Abstract
Transcription in eukaryotes is a multistep process involving the assembly and disassembly of numerous inter- and intramolecular interactions between transcription factors and nucleic acids. The roles of each of these interactions and the regions responsible for them have been identified and studied primarily by the use of mutants, which destroy the inherent properties of the interacting surface. A less intrusive but potentially effective way to study the interactions as well as the surfaces responsible for them is the use of RNA aptamers that bind to the interacting factors. Here, we report the isolation and characterization of high-affinity RNA aptamers that bind to the yeast general transcription factor TFIIB. These aptamers fall into two classes that interfere with TFIIB's interactions with either TBP or RNA polymerase II, both of which are crucial for transcription in yeast. We demonstrate the high affinity and specificity of these reagents, their effect on transcription and preinitiation complex formation and discuss their potential use to address mechanistic questions in vitro as well as in vivo.
INTRODUCTION
Cellular architecture and function depend critically upon a myriad of specific macromolecular interactions. These include contacts involving proteins with other proteins, nucleic acids and small molecules. Protein function has been traditionally analyzed by inactivating or inhibiting the protein itself using conditional and other mutations, small molecule inhibitors (drugs), and RNAi. Since many proteins have distinct surfaces that are responsible for independent interactions, the ability to interfere with these regions individually is extremely valuable in evaluating the role of specific macromolecular interactions. Aptamers, which are single-stranded oligonucleotides that bind with high affinity and specificity to a target molecular surface, are well suited for this purpose (1). Aptamers are a powerful tool for mechanistic experiments because (i) they do not irreversibly alter the protein they bind, (ii) they interact with and sterically block a specific surface on the protein in much the same way as a natural interacting partner would, and (iii) they can be expressed in a rapid yet inducible manner in vivo, thus avoiding secondary (plieotropic) effects and allowing kinetic analysis of the consequences of the perturbation.
RNA aptamers are isolated by a process of iterative selection called SELEX (systematic evolution of ligands by exponential enrichment), from a large combinatorial starting library (2,3). The process involves repeated cycles of binding to the target, partitioning and amplification of ‘binders’, until high-affinity aptamers are isolated. Aptamers have been generated to bind a wide variety of targets such as small organic molecules, peptides, proteins, cells and even virus particles (4). They have the high affinity and specificity that is desired in drugs. Additionally, they can be efficiently and cost effectively mass-produced as well as expressed in vivo in a functional form in several model organisms (5).
Transcription initiation in yeast is an orchestrated event that involves interactions of hundreds of polypeptides that each has to be recruited to the promoter in a coordinated and timely manner (6). The process starts with the assembly of the preinitiation complex (PIC) at the promoter (7). TATA-binding protein (TBP) binds to the promoter element, often as part of a bigger complex such as TFIID or SAGA. This binding is stabilized by two general transcription factors (GTFs), TFIIA and TFIIB. TFIIB properly orients the TBP–DNA complex and facilitates the recruitment of the RNA polymerase (Pol) II–TFIIF complex by directly binding to Pol II (8). PIC formation is complete with the entry of TFIIE and TFIIH, factors involved in promoter melting and open complex formation.
Once the PIC is assembled, DNA at the promoter is melted, an open complex is formed and transcription initiates. Upon transition into productive elongation, several of the contacts established during PIC formation are lost and new ones are made with elongation factors that are recruited to the now elongating Pol II (9). Thus, the entire process comprises a dynamic landscape of interactions between the players involved. The general transcription factors are at the heart of this interactome, with each GTF forming a ‘node’, in contact with multiple other factors. Our goal is to use RNA aptamers to disable each of these ‘connections’ and thus decipher the contribution of that interaction in the process of initiation.
In our past reports, we have described the selection of two classes of RNA aptamers that bind to yeast TBP, where one class interferes with the TBP–DNA interaction and the other interferes with the TBP–TFIIA interaction (10,11). We have also shown that while the loss of either molecular interaction surface blocks RNA synthesis in vitro in extracts, PIC formation is affected in mechanistically distinct ways (10). In this study, our focus is the general transcription factor TFIIB. TFIIB in yeast consists of several domains: the N-terminal Zn ribbon domain interacts with the dock region of Pol II; the B finger domain inserts itself into the RNA exit channel and is in direct contact with the active site of Pol II, wherein it influences start site selection along with Pol II and TFIIF; the C terminal core domain consists of two imperfect direct repeats that are responsible for TBP and DNA binding (8,12). In addition to these contacts, TFIIB is also thought to be a direct target of some acidic activators and interacts with a Pol II C-terminal domain phosphatase, Ssu72 (13,14). TFIIB is therefore central to PIC assembly, participates in several crucial interactions and is in direct contact with TBP.
In this article, we report the isolation and characterization of aptamers that bind with high affinity and specificity to yeast TFIIB. We demonstrate that these aptamers are potent transcription inhibitors in vitro that block PIC assembly in a manner distinct from each other and either of the TBP aptamers. We anticipate that the TBP and TFIIB aptamers will serve as specific inhibitors to address questions concerning basic transcription mechanisms that have been difficult to address with existing reagents.
MATERIALS AND METHODS
Plasmid constructs
The deletion constructs of yeast TFIIB (except ΔB Finger) were made by amplifying the appropriate regions from the full-length clone by PCR using primers that added an NcoI site and a 6-His tag on the N-terminal end and an XhoI site on the C-terminal end. For the ΔB Finger construct, the DNA fragment encoding the Zn ribbon domain was amplified with the addition of 5′ NcoI and 3′ NarI sites and the DNA fragment encoding the core domain was amplified with the addition of 5′ NarI and 3′ XhoI sites. These were then digested with NarI and ligated first and subsequently digested with NcoI and XhoI to insert into pET16b. The PCR products were then inserted into the bacterial expression pET16b using the NcoI and XhoI (the nucleotide positions for each of the deletion constructs are as follows: Core, 358–1035 bp; ΔZn, 165–1035 bp; ΔB, 1–165 + 358–1035; NTD, 1–267 bp).
The heart muscle kinase (HMK) tag was inserted at the C-terminal end of the core IIB construct described earlier. First, PCR was carried out using the DNA fragment encoding core IIB with a forward primer containing the NcoI site and a 6-His tag and a reverse primer that did not include the Stop codon. This was followed by a second PCR with the same forward primer and a reverse primer that added the HMK sequence, a Stop codon and an XhoI site. This fragment was then inserted into pET16b as described earlier. The sequence for the HMK tag was obtained from (15). The constructs for TFIIB mutants K190E, K201E, K205E and the double mutants K190E/K201E, K201E/K205E were a gift from Dr A. Ponticelli.
Protein purification
Full-length His-tagged versions of yeast TFIIB, TBP, the deletion constructs of TFIIB and HMK-tagged core IIB were all purified from BL21-DE3 cells according to a standard His-tagged protein purification protocol. The single- and double-point mutants were purified from JM109 cells by the same protocol. Yeast TFIIA was purified by using a protocol obtained from S. Hahn (Fred Hutchinson Cancer Research Center, Seattle, USA), described earlier (11).
In vitro evolution
SELEX was carried out as described earlier (11). The template DNA library (previously described in ref. 16) contains on the order of 2 × 1015 sequences. It consists of a central 50-bp long randomized region flanked by two constant regions that contain the 5′T7 promoter and facilitate amplification by PCR. RNA was made from this DNA library by in vitro transcription with T7 RNAP and eight rounds of selection were carried out with bacterially expressed yeast TFIIB protein. For each cycle, the RNA protein mixture was incubated in 1× binding buffer (12 mM HEPES, pH 7.9, 150–200 mM NaCl, 1–10 mM MgCl2, 1 mM DTT), partitioned using a nitrocellulose filter, the bound RNA recovered by extraction with hot phenol and amplified to yield an enriched pool for the next cycle. The concentrations of NaCl, MgCl2 and TFIIB were decreased gradually with each cycle to increase the stringency of the selection. Negative selection (without protein) was included with every cycle after the third. The final DNA pool was cloned into the pSTBlue-1 vector and 70 individual clones were sequenced.
Electrophoretic mobility shift (EMS) assay
RNA-binding experiments were carried using radiolabeled RNA synthesized by internally radiolabeling RNA (with α-P32 UTP) during in vitro transcription by T7 RNA polymerase from the template DNA, using the Ambion MegaShortscript kit. Radiolabeled RNA, present at a final concentration of 0.5 nM (0.2 nM in Figure 1B) was incubated with indicated amounts of protein in 1× B binding buffer (12 mM HEPES, pH 7.9, 150 mM NaCl, 2 mM MgCl2, 1 mM DTT, 5 ng/μl yeast tRNA, 0.1 μg/μl BSA, 10% glycerol) for 30 min at room temperature and loaded on a native polyacrylamide gel (0.5× TBE, 6% acrylamide with 1.5 mM MgCl2 and 1 mM DTT). Complexes were resolved at 180 V for 2 h, gels were dried and exposed to a phosphorimager screen overnight. The images were scanned and analyzed with the ImageQuant (5.2) software. EMS assays with labeled TATA DNA were carried out as described in ref. (11). Labeled TATA DNA was present at 0.5 nM, TBP at 20 nM, TFIIA at 0.5 nM, TFIIB at 200 nM and cold aptamer RNAs or yeast tRNA at 800 nM. For the reverse bandshift experiment in Figures 1C and S4, the HMK-tagged core IIB protein was radiolabeled using HMK (see the subsequent section: ‘protease protection assay’). The binding experiment was carried out in 1× B binding buffer with 70 nM core IIB and 700 nM of indicated cold aptamer RNA. The mixture was incubated for 30 min at room temperature, resolved on a 0.5 × TBE, 6% native PAGE with 1.5 mM MgCl2, and processed as described for the experiments using radiolabeled RNA.
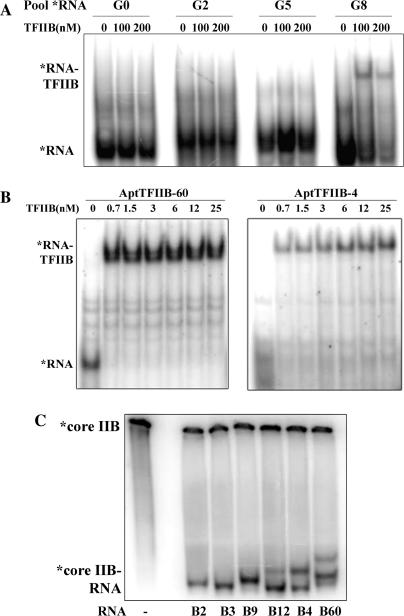
Figure 1.
RNA aptamers to TFIIB selected from a starting pool of 2 × 1015 sequences. (A) Band shift assay with labeled RNA obtained from the starting pool (G0), and after two (G2), five (G5) and eight (G8) cycles of SELEX was performed with increasing concentrations of yeast TFIIB (0, 100, 200 nM). (B) Band shift assay with labeled candidate aptamers B60 and B4 was carried out using indicated amounts of recombinant TFIIB protein. The KD for binding was determined to be on the order of 1 nM or less. (C) Band shift assay carried out with radiolabeled core domain of TFIIB (70 nM) and indicated cold aptamer RNA (700 nM).
In vitro transcription
The in vitro transcription experiments were carried out as described in ref. (17). Yeast whole-cell extract was made from W3031A. A plasmid template pSH515-Gless was used which contained one Gal4-binding site, the HIS4 core promoter (−141 to +3) and a 390-nt G-less cassette [made by insertion of the G-less cassette from pG5MLT into pSH515 (9) downstream of the promoter sequence]. The aptamers were added at 50 nM along with the extract and transcription was allowed to proceed for 30 min for assaying multiple-round transcription. For the single-round experiments, aptamers were added at 50 nM after PIC formation, transcription was initiated by the addition of NTPs for 45 s, and transcription was stopped by the addition of heparin. For the TFIIB-rescue experiments, aptamers at 30 nM and excess TFIIB at 300 nM were added along with the template to the extract.
Immobilized template assay
These experiments were carried out as described in ref. (10). The wash buffer included nonspecific competitor DNA at 10 ng/μl and NP-40 at 0.1% final concentration.
Protease protection assay
C-terminally HMK-tagged core IIB protein (0.3 nmol) was radiolabeled using 2-μl HMK (5 mg/ml), 5 μl γ-P32ATP (16.5 pmol) in 30 μl 1× B binding buffer for 1 h at 37°C. The unincorporated ATP was removed by purification on a P6 column (BioRAD, Hercules, CA). Labeled core IIB measuring 7.8 pmol was incubated with 78 pmol of yeast tRNA (control), AptIIB-60 or AptIIB-4 RNA in 1× B binding buffer for 15 min to allow RNA–protein complexes to form. Partial protease digestion was then carried out by addition of 300 ng, 100 ng, 30 ng or 10 ng of chymotrypsin to the mixture in a final volume of 10 μl. After a 7-min incubation, PMSF was added to a final concentration of 10 mM to stop the protease digestion, SDS loading dye was added, the samples were boiled at 95°C for 3 min and resolved on a 15% Tricine–SDS–PAGE overnight at 4°C (18). The gel was dried and exposed overnight on a phosphorimager screen. Analysis of the image was done using ImageQuant (5.2) software.
RESULTS
Selection of RNA aptamers to yeast TFIIB
To isolate high-affinity RNA aptamers that bound to yeast TFIIB (IIB), we screened a randomized DNA library that contains on the order of 2 × 1015 random sequences [described previously, (11,16)] by SELEX. Briefly, for each round of selection, the template DNA was transcribed using T7 RNA polymerase to generate an RNA pool, which was incubated with the bacterially expressed and purified TFIIB protein. The protein-bound RNA was partitioned away from the free RNA by nitrocellulose filtration and RNA was recovered from filter-bound complexes by extraction with hot phenol. RNAs were reverse transcribed and amplified to yield an enriched DNA pool to be used in the subsequent round of selection. Affinity of the pool RNA for TFIIB was monitored by a band shift assay with labeled RNA after every third round of selection (Figure 1A). After eight rounds of selection, a significant fraction of the RNA pool bound to IIB, the enriched DNA pool was cloned, and 70 randomly picked clones were sequenced to identify individual candidates.
The 43 unique candidates identified were aligned by the ClustalW multiple sequence alignment program (19) and classified into 12 families based on sequence information. We tested one representative member of each family for binding to TFIIB protein and nine out of the 12 showed strong TFIIB binding activity (Supplementary Data, Figure S1). In addition, we performed EMS assays with each of the candidate aptamers to determine their affinity to IIB. Figure 1B shows representative EMS experiments with two radiolabeled aptamers, AptTFIIB-4 and AptTFIIB-60. There is a single shifted band in the IIB containing lanes that corresponds to the aptamer RNA–TFIIB complex, and the KD calculated from the binding curve is on the order of 1 nM as measured by this assay (Figure S2). We also determined that the aptamer protein interaction is stable for >30 min by carrying out a similar EMS experiment in the presence of excess cold aptamer RNA (Figure S3).
In order to assess whether the aptamers formed conformationally distinct complexes with IIB, we performed a band shift assay with the radiolabeled protein. The core domain of TFIIB (residues 124–345) (12), was C-terminally tagged with the HMK recognition site and radiolabeled using recombinant HMK (Sigma, St. Loius, MO). The labeled protein was incubated with the indicated cold RNA aptamer in molar excess and loaded onto a native gel (Figure 1C). Since the RNA is not limiting in this assay, the sensitivity is higher and all forms of RNA–protein complexes are detected. In addition, the unbound protein is positively charged at the pH of the gel (7.5) and therefore remains at the well unless bound to RNA. This allows for the gel to be run much longer and the distinct complexes are more resolved relative to a conventional band shift assay performed with labeled RNA. All of the RNA aptamers shown in Figure 1C are precisely the same length (100 nt) except for AptTFIIB-2, which is 10-nt shorter (90 nt); however, complexes with distinct mobilities appear to be formed with some of these aptamers. The overall shape of each aptamer–TFIIB complex is determined to a large extent by the conformation(s) adopted by the RNA molecule since the protein component is the same. Therefore, the differences in mobilities observed could be attributed to the differences between aptamers in their secondary and tertiary structures. However, complexes containing different RNA aptamers appear to have nearly identical mobility on the native gel (B2, B3 and B9; B4 and B12). On the other hand, complexes formed with B60 are distinct from those with B4 and B12, and from the rest. This experiment serves as a demonstration of a complementary approach to characterizing aptamer–protein complexes by EMSA, which provides information that is not obtained in a normal *RNA EMSA. In previous studies with TBP aptamers from our lab [(10) and our unpublished data], we have observed that aptamers that form complexes with distinct mobilities on native gels (i) often bind to discrete surfaces on the target and (ii) have disparate inhibitory characteristics in functional assays. Taken together with Figure 1C, these results lend support to the possibility that these reagents may exert different effects on various TFIIB interactions and hence affect TFIIB function in transcription in mechanistically diverse ways.
Effect of TFIIB aptamers on the TFIIB–TBP interaction
In eukaryotic mRNA transcription, TBP nucleates the assembly of PIC formation by binding to DNA at the promoter (6). TFIIB is then thought to bind and stabilize the TBP–DNA interaction as well as facilitate Pol II recruitment via its N-terminal domain (20). Because the contacts with TBP and Pol II are functionally the most important of TFIIB's interactions, we set out to determine if the TFIIB aptamers interfered with either of these interactions.
We tested the TFIIB aptamers both for their abilities to interfere with the formation of a TFIIB–TFIIA–TBP–TATA (DAB) complex assembled in vitro and to disrupt a preformed complex. Native gel conditions that are required to visualize the TFIIB–TBP–DNA (DB) complex do not resolve the TBP–DNA complex as distinct from the larger complex; therefore, TFIIA, which stabilizes the TBP–DNA interaction but does not appear to affect the TFIIB–TBP interaction directly in any manner (21), was included to overcome this technical limitation (the TFIIB aptamers do not bind to TFIIA or to TBP, data not shown). First, the direct ‘competition’ experiment was designed to reveal if the aptamer interfered with the formation of the quaternary complex (DAB). The reaction was allowed to proceed to equilibrium and complexes were resolved by native PAGE. Figure 2A shows that in the presence of a molar excess of cold RNA aptamer (over IIB), some of the aptamers interfere with formation of the DAB complex but not the DA complex. These results indicate that these aptamers are able to compete with TBP for binding to IIB. All of the tested aptamers except AptTFIIB-4, 9, 12 and 27, and the control tRNA, were able to compete to a large extent. Based on their ability to, or not to, interfere with the TBP–TFIIB interaction we divided these RNA aptamers into two functionally distinct classes.
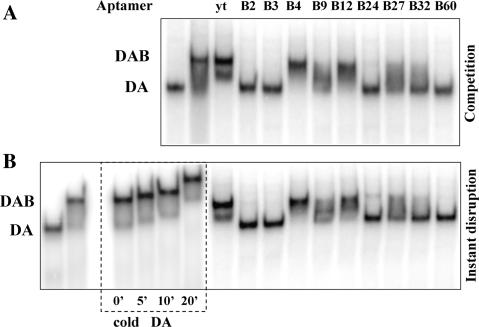
Figure 2.
The TFIIB aptamers fall into two classes based on their ability to interfere with TFIIB–TBP interaction. (A) Complexes were assembled on labeled TATA DNA with yeast TBP (20 nM), TFIIA (0.5 nM) and TFIIB (200 nM) in the presence or absence of the TFIIB aptamers (800 nM). Class I aptamers (B2, B60, etc.) compete effectively and prevent formation of the TATA–TBP–TFIIA–TFIIB (DAB) complex, whereas Class II aptamers (B4, etc.) were not as efficient. The lighter band seen migrating slightly faster than the DAB complex in the ytRNA lane is most likely due to dissociation of the complex during electrophoresis. (B) The Class I aptamers instantaneously disrupt the preformed DAB complex by removing TFIIB (leaving the DA complex intact), whereas the Class II aptamers had no such effect. The ‘yt’ represents a yeast tRNA which is used as a control RNA. In the lanes marked by the box, excess cold DA was added to the DAB complex to verify that the DAB complex is stable for the time frame of the disruption and competition experiments.
The second assay was a ‘disruption’ experiment, which allowed us to assess whether the aptamers that do interfere with this interaction could only do so passively by competition or if they were able to actively dislodge the competitor from a preformed complex. DAB complexes were first assembled on TATA DNA and allowed to reach equilibrium. A molar excess of RNA aptamer or control yeast tRNA was added subsequently to the complex and the reaction was immediately loaded on a native gel and resolved. The stability of the DAB complex was also simultaneously assayed by incubating the complex in the presence of excess cold DA for increasing amounts of time. In this instantaneous ‘disruption’ experiment (Figure 2B), it was clear that all the aptamers that were able to compete were also able to instantaneously disrupt the TFIIB–TBP interaction and remove TFIIB from the complex. It is also apparent that this disruption could not be explained by the instability of TFIIB binding because incubation with excess cold DA for the same amount of time did not lead to a loss of TFIIB from the DAB complex. Thus, the TFIIB aptamers fall into two classes—class I includes the majority of the aptamers (such as AptTIIB-60) which interfere with the TFIIB–TBP interaction and class II aptamers (such as AptTFIIB-4) which bind with comparable affinity, but do not affect this interaction.
Because the class II aptamers do not interfere with the TBP–TFIIB interaction, they would be expected to bind this complex. The smears seen between the DA and DAB complex in the lanes containing the class II aptamers in Figure 2A and B could contain a faster moving, RNA containing DAB–aptamer complex, as seen previously with other RNA–protein interactions (22). However, we were unable to detect a new distinct band corresponding to this new DAB–RNA complex in our experiments (Figures 2A and S7). Additionally, it is evident that the class I aptamers not only bind with high affinity but are also able to dramatically disrupt a physiological TBP interaction as a consequence of this binding. This property of the aptamers makes them very valuable inhibitors for use in a physiological setting, because a large proportion of TFIIB in the cell likely resides in macromolecular assemblies (and not as a free entity in solution), in which case a passive competitor would be unable to effect this inhibition.
The TFIIB aptamers inhibit transcription, in vitro
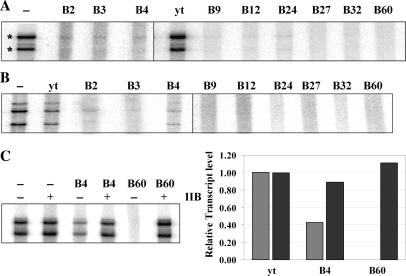
The TFIIB–TBP interaction is believed to be essential for PIC formation and therefore for transcription by Pol II. Therefore, the class I aptamers that disrupt this interaction when a complex is assembled from purified proteins, would be expected to prevent Pol II transcription, if they were functional in the context of a complex milieu of proteins such as a whole-cell extract. To test this, we performed an in vitro transcription assay using a plasmid template, which contained Gal4-binding sites and the yeast HIS4 promoter followed by a G-less cassette, in the presence of whole-cell extract from Saccharomyces cerevisiae (yeast). Aptamer RNA, or control yeast tRNA (or G0 RNA, Figure S6) was added to the extract along with the template (Figure 3A) or after PIC formation (Figure 3B), mimicking the competition and disruption experiments described above [the aptamers are stable in the whole-cell extract in the presence of RNase Inhibitor SuperaseIn (Ambion, Austin, Texas), for the duration of the experiment, data not shown]. Transcription was initiated by the addition of a nucleotide mix containing one radiolabeled NTP and allowed to proceed for 30 min (multiple rounds, Figure 3A) or 45 s (single round, Figure 3B). The radiolabeled transcript synthesized was run out on a denaturing gel and analyzed. The large reduction in the intensity of the bands corresponding to the full-length transcript in every lane that contained aptamer RNA, in both of these experiments, leads to the following conclusions: (i) that all the aptamers, irrespective of whether they are class I or class II, inhibit transcription both when added before (Figure 3A) as well as after PIC formation (Figure 3B); and (ii) they appear to work in the context of an extract very efficiently at only 2–3× molar excess of their target (TFIIB concentration in the reaction mix ∼20 nM as determined by western analysis, data not shown). In order to confirm that this inhibition was because of the aptamers acting through TFIIB and not an indirect effect of addition of exogenous RNA, two controls were performed. First, addition of yeast tRNA or G0 RNA (Figure S6) at a similar concentration did not inhibit transcription in any experiment (Figures 3A and B, and S6). Second, when the extract was supplemented with molar excess of recombinant TFIIB, prior to addition of aptamer RNA (or after aptamer addition, Figure S6), transcription activity was unaffected (Figure 3C). For the experiment in Figure 3C, the aptamers were used at 30 nM (not 50 nM as in Figure 3A and B) so as to enable add-back of exogenous TFIIB at 300 nM (which is the maximum TFIIB that could be added without any significant effect on the control lanes). The inhibitory effect of B4 at this concentration is slightly less than when added at 50 nM. The fact that excess TFIIB is able to suppress the transcription inhibition mediated by the aptamer supports the idea that the transcription inhibition observed in the presence of the aptamer is due to the unavailability, specifically, of TFIIB, because when excess TFIIB is provided, this inhibition is reversed.
Figure 3.
The TFIIB aptamers inhibit basal transcription in vitro from a Pol II promoter in a yeast whole-cell extract. (A) The aptamers were added at 50 nM along with the extract to a plasmid template containing the HIS4 promoter fused to a G-less cassette and transcription was allowed to proceed for 30 min. The correct transcripts are indicated (asterisks). (B) Aptamers were added at 50 nM after PIC formation, and single-round transcription was assayed by incubating with NTPs for 45 s, and transcription was stopped by the addition of heparin. (C) Transcription inhibition with the aptamers at 30 nM (lanes 3 and 5), (hatched bars) and with addition of recombinant yeast TFIIB (lanes 2, 4 and 6) to the extract (black bars) were quantified and plotted. Excess TFIIB was able to completely restore transcription in the presence of the aptamers.
In conclusion, the RNA aptamers are effective inhibitors of Pol II driven transcription in a yeast whole-cell extract. However, given their distinct effects on DAB complex formation, it seems likely that the mechanism of the inhibition achieved is different for each aptamer class.
Aptamers affect PIC formation at discrete steps
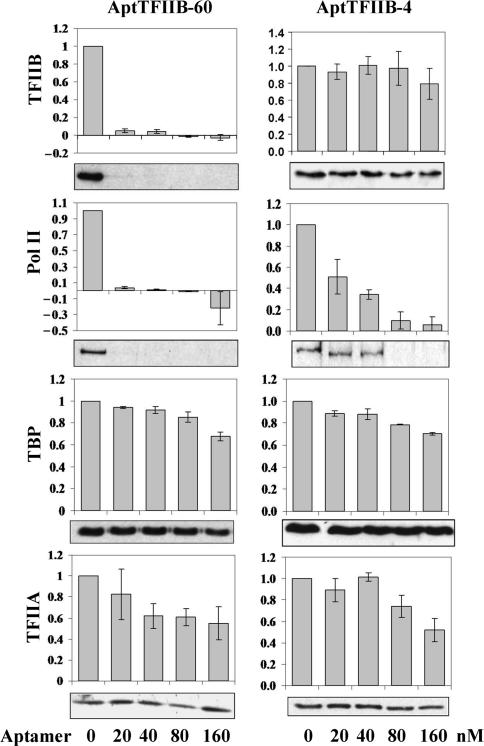
To evaluate the mechanistic differences among the different classes of aptamer RNAs, we devised an immobilized promoter template assay to monitor PIC assembly in the presence of each of the representative aptamers (AptTFIIB-4 and AptTFIIB-60). A DNA template containing the HIS4 promoter and Gal4-binding sites was immobilized onto magnetic beads and PICs were assembled on the promoter using yeast whole-cell extract as a source for transcription machinery. The unbound proteins were then washed away and the composition of the assembled PIC was ascertained by SDS–PAGE and western analysis. Occupancy of four components of the PIC, namely, TBP, TFIIB, TFIIA and Pol II were monitored using specific antibodies. When a class I aptamer such as B60 was added along with the extract to the template, PIC formation appeared to be severely compromised (Figure 4). TBP and TFIIA levels at the promoter were not affected significantly throughout the range of aptamer concentrations explored; however, even at the lowest aptamer concentration, TFIIB and Pol II were reduced to undetectable levels. This is consistent with the observation that B60 interferes with the TFIIB–TBP interaction. Therefore, in the presence of B60, TBP and TFIIA assemble at the promoter, but TFIIB, which is now in complex with B60, is unable to do so. Also, because the N-terminal domain of TFIIB, specifically the Zn ribbon, is known to be required to recruit Pol II (8) to the PIC, the absence of TFIIB results in a loss of Pol II recruitment at the promoter.
Figure 4.
Pre-initiation complex (PIC) formation at the HIS4 promoter was monitored in an immobilized template assay. Bead bound templates were incubated with yeast whole-cell extract in the presence of the TFIIB aptamers B60 or B4 at 50 nM for 30 min. The templates were then washed and promoter bound factors were analyzed by western blots using specific antibodies [anti-TFIIB, anti-Rpb3 (PolII), anti-TBP and anti-Toa2 (TFIIA)]. The results were quantified and the values normalized to the no aptamer sample are plotted. Error bars represent the SEM for three independent experiments.
In the presence of AptTFIIB-4, a class II aptamer, TFIIB occupancy at the PIC is nearly unchanged, as are TBP and TFIIA levels, even at the highest aptamer concentration tested (Figure 4). Nonetheless, the level of Pol II at the promoter decreased proportionally with increasing aptamer concentration, suggesting that B4 binds to TFIIB and this binding affects the TFIIB–Pol II interaction. Thus, although both classes of aptamers result in the same ‘phenotype’ i.e. transcription inhibition, they do so by disconnecting discrete links in the transcription circuit.
Protease footprinting reveals the contact sites of the aptamers on TFIIB
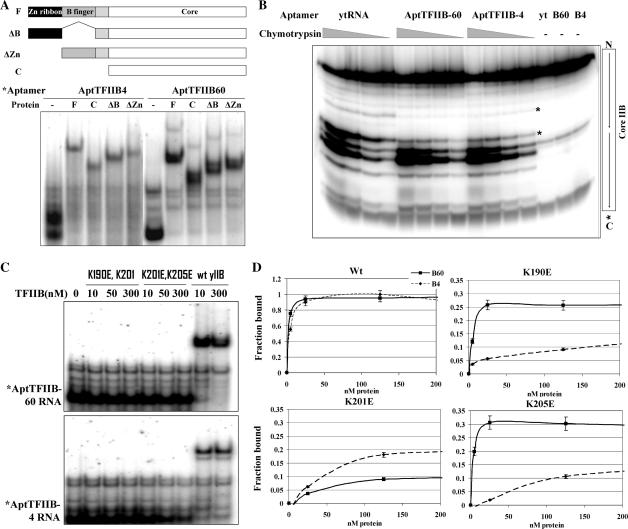
The stark difference in the way the two classes of aptamers inhibited PIC formation lead to the possibility that they might bind to distinct molecular surfaces on TFIIB. In an attempt to map the region of the protein bound by each of these aptamers, we generated a series of deletions in yeast TFIIB. Figure 5A illustrates these deletions in the Zn ribbon (ΔZn), B finger (ΔB), Zn ribbon and B finger (core) and the core domain (NTD). All the purified Escherichia coli expressed proteins except the core domain deletion bound AptTFIIB-4 as well as AptTFIIB-60 with affinity (KD) similar to that of full-length TFIIB (Figure 5A). Thus, the binding site for both of the aptamers (and that of all the other aptamers mentioned previously in this study, data not shown) resides in the core domain of TFIIB (the minimal domain tested).
Figure 5.
Defining the RNA aptamer contact sites on TFIIB. (A) Bandshift assay with the full length (F), core domain (C), delta Zn ribbon (ΔZn) and delta B-finger (ΔB) versions of yeast TFIIB. The labeled aptamer RNA (∼1 nM) was incubated with the indicated protein (50 nM) and loaded on a native gel. Schematic showing the deletion constructs used in the experiment. (B) C-terminally kinase-labeled core domain of TFIIB (0.6 uM) was subjected to digestion with increasing amounts of chymotrypsin in the presence of aptamers B4 or B60 or yeast tRNA (3 uM) for 7 min. Products were resolved on a Tricine–SDS 15% polyacrylamide gel. Asterisk indicates the region where protection is seen in the presence of the aptamers. (C) Bandshift assay with the two double mutants, K190E K201E and K201E K205E, as well as wt yeast TFIIB. Labeled B60 or B4 RNA was incubated with indicated amounts of the mutant or wt yeast TFIIB for 30 min and loaded on a native gel. (D) Bandshift assay with K190E, K201E and K205E as well as wt TFIIB. Labeled B4 or B60 was incubated with indicated amounts of each protein for 30 min and loaded on a native gel. Graphs are plotted as % of total counts bound to protein (n = 2).
Protease footprinting provided a higher resolution mapping. This technique has previously been used to map protein–protein interaction sites (15,23). The digestion of an end-labeled protein with limiting amounts of a specific protease gives rise to a pattern of bands corresponding to the cleavage products. The protection of a protease site by another factor (protein or RNA) obscures access of the protease and causes the reduction in the intensity of the corresponding band(s).
The core domain of TFIIB was C-terminally tagged with the HMK recognition site and radiolabeled using recombinant HMK (Sigma). This protein was allowed to bind to each of the aptamers or yeast tRNA as a control, digested with increasing amounts of chymotrypsin (or V8 protease, Figure S5) and analyzed by high-resolution Tricine–SDS–PAGE (we ensured that all of the protein was RNA-bound, Figure S4). Sites on the protein that were bound by the aptamer were protected from digestion and did not give rise to bands on the denaturing gel. There were several distinguishable bands corresponding to cut sites on core IIB, the most predominant of these corresponded to the linker region that connected the two direct repeats within the core (Figure 5B) (a similar experiment carried out with 350 nM aptamer RNA and 70 nM labeled TFIIB shows comparable protection pattern, data not shown). Surprisingly, in the presence of either aptamer, the pattern was remarkably similar—showing protection at residues just preceding the linker region on the first direct repeat. A stretch of basic residues in the last two helices of the first repeat of the core domain (helices D1 and E1) provide potential targets for RNA interactions.
Previously, mutations of specific lysine residues in these last two helices of the first repeat (K190, K201 and K205) have been shown to affect basal (in yeast) or activated (in human) Pol II transcription (24,25). Therefore, we tested the ability of the aptamers to bind to double mutant IIB with each pair of lysine residues mutated to glutamic acid (K190E, K201E and K201E, K205E). In the EMS assay shown in Figure 5C, neither aptamer was able to bind any of the mutants indicating that both aptamers indeed contacted this region within the core domain of TFIIB (the double radical substitution mutants are properly folded and support partial basal transcription from a whole-cell extract in vitro, Figure S6). Overall, it appeared that both classes of aptamers contacted at least a few of the same residues in the core domain. However, when we carried out binding assays with the single mutants in the same region (K190E, K201E and K205E), we observed differences in the binding characteristics of the two aptamers. While B4 is unable to bind well to all three single mutants, B60 appears to be severely compromised in forming a complex mainly with the K201E mutant (Figure 5D). Because the B4 and B60 aptamers are at least as big as the protein in overall size, we propose that they both depend on contact with the end of the first repeat in the core IIB, but the molecular interaction surface for each aptamer–protein complex is likely to be distinct, leading to their differential effects on TFIIB containing complexes.
DISCUSSION
RNA aptamers have been selected using SELEX procedures to bind a variety of protein targets, the vast majority made with therapeutic applications in mind (26–30). However, with this study and our previously published studies (10,17,31), we are illustrating the potential of these reagents as tools to probe macromolecular interactions in basic research as well. These RNAs can bind with high affinity to a protein surface and thereby effectively prevent specific interactions of that surface with other particular proteins or nucleic acids. This allows the assessment of that interaction without necessarily interfering with the overall integrity of the targeted protein or its other interaction surfaces. Our goal is to develop a collection of highly specific reagents targeting key regulatory transcription factors to dissect the role of particular factor interactions in transcription and its regulation both in vitro and in vivo.
Here, we isolate and characterize RNA aptamers that bind to yeast general transcription factor TFIIB. Although several families of aptamer RNAs were isolated, we have focused mainly on two representative molecules, B60 and B4. These aptamer RNA molecules display extremely high affinity to their target with dissociation constants (KD) of <1 nM. We believe that extensive protein–RNA interaction interfaces, as well as a stable RNA structure, contribute to this high affinity. It has previously been observed that sequences with higher information content, as judged from the complexity of the structure of these folded RNAs, tend to be the ones with the highest affinity to the target (32). In agreement with this, many of the aptamers isolated in the selection appear to include three-way junctions and pseudoknots in their secondary and tertiary structure as predicted by the RNA structure prediction program, Kinefold (33).
Although high-affinity aptamers do not guarantee a high degree of specificity in binding to small molecules (34), this is the exception rather than the rule with protein-binding aptamers. The specificity arises from the extensive contacts that the RNA makes on the structured surface of the protein, which is unlikely to be duplicated exactly in another unrelated protein (25). In the case of the TFIIB aptamers, we know that B60 as well as B4 are unable to bind to human TFIIB, which is about 80% similar to the yeast TFIIB in its C terminal domain. The region on the protein that is contacted by the RNAs is highly conserved in both yeast and human TFIIB, but this region alone is probably not sufficient for aptamer binding. The RNA molecules, which are reasonably large (∼30 kDa), recognize a molecular ‘surface’, only a part of which is highly conserved, and the entire surface is not identical in the two orthologs. The fact that a large molecular surface is probably buried in the RNA–protein complex then provides the basis for the spectacular affinity and specificity of this interaction. An additional proof for this specificity comes from the observation that we are able to reverse the aptamer-mediated inhibition of transcription from extracts in vitro by addition of molar excess of recombinant TFIIB.
Aptamers, in general, tend to bind to the functionally important epitopes on the target. TFIIB makes crucial contacts with TBP as well as Pol II, and so the surfaces involved in these interactions are the important ‘functional’ epitopes in the protein. From the protease mapping experiments, we find that both aptamers contact a helix located at the end of the first repeat in the core domain of TFIIB. This region contains a high density of positively charged residues on one face of the helix, which explains why it seems to be a preferred nucleic acid-binding site. Interestingly, although both B60 and B4 aptamers have a common region of contact they interfere in distinct ways with the formation of the PIC with B60 blocking TFIIB's binding to TBP and B4 blocking interaction with Pol II. This common region in the TFIIB core domain is also adjacent to the linker that connects the two repeats in the core, and has been shown to influence the ‘closed’ to ‘open’ conformational change that is required for hTFIIB incorporation into the PIC (8,35). This suggests that RNAs that bind in the vicinity of the linker could interfere with TBP or Pol II binding directly by steric hindrance. Alternatively, they could also affect TFIIB conformation and therefore indirectly influence one or both functional interactions. We favor the former model because the pattern of protease cleavage of IIB is altered in a very localized manner by addition of either RNA aptamer.
Another remarkable property of these RNAs is their ability to disrupt a preformed complex containing TFIIB, be it a TFIIB–TBP–TATA complex assembled from purified proteins or a complete PIC formed in the context of a whole-cell extract, with multiple other proteins in contact with TFIIB. We have observed this property in some of our TBP aptamers as well (11). This is an especially important feature for any reagent meant for use in vivo as all of the target protein cannot be expected to be freely available in solution for binding. At least in one case, with AptTBP-12, trimming the RNA sequence can lead to a selective loss of just the ability to disrupt (our unpublished data) complexes making the aptamer a passive competitor. It is therefore tempting to speculate that the structured RNA uses one of its ‘arms’ as a foothold to bind first to an exposed surface on the target and subsequently gain access to the ‘functional’ epitope that forms the higher-affinity binding site.
Finally, the fact that the RNA aptamers are effective in whole-cell extracts from yeast bodes well for their application in vivo. In the future, we plan to express these RNAs in a regulated manner in vivo in yeast to quantitatively inhibit PIC formation at specific stages. The use of the four classes of aptamers that target the TBP–DNA, TBP–TFIIA, TFIIB–TBP and TFIIB–PolII interactions specifically will allow us to dissect the role of each of these contacts in the transcriptional program of yeast.
From previous studies, we know that RNA aptamers can be produced in a controlled manner in vivo by constructing regulated genes encoding the aptamer RNAs and expressing them in a cell type and developmental stage-specific manner in higher eukaryotes as well (31,36). These RNAs can then selectively inhibit a specific surface on the target protein rapidly and completely in vivo, and the consequences can be assayed almost immediately at the genomic level. We hope that this strategy will prove to be generally applicable and provide a finer tool to dissect the complex network of macromolecular interactions that are key to transcription initiation as well as other cellular processes in higher organisms.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
AKNOWLEDGEMENTS
We are very grateful to Drs A. Ponticelli, J. Fu, M.H. Suh and S. Hahn for kindly providing strains and plasmids; to M. Spatz, M. Guertin and Drs K. Adelman and X. Fan for advice and technical assistance; and to H. Salamanca and Dr J. Waterfall for comments on the article. This work was supported by National Institutes of Health grant (GM40918) to J.T.L. Funding to pay the Open Access publication charges for this article was provided by National Institutes of Health grant (GM40918) to J.T.L.
Conflict of interest statement. None declared.
REFERENCES
- 1.Famulok M, Mayer G. Intramers and aptamers: Applications in protein-function analyses and potential for drug screening. Chembiochem. 2005;6:19–26. doi: 10.1002/cbic.200400299. [DOI] [PubMed] [Google Scholar]
- 2.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 3.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 4.Gold L, Polisky B, Uhlenbeck O, Yarus M. Diversity of oligonucleotide functions. Annu. Rev. Biochem. 1995;64:763–797. doi: 10.1146/annurev.bi.64.070195.003555. [DOI] [PubMed] [Google Scholar]
- 5.Famulok M, Blind M, Mayer G. Intramers as promising new tools in functional proteomics. Chem. Biol. 2001;8:931–939. doi: 10.1016/s1074-5521(01)00070-9. [DOI] [PubMed] [Google Scholar]
- 6.Hahn S. Structure and mechanism of the RNA polymerase II transcription machinery. Nat. Struct. Mol. Biol. 2004;11:394–403. doi: 10.1038/nsmb763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thomas MC, Chiang CM. The general transcription machinery and general cofactors. Crit. Rev. Biochem. Mol. Biol. 2006;41:105–178. doi: 10.1080/10409230600648736. [DOI] [PubMed] [Google Scholar]
- 8.Deng W, Roberts SG. TFIIB and the regulation of transcription by RNA polymerase II. Chromosoma. 2007;116:417–429. doi: 10.1007/s00412-007-0113-9. [DOI] [PubMed] [Google Scholar]
- 9.Yudkovsky N, Ranish JA, Hahn S. A transcription reinitiation intermediate that is stabilized by activator. Nature. 2000;408:225–229. doi: 10.1038/35041603. [DOI] [PubMed] [Google Scholar]
- 10.Shi H, Fan X, Sevilimedu A, Lis JT. RNA aptamers directed to discrete functional sites on a single protein structural domain. Proc. Natl Acad. Sci. USA. 2007;104:3742–3746. doi: 10.1073/pnas.0607805104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan X, Shi H, Adelman K, Lis JT. Probing TBP interactions in transcription initiation and reinitiation with RNA aptamers that act in distinct modes. Proc. Natl Acad. Sci. USA. 2004;101:6934–6939. doi: 10.1073/pnas.0401523101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pinto I, Ware DE, Hampsey M. The yeast SUA7 gene encodes a homolog of human transcription factor TFIIB and is required for normal start site selection in vivo. Cell. 1992;68:977–988. doi: 10.1016/0092-8674(92)90040-j. [DOI] [PubMed] [Google Scholar]
- 13.Wu WH, Pinto I, Chen BS, Hampsey M. Mutational analysis of yeast TFIIB. A functional relationship between Ssu72 and Sub1/Tsp1 defined by allele-specific interactions with TFIIB. Genetics. 1999;153:643–652. doi: 10.1093/genetics/153.2.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grossmann JG, Sharff AJ, O’Hare P, Luisi B. Molecular shapes of transcription factors TFIIB and VP16 in solution: implications for recognition. Biochemistry. 2001;40:6267–6274. doi: 10.1021/bi0028946. [DOI] [PubMed] [Google Scholar]
- 15.Hori R, Carey M. Protease footprinting analysis of ternary complex formation by human TFIIA. J. Biol. Chem. 1997;272:1180–1117. doi: 10.1074/jbc.272.2.1180. [DOI] [PubMed] [Google Scholar]
- 16.Shi H, Hoffman BE, Lis JT. A specific RNA hairpin loop structure binds the RNA recognition motifs of the drosophila SR protein B52. Mol. Cell. Biol. 1997;17:2649–2657. doi: 10.1128/mcb.17.5.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao X, Shi H, Sevilimedu A, Liachko N, Nelson HC, Lis JT. An RNA aptamer that interferes with the DNA binding of the HSF transcription activator. Nucleic Acids Res. 2006;34:3755–3761. doi: 10.1093/nar/gkl470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schagger H. Tricine-SDS-PAGE. Nat. Protoc. 2006;1:16–22. doi: 10.1038/nprot.2006.4. [DOI] [PubMed] [Google Scholar]
- 19.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pardee TS, Bangur CS, Ponticelli AS. The N-terminal region of yeast TFIIB contains two adjacent functional domains involved in stable RNA polymerase II binding and transcription start site selection. J. Biol. Chem. 1998;273:17859–17864. doi: 10.1074/jbc.273.28.17859. [DOI] [PubMed] [Google Scholar]
- 21.Buratowski S, Hahn S, Guarente L, Sharp PA. Five intermediate complexes in transcription initiation by RNA polymerase II. Cell. 1989;56:549–561. doi: 10.1016/0092-8674(89)90578-3. [DOI] [PubMed] [Google Scholar]
- 22.Espinoza CA, Allen TA, Hieb AR, Kugel JF, Goodrich JA. B2 RNA binds directly to RNA polymerase II to repress transcript synthesis. Nat. Struct. Mol. Biol. 2004;11:822–829. doi: 10.1038/nsmb812. [DOI] [PubMed] [Google Scholar]
- 23.Hori R, Pyo S, Carey M. Protease footprinting reveals a surface on transcription factor TFIIB that serves as an interface for activators and coactivators. Proc. Natl Acad. Sci. USA. 1995;92:6047–6051. doi: 10.1073/pnas.92.13.6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bangur CS, Pardee TS, Ponticelli AS. Mutational analysis of the D1/E1 core helices and the conserved N-terminal region of yeast transcription factor IIB (TFIIB): Identification of an N-terminal mutant that stabilizes TATA-binding protein-TFIIB-DNA complexes. Mol. Cell. Biol. 1997;17:6784–6793. doi: 10.1128/mcb.17.12.6784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roberts SG, Ha I, Maldonado E, Reinberg D, Green MR. Interaction between an acidic activator and transcription factor TFIIB is required for transcriptional activation. Nature. 1993;363:741–744. doi: 10.1038/363741a0. [DOI] [PubMed] [Google Scholar]
- 26.White RR, Sullenger BA, Rusconi CP. Developing aptamers into therapeutics. J. Clin. Invest. 2000;106:929–934. doi: 10.1172/JCI11325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gold L. Oligonucleotides as research, diagnostic, and therapeutic agents. J. Biol. Chem. 1995;270:13581–13584. doi: 10.1074/jbc.270.23.13581. [DOI] [PubMed] [Google Scholar]
- 28.Miyakawa S, Fujiwara M, Nakamura Y. Aptamer therapeutics. Tanpakushitsu Kakusan Koso. 2006;51:2521–2527. [PubMed] [Google Scholar]
- 29.Que-Gewirth NS, Sullenger BA. Gene therapy progress and prospects: RNA aptamers. Gene Ther. 2007;14:283–291. doi: 10.1038/sj.gt.3302900. [DOI] [PubMed] [Google Scholar]
- 30.Eckstein F. The versatility of oligonucleotides as potential therapeutics. Expert Opin. Biol. Ther. 2007;7:1021–1034. doi: 10.1517/14712598.7.7.1021. [DOI] [PubMed] [Google Scholar]
- 31.Shi H, Hoffman BE, Lis JT. RNA aptamers as effective protein antagonists in a multicellular organism. Proc. Natl Acad. Sci. USA. 1999;96:10033–10038. doi: 10.1073/pnas.96.18.10033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carothers JM, Oestreich SC, Davis JH, Szostak JW. Informational complexity and functional activity of RNA structures. J. Am. Chem. Soc. 2004;126:5130–5137. doi: 10.1021/ja031504a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xayaphoummine A, Bucher T, Isambert H. Kinefold web server for RNA/DNA folding path and structure prediction including pseudoknots and knots. Nucleic Acids Res. 2005;33:W605–W610. doi: 10.1093/nar/gki447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carothers JM, Oestreich SC, Szostak JW. Aptamers selected for higher-affinity binding are not more specific for the target ligand. J. Am. Chem. Soc. 2006;128:7929–7937. doi: 10.1021/ja060952q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roberts SG, Green MR. Activator-induced conformational change in general transcription factor TFIIB. Nature. 1994;371:717–720. doi: 10.1038/371717a0. [DOI] [PubMed] [Google Scholar]
- 36.Burke DH, Nickens DG. Expressing RNA aptamers inside cells to reveal proteome and ribonome function. Brief Funct. Genomic Proteomic. 2002;1:169–188. doi: 10.1093/bfgp/1.2.169. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.