Abstract
In this article, we characterize histone demethylase activity of the entire family of JmjC+N proteins of Drosophila melanogaster. Our results show that Lid (little imaginal discs), which is structurally homologous to JARID1, demethylates H3K4me3. However, contrary to what would be inferred from its demethylase activity, lid contributes to the establishment of transcriptionally competent chromatin states as: (i) is required for histone H3 acetylation; (ii) contributes to expression of the homoeotic gene Ultrabithorax (Ubx); and (iii) antagonizes heterochromatin-mediated gene silencing (PEV). These results, which are consistent with the identification of lid as a trithorax group (trxG) gene, are discussed in the context of current models for the contribution of H3K4me3 to the regulation of gene expression. Here, we also show that the two Drosophila JMJD2 homologues, dJMJD2(1)/CG15835 and dJMJD2(2)/CG33182, are capable of demethylating both H3K9me3 and H3K36me3. dJMJD2(1)/CG15835 regulates heterochromatin organization, as its over-expression induces spreading of HP1, out of heterochromatin, into euchromatin, without affecting the actual pattern of histone modifications of heterochromatin. dJMJD2(1)/CG15835 is excluded from heterochromatin and localizes to multiple euchromatic sites, where it regulates H3K36 methylation. These results indicate that dJMJD2(1)/CG15835 contributes to delimit hetero- and euchromatic territories through the regulation of H3K36 methylation in euchromatin. On the other hand, dJARID2/CG3654 shows no demethylase activity on H3K4me3, H3K9me3, H3K27me3, H3K36me3 and H4K20me3.
INTRODUCTION
Histones are subjected to a number of post-translational modifications, such as methylation, acetylation, phosphorylation, sumoylation, ubiquitinylation and ADP ribosylation. Covalent modifications of core histones are known to play important genomic functions as they are recognized by regulatory proteins that bind chromatin and modify its functional state (1). In this context, methylation at lysine residues appears to play a central role, as it is involved in regulating a wide range of genomic functions including heterochromatin formation, dosage compensation, gene expression and cell memory (2). Moreover, several protein domains are known to specifically recognize methylated lysines. These include the chromo, tudor, WD40 repeat and PHD finger domains (2–4). Chromatin-binding proteins containing these domains have been shown to bind histones methylated at specific lysine residues. Some well-established examples are the BPTF component of the remodelling complex NURF that, through its PHD finger, binds H3K4me3 (5), or the silencing proteins HP1 and Polycomb, which contain N-terminal chromo domains that recognize H3K9me2,3 and/or H3K27me2,3 (6–9).
Key for regulation is the possibility to revert the modification. The existence of lysine-specific histone methyl-transferases (HMTs) has been known for some years (10,11). But, on the other hand, it was not until recently that enzymes capable of antagonizing lysine-methylation were identified. The first lysine-specific histone demethylase identified was LSD1, which is capable to demethylate mono- and di-, but not tri-, methylated H3K4 or H3K9 (12,13). More recently, proteins containing the JumonjiC (JmjC) domain were found to be capable of acting on trimethylated H3K9 and/or H3K36 (14), H3K4 (15–23) and H3K27 (24–27).
JmjC-containing proteins constitute an extensive phylogenetic family with multiple members in all eukaryotic species analysed to date, from yeasts to humans (14). Little is known, however, about the actual histone demethylase activity of most of them. In Drosophila melanogaster, homology search identified up to 13 JmjC-containing proteins (14), whose enzymatic activity remains largely uncharacterized. In general, the JmjC domain is found in combination with other protein domains. In particular, a subclass of JmjC proteins contain, in addition to the catalytic JmjC domain, a second highly conserved N-terminal domain (JmjN), which is also required for enzymatic activity (14). JmjC+N proteins are known to play important regulatory roles during development and cell cycle progression, and are frequently deregulated in cancer (14). Drosophila contains four JmjC+N proteins: Lid (little imaginal discs), CG3654, CG15835 and CG33182. Here, we have characterized their histone-demethylase activity in vivo. Our results indicate that Lid is capable of demethylating H3K4me3. Similar results were recently reported by others (15,17,23). In addition, we also show that, opposite to what would be expected from its enzymatic activity, lid antagonizes gene silencing being required for acetylation of histone H3. On the other hand, CG15835 and CG33182 are able to demethylate H3K9me3 and H3K36me3. Moreover, over-expression of CG15835 results in spreading of HP1 into euchromatin. Finally, over-expression of CG3654 shows no significant effect on the levels of H3K4me3, H3K9me3, H3K27me3, H3K36me3 and H4K20me3.
MATERIALS AND METHODS
DNAs
cDNAs of the genes encoding JmjC+N proteins were obtained from Drosophila Genomics Resource Center: GH09982 (CG3654); LD33386 (CG15835) and LD40310 (lid). GH09982 corresponds to a truncated form missing part of the N-terminal region but carrying the complete C-terminal part containing the JmjN, JmjC and ARID domains (aa positions 1521 to 2351). For dJMJD2(1)/CG15835, a H195A substitution was introduced into pActPPA-CG15835-Flag by site-directed mutagenesis using the QuickChange II mutagenesis kit (Stratagene) and verified by DNA sequencing.
Fly strains
EP-line P{EPgy2}CG33182EY10737, lid12367 and lid10403 mutants and the white inversion line In(1)wm4h were obtained from the Bloomington Stock Center (USA). The ‘engrailed’-GAL4 (en-GAL4) and UAS-GFP lines were provided by Dr Jean-Paul Vincent (MRC, London, UK). lid12367 and lid10403 mutants are described in ref. (15,28). In(1)wm4h is described in ref. (29). To over-express dJMJD2(1)/CG15835 in flies, transgenic lines carrying a UASGAL4-CG15835-Flag construct were obtained. For that, CG15835-Flag was cloned into in the pUAST vector (30). Transgenic flies for UAS constructs were generated using a w1118 strain as a recipient stock.
In silico analysis
Protein sequences from Drosophila JmjC+N proteins (CG15835, CG33182, CG3654 and CG9088) were retrieved from Ensembl release 43 (February 2007) and human protein sequences (JARID1A-D, JARID2 and JMJD2A-D) were retrieved from Ensembl release 45 (June 2007) (31). Multiple sequence alignment was performed using ClustalW (32). Protein domains were retrieved from PFAM (33) and SMART databases (34,35). Percentage identity was calculated using the program needle (36–38) at the EBI webpage (www.ebi.ac.uk).
Over-expression in S2 cells
For over-expression in S2 cells, cDNAs were Flag-tagged at C-terminal, cloned into the Drosophila expression vector pActPPA, where expression is driven by the actin5C promoter, and transfected (15 μg) into S2 cells by the calcium-phosphate method (39). Immunolocalization experiments were performed 48 h after transfection according to standard procedures using antibodies against H3K4me3 (Abcam), H3K4me2 (Upstate), H3K9me3 (Upstate), H3K9me2 (Upstate), H3K27me3 (Upstate) and H3K36me3 (Abcam). αFlag antibodies were FITC-conjugated monoclonal M2 (Upstate) or rabbit polyclonal (Sigma), and were added at the same time than the secondary antibody at a 1/200 and 1/500 dilution, respectively. αHP1 antibodies were raised in rat and used at a 1/200 dilution. Cy3-conjugated anti-rat, Cy3-conjugated anti-rabbit and Cy2-conjugated anti-rabbit secondary antibodies were obtained from Jackson ImmunoResearch. For fluorescence microscopy analysis, samples were mounted in Vectashield mounting medium (Vector Laboratories) with 1.5 μg/ml DAPI (4,6-diamidino-2-phenylindole) and visualized in an Eclipse E-1000 (Nikon) fluorescence microscope equipped with a CoolSnapfx camera (Photometrics) and Metamorph software (version 6.3r1).
Over-expression in flies
For over-expression in flies the GAL4-UAS system was used (30). In these experiments, UASGAL4-CG15835-Flag lines and the EP_line P{EPgy2}CG33182EY10737 were crossed to either an ‘engrailed’-GAL4 (en-GAL4); UAS-GFP line, when over-expression was analysed in imaginal discs, or an actin5C-GAL4 line, when over-expression was analysed in polytene chromosomes.
Immunostaining of imaginal discs
Dissection and immunostaining of imaginal discs obtained from third instar larvae was performed as described elsewhere (40), using αH3K4me3 (Abcam), αH3K9me3 (Upstate), αH3K27me3 (Upstate) and αH3K36me3 (Abcam). Secondary Cy3-conjugated antibodies (Jackson Laboratories) were used at a 1/200 dilution. GFP fluorescence was visualized directly. Preparations were mounted in Vectashield mounting medium (Vector Laboratories) with 1.5 μg/ml DAPI (4,6-diamidino-2-phenylindole) and visualized in a Leica TCS/SPE confocal microscope equipped with LAS/AF software (version v.1.6.3).
Immunostaining of polytene chromosomes
Polytene chromosomes were obtained from third instar larvae raised at 25°C. Dissection of salivary glands and immunostaining were performed as described in ref. (41), using αH3K4me3 (Abcam), αH3K9me3 (Upstate), αH3K9me2 (Upstate), αH3K36me3 (Abcam), αFlag_M2 monoclonal (Upstate), αH3K9Ac (Upstate), αH3K9/K14Ac (αH3Ac) (Upstate) and rat αHP1. Secondary antibodies Cy3- and Cy2-conjugated antibodies (Jackson Laboratories) were used at a 1/200 dilution. Preparations were mounted in Vectashield mounting medium (Vector Laboratories) with 1.5 μg/ml DAPI (4,6-diamidino-2-phenylindole) and visualized in an Eclipse E-1000 (Nikon) fluorescence microscope equipped with a CoolSnapfx camera (Photometrics) and Metamorph software (version 6.3r1).
Analysis of Ubx expression
When the effect of lid mutations on Ubx expression was determined, imaginal discs obtained from wild-type, homozygous lid10403 or lid12367, or trans-heterozygous lid12367/lid10403 mid-third instar larvae were immunostained with αUbx antibodies (42), used at a 1/20 dilution. Immunostaining and visualization were performed as described above.
PEV analysis
To analyse the effect of lid mutations on PEV, the white inversion In(1)wm4h line was used. In these experiments, w1118; lid12367/CyO [w-] females were crossed with In(1)wm4h/Y; +/+ males and the eye phenotype of heterozygous lid12367/+ females was compared to that of siblings ‘wt’ for this ‘locus’. Similar results were obtained when the reverse cross was performed.
RESULTS
Domain organization and sequence homology analysis of JmjC+N proteins of D. melanogaster
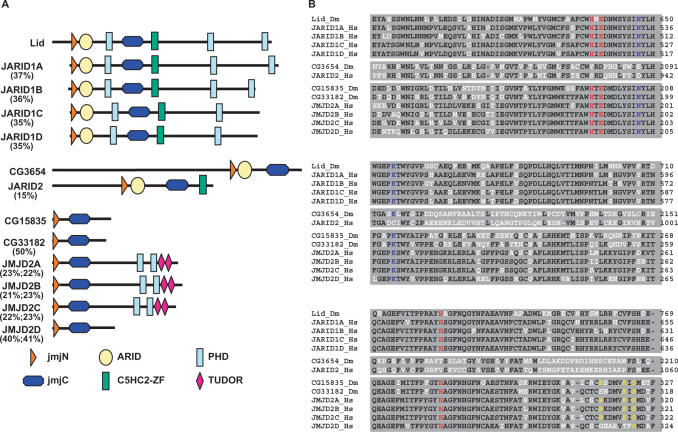
JmjC+N proteins are divided into two evolutionarily conserved groups showing important differences in their domain structure and organization, the JARID family and the JMJD2 family (14) (Figure 1A). JARID proteins contain, in addition to the JmjN and JmjC domains, ARID and C5HC2-zinc finger domains, which mediate DNA binding. JARID proteins are, in turn, divided into two subgroups according to the presence, JARID1, or not, JARID2, of chromatin-binding PHD domains (Figure 1A). On the other hand, JMJD2 proteins contain several PHD and/or tudor domains, for binding to chromatin, but no DNA-binding domains (Figure 1A). JMJD2D constitutes an exception, as it does not contain any known chromatin- or DNA-binding domains.
Figure 1.
Drosophila contains four JmjC+N proteins that are structurally homologous to mammalian JARID1 (Lid), JARID2 (CG3654) and JMJD2 (CG15835 and CG33182). (A) The domain structure of the four Drosophila JmjC+N proteins is compared to those corresponding to mammalian JARID1A-D, JARID2 and JMJD2A-D proteins. The positions of the JumonjiC (JmjC) and JumonjiN (JmjN) domains are indicated. DNA-binding domains, ARID and C5HC2-zinc fingers and chromatin-binding domains, PHD and tudor, are also indicated. Numbers indicate percentage identity with respect to the corresponding Drosophila homologue. (B) Sequence alignment of the catalytic JmjC domains of JmjC+N proteins from Drosophila (Dm) and humans (Hs). For each group, identical residues are shown in black. Catalytic residues involved in binding of co-factors are shown in red [Fe(II)] and blue (αKG). Shown in yellow are residues involved in differential binding of H3K36me3 and H3K9me3 peptides. Numbers on the right indicate amino acid positions on the corresponding proteins.
In Drosophila, Lid and CG3654 are JARID proteins (Figure 1A). Lid is structurally homologous to mammalian JARID1 as, in addition to the JmjN and JmjC domains, it contains an ARID domain, a C5HC2-zinc finger domain and three PHD domains. In mammals, there are four JARID1 isoforms (Figure 1A) (14). JARID1D and JARID1C, which are closely related (84% identity), contain only two PHD fingers. On the other hand, JARID1A and JARID1B contain three PHD fingers, like Lid, and they are more distantly related to JARID1D (45% identity). Hence, from the point of view of its domain organization, Lid is more closely related to JARID1A/B than to JARID1C/D, though identity is similar (37–35%). On the other hand, CG3654 is the structural homologue of mammalian JARID2 (Figure 1A), as it does not contain any chromatin-binding domains. In this case, however, identity is lower (15%) and, in addition, CG3654 is missing the C5HC2-zinc finger domain present in mammalian JARID2.
CG15835 and CG33182 are JMJD2 proteins (Figure 1A). In mammals, the JMJD2 family consists of four closely related isoforms (14). Three of these isoforms (JMJD2A-C) contain chromatin-binding domains (PHD fingers and tudor domains), which are missing in JMJD2D (43,44). In Drosophila, CG15835 and CG33182, which are closely related to each other (50% identity), do not contain any known DNA- or chromatin-binding domains being, therefore, structurally homologous to mammalian JMJD2D. Actually, both CG15835 and CG33182 show higher identity to mammalian JMJD2D (40%) than to any of the other mammalian JMJD2 isoforms (22%).
At the level of the catalytic JmjC domain, Lid shows high identity to mammalian JARID1 isoforms (77–75%) (Figure 1B). Similarly, the JmjC domains of CG15835 and CG33182 are closely related to those of the mammalian JMJD2 isoforms (63–71% identity) (Figure 1B). On the other hand, in the case of the JARID2 homologue of Drosophila, CG3654, conservation of the JmjC domain is lower (40% identity to mammalian JARID2).
Determination of histone demethylase activity of Drosophila JmjC+N proteins
Histone demethylase activity of the Drosophila JmjC+N proteins was analysed through over-expression experiments performed in Drosophila cultured S2 cells and/or in flies.
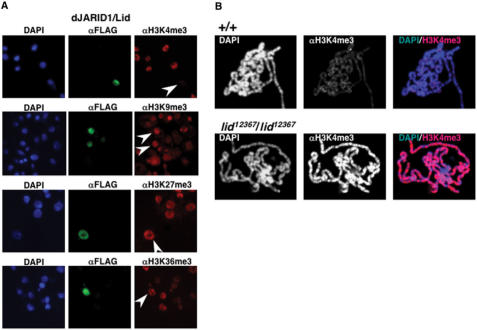
To determine the potential histone demethylase activity of dJARID1/Lid, a dJARID1/Lid-Flag fusion protein was transiently expressed in S2 cells and after transfection, cells were stained with αFlag antibodies, to identify cells expressing the fusion protein, and with antibodies that specifically recognize H3K4me3, H3K9me3, H3K27me3 and H3K36me3. Over-expression of dJARID1/Lid results in a strong reduction in the overall levels of H3K4me3. On the other hand, no significant effects on the levels of H3K9me3, H3K27me3 and H3K36me3 were detected. These results indicate that dJARID1/Lid is capable to specifically demethylate H3K4me3. Consistent with this hypothesis, a strong increase in H3K4me3 is detected in polytene chromosomes from homozygous lid12367 mutants (Figure 2B). In the course of these experiments, others reported a similar demethylase activity for several members of the JARID1 family of JmjC+N proteins, including Lid (15–23).
Figure 2.
dJARID1/Lid demethylates H3K4me3. (A) A Lid-Flag fusion protein was over-expressed in Drosophila S2-cells and transfected cells were stained with αFlag (shown in green), and αH3K4me3, αH3K9me3, αH3K27me3 or αH3K36me3 antibodies (shown in red). DNA was stained with DAPI (shown in blue). Arrows indicate cells over-expressing dJARID1/Lid. (B) Polytene chromosomes from control wild-type larvae (+/+) and homozygous lid12367/lid12367 mutant larvae stained with αH3K4me3 are shown. DNA was stained with DAPI.
In Drosophila, there are two JMJD2 isoforms, dJMJD2(1)/CG15835 and dJMJD2(2)/CG33182 (Figure 1A). To analyse the potential histone demethylase activity of dJMJD2(1)/CG15835, we performed similar over-expression experiments in S2 cells as those described above for dJARID1/Lid (Figure 2A). Over-expression of dJMJD2(1)/CG15835 results in a strong decrease on the levels of H3K9me3 and H3K36me3 (Figure 3A). On the other hand, the levels of H3K4me3 and H3K27me3 are not significantly altered. Demethylase activity of dJMJD2(1)/CG15835 depends on the JmjC domain, as it is abolished by mutations that affect its catalytic activity. JmjC proteins used Fe(II) and α-ketoglutarate (αKG) as co-factors to carry out the demethylating reaction (44,45). Structural studies allowed the identification of residues within the JmjC domain involved in binding of Fe(II) and αKG (Figure 1B, shown in red and blue, respectively) (46). In the case of dJMJD2(1)/CG15835, co-ordination of Fe(II) involves residues H195, E197 and H223 (Figure 1B). As shown in Figure 3B, a single-point mutation involving one of these residues, H195A, abolishes demethylase activity of dJMJD2(1)/CG15835.
Figure 3.
dJMJD2(1)/CG15835 demethylates H3K9me3 and H3K36me3. (A) A CG15835-Flag fusion protein was over-expressed in Drosophila S2 cells and transfected cells were stained with αFlag (shown in green), and αH3K4me3, αH3K9me3, αH3K27me3 or αH3K36me3 antibodies (shown in red). DNA was stained with DAPI (shown in blue). Arrows indicate cells over-expressing CG15835. (B) Similar experiments as those described in (A), but with a mutated CG15835 form carrying a single-point mutation at the catalytic JmjC domain, H195A, which perturbs co-ordination of Fe(II) and impairs enzymatic activity.
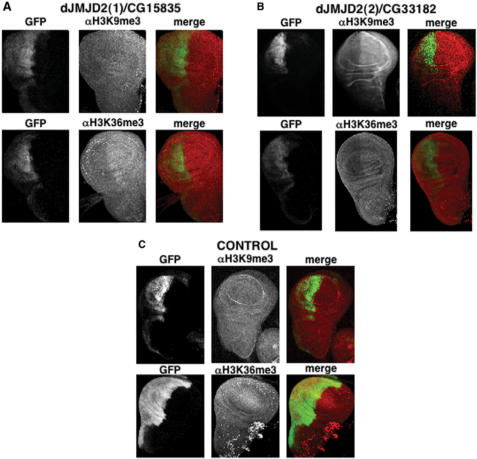
Demethylase activity of dJMJD2(1)/CG15835 was also analysed in flies (Figure 4). For this purpose, we obtained transgenic lines carrying a UASGAL4-CG15835-Flag construct, where expression of dJMJD2(1)/CG15835 is under the control of the yeast activator GAL4, allowing its over-expression upon crossing with lines expressing GAL4. In these experiments, we used a line carrying an en-GAL4 transgen, where GAL4 is expressed under the control of the ‘engrailed’ promoter, and, in addition, a second UAS-GFP transgen, where GFP is also expressed in response to GAL4, allowing identification of the domains of expression of en-GAL4. Then, the effects of dJMJD2(1)/CG15835 over-expression were determined in wing imaginal discs, where the ‘engrailed’ promoter is specifically active in the posterior compartment. In this system, the en-GAL4 driver induces both GFP expression and specific over-expression of dJMJD2(1)/CG15835 only in the posterior wing-disc compartment. Under these conditions, cells of the posterior compartment show a reduced reactivity with αH3K36me3 antibodies when compared to cells of the anterior compartment, where no over-expression takes place (Figure 4A). A weaker, but reproducible, effect on the levels of H3K9me3 is also detected (Figure 4A). On the other hand, no effect on H3K9me3 or H3K36me3 is observed in the posterior compartment of flies expressing en-GAL4; UAS-GFP alone (Figure 4C). A similar approach was used to analyse demethylase activity of dJMJD2(2)/CG33182 in flies (Figure 4B). In this case, we took advantage of a P-element insertion at the 5′-UTR of CG33182, P{EPgy2}CG33182EY10737, which contains GAL4-binding sites and, therefore, allows its over-expression upon crossing to the en-GAL4 line. In this case, a strong decrease in reactivity with αH3K9me3 and αH3K36me3 antibodies is observed in cells of the posterior compartment (Figure 4B), indicating that dJMJD2(2)/CG33182 is also capable of demethylating H3K9me3 and H3K36me3. On the other hand, over-expression of dJMJD2(2)/CG33182 shows no significant effect on the levels of H3K4me3 and H3K27me3 (Supplementary Data, Figure S1).
Figure 4.
In flies, dJMJD2(1)/CG15835 and dJMJD2(2)/CG33182 demethylate H3K9me3 and H3K36me3. (A) To over-express CG15835, flies carrying a transgenic UASGAL4-CG15835-Flag construct, where expression of dJMJD2(1)/CG15835 is under the control of GAL4, were crossed to en-GAL4; UAS-GFP flies (see text for details). The effects on the levels of H3K9me3 and H3K36me3 were determined by staining wing imaginal discs from en-GAL4; UAS-GFP; UASGAL4-CG15835-Flag larvae with αH3K9me3 and αH3K36me3 antibodies. In this system, expression of the GFP reporter labels the posterior wing-disc compartment, where over-expression of CG15835 is specifically induced. (B) Similar experiments as those described in (A), but performed with flies carrying a P-element insertion at the 5'-UTR of CG33182, P{EPgy2}CG33182EY10737, that contains GAL4-binding sites and, therefore, allows over-expression of dJMJD2(2)/CG33182 at the posterior wing-disc compartment upon crossing to the en-GAL4; UAS-GFP line. (C) Similar experiments as those described in (A) and (B), but performed with control en-GAL4; UAS-GFP flies, where only the GFP reporter is expressed at the posterior wing-disc compartment.
Similar over-expression experiments, performed both in S2 cells and flies, failed to detect any demethylase activity of dJARID2/CG3654 on H3K4me3, H3K9me3, H3K27me3, H3K36me3 and H4K20me3 (data not shown).
dJARID1/Lid antagonizes gene silencing
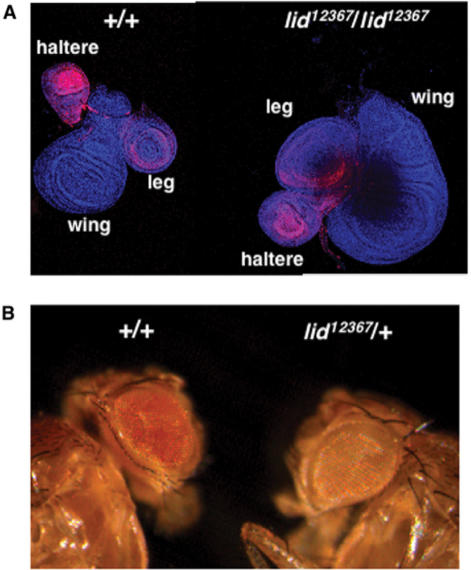
lid was first identified as a member of the trithorax group of genes (trxG), as lid mutants enhance the homoeotic transformations associated to mutations in other trxG genes (28). In fact, lid interacts genetically with the major HMTs of H3K4 of Drosophila, trithorax (trx) and ash1, which are also trxG genes (28). These observations suggest that lid is required to maintain expression of the homoeotic genes, which is opposite to what would be inferred from its demethylase activity as, in general, H3K4me3 is an epigenetic mark that correlates with transcriptionally active genes (47–51). Therefore, demethylation of H3K4 would be expected to contribute to gene silencing rather than to activation. These considerations prompted us to investigate the actual contribution of lid to expression of the homoeotic gene Ultrabithorax (Ubx) (Figure 5A). In these experiments, we analysed Ubx expression in imaginal discs from lid12367 or lid10403 mutant larvae by immunostaining with specific αUbx antibodies (42). It was reported that lid mutations result in small imaginal discs (28). Actually, it was recently shown that, independent of its demethylase activity, lid regulates cell proliferation through its interaction with myc (15). This phenotype occurs, however, at a very low frequency (<2%), so that most homozygous mutant larvae contain discs of normal size. In a wild-type condition, Ubx is strongly expressed in haltere, much less expressed in leg and not expressed at all in wing discs (Figure 5A, panel +/+). In homozygous lid12367 larvae, the pattern of Ubx expression is not altered but expression in haltere discs is reduced being similar to that observed in leg discs (Figure 5A, panel lid12367/lid12367), which is in contrast with the much higher expression in haltere versus leg discs observed in wild-type larvae. Similar results were obtained with homozygous lid10403 or trans-heterozygous lid12367/lid10403 mutant larvae. A similar effect on Ubx expression was reported in S2 cells (23).
Figure 5.
lid antagonizes gene silencing. (A) Expression of the homoeotic gene Ultrabithorax (Ubx) was determined by immunostaining with αUbx antibodies in imaginal discs from control wild-type larvae (+/+) and homozygous lid12367/lid12367 mutant larvae. Haltere, wing and leg discs are indicated. (B) The effect of lid mutations on the PEV of In(1)wm4 is shown. The eye phenotype of heterozygous lid12367/+ flies is compared to that of control siblings wild type for the ‘locus’ (+/+).
The contribution of lid to heterochromatin-dependent gene silencing (PEV) was also analysed (Figure 5B). In these experiments, we determined the effects of lid12367 on silencing of the white gene in In(1)wm4. Heterozygous lid12367 flies show strong enhancement of PEV when compared to control siblings, indicating that lid12367 is a strong dominant E(var), which is in contrast with the suppressor effect associated to mutants of Drosophila LSD1/Su(var)3-3 that demethylates H3K4me1,2 but not H3K4me3 (52).
These results indicate that lid antagonizes gene silencing in two different contexts, expression of the homoeotic gene Ubx and heterochromatin-dependent gene silencing, and suggest a role of lid in maintaining transcriptionally competent chromatin states. Consistent with this hypothesis, lid is required to maintain the pattern of acetylation of histone H3 (Figure 6), a modification that is characteristic of open chromatin domains (53,54). In these experiments, the levels of H3K9Ac were found to be strongly reduced in polytene chromosomes from homozygous lid12367 mutants when compared to control polytenes obtained from wild-type larvae (Figure 6A). Similar results were obtained when αH3Ac antibodies, which recognize histone H3 acetylated at K9 and K14, were used (Figure 6B).
Figure 6.
lid is required for histone H3 acetylation. Polytene chromosomes obtained from control wild-type larvae (+/+) and homozygous lid12367/lid12367 mutant larvae were stained with antibodies that specifically recognize histone H3 acetylated at K9, αH3K9, (A) or at K9 and K14, αH3Ac, (B). DNA was stained with DAPI.
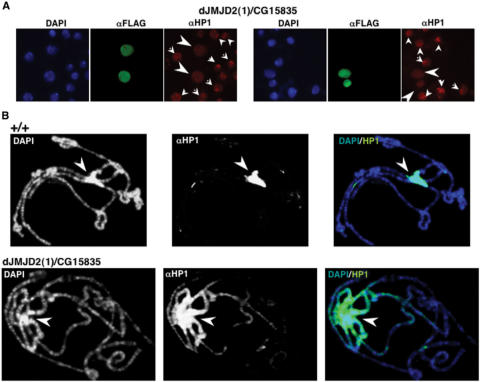
dJMJD2(1)/CG15835 regulates spreading of HP1
As shown above, dJMJD2 isoforms demethylate H3K9me3 and H3K36me3. In this context, we rationalize that histone demethylases capable of removing H3K9me3 are likely to regulate HP1 binding to chromatin and, therefore, contribute to the structural organization of heterochromatin. To address this question, dJMJD2(1)/CG15835-Flag was over-expressed in S2 cells, and the pattern of HP1 localization was determined using specific αHP1 antibodies. As shown in Figure 7A, most untransfected cells show a bright αHP1 spot, which reflects localization of HP1 to the heterochromatic chromocentre. On the other hand, cells over-expressing dJMJD2(1)/CG15835 show a diffuse pattern of HP1 localization with no preferential localization at heterochromatin, indicating mislocalization throughout euchromatin. Similar results were obtained when over-expression was performed in flies (Figure 7B). In these experiments, transgenic flies carrying the UASGAL4-CG15835-Flag construct were crossed to an act5C-GAL4 line, where GAL4 is expressed under the control of the constitutive act5C-promoter and, therefore, CG15835 is ubiquitously expressed. Then, the pattern of HP1 localization was determined in polytene chromosomes, where HP1 localizes to the heterochromatic chromocentre in control wild-type flies (Figure 7B, panels +/+). However, upon over-expression of dJMJD2(1)/CG15835, HP1 spreads out of the chromocentre invading the euchromatic chromosome arms (Figure 7B, panels dJMJD2(1)/CG15835). Actually, under these conditions, HP1 concentration along the chromosome arms decreases progressively as distance to the chromocentre increases. These results indicate that, rather than delocalization, over-expression of dJMJD2(1)/CG15835 induces spreading of HP1 into euchromatin.
Figure 7.
Over-expression of dJMJD2(1)/CG15835 induces spreading of HP1 into euchromatin. (A) S2 cells transfected with CG15835-Flag were stained with αFlag (shown in green) and αHP1 antibodies (shown in red). DNA was stained with DAPI (shown in blue). In untransfected cells, HP1 localizes mostly to a single intense spot in the nuclei (small arrow heads), which corresponds to the heterochromatic chromocentre. Cells over-expressing CG15835 (big arrow heads) show a diffuse nuclear distribution of HP1 with no specific enrichment at the chromocentre. (B) Polytene chromosomes from UASGAL4-CG15835-Flag; act5C-GAL4 larvae, where CG15835 is ubiquitously expressed (see text for details) (panels dJMJD2(1)/CG15835), and control wild-type larvae (panels +/+), were stained with specific αHP1 antibodies. DNA was stained with DAPI. Arrows indicate the position of the heterochromatic chromocentre.
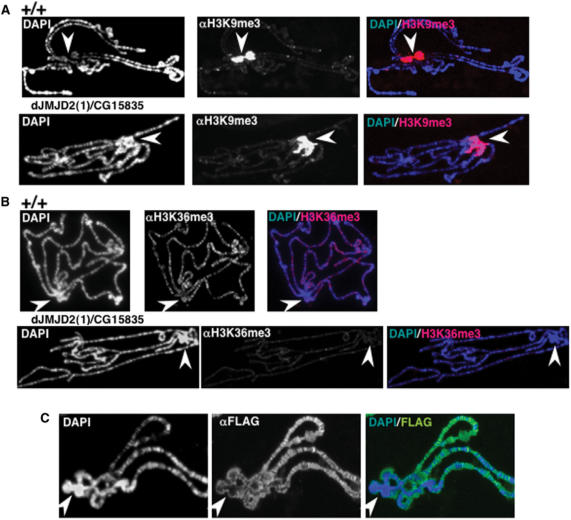
HP1 is known to recognize H3K9me2,3, a modification that is specifically enriched at heterochromatin (6,7,9). Therefore, spreading of HP1 observed upon over-expression of dJMJD2(1)/CG15835 might simply reflect decreased H3K9me2,3 at heterochromatin, which will result on its mobilization from the chromocentre. Over-expression of dJMJD2(1)/CG15835, however, does not show any significant effect on the extent of H3K9me3 (Figure 8A) and H3K9me2 (Supplementary Data, Figure S2), detected at the chromocentre. Actually, the effect on H3K9me3 observed in imaginal discs is only weak (Figure 4A), and, in polytene chromosomes, decreased H3K9me3 can be detected only at euchromatin, where it localizes to a few sites (Figure 8A). Moreover, spreading of HP1 into euchromatin is not accompanied by an equivalent spreading of H3K9me2,3 (Figures 8A and S2). On the other hand, as observed in imaginal discs (Figure 4A), over-expression of dJMJD2(1)/CG15835 shows a strong effect on H3K36me3 in polytene chromosomes (Figure 8B), where this modification is restricted to the euchromatic chromosome arms. In fact, H3K36me3 is a modification that correlates with active chromatin domains (55–58). These observations strongly suggest that dJMJD2(1)/CG15835 acts at euchromatin but not at heterochromatin. Consistent with this interpretation, in polytene chromosomes, over-expressed dJMJD2(1)/CG15835-Flag localizes to multiple sites on the euchromatic chromosome arms, being excluded from heterochromatin (Figure 8C).
Figure 8.
dJMJD2(1)/CG15835 localizes at euchromatin and regulates H3K36me3. (A) Polytene chromosomes from UASGAL4-CG15835-Flag; act5C-GAL4 larvae, where CG15835-Flag is ubiquitously expressed, (panels dJMJD2(1)/CG15835), and control wild-type larvae (panels +/+), were stained with αH3K9me3 antibodies. (B) Similar experiments as those described in (A), but polytene chromosomes were stained αH3K36me3 antibodies. (C) Polytene chromosomes from UASGAL4-CG15835-Flag; act5C-GAL4 larvae were stained with αFlag antibodies to determine the localization of the CG15835-Flag fused protein. DNA was stained with DAPI. Arrows indicate the position of the heterochromatic chromocentre.
DISCUSSION
Here, we have characterized the entire family of JmjC+N histone demethylases of D. melanogaster, which consists of four members: dJARID1/Lid, dJARID2/CG3654, dJMJD2(1)/CG15835 and dJMJD2(2)/CG33182. JmjC-demethylases are characterized by their ability to demethylate trimethyllysine residues (14). Our results indicate that dJARID1/Lid demethylates H3K4me3, which is in agreement with recently reported results by others (15,17,23). On the other hand, dJMJD2(1)/CG15835 and dJMJD2(2)/CG33182 demethylate both H3K9me3 and H3K36me3. Our results strongly suggest that these enzymes are also capable to demethylate dimethyllysine residues, as decreased trimethylation observed when they are over-expressed is not accompanied by increased dimethylation (Supplementary Data, Figure S3). Finally, no demethylase activity on H3K4me3, H3K9me3, H3K27me3, H3K36me3 and H4K20me3 could be attributed to dJARID2/CG3654. Interestingly, residues within the JmjC domain involved in binding Fe(II) and αKG (Figure 1B, shown in red and blue, respectively) (46), which are highly conserved in all JmjC proteins showing histone demethylase activity (14), are present in dJARID1/Lid, dJMJD2(1)/CG15835 and dJMJD2(2)/CG33182, but not in dJARID2/CG3654. In dJARID2/CG3654, two of the three residues involved in co-ordination of Fe(II) are not conserved (Figure 1B), indicating that binding of Fe(II) should be strongly perturbed. Similarly, binding of αKG is likely to be perturbed as one of the two essential JmjC domain residues involved in this interaction is not conserved (Figure 1B). A similar situation is observed in mammalian JARID2, where residues involved in binding αKG and Fe(II) are not conserved either (Figure 1B). Altogether, these observations suggest that the JmjC domain of JARID2 proteins might not be enzymatically active. Whether, they are capable at all of carrying out any demethylating reaction remains, however, to be determined.
dJMJD2(1)/CG15835 and dJMJD2(2)/CG33182, which demethylate both H3K9me3 and H3K36me3, are structurally homologous to mammalian JMJD2D (Figure 1A). Mammalian JMJD2D, however, demethylates H3K9me3 but is not capable to demethylate H3K36me3 (44). On the other hand, similar to the Drosophila JMJD2 proteins, mammalian JMJD2A and JMJD2C isoforms demethylate both H3K9me3 and H3K36me3 (44). Actually, though missing any DNA/chromatin-binding domains, the catalytic JmjC domains of CG15835 and CG33182 show slightly higher homology to those of mammalian JMJD2A-C (68–71% identity) than to that of JMJD2D (63–66% identity) (Figure 1B). Recent structural studies identified residues within the JmjC domain of mammalian JMJD2A involved in binding H3K9me3 and H3K36me3 peptides (59–61). In both cases, similar interactions help to bring the methyllysine residue close to the active site of the enzyme. However, significant differences are observed at the regions where the peptides enter and exit the JmjC domain. For the H3K36me3 peptide, these interactions involved the C-terminal region of the JmjC domain and, in particular, residues R309, K314 and S316 that, in the case of H3K9me3, are not involved in peptide binding. Interestingly, this region is conserved in the Drosophila JMJD2 isoforms, as well as in mammalian JMJD2C, but not in mammalian JMJD2D (Figure 1B).
Our results also show that, despite its ability to demethylate H3K4me3, dJARID1/Lid antagonizes silencing both of the homoeotic gene Ubx as well as heterochromatin-dependent gene silencing. Moreover, consistent with a contribution to gene activation, dJARID1/Lid regulates acetylation of histone H3. These observations are surprising, as, on the basis of its histone demethylase activity, the opposite effects would be expected since H3K4me3 correlates with transcriptionally active genes. Actually, more in agreement with its enzymatic activity, mammalian JARID1D was reported to interact with the polycomb (PcG) protein RING6a/MBLR and to prevent access of the basal transcription machinery to the human ‘engrailed’ promoter (16). Whether dJARID1/Lid, which is more closely related to JARID1A/B than to JARID1D (Figure 1A), interacts with PcG proteins is not known. It is possible that the various JARID1 isoforms would perform different non-redundant functions. It is also possible that, depending on the actual context, dJARID1/Lid would favour transcription activation or repression. Actually, the fission yeast orthologue (SpLid2C) is a component of a multi-protein complex containing Ash2 (62), an evolutionarily conserved TrxG protein involved both in gene activation and silencing. Interestingly, the same complex contains SpEcm5, which is also a JmjC+N protein of the JARID1 family (62). Whether these interactions are conserved in Drosophila, and if they contribute to the functional properties of dJARID1/Lid, remains to be determined. Finally, the overall increase on H3K4me3 that occurs in the absence of dJARID1/Lid might alter binding to chromatin of transcription activators recognizing H3K4me3. Actually, slight changes in the abundance of the chromo-helicase Chd1, which binds H3K4me3 and associates with transcriptionally active loci, were reported at the Sgs4 ‘locus’ in polytene chromosomes from RNAilid knockdown larvae (17). At this respect, it must be noticed that the pattern of H3K4me3 of the Ubx ‘locus’ is finely regulated (63). In haltere/leg discs, where Ubx is highly expressed, trimethylation of H3K4 is constrained to a relatively short region spanning the transcription start site. On the other hand, in wing discs, where Ubx is not expressed, no significant enrichment in H3K4me3 is detected throughout the ‘locus’. Various HMTs contribute to establishment/maintenance of this pattern of H3K4me3. It is known that, in haltere/leg discs, ash1 is required to maintain high levels of H3K4me3 at the transcription start site (63). On the other hand, E(z) is required to prevent H3K4 trimethylation in the wing disc (63). lid, which is the only H3K4me3 demethylase of Drosophila, is likely to contribute also to establishment/maintenance of the pattern of H3K4me3 of the Ubx gene. Further work is required to determine the precise molecular mechanisms of the contribution of dJARID1/Lid to Ubx activation. However, it is possible that, in its absence, H3K4me3 would spread throughout the ‘locus’ altering loading/assembly of the initiation complex at the transcription start site. Actually, it was recently shown that H3K4me3 regulates binding of the general transcription factor TFIID (64).
Here, we have also shown that dJMJD2(1)/CG15835 influences heterochromatin organization, as its over-expression induces spreading of HP1 into euchromatin. dJMJD2(1)/CG15835, however, is excluded from heterochromatin and, consistent with this observation, over-expression of dJMJD2(1)/CG15835 does not affect the pattern of H3K9me2,3 at heterochromatin. On the contrary, dJMJD2(1)/CG15835 localizes to multiple euchromatic sites, where it mostly regulates H3K36me3, as its over-expression results in a strong decrease in the levels of H3K36me3. At euchromatin, dJMJD2(1)/CG15835 also demethylates H3K9me3, but to a lesser extent. Altogether, these observations indicate that H3K36me3 acts as a barrier that prevents spreading of HP1 into euchromatin. It was shown earlier that methylation of H3K4 also prevents spreading of heterochromatin (52). Interestingly, both H3K36 and H3K4 methylation associate to actively transcribed genes, suggesting that gene activity is a main determinant to delimit hetero- and euchromatic territories. Specific recognition of H3K9me2,3 is known to regulate binding of HP1 to chromatin (6–9). However, spreading of HP1 observed upon over-expression of dJMJD2(1)/CG15835 is not accompanied by a parallel spreading of H3K9me2,3, indicating that it involves additional mechanisms. Actually, HP1 is also known to be capable of binding directly to DNA, RNA as well as unmodified histones (65–67).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We are thankful to J. Font, and Drs J. Bernués, M. Milán and J.P. Vincent, for antibodies, plasmids and fly stocks. We are highly thankful to E. Fuentes for technical assistance. M.L-.L. and C.C. acknowledge receipt of a I3P pre-doctoral fellowship and a I3P post-doctoral contract from CSIC, respectively. C.C. also acknowledges receipt of an ARC fellowship. A.V. acknowledges receipt of an ICREA junior contract from the Generalitat de Catalunya. This work was supported by grants from the MEC (BMC2006-1627; CSD2006-00049), the CIRIT (2001SGR00344) and the EU (LSHB-CT-2004-511965). This work was carried out within the framework of the CeRBa of the Generalitat de Catalunya. Funding to pay the Open Access publication charges for this article was provided by MEC grant (BMC-2006-1627).
Conflict of interest statement. None declared.
REFERENCES
- 1.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 2.Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell Biol. 2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- 3.Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol. Cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 4.Mellor J. It takes a PHD to read the histone code. Cell. 2006;126:22–24. doi: 10.1016/j.cell.2006.06.028. [DOI] [PubMed] [Google Scholar]
- 5.Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 6.Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 7.Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 8.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 9.Fischle W, Wang Y, Jacobs SA, Kim Y, Allis CD, Khorasanizadeh S. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 domains. Genes Dev. 2003;17:1870–1881. doi: 10.1101/gad.1110503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y, Reinberg D. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev. 2001;15:2343–2360. doi: 10.1101/gad.927301. [DOI] [PubMed] [Google Scholar]
- 11.Lachner M, Jenuwein T. The many faces of histone lysine methylation. Curr. Opin. Cell Biol. 2002;14:286–298. doi: 10.1016/s0955-0674(02)00335-6. [DOI] [PubMed] [Google Scholar]
- 12.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 13.Metzger E, Wissmann M, Yin N, Müller JM, Schneider R, Peters AH, Günther T, Buettner R, Schüle R. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- 14.Klose RJ, Kallin EM, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nat. Rev. Genet. 2006;7:715–727. doi: 10.1038/nrg1945. [DOI] [PubMed] [Google Scholar]
- 15.Secombe J, Li L, Carlos L, Eisenman RN. The Trithorax group protein Lid is a trimethyl histone H3K4 demethylase required for dMyc-induced cell growth. Genes Dev. 2007;21:537–551. doi: 10.1101/gad.1523007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee MG, Norman J, Shilatifard A, Shiekhattar R. Physical and functional association of a trimethyl H3K4 demethylase and Ring6a/MBLR, a polycomb-like protein. Cell. 2007;128:877–887. doi: 10.1016/j.cell.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 17.Eissenberg JC, Lee MG, Schneider J, Ilvarsonn A, Shiekhattar R, Shilatifard A. The trithorax-group gene in Drosophila little imaginal discs encodes a trimethylated histone H3 Lys4 demethylase. Nat. Struct. Mol. Biol. 2007;14:344–346. doi: 10.1038/nsmb1217. [DOI] [PubMed] [Google Scholar]
- 18.Yamane K, Tateishi K, Klose RJ, Fang J, Fabrizio LA, Erdjument-Bromage H, Taylor-Papadimitriou J, Tempst P, Zhang Y. PLU-1 is an H3K4 demethylase involved in transcriptional repression and breast cancer cell proliferation. Mol. Cell. 2007;25:801–812. doi: 10.1016/j.molcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 19.Klose RJ, Yan Q, Tothova Z, Yamane K, Erdjument-Bromage H, Tempst P, Gilliland DG, Zhang Y, Kaelin W.GJ. The Retinoblastoma Binding Protein RBP2 is an H3K4 demethylase. Cell. 2007;128:889–900. doi: 10.1016/j.cell.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 20.Tahiliani M, Mei P, Fang R, Leonor T, Rutenberg M, Shimizu F, Li J, Rao A, Shi Y. The histone H3K4 demethylase SMCX links REST target genes to X-linked mental retardation. Nature. 2007;447:601–605. doi: 10.1038/nature05823. [DOI] [PubMed] [Google Scholar]
- 21.Liang G, Klose RJ, Gardner KE, Zhang Y. Yeast Jhd2p is a histone H3 Lys4 trimethyl demethylase. Nat. Struct. Mol. Biol. 2007;14:243–245. doi: 10.1038/nsmb1204. [DOI] [PubMed] [Google Scholar]
- 22.Seward DJ, Cubberley G, Kim S, Schonewald M, Zhang L, Tripet B, Bentley DL. Demethylation of trimethylated histone H3 Lys4 in vivo by JARID1 JmjC proteins. Nat. Struct. Mol. Biol. 2007;14:240–242. doi: 10.1038/nsmb1200. [DOI] [PubMed] [Google Scholar]
- 23.Lee N, Zhang J, Klose RJ, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. The trithorax-group protein Lid is a histone H3 trimethyl-Lys4 demethylase. Nat. Struct. Mol. Biol. 2007;14:341–343. doi: 10.1038/nsmb1216. [DOI] [PubMed] [Google Scholar]
- 24.Agger K, Cloos P.AC, Christeen J, Pasini D, Rose S, Rappsilber J, Issaeva I, Canaani E, Salcini AE, Helin K. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- 25.De Santa F, Totaro MG, Prosperini E, Notarbartolo S, Testa G, Natoli G. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell. 2007;130:1–12. doi: 10.1016/j.cell.2007.08.019. [DOI] [PubMed] [Google Scholar]
- 26.Lan F, Bayliss PE, Rinn JL, Whetstine JR, Wang JR, Chen S, Iwase S, Alpatov R, Issaeva I, Canaani E, et al. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature. 2007;449:689–694. doi: 10.1038/nature06192. [DOI] [PubMed] [Google Scholar]
- 27.Lee MG, Villa R, Trojer P, Norman J, Yan K.-P, Reinberg D, Di Croce L, Shiekhattar R. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science. 2007;318:447–450. doi: 10.1126/science.1149042. [DOI] [PubMed] [Google Scholar]
- 28.Gildea JJ, Lopez R, Shearn A. A screen for new trithorax group genes identified little imaginal dics, the Drosophila melanogaster homologue of human retinoblastoma binding protein 2. Genetics. 2000;156:645–663. doi: 10.1093/genetics/156.2.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tartof KD, Hobbs C, Jones M. A structural basis for variegating position effects. Cell. 1984;37:869–878. doi: 10.1016/0092-8674(84)90422-7. [DOI] [PubMed] [Google Scholar]
- 30.Brand AH, Perrimon N. Targeted expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 31.Hubbard TJ, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, Cutts T, et al. Ensembl 2007. Nucleic Acids Res. 2007;35:D610–D617. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, Khanna A, Marshall M, Moxon S, Sonnhammer EL, et al. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl Acad. Sci. USA. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Letunic I, Copley RR, Pils B, Pinkert S, Schultz J, Bork P. SMART 5: domains in the context of genomes and networks. Nucleic Acids Res. 2006;34:D257–D260. doi: 10.1093/nar/gkj079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Needleman SB, Wunsch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 37.Kruskal JB. An overview of sequence comparison. In: Sankoff D, Kruskal JB, editors. Time Warps, String Edits and Macromolecules: The Theory and Practice of Sequence Comparison. Massachussetts: Addison Wesley; 1983. pp. 1–33. [Google Scholar]
- 38.Smith TF, Waterman MS. Identification of common molecular subsequences. J. Mol. Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- 39.Di Nocera PP, Dawid IB. Transient expression of genes introduced into cultured cells of Drosophila. Proc. Natl Acad. Sci. USA. 1983;80:7095–7098. doi: 10.1073/pnas.80.23.7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dequier E, Souid S, Pál M, Mároy P, Lepesant JA, Yanicostas C. Top-DER- and Dpp-dependent requirements for the Drosophila fos/kayak gene in follicular epithelium morphogenesis. Mech. Dev. 2001;106:47–60. doi: 10.1016/s0925-4773(01)00418-x. [DOI] [PubMed] [Google Scholar]
- 41.Cortés A, Huertas D, Fanti L, Pimpinelli S, Marsellach FX, Piña B, Azorín F. DDP1, a single-stranded nucleic acid-binding protein of Drosophila, associates with pericentric heterochromatin and is functionally homologous to the yeast Scp160p, which is involved in the control of cell ploidy. EMBO J. 1999;18:3820–3833. doi: 10.1093/emboj/18.13.3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.White RA, Wilcox M. Protein products of the bithorax complex in Drosophila. Cell. 1984;1984:163–171. doi: 10.1016/0092-8674(84)90202-2. [DOI] [PubMed] [Google Scholar]
- 43.Katoh M, Katoh M. Identification and characterization of JMJD2 family genes in silico. Int. J. Oncol. 2004;24:1623–1628. [PubMed] [Google Scholar]
- 44.Whetstine JR, Nottke A, Lan F, Huarte M, Smolikov S, Chen Z, Spooner E, Li E, Zhang G, Colaiacovo M, et al. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell. 2006;125:1–15. doi: 10.1016/j.cell.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 45.Tsukada Y-I, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 46.Chen Z, Zang J, Whetstine JR, Hong X, Davrazou F, Kutateladze TG, Simpson M, Mao Q, Pan C.-H, Dai S, et al. Structural insights into histone demethylation by JMJD2 family members. Cell. 2006;125:691–702. doi: 10.1016/j.cell.2006.04.024. [DOI] [PubMed] [Google Scholar]
- 47.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 48.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 49.Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ, Gingeras TR, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 50.Wirbelauer C, Bell O, Schubeler D. Variant histone H3.3 is deposited at sites of nucleosomal displacement throughout transcribed genes while active histone modifications show a promoter-proximal bias. Genes Dev. 2005;19:1761–1766. doi: 10.1101/gad.347705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell. 2003;11:709–719. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 52.Rudolph T, Yonezawa M, Lein S, Heidrich K, Kubicek S, Schäfer C, Phalke S, Walther M, Schmidt A, Jenuwein T, et al. Heterochromatin formation in Drosophila is initiated through active removal of H3K4 methylation by the LSD1 homolog SU(VAR)3-3. Mol. Cell. 2007;26:103–115. doi: 10.1016/j.molcel.2007.02.025. [DOI] [PubMed] [Google Scholar]
- 53.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 54.Shahbazian MD, Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 2007;76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114. [DOI] [PubMed] [Google Scholar]
- 55.Bell O, Wirbelauer C, Hild M, Scharf AN, Schwaiger M, MacAlpine DM, Zilbermann F, van Leeuwen F, Bell SP, Imhof A, et al. Localized H3K36 methylation states define histone H4K16 acetylation during transcriptional elongation in Drosophila. EMBO J. 2007;26:4974–4984. doi: 10.1038/sj.emboj.7601926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Eissenberg JC, Shilatifard A. Leaving a mark: the many footprints of the elongating RNA polymerase II. Curr. Opin. Genet. Dev. 2006;16:184–190. doi: 10.1016/j.gde.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 57.Kizer KO, Phatnani HP, Shibata Y, Hall H, Greenleaf AL, Strahl BD. A novel domain in Set2 mediates RNA polymerase II interaction and couples histone H3 K36 methylation with transcript elongation. Mol. Cell. Biol. 2005;25:3305–3316. doi: 10.1128/MCB.25.8.3305-3316.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Krogan NJ, Kim M, Tong A, Golshani A, Cagney G, Canadien V, Richards DP, Beattie BK, Emili A, Boone C, et al. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 2003;23:4207–4218. doi: 10.1128/MCB.23.12.4207-4218.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ng SS, Kavanagh KL, McDonough MA, Butler D, Pilka ES, Lienard B.MR, Bray JE, Savitsky P, Gileadi O, von Delft F, et al. Crystal structures of histone demethylase JMJD2A reveal basis for substrate specificity. Nature. 2007;448:87–91. doi: 10.1038/nature05971. [DOI] [PubMed] [Google Scholar]
- 60.Chen Z, Zang J, Kappler J, Hong X, Crawford F, Wang Q, Lan F, Jiang C, Whetstine J, Dai S, et al. Structural basis of the recognition of a methylated histone tail by JMJD2A. Proc. Natl Acad. Sci. USA. 2007;104:10818–10823. doi: 10.1073/pnas.0704525104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Couture J-F, Collazo E, Ortiz-Tello PA, Brunzelle JS, Trievel RC. Specificity and mechanism of JMJD2A, a trimethyllysine-specific histone demethylase. Nat. Struct. Mol. Biol. 2007;14:689–695. doi: 10.1038/nsmb1273. [DOI] [PubMed] [Google Scholar]
- 62.Roguev A, Schaft D, Shevchenko A, Aasland R, Shevchenko A, Stewart AF. High conservation of the Set1/Rad6 axis of histone 3 lysine 4 methylation in budding and fission yeasts. J. Biol. Chem. 2003;278:8487–8493. doi: 10.1074/jbc.M209562200. [DOI] [PubMed] [Google Scholar]
- 63.Papp B, Müller J. Histone trimethylation and the maintenance of transcriptional ON and OFF states by trxG and PcG proteins. Genes Dev. 2006;20:2041–2054. doi: 10.1101/gad.388706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vermeulen M, Mulder KW, Denissov S, Pijnappel W.W.MP, van Schaik F.MA, Varier RA, Baltissen M.PA, Stunnenberg HG, Mann M, Timmers H.TM. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell. 2007;131:58–69. doi: 10.1016/j.cell.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 65.Muchardt C, Guillemé M, Seeler J-S, Trouche D, Dejean A, Yaniv M. Coordinated methyl and RNA binding is required for heterochromatin localization of mammalian HPIα. EMBO Rep. 2002;3:975–981. doi: 10.1093/embo-reports/kvf194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nielsen AL, Oulad-Abdelghani M, Ortiz JA, Remboutsika E, Chambon P, Losson R. Heterochromatin formation in mammalian cells: interaction between histones and HP1 proteins. Mol. Cell. 2001;7:729–739. doi: 10.1016/s1097-2765(01)00218-0. [DOI] [PubMed] [Google Scholar]
- 67.Zhao T, Heyduk T, Allis CD, Eissenberg JC. Heterochromatin protein 1 binds to nucleosomes and DNA in vitro. J. Biol. Chem. 2000;275:28332–28338. doi: 10.1074/jbc.M003493200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.