1 × 105) were transduced with the pLV vector expressing an unrelated miRNA N (Invitrogen) or the antiviral miRNAs Apol47, Bpol1, Cgag5, Dr/t5, Eldr9 and ACDE at a multiplicity of infection (MOI) of 0.15 as described previously (32). The pLV vector contains the blasticidin resistance gene, which allows the selection of transduced cells using 3.5 μg/ml blasticidin. The GFP marker encoded by the miRNA cassette was used to select GFP+ cells by FACS sorting approximately 10 days post-transduction.
RESULTS
Design of antiviral miRNAs
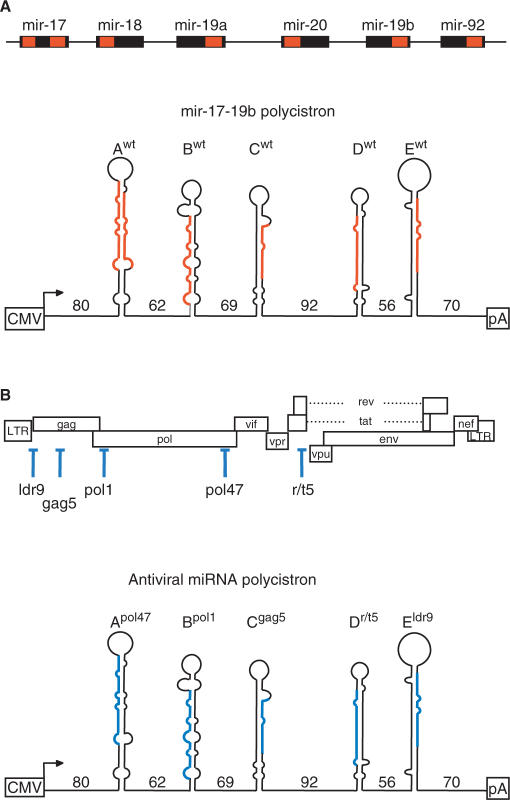
The human mir-17-92 cluster on chromosome 13 encodes a 1 kb pri-miRNA polycistronic transcript with six pre-miRNAs that produces seven mature miRNAs (Figure 1A, upper panel) (15). The pre-miR-17 hairpin encodes two miRNAs, one on the 5′ side of the hairpin (miR-17-5p) and one on the 3′ side (miR-17-3p). We amplified the sequences encoding the first five pri-miRNAs from the mir-17-92 polycistron and cloned it under the control of the cytomegalovirus (CMV) immediate early promoter (Figure 1A, lower panel). We subcloned each individual pre-miRNA with at least 40-nt flanks on each side of the hairpin and systematically replaced the mature miRNA sequences with antiviral sequences as explained in detail in the Supplementary Figure 1. The original miRNA names were replaced with letters A-E and we inserted 19- to 24-nt antiviral sequences against five HIV-1 genes (Figure 1B, upper panel). The HIV-1 targets represent highly conserved sequences to which we successfully raised potent shRNA inhibitors (32). We set out to combine 2, 3, 4 or 5 antiviral miRNAs, which will eventually result in an antiviral pri-miRNA polycistron (Figure 1B, lower panel).
Figure 1.
Schematic of the mir-17-92 cluster and design of the antiviral miRNAs. (A) Genomic organization of the mir-17-92 polycistron (upper panel) and predicted secondary structure of the mir-17-19b transcript (Awt-Ewt, lower panel). Mature miRNAs are indicated in red; CMV, cytomegalovirus immediate early promoter; pA, SV40 polyadenylation signal. (B) Scheme of the HIV-1 proviral genome and position of the target sequences (upper panel) used for the design of the antiviral miRNA polycistron (lower panel). Guide strands of the siRNA against HIV-1 are blue. Numbers between the pre-miRNAs represent the length of the flanks (in nucleotides). Original miRNA names were replaced with the letters A–E and target genes are indicated in superscript.
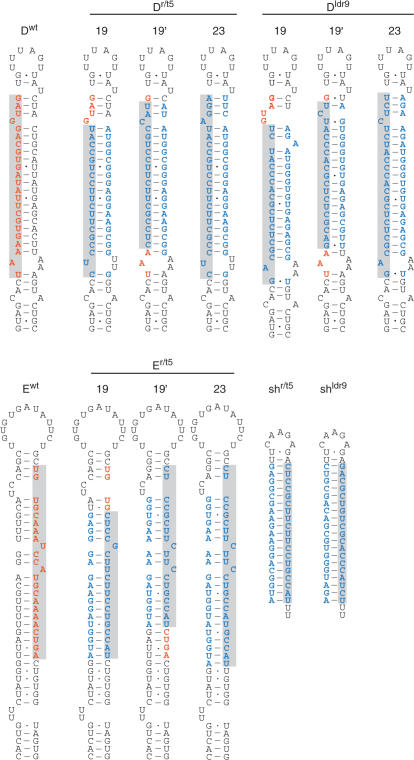
We first determined if we could produce an optimal antiviral miRNA construct by varying the position and length of the anti-HIV sequence. For the initial experiments we chose miRNAs miR-20 and miR-19b (Dwt and Ewt) because they contain the smallest number of bulges and thus most resemble the original shRNA structure (Figure 2). The original mature miRNA and the antiviral miRNA strand are boxed. We modified the passenger strand of the basepaired stem to mimic structural features (mismatches, bulges and thermodynamic stability) of the natural pre-miRNA. We constructed a series of antiviral miRNAs with the effective r/t5 and ldr9 siRNAs in the Dwt backbone, which produces the antiviral guide strand from the 5′ side of the hairpin duplex. The 19-nt antiviral siRNAs were positioned either at the 5′ (Dr/t5-19′ and Dldr9-19′) or 3′-end (Dr/t5-19′and Dldr9-19′) of the original miRNA sequence (Figure 2, upper panel). Additional constructs were made in which we extended the antiviral siRNAs at the 3′-end to 23-nt, which is the actual size of the wild-type mature miRNA (Dr/t5-23 and Dldr9-23) (Figure 2, Table 1). We repeated this strategy for the r/t5 inhibitor in the Ewt hairpin, which produces the antiviral guide strand from the 3′ side of the hairpin stem (Figure 2, lower panel). We designed a similar set of constructs in which the 19-nt r/t5 siRNA was positioned at the 5′ (Er/t5-19′) or 3′-end (Er/t5-19′) of the wild-type miRNA and an extended 23-nt version (Er/t5-23).
Figure 2.
Structure of the antiviral miRNAs based on pre-miRNAs Dwt and Ewt. Upper panel: The r/t5 and ldr9 siRNAs were incorporated into the Dwt backbone, which produces the guide strand from the 5′-side of the hairpin. The guide strand is marked in grey. We modified the passenger strand to mimic structural features (mismatches, bulges and thermodynamic stability) of the natural pre-miRNA. The 19-nt antiviral siRNAs were positioned either at the 5′ (Dr/t5-19 and Dldr9-19) or 3′-end (Dr/t5-19' and Dldr9-19') of the original miRNA and the length of the siRNAs was extended at the 3′-end to 23-nt (Dr/t5-23 and Dldr9-23). Lower panel: The r/t5 siRNA was similarly incorporated into the Ewt pre-miRNA, which produces the guide strand from the 3′-side of the hairpin. The structure of the original shRNAs shr/t5 and shldr9 are presented. HIV-1 sequences are blue, mature wild-type miRNA sequences are red, pre-miRNA sequences are black, Watson–Crick base pairs are shown with dashes and G•U wobbles with dots.
Table 1.
Structure and properties of the anti-HIV miRNAs
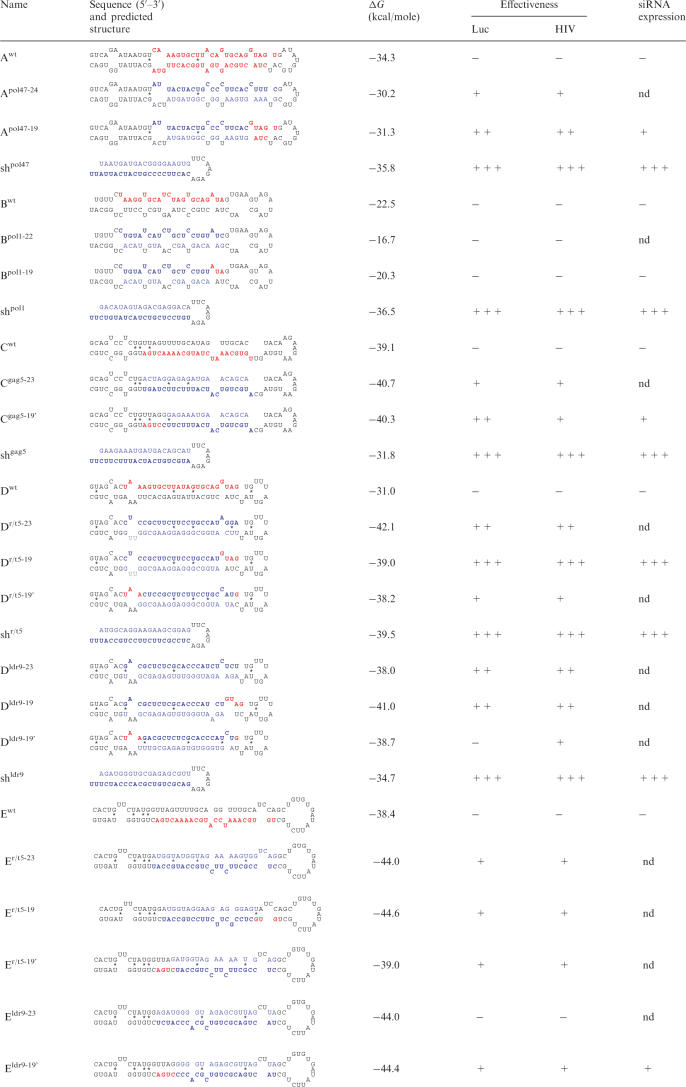 |
HIV-1 sequences are blue; pre-miRNA sequences are black; mature miRNA sequences are red; guide strand is bold; −, no; +, 10–30%; ++, 30–70%; +++, 70–100%; nd, not determined.
Activity of the antiviral miRNAs
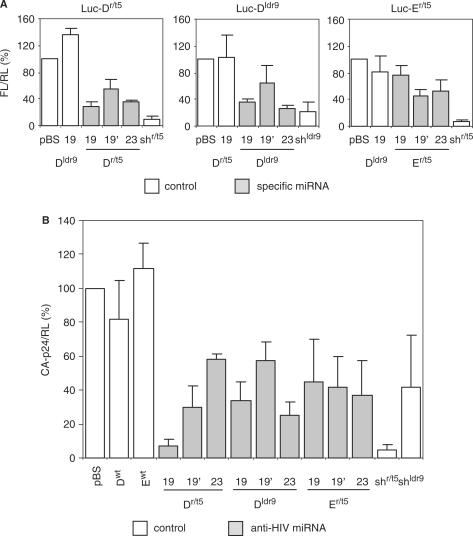
To determine the inhibitory activity of the miRNA constructs, we co-transfected 293T cells with the inhibitors and luciferase reporter constructs containing either the 50-nt r/t5 target (Luc-Dr/t5 and Luc-Er/t5) or the 50-nt ldr9 target (Luc-Dldr9). A plasmid encoding renilla luciferase (pRL) was included to correct for transfection variation and to monitor for cell viability that may be affected by off-target effects of the modified miRNAs and/or the antiviral siRNAs. Firefly and renilla luciferase expression was measured 48 h post-transfection. Firefly luciferase expression was normalized to the control renilla luciferase expression. Firefly luciferase expression in the presence of pBluescript (pBS) was set at 100% (Figure 3A). A common pattern was observed in that most efficient inhibition was scored for the 19- and 23-nt siRNA positioned at the 5′-end of the original miRNA sequence, which are the 19 and 23 variants of the D hairpin and the 19′ and 23 variants of the E hairpin.
Figure 3.
Inhibition of luciferase reporters and HIV-1 production by individual antiviral miRNAs. (A) Luciferase reporters (100 ng) were co-transfected in 293T cells with 20 ng antiviral miRNA construct and 1 ng pRL. The original shRNAs shr/t5 and shldr9 were used as positive controls and non-matching miRNAs as negative controls. Firefly and renilla luciferase expression was measured 2 days post-transfection. Normalized firefly luciferase expression in the presence of pBS was set at 100%. Mean values obtained in three independent experiments are shown (±SE). FL, firefly luciferase; RL, renilla luciferase. (B) 239T cells were co-transfected with 150 ng HIV-1 molecular clone LAI, 1 ng pRL and 20 ng of the indicated antiviral miRNA constructs. shr/t5 and shldr9 constructs were used as positive controls, Dwt and Ewt as negative controls. CA-p24 levels in the culture supernatant and renilla luciferase expression were measured 2 days post-transfection. Normalized CA-p24 expression in the presence of pBS was set at 100%. Error bars represent the standard deviation in four independent experiments.
A similar trend was observed in HIV-1 inhibition studies (Figure 3B). We co-transfected the HIV-1 molecular clone LAI and the miRNA constructs into 293T cells and virus production was measured as the CA-p24 level in the culture supernatant at 2 days post-transfection. We observed the strongest inhibition when the siRNA inhibitor was situated at the 5′-end of the miRNA sequence. The optimized hairpins have a similar efficiency of inhibiting HIV-1 production as the shRNA constructs that were used as positive controls.
Based on these initial findings, we designed additional miRNA constructs with 19–24 nt antiviral sequences at the 5′-end of the miRNA: Apol47-24, Apol47-19, Bpol1-22, Bpol1-19, Cgag5-23, Cgag5-19', Eldr9-23 and Eldr9-19' (Table 1). Of the original Dr/t5, Dldr9 and Er/t5 constructs, we selected Dr/t5-19 because it is the best inhibitor. As a consequence, the ldr9 inhibitor was introduced in the E hairpin. We tested the effectiveness of all new constructs against luciferase reporters and HIV-1 (Table 1). The observed inhibition is sequence-specific because non-matching miRNAs did not have any effect. Furthermore, the expression of the control renilla reporter was stable in all experiments. The miRNA constructs with 19-nt anti-HIV sequences showed a somewhat higher activity than the extended versions. We therefore selected the 19-nt inhibitors for the construction of antiviral miRNA polycistrons. For simplicity, we removed the 19 indications from the miRNA names (e.g. Apol47-19 becomes Apol47).
Expression and processing of an antiviral miRNA
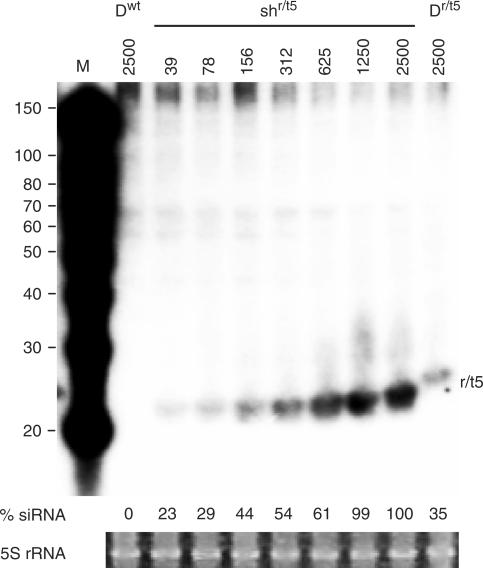
The original shRNA inhibitor shr/t5 seems slightly more active than the optimized Dr/t5 miRNA molecule (Figure 3). This could be due to the use of different promoters (polymerase III U6 versus polymerase II CMV, respectively), but may also be due to differential RNA processing efficiencies. We therefore wanted to compare the siRNA level expressed from each construct. We titrated 39–2500 ng shr/t5 construct in 293T cells and compared that with 2500 ng Dr/t5 construct. Two days post-transfection, total cellular RNA was isolated from the transfected cells and analyzed on a Northern blot with an r/t5 probe that detects the guide (antisense) strand (Figure 4). The original Dwt hairpin was used as a negative control. Quantification of the RNA bands showed that expression of the antiviral 19-nt siRNA by construct Dr/t5 is about 15–30-fold lower than the expression by construct shr/t5 (Figure 4, see numbers below the blot). Although the siRNA expression level of Dr/t5 is significantly lower than that of shr/t5, the inhibitory effect measured in the luciferase and HIV-1 inhibition assays is comparable. This result suggests that the intrinsic inhibitory capacity of Dr/t5 is in fact much greater than that of shr/t5.
Figure 4.
Expression and processing of siRNA from a miRNA versus an shRNA construct. We titrated 39–2500 ng shr/t5 construct in 293T cells and compared that with 2500 ng Dr/t5 construct. Northern blot analysis was performed on total cellular RNA. M, RNA size marker (nt) is shown on the left. Expression of siRNAs by 2500 ng shr/t5 was set at 100% and relative siRNA expression levels are indicated below the blot. Ethidium bromide staining of the 5S rRNA serves as sample loading control.
Multimerization of miRNAs increases the inhibitory activity
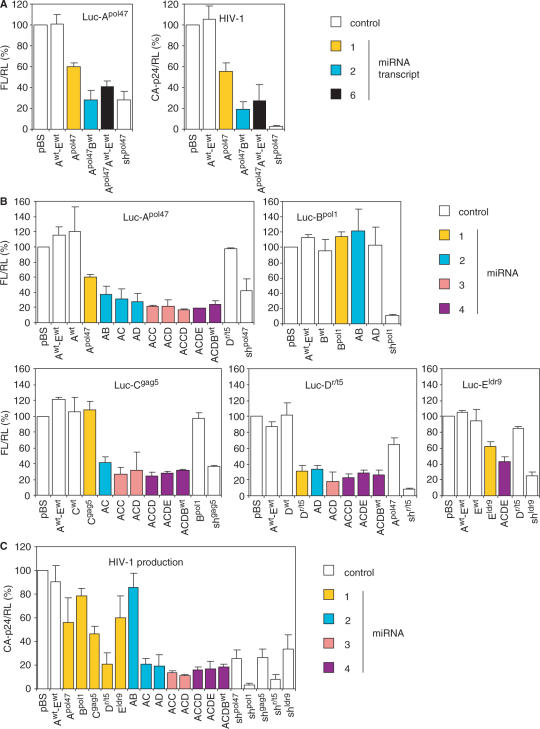
To address the effect of chaining of different hairpins for the silencing activity of the individual hairpins, we coupled the Apol47 inhibitor to wild-type pri-miRNAs. We constructed two variants of Apol47 with two or six hairpins in a single transcript: Apol47Bwt and Apol47Awt-Ewt (Awt-Ewt represents the complete wild-type mir-17-19b polycistron). We tested the ability of these hairpins to inhibit the Luc-Apol47 reporter (Figure 5A, left). Firefly luciferase expression was normalized to the renilla luciferase expression from the co-transfected pRL plasmid. We set the luciferase expression in the presence of pBS at 100%. The knockdown efficiency of the single Apol47 hairpin was ∼40%, but chaining it with the Bwt hairpin or the Awt-Ewt cluster enhanced the silencing activity. As a negative control, we included the Awt-Ewt construct. As a positive control, we included the original shRNA. A similar pattern was observed for inhibition of HIV-1 production (Figure 5A, right). Thus, expression of a miRNA inhibitor as part of a miRNA polycistronic transcript enhances the silencing activity.
Figure 5.
Inhibition of luciferase reporters and HIV-1 production by antiviral polycistronic miRNAs. (A) Knockdown efficiencies of the antiviral miRNA Apol47 expressed from 1-, 2- and 6-hairpin transcripts: Apol47, Apol47Bwt and Apol47Awt-Ewt. Left panel: 293T cells were co-transfected with 100 ng Luc-Apol47, 10 ng of different hairpin RNA constructs and 1 ng pRL. Two days post-transfection, firefly and renilla luciferase expression was measured. Normalized firefly luciferase expression with pBS was set at 100%. The Awt-Ewt construct was used as negative control and shpol47 as positive control. Right panel: 293T cells were co-transfected with 150 ng pLAI, 1 ng pRL and 5 ng of the hairpin constructs. Normalized CA-p24 level in the absence of inhibitor was set at 100%. FL, firefly luciferase; RL, renilla luciferase. (B) Luciferase knockdown efficiencies of 1, 2, 3 and 4 antiviral miRNA transcripts. 293T cells were co-transfected with 100 ng of the indicated luciferase reporter, 10 ng miRNA construct and 1 ng pRL. Two days post-transfection, firefly and renilla luciferase expression was measured. The Awt-Ewt construct and a non-matching miRNA were used as negative controls. The original shRNA constructs shpol47, shpol1, shgag5, shr/t5 and shldr9 were used as positive controls. (C) Inhibition of HIV-1 production by the miRNA constructs. 293T cells were co-transfected with 150 ng pLAI, 1 ng pRL and 5 ng 1, 2, 3 or 4 miRNA constructs. Two days post-transfection, CA-p24 levels in the culture supernatant and renilla luciferase expression were measured. Normalized CA-p24 level in the absence of inhibitor was set at 100%. Mean values obtained in four independent experiments are shown (±SE).
Next, we constructed polycistronic hairpin constructs with different combinations of two, three or four hairpins of the miRNA inhibitors Apol47, Bpol1, Cgag5, Dr/t5 and Eldr9: AB, AC, AD, ACC, ACD, ACCD, ACDE and ACDBwt. We first determined the knockdown efficiency of each individual hairpin within the multiplex transcripts by co-transfection with the corresponding luciferase reporter into 293T cells (Figure 5B, Supplementary Table 2). Constructs encoding the Awt-Ewt cistron and individual wild-type or non-matching miRNAs were used as negative controls. For inhibitors Apol47, Cgag5 and Eldr9, we observed a remarkable enhancement of silencing activity when chained to other hairpins. For instance, construct Cgag5 is poorly active compared to construct AC or ACDE (Figure 5B). Construct Bpol1 does not exhibit any inhibitory activity, either alone or when combined with another hairpin as in AB. Since the Bpol1 hairpin structure is rather unstable as predicted by the Mfold algorithm, this could be due to misfolding of the hairpin RNA. We therefore will not use hairpin Bpol1 in the final polycistron construct. Hairpin Dr/t5 does not benefit from linkage to other hairpins, but this is likely due to the high inhibitory activity of the individual Dr/t5 inhibitor. Strong inhibition of Luc-Dr/t5 was observed with Dr/t5, AD, ACD, ACCD and ACDE, indicating that generation of effective siRNAs from the multiplex hairpin transcripts does not depend on the miRNA position in the polycistron.
We further tested the ability of the antiviral miRNAs to inhibit HIV-1 by co-transfection with the HIV-1 molecular clone LAI (Figure 5C, Supplementary Table 2). Consistent with the luciferase results, we observed a moderate HIV-1 inhibition by the single miRNA constructs, except for construct Bpol1 (inactive) and Dr/t5 (highly active) (Figure 5C). Multimerization of the hairpins strongly enhanced the inhibition of HIV-1 production, which is due both to enhancement of the silencing activity of the individual hairpins and to the presence of multiple antiviral siRNAs. The Awt-Ewt construct was used as a negative control and the shRNA constructs were used as positive controls.
Increased siRNA expression from polycistronic miRNA transcripts
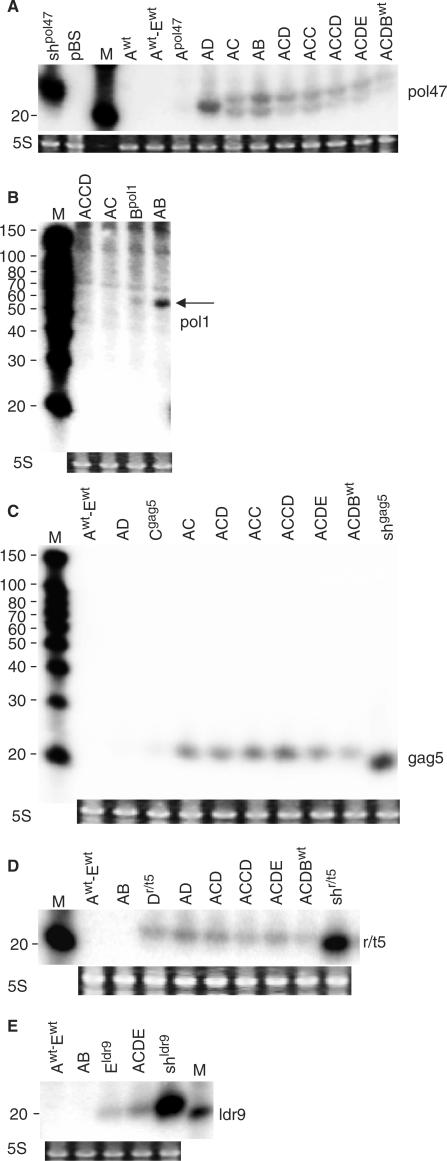
We next performed Northern blot analysis of the antiviral siRNAs made by the different polycistron constructs (Figure 6). The results are remarkably similar to the activity data in Figure 5. For instance, siRNA expression of Apol47 is greatly increased by linkage to another hairpin as in AD (Figure 6A). The Northern blot analysis of the Bpol1 and the AB inhibitors provides an explanation for its inactivity as only the ∼60-nt Bpol1 precursor is observed (Figure 6B, indicated by an arrow). To study whether the passenger siRNA strand of Bpol1 is made, we performed a Northern blot analysis with the corresponding probe, but failed to detect any passenger siRNAs (results not shown). In contrast, properly processed siRNAs are produced from hairpin Apol47 in construct AB, indicating that the inactive B unit in the two-hairpin transcript does not negatively influence the active A unit (Figure 6A and B). In addition, siRNA production from the A hairpin is boosted for the polycistronic transcripts compared to the single Apol47 hairpin transcript. For inhibitor Cgag5, we also observed a strong increase in siRNA production when the hairpin is combined with other hairpins in a polycistronic transcript (Figure 6C). The Dr/t5 inhibitor is expressed individually and does not benefit from chaining to other hairpins (Figure 6D), which correlates nicely with the luciferase results (Figure 5B). For inhibitor Eldr9, we observed increased siRNA levels for ACDE compared to the single hairpin (Figure 6E). The combined luciferase inhibition results and the Northern blot analyses demonstrate that the silencing activity of an individual hairpin RNA can be significantly enhanced when expressed in a polycistronic transcript.
Figure 6.
Northern blot analyses of siRNAs derived from antiviral miRNA constructs. 293T cells were transfected with the indicated 1, 2, 3 and 4 antiviral miRNA constructs and siRNAs were detected in total cellular RNA using 19-nt complementary LNA oligonucleotide probes. Different miRNA transcripts were analysed: (A) Apol47, (B) Bpol1, (C) Cgag5, (D) Dr/t5 and (E) Eldr9. As negative controls, pBS or miRNA constructs targeting another gene were used. The original shRNA constructs were used as positive controls, showing the mature siRNAs. Ethidium bromide staining of 5S rRNA serves as sample loading control. M, RNA size marker (nt) is shown on the left, the probe used, is shown on the right.
HIV-1 replication is suppressed in cells stably expressing an antiviral miRNA polycistron
We next created stably transduced SupT1 cells with a lentiviral vector (pLV) expressing the individual miRNA constructs Apol47, Bpol1, Cgag5, Dr/t5, Eldr9 and the polycistronic ACDE construct. pLV expressing the control miRNA N (Invitrogen) was used as negative control. To study the impact of the antiviral miRNAs on SupT1 cell viability, we set up a sensitive toxicity screen for cells transduced with N, Apol47 and ACDE. We cultured the cells for 36 days after lentiviral transduction and followed the percentage of GFP+ (transduced) and GFP- (untransduced) cells by FACS (Table 2). We did not observe a decrease in the fraction of transduced cells, indicating a similar growth rate as untransduced cells.
Table 2.
Toxicity screen of antiviral miRNA constructs
| Construct | Percentage of GFP + cells after transduction | |||
|---|---|---|---|---|
| Day 0 | Day 4 | Day 20 | Day 36 | |
| pLV | 0 | 0 | 0 | 0 |
| N | 2.8 | 3 | 1.5 | 4.5 |
| Apol47 | 6 | 6 | 6 | 4.8 |
| ACDE | 2.4 | 3 | 1.5 | 2.9 |
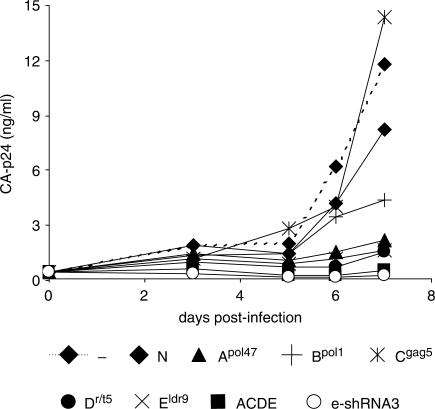
The stably transduced SupT1 cells were selected by FACS sorting and subsequently infected with HIV-1 (0.5 ng of CA-p24). Virus replication was followed for 7 days by measuring the CA-p24 level in the culture supernatant. Fast virus replication and virus-induced cytopathic effects were observed in cells expressing miRNAs N, Bpol1, Cgag5 and untransduced SupT1 cells (Figure 7). HIV-1 replication was inhibited by the individual hairpins Apol47, Dr/t5 and Eldr9. Virus replication was profoundly inhibited in SupT1 cells expressing the polycistronic miRNA construct ACDE and control cells that express an extended triple shRNA construct (e-shRNA 3) (manuscript in preparation).
Figure 7.
Inhibition of HIV-1 replication in SupT1 cells transduced with lentiviral vectors expressing antiviral miRNAs Apol47, Bpol1, Cgag5, Dr/t5, Eldr9 and ACDE. Untransduced SupT1 cells and cells transduced with the unrelated miRNA N were used as negative controls. Cells expressing an extended triple shRNA construct (e-shRNA 3) serve as positive control. SupT1 cells stably expressing miRNAs Apol47, Bpol1, Cgag5, Dr/t5, Eldr9 and ACDE were infected with HIV-1 and virus replication was monitored for 7 days by measuring CA-p24 in the culture supernatant. One representative experiment is shown, similar results were obtained in three independent experiments.
DISCUSSION
In this study, we designed a combinatorial RNAi approach against HIV-1 using the human mir-17-92 polycistron. We first constructed individual pri-miRNA transcripts against five conserved regions of HIV-1 under the control of a CMV promoter. We maintained the secondary structure of the original pre-miRNAs and included the single-stranded flanks because these are important for proper miRNA processing and subsequent RISC loading (12). We used the CMV promoter to express the transcripts because most primary miRNAs are transcribed by RNA polymerase II (7), which also allows inducible or tissue-specific miRNA expression (22,53,54). Previously, several reports have demonstrated effective gene knockdown in mammalian cells with siRNAs derived from miRNA precursors (17,18,22,53). However, none of these studies addressed the issue that a mature miRNA is typically 22–24 nt, whereas an siRNA is only 19-nt in length. Here, we demonstrate that positioning of 19–24 nt antiviral siRNA sequences at the 5′-end of the pre-miRNA hairpin stem results in optimal HIV-1 inhibition.
Despite the optimization of the miRNA-like inhibitors, their activity is less than that of the original shRNA antivirals that were used to design the miRNAs. We therefore addressed whether the miRNA-like inhibitor is correctly processed and compared the amount of siRNA produced from the miRNA versus shRNA constructs. The siRNA level produced by the shRNA construct shr/t5 is 15–30-fold higher than the Dr/t5 miRNA, which is likely due to the use of different promoters (RNA polymerase III versus RNA polymerase II). Interestingly, the shRNA inhibitor did only show marginally higher inhibitory activity, suggesting that the intrinsic inhibitory activity of the miRNA-like inhibitor is in fact much greater than the shRNA variant. Consistent with these results, vectors encoding hairpin structures that closely resemble a natural pre-miRNA produced ∼12-fold more mature siRNAs than vectors encoding simple hairpin structures (21). The superior activity of natural miRNA-like inhibitors is likely attributed by the intrinsic properties of a miRNA, which is processed in the nucleus by Drosha, exported to the cytoplasm, processed further by Dicer and loaded into RISC. In the case of an shRNA, the Drosha step is bypassed, which could provide a less-efficient entry into the RNAi pathway.
Moderate HIV-1 inhibitory activity was observed with constructs expressing a single miRNA hairpin. Interestingly, we demonstrate that co-expression of two or more hairpins in a single transcript greatly enhanced the silencing activity of each individual hairpin in the transcript. Northern blot analyses showed that the increased inhibitory activity correlates with higher siRNA expression levels. Multimerization of different miRNA hairpins is of particular interest for targeting of RNA viruses such as HIV-1 and HCV because of their extreme genetic diversity and potential for mutational escape. Two recent papers presented the potential of combinatorial RNAi using two miRNA-30 hairpins (55,56). In agreement with these studies, we showed an increase in RNAi activity upon multimerization of 2, 3, 4 or 6 hairpins in a single transcript. Another study used the miR-30 backbone to multiplex artificial miRNAs and reported decreased RNAi activity for the tandem plasmid, which may be due to miRNA processing problems (57). Thus, chaining of miRNAs needs a very careful transcript design. Most studies have used either the miR-30 or miR-155 backbone because these molecules have been studied extensively. Our study focuses on the natural mir-17-92 polycistron and we demonstrate that insertion of four antiviral siRNAs creates a transcript that is properly processed into functional antiviral miRNAs that effectively inhibit HIV-1 production and replication.
A major concern of the miRNA approach is the off-target effect on cellular transcripts with partial sequence complementarity. Such an off-target effect may require only a complementarity of 6–8 nt between the seed region of the miRNA and the target (58,59). Such a weak restraint results in numerous potential off-target genes for any miRNA. When multiple miRNAs are used, the number of potential off-targets will increase, increasing the chance of a negative effect on the treated cells. However, we observed no obvious cellular changes in a sensitive toxicity screen. Furthermore, the observed inhibition of firefly luciferase reporters clearly showed sequence-specificity as non-matching miRNAs did not have any effect and the expression of the control renilla reporter was not affected. Another study, in which artificial miRNAs were expressed in Arabidopsis thaliana, conferred viral resistance without cellular alterations, suggesting that off-target effects are not significant (60). Furthermore, recently evidence emerges that siRNA sequences inserted in a miRNA backbone do not compete for transport and incorporation into RISC, while competition was observed when the same siRNA sequences were presented as synthetic siRNAs or shRNAs (61). Nonetheless, off-targeting is a genuine concern for the development of any RNAi-based gene therapy against HIV-1 and the potential risk should be assessed properly in relevant in vivo models prior to an eventual clinical application (62).
We have shown in stably transduced T-cell lines that multiple effective miRNAs inhibit HIV-1 replication much stronger than a single miRNA. These data, together with our previous shRNA studies (32,63), indicate that a combinatorial RNAi approach against HIV-1 results in an increased magnitude of inhibition and consequently a restriction of viral escape. Current strategies combine multiple polymerase III shRNA expression cassettes (63), which results in high expression levels. This may not always be desired in a gene therapy setting because of increased toxicity due to saturation of the RNAi machinery with siRNAs (46). The use of polymerase II promoters to express the miRNA polycistron will reduce this risk because of lower expression levels. In addition, polymerase II cassettes allow expression in a tissue-specific manner and inducible gene expression, which increases the flexibity for gene therapy and functional genomic applications (22,54).
For HIV-1 gene therapy applications the use of a hematopoietic or T-cell-specific promoter can increase the target cell specificity. An interesting candidate is the WAS promoter that is active in human hematopoietic precursor cells (CD34+) and T lymphocytes, B cells and dendritic cells (64). Another intriguing possibility is to use the HIV-1 LTR promoter to express the miRNA polycistron. Transcriptional activation of the HIV-1 LTR requires the viral Tat protein, which is produced only in HIV-1-infected cells, thereby allowing exquisite target cell specificity. This approach has previously been employed for shRNA expression (65). Thus, several approaches can be tested experimentally in order to optimize the antiviral miRNA polycistron strategy for HIV-1 inhibition.
In summary, one can effectively combat HIV-1 with multiple miRNA effector molecules transcribed from a single polycistronic transcript. We showed that expression of the miRNA polycistron results in the production of functional mature miRNAs that can efficiently and selectively inhibit HIV-1. Further optimization of this construct by increasing the target cell specificity and inducibility will be a further step towards a gene therapeutic approach against HIV-1.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
HIV-1 RNA research in the Berkhoutlab is sponsored by ZonMw (VICI grant) and NWO-CW (TOP grant). We thank Stephan Heynen for performing CA-p24 ELISA and Jens Gruber for useful discussions. Funding to pay the Open Access publication charges for this article was provided by ZonMw and NWO-CW.
Conflict of interest statement. None declared.
REFERENCES
- 1.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- 3.Zamore PD, Tuschl T, Sharp PA, Bartel DP. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- 4.Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- 5.Hammond SM, Caudy AA, Hannon GJ. Post-transcriptional gene silencing by double-stranded RNA. Nat. Rev. Genet. 2001;2:110–119. doi: 10.1038/35052556. [DOI] [PubMed] [Google Scholar]
- 6.Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 10.Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 11.Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeng Y, Yi R, Cullen BR. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. EMBO J. 2005;24:138–148. doi: 10.1038/sj.emboj.7600491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cullen BR. Transcription and processing of human microRNA precursors. Mol. Cell. 2004;16:861–865. doi: 10.1016/j.molcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 14.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat. Rev. Mol Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 15.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suh MR, Lee Y, Kim JY, Kim SK, Moon SH, Lee JY, Cha KY, Chung HM, Yoon HS, Moon SY, et al. Human embryonic stem cells express a unique set of microRNAs. Dev. Biol. 2004;270:488–498. doi: 10.1016/j.ydbio.2004.02.019. [DOI] [PubMed] [Google Scholar]
- 17.Boden D, Pusch O, Silbermann R, Lee F, Tucker L, Ramratnam B. Enhanced gene silencing of HIV-1 specific siRNA using microRNA designed hairpins. Nucleic Acids Res. 2004;32:1154–1158. doi: 10.1093/nar/gkh278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeng Y, Wagner EJ, Cullen BR. Both natural and designed micro RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol. Cell. 2002;9:1327–1333. doi: 10.1016/s1097-2765(02)00541-5. [DOI] [PubMed] [Google Scholar]
- 19.Zeng Y, Yi R, Cullen BR. MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc. Natl Acad. Sci. USA. 2003;100:9779–9784. doi: 10.1073/pnas.1630797100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeng Y, Cai X, Cullen BR. Use of RNA polymerase II to transcribe artificial microRNAs. Methods Enzymol. 2005;392:371–380. doi: 10.1016/S0076-6879(04)92022-8. [DOI] [PubMed] [Google Scholar]
- 21.Silva JM, Li MZ, Chang K, Ge W, Golding MC, Rickles RJ, Siolas D, Hu G, Paddison PJ, Schlabach MR, et al. Second-generation shRNA libraries covering the mouse and human genomes. Nat. Genet. 2005;37:1281–1288. doi: 10.1038/ng1650. [DOI] [PubMed] [Google Scholar]
- 22.Stegmeier F, Hu G, Rickles RJ, Hannon GJ, Elledge SJ. A lentiviral microRNA-based system for single-copy polymerase II-regulated RNA interference in mammalian cells. Proc. Natl Acad. Sci. USA. 2005;102:13212–13217. doi: 10.1073/pnas.0506306102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dickins RA, Hemann MT, Zilfou JT, Simpson DR, Ibarra I, Hannon GJ, Lowe SW. Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nat. Genet. 2005;37:1289–1295. doi: 10.1038/ng1651. [DOI] [PubMed] [Google Scholar]
- 24.Haasnoot PCJ, Cupac D, Berkhout B. Inhibition of virus replication by RNA interference. J. Biomed. Sci. 2003;10:607–616. doi: 10.1159/000073526. [DOI] [PubMed] [Google Scholar]
- 25.Haasnoot PCJ, Berkhout B. Handbook of Experimental Pharmacology. Vol. 173. Berlin, Heidelberg: Springer-Verlag; 2006. RNA interference: its use as antiviral therapy; pp. 117–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anderson J, Banerjea A, Akkina R. Bispecific short hairpin siRNA constructs targeted to CD4, CXCR4, and CCR5 confer HIV-1 resistance. Oligonucleotides. 2003;13:303–312. doi: 10.1089/154545703322616989. [DOI] [PubMed] [Google Scholar]
- 27.Das AT, Brummelkamp TR, Westerhout EM, Vink M, Madiredjo M, Bernards R, Berkhout B. Human immunodeficiency virus type 1 escapes from RNA interference-mediated inhibition. J. Virol. 2004;78:2601–2605. doi: 10.1128/JVI.78.5.2601-2605.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lo HL, Chang T, Yam P, Marcovecchio PM, Li S, Zaia JA, Yee JK. Inhibition of HIV-1 replication with designed miRNAs expressed from RNA polymerase II promoters. Gene Ther. 2007;14:1503–1512. doi: 10.1038/sj.gt.3303011. [DOI] [PubMed] [Google Scholar]
- 29.Boden D, Pusch O, Lee F, Tucker L, Ramratnam B. Human immunodeficiency virus type 1 escape from RNA interference. J. Virol. 2003;77:11531–11535. doi: 10.1128/JVI.77.21.11531-11535.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Westerhout EM, Ooms M, Vink M, Das AT, Berkhout B. HIV-1 can escape from RNA interference by evolving an alternative structure in its RNA genome. Nucleic Acids Res. 2005;33:796–804. doi: 10.1093/nar/gki220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berkhout B. RNA interference as an antiviral approach: targeting HIV-1. Curr. Opin. Mol. Ther. 2004;6:141–145. [PubMed] [Google Scholar]
- 32.Ter Brake O, Konstantinova P, Ceylan M, Berkhout B. Silencing of HIV-1 with RNA interference: a multiple shRNA approach. Mol. Ther. 2006;14:883–892. doi: 10.1016/j.ymthe.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 33.Lisziewicz J, Sun D, Klotman M, Agrawal S, Zamecnik P, Gallo R. Specific inhibition of human immunodeficiency virus type 1 replication by antisense oligonucleotides: an in vitro model for treatment. Proc. Natl Acad. Sci. USA. 1992;89:11209–11213. doi: 10.1073/pnas.89.23.11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bertrand E, Pictet R, Grange T. Can hammerhead ribozymes be efficient tools to inactivate gene function? Nucleic Acids Res. 1994;22:293–300. doi: 10.1093/nar/22.3.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohkawa J, Yuyama N, Takebe Y, Nishikawa S, Taira K. Importance of independence in ribozyme reactions: kinetic behavior of trimmed and of simply connected multiple ribozymes with potential activity against human immunodeficiency virus. Proc. Natl Acad. Sci. USA. 1993;90:11302–11306. doi: 10.1073/pnas.90.23.11302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ter Brake O, Berkhout B. Lentiviral vectors that carry anti-HIV shRNAs: problems and solutions. J. Gene Med. 2007;9:743–750. doi: 10.1002/jgm.1078. [DOI] [PubMed] [Google Scholar]
- 37.Ter Brake O, Berkhout B. A novel approach for inhibition of HIV-1 by RNA interference: counteracting viral escape with a second generation of siRNAs. J. RNAi Gene Silencing. 2005;1:56–65. [PMC free article] [PubMed] [Google Scholar]
- 38.Liu YP, Haasnoot J, Berkhout B. Design of extended short hairpin RNAs for HIV-1 inhibition. Nucleic Acids Res. 2007;35:5683–5693. doi: 10.1093/nar/gkm596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Konstantinova P, de Vries W, Haasnoot J, Ter Brake O, de Haan P, Berkhout B. Inhibition of human immunodeficiency virus type 1 by RNA interference using long-hairpin RNA. Gene Ther. 2006;13:1403–1413. doi: 10.1038/sj.gt.3302786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Novina CD, Murray MF, Dykxhoorn DM, Beresford PJ, Riess J, Lee SK, Collman RG, Lieberman J, Shankar P, Sharp PA. siRNA-directed inhibition of HIV-1 infection. Nat. Med. 2002;8:681–686. doi: 10.1038/nm725. [DOI] [PubMed] [Google Scholar]
- 41.Martinez MA, Gutierrez A, Armand-Ugon M, Blanco J, Parera M, Gomez J, Clotet B, Este JA. Suppression of chemokine receptor expression by RNA interference allows for inhibition of HIV-1 replication. AIDS. 2002;16:2385–2390. doi: 10.1097/00002030-200212060-00002. [DOI] [PubMed] [Google Scholar]
- 42.Zhou N, Fang J, Mukhtar M, Acheampong E, Pomerantz RJ. Inhibition of HIV-1 fusion with small interfering RNAs targeting the chemokine coreceptor CXCR4. Gene Ther. 2004;11:1703–1712. doi: 10.1038/sj.gt.3302339. [DOI] [PubMed] [Google Scholar]
- 43.Jacque JM, Stevenson M. The inner-nuclear-envelope protein emerin regulates HIV-1 infectivity. Nature. 2006;441:641–645. doi: 10.1038/nature04682. [DOI] [PubMed] [Google Scholar]
- 44.Nguyen DG, Wolff KC, Yin H, Caldwell JS, Kuhen KL. “UnPAKing” human immunodeficiency virus (HIV) replication: using small interfering RNA screening to identify novel cofactors and elucidate the role of group I PAKs in HIV Infection. J. Virol. 2006;80:130–137. doi: 10.1128/JVI.80.1.130-137.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qu J, Ye J, Fang R. Artificial microRNA-mediated virus resistance in plants. J. Virol. 2007;81:6690–6699. doi: 10.1128/JVI.02457-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, Marion P, Salazar F, Kay MA. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 47.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peden K, Emerman M, Montagnier L. Changes in growth properties on passage in tissue culture of viruses derived from infectious molecular clones of HIV-1LAI, HIV-1MAL, and HIV-1ELI. Virology. 1991;185:661–672. doi: 10.1016/0042-6822(91)90537-l. [DOI] [PubMed] [Google Scholar]
- 49.Jeeninga RE, Hoogenkamp M, Armand-Ugon M, de Baar M, Verhoef K, Berkhout B. Functional differences between the long terminal repeat transcriptional promoters of HIV-1 subtypes A through G. J. Virol. 2000;74:3740–3751. doi: 10.1128/jvi.74.8.3740-3751.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ruijter JM, Thygesen HH, Schoneveld OJ, Das AT, Berkhout B, Lamers WH. Factor correction as a tool to eliminate between-session variation in replicate experiments: application to molecular biology and retrovirology. Retrovirology. 2006;3:1–8. doi: 10.1186/1742-4690-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kotsopoulou E, Kim VN, Kingsman AJ, Kingsman SM, Mitrophanous KA. A Rev-independent human immunodeficiency virus type 1 (HIV-1)-based vector that exploits a codon-optimized HIV-1 gag-pol gene. J. Virol. 2000;74:4839–4852. doi: 10.1128/jvi.74.10.4839-4852.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zufferey R, Dull T, Mandel RJ, Bukovsky A, Quiroz D, Naldini L, Trono D. Self-inactivating lentivirus vector for safe and efficient in vivo gene delivery. J. Virol. 1998;72:9873–9880. doi: 10.1128/jvi.72.12.9873-9880.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chung KH, Hart CC, Al Bassam S, Avery A, Taylor J, Patel PD, Vojtek AB, Turner DL. Polycistronic RNA polymerase II expression vectors for RNA interference based on BIC/miR-155. Nucleic Acids Res. 2006;34:e53. doi: 10.1093/nar/gkl143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shin KJ, Wall EA, Zavzavadjian JR, Santat LA, Liu J, Hwang JI, Rebres R, Roach T, Seaman W, Simon MI, et al. A single lentiviral vector platform for microRNA-based conditional RNA interference and coordinated transgene expression. Proc. Natl Acad. Sci. USA. 2006;103:13759–13764. doi: 10.1073/pnas.0606179103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sun D, Melegari M, Sridhar S, Rogler CE, Zhu L. Multi-miRNA hairpin method that improves gene knockdown efficiency and provides linked multi-gene knockdown. Biotechniques. 2006;41:59–63. doi: 10.2144/000112203. [DOI] [PubMed] [Google Scholar]
- 56.Xia XG, Zhou H, Xu Z. Multiple shRNAs expressed by an inducible pol II promoter can knock down the expression of multiple target genes. Biotechniques. 2006;41:64–68. doi: 10.2144/000112198. [DOI] [PubMed] [Google Scholar]
- 57.Zhou H, Xia XG, Xu Z. An RNA polymerase II construct synthesizes short-hairpin RNA with a quantitative indicator and mediates highly efficient RNAi. Nucleic Acids Res. 2005;33:e62. doi: 10.1093/nar/gni061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 59.Birmingham A, Anderson EM, Reynolds A, Ilsley-Tyree D, Leake D, Fedorov Y, Baskerville S, Maksimova E, Robinson K, Karpilow J, et al. 3′ UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat. Methods. 2006;3:199–204. doi: 10.1038/nmeth854. [DOI] [PubMed] [Google Scholar]
- 60.Niu QW, Lin SS, Reyes JL, Chen KC, Wu HW, Yeh SD, Chua NH. Expression of artificial microRNAs in transgenic Arabidopsis thaliana confers virus resistance. Nat. Biotechnol. 2006;24:1420–1428. doi: 10.1038/nbt1255. [DOI] [PubMed] [Google Scholar]
- 61.Castanotto D, Sakurai K, Lingeman R, Li H, Shively L, Aagaard L, Soifer H, Gatignol A, Riggs A, Rossi JJ. Combinatorial delivery of small interfering RNAs reduces RNAi efficacy by selective incorporation into RISC. Nucleic Acids Res. 2007;35:5154–5164. doi: 10.1093/nar/gkm543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Haasnoot J, Westerhout EM, Berkhout B. RNA interference against viruses: strike and counterstrike. Nat. Biotechnol. 2007;25:1435–1443. doi: 10.1038/nbt1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ter Brake O, 't Hooft K, Liu YP, Centlivre M, von Eije KJ, Berkhout B. Lentiviral vector design for multiple shRNA expression and durable HIV-1 inhibition. Mol. Ther. 2008;16:557–564. doi: 10.1038/sj.mt.6300382. [DOI] [PubMed] [Google Scholar]
- 64.Charrier S, Dupre L, Scaramuzza S, Jeanson-Leh L, Blundell MP, Danos O, Cattaneo F, Aiuti A, Eckenberg R, Thrasher AJ, et al. Lentiviral vectors targeting WASp expression to hematopoietic cells, efficiently transduce and correct cells from WAS patients. Gene Ther. 2007;14:415–428. doi: 10.1038/sj.gt.3302863. [DOI] [PubMed] [Google Scholar]
- 65.Unwalla HJ, Li MJ, Kim JD, Li HT, Ehsani A, Alluin J, Rossi JJ. Negative feedback inhibition of HIV-1 by TAT-inducible expression of siRNA. Nat. Biotechnol. 2004;22:1573–1578. doi: 10.1038/nbt1040. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.