Abstract
Nucleotide composition varies greatly among DNA viruses of animals, yet the evolutionary pressures and biological mechanisms driving these patterns are unclear. One of the most striking discrepancies lies in the frequency of CpG (the dinucleotide CG, linked by a phosphate group), which is underrepresented in most small DNA viruses (those with genomes below 10 kb) but not in larger DNA viruses. Cytosine methylation might be partially responsible, but research on this topic has focused on a few virus groups. For several viruses that integrate their genome into the host genome, the methylation status during this stage has been studied extensively, and the relationship between methylation and viral-induced tumor formation has been examined carefully. However, for actively replicating viruses—particularly small DNA viruses—the methylation status of CpG motifs is rarely known and the effects on the viral life cycle are obscure. In vertebrate host genomes, most cytosines at CpG sites are methylated, which in vertebrates acts to regulate gene expression and facilitates the recognition of unmethylated, potentially pathogen-associated DNA. Here we briefly introduce cytosine methylation before reviewing what is currently known about CpG methylation in DNA viruses.
INTRODUCTION
CpG underrepresentation in vertebrate genomes
The denotation ‘CpG’ is shorthand for the occurrence of a cytosine linked, through a phosphate bond, to a guanine. CpGs are underrepresented in most eukaryote genomes, but the frequency varies widely among species and is negatively correlated with the presence and extend of cytosine methylation in the genome (1,2). In vertebrate genomes, CpGs are present at one-third to one-fourth of the expected frequency, yet the reasons are disputed (1–4). Cytosine and guanine tend to have higher stacking energies than adenine and thymine, so structural constrains may be important in CpG avoidance (5). In many species, the proportion of tRNAs containing CpG in their anticodons is lower than that of tRNAs with other dinucleotides; therefore, the transcription efficiency might be higher for codons not containing CpGs (6). Another explanation may lie in the fact that unmethylated CpGs can stimulate innate immune responses, potentially resulting in autoimmune reactions (7–9); therefore, large numbers of CpGs may be detrimental if not all are methylated. Finally, methylated cytosines have a tendency for spontaneous deamination, which may also account for the CpG depletion (10–12). While the deamination of unmethylated cytosines leads to uracil and can be corrected by cellular DNA repair machinery, the transition of methylated cytosines to thymines is irreversible, leading to elevated mutation frequencies in highly methylated genomes.
In general, vertebrate genomic CpGs are highly methylated. Sixty to 90% of genomic CpGs are thought to be in a methylated state (13,14), but both CpG frequency and methylation patterns can vary widely across a single vertebrate genome. Notably, some regions are CpG-enriched yet practically devoid of methylation. These sequence stretches, termed ‘CpG islands’, are >500 bp in length and comprise ∼1% of total genomic DNA [e.g. the human genome contains more than 29 000 such islands, with estimates reaching as high as 45 000 islands per haploid genome (15,16)]. The islands are often associated with 5′ promoter regions of housekeeping genes [for reviews see, among others, (15,17,18)].
Vertebrate CpG methyltransferases
Vertebrate genomes are methylated by DNA methyltransferases (DNMTs) which convert cytosine to 5-methylcytosine (5Me-cytosine). DNMTs are functionally divided into ‘de novo’ (DNMT3a and DNMT3b) and ‘maintenance’ (DNMT1) methyltransferases (Table 1). Their catalytic domains appear highly conserved across species and S-adenosyl methionine (SAM) appears to function as the only methyl donor [reviews of mammalian DNA methyltransferases can be found in (19–22)]. DNMT expression levels in nontransformed tissues have rarely been studied explicitly. The available data indicates the translation of all three DNMTs in the majority of tissues, but translation levels seem to vary between tissues, cell differentiation levels, host developmental stages and potentially host species. Robertson et al. (23) analyzed DNMT mRNA levels in various human tissues and found DNMT1, 3a and 3b expressed in nearly all analyzed fetal and adult tissues. In adult tissues, DNMT1 appeared to be generally more highly expressed than DNMT3a and 3b, and DNMT1 mRNA was detected in all analyzed tissues except the small intestine. DNMT3a and 3b mRNAs were detected in large quantities in heart, skeletal muscle, thymus, kidney, liver and peripheral blood mononuclear cells, but were present at low levels in all other analyzed tissues. DNMT1 and 3a seemed to be expressed at high levels in all analyzed fetal tissues, while DNMT 3b was expressed at high levels only in fetal liver. The authors proposed fetal hepatic hematopoiesis as the reason for the observed high levels of DNMT3b in the fetal liver. In bovine fetuses, Golding et al. (24) found DNMT3b highly expressed in rumen, kidney, testes and lung while the highest mRNA levels were again detected in the liver. MessengerRNAs of all three DNMTs were again detected in all of the adult bovine tissues analyzed, but highest levels were detected in kidney, brain and testes. Mizuno et al. (25) detected mRNAs for all three DNMTs in human neutrophils, monocytes, T lymphocytes, bone marrow cells and CD34-positive immune cells, but mRNA levels varied between cell types. DNMT1 appeared to be expressed at high levels in all cell types except neutrophiles, while DNMT3a mRNA levels in neutrophiles and T lymphocytes appeared high. DNMT3b mRNA levels were low in differentiated cells such as neutrophiles, but high in bone marrow and particularly CD34-positive cells.
Table 1.
Overview of vertebrate DNA methyltransferases and their functions
| DNMT | Functional role | Reference |
|---|---|---|
| De-novo DNMTs | ||
| DNMT 3a | Embryonic development, methylation of CpG sites, meiosis induction in sperm(?) | (110–112) |
| DNMT 3b | Embryonic development, spermatogenesis(?) | (110,113) |
| DNMT 3L | Maternal genomic imprinting, silencing of retrotransposons in spermatogonial stem cells | (114) |
| Maintenance DNMTs | ||
| DNMT 1 | Cellular maintenance methylation, maintenance of imprinting, silencing of mobile elements during genomic demethylation, contribution to histone deacetylases | (115–118) |
| DNMT 2 | Unclear. Methylation in Drosophila melanogaster(?) | (119) |
The enzymatic functions attributed to known DNMTs are summarized here, and some key references are provided. Some simplifications were made for the purpose of clarity, and the reader is referred to specific reviews of DNMTs, indicated in the text, for more detail. Cases where the functional role has been proposed, but not yet established conclusively, are indicated by question marks.
Methylation patterns are generally stable, sustained by DNMT1 and are inherited by both daughter DNA molecules during mitosis. However, during meiosis both parental genomes are demethylated prior to fertilization, and cellular methylation patterns become established de novo during embryogenesis. This process usually starts during implantation and is finished by the end of gastrulation (26). The development of site-specific methylation patterns is crucial for normal embryonic development (27–29) and is responsible for genomic imprinting and for X chromosome silencing (30,31).
Methylation-induced gene silencing
CpG methylation acts to suppress transcription in several ways (32). It can directly prevent the binding of transcription factors to the promoter regions of genes, most likely due to steric hindrance. Alternatively, a number of proteins, known as methyl-CpG binding (MeCP) proteins, can selectively bind methylated CpG sites. One of these, Kaiso, recognizes methylated CpG sites through a zinc-finger motif, while another subgroup shares a conserved methyl-CpG-binding domain (MBD). The MBD proteins MBD 1, 2 and MeCP2, as well as Kaiso, all function as transcriptional repressors. MBD 3, however, associates with the nucleosome remodeling and histone deacetylation (NuRD) complex, a co-repressor complex containing histone deacetylases. Deacetylation of histones increases their affinity for DNA, leading to the formation of inactive heterochromatin (30,33,34).
Methylation-induced silencing of foreign DNA
Methylation is involved in the inactivation of integrated foreign DNA, such as retrotransposons, proviral sequences and other transposable elements (TEs) (33). These ‘parasitic’ DNAs constitute a quantitatively significant portion of mammalian genomes, comprising, for example, ∼40% of the human genome. They are notably GC rich and, within mammalian genomes, are hypermethylated at CpG motifs. In several studies, the inhibition of de novo or maintenance methylation was associated with increased transcription levels of TEs, indicating that effective TE silencing requires both de novo and maintenance methylation, likely catalyzed by DNMT3L and DNMT1, respectively (35,36).
Methylation and innate immunity
CpG methylation also plays an important role in immune surveillance and the detection of pathogens, as unmethylated CpGs are a signature of bacterial and other pathogen-associated DNA. CpG-containing oligodeoxynucleotides (ODNs), even as short as 6 nt in length, can be potent stimulators of the vertebrate immune system, resulting in the activation of a wide variety of immune cells such as B cells, NK cells, monocytes/macrophages and dendritic cells (DCs). Stimulation with CpG ODNs directly activates plasmacytoid dendritic cells (PDCs) and B cells. The PDCs secrete large amounts of IFN-α and IL-12 which subsequently stimulate monocytes, myeloid DCs, T cells and type-1 NK cells (37–39). One pathway through which unmethylated CpGs trigger innate immune responses is by binding a member of the Toll-like receptor (TLR) family, TLR9 [see, as one example, (40)]. TLR9 is expressed at high levels on subsets of human and mouse DCs and gene homologs have been found in many other mammalian species. The degree to which particular CpG sequences stimulate immune responses depends upon their flanking nucleotide sequences, and motifs which confer maximal stimulation are at least partially species-specific (41). For example, ODNs containing the motif GACGTT very efficiently stimulate murine and rabbit B cells, but only weakly stimulate human B cells, which are most effectively stimulated by GTCGTT (42). Some similarities in recognition sequences among related species appear likely. For example, GTCGTT motifs are highly immune stimulatory in a variety of vertebrates including humans, dogs, cats, cows, chicken and nonhuman primates (41). ODNs that are thymine rich at the 3′ end and contain a TpC dinucleotide at their 5′ end appear to be generally potent immune stimulators, while those that contain CpG motifs toward the 3′ end appear to be less immunogenic (41). Generalities, however, may be misleading in some cases, as resulting immune effects are thought to be highly cell-type and species specific (43–45).
CpG dinucleotides in vertebrate genomes are most commonly preceded by a C and followed by a G (46), a signature which does not appear to be immune stimulatory. Motifs such as TTCCGA, on the other hand, appear to be potent stimulators of the human immune system and may play a role in autoimmune diseases such as lupus erythematosus. Other CpG motifs, for example those containing poly(G) sequences, are thought to inhibit NF-κB-induced immune stimulation (47). The relatively low occurrence of stimulatory motifs in mammalian DNA might explain the lower immunogenicity of unmethylated vertebrate DNA compared to bacterial DNA.
Stimulatory motifs in DNA virus genomes
The genomes of some, but not all, viruses are also lacking many such stimulatory sequences. For example, likely stimulatory sequences are highly underrepresented in the genomes of adenovirus serotypes 2 and 5, but not in serotype 12. Adenovirus type 2 and 5 can cause persistent infections, their DNA is not immune stimulatory and it might even suppress immune stimulation by DNA from other sources. Type 12, however, does not cause persistent infections, and its DNA appears highly immune stimulatory. Experimental evidence for a correlation between the stimulatory potential of the viral DNA and the number of supposedly stimulatory motifs has been presented (48).
Table 2 summarizes the frequencies of certain putatively stimulatory and nonstimulatory motifs in genomes from several members of different viral families. General conclusions are difficult to draw since the biological effects of individual motifs likely differ between virus families and host species. Moreover, the limited genome size and CpG underrepresentation complicates inference for small DNA viruses. However, certain patterns appear noticeable even without using formal statistical inference. The ratio of total stimulatory to total nonstimulatory motifs is higher than expected based on the viral nucleotide composition for Frog adenovirus, Frog virus 3 and herpes simplex virus type-1 (HSV1), while it is lower than expected for all other viruses for which this value can be calculated. Putatively stimulatory motifs are rarely present more frequently than expected based on the viral nucleotide composition, with the exception of HSV1 and Frog virus 3. Putatively nonstimulatory motifs, on the other hand, are present at higher frequency than expected in several cases, as seen for several adeno- and herpesviruses. The driving forces determining this nucleotide composition are difficult to disentangle. HSV1 induces mainly subclinical, persistent and recurrent infections but HSV1 DNA has been shown to efficiently stimulate TLR9 responses (49). The mammalian genomes examined contain a functional TLR9 ortholog, but chickens lack an orthologous TLR9 gene, and the same is likely true for most if not all avian species. Unfortunately, data on other avian species are thus far limited (50) and it is not known, in most cases, whether amphibian species have a functional TLR9. Several other factors likely influence presence and distribution of CpG-containing motifs, but many of these remain to be understood.
Table 2.
Frequency of TLR9 stimulatory and non-stimulatory/inhibitory sequences
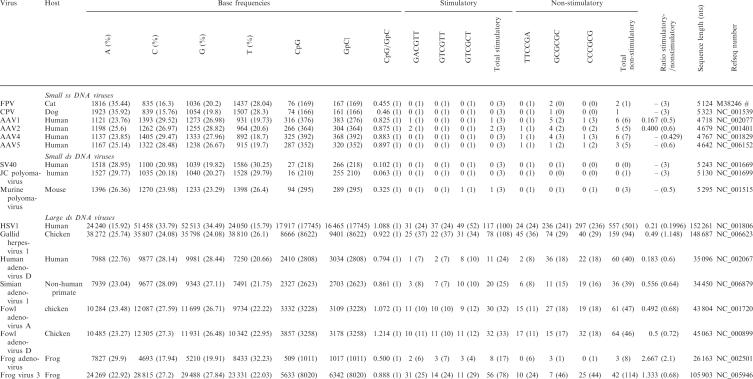 |
GpC and CpG contents, as well as individual nucleotide and motif frequencies, were determined from the publicly available reference sequences (RefSeq) in Genbank (MatLab script available from the authors upon request). Sequence length, frequencies and percentages of individual nucleotides, GpC and CpG content, as well as the frequencies of potentially stimulating and non-stimulating motifs, are provided. Potentially stimulatory sequences are those described by Rankin et al. (120); values represent the total number of the respective motif present in the genome. The number in parentheses indicates the number of motifs expected in the sequence, given the individual nucleotide composition. This was calculated using the following formula: E(UVWXYZ) = 10−12* sequence length*(%U*%V*%W*%X*%Y*%Z), where E(UVWXYZ) = expected value of the motif UVWXYZ; U,V,W,X,Y,Z = nucleotides of which the motif consists, and %U = % of viral sequence consisting of base U. Expected values were rounded to the nearest integer. Likely non-stimulatory and potentially inhibitory sequences are those described by Krieg; values again represent the total number of the respective motif present in the genome, and expected values were calculated as describe above. ‘Total stimulatory’ or ‘total non-stimulatory’ values represents the sum of all putatively stimulatory or non-stimulatory motifs in the sequence. ‘Ratio of stimulatory/non-stimulatory sequences’ represent the fraction of the total number of stimulatory divided by the total number of non-stimulatory sequences. ‘CpG’ or ‘GpC’ represents the total number of the respective dinucleotide CpG or GpC in the viral DNA sequence. ‘CpG/GpC’ represents fraction of the number of CpG motifs divided by the number of GpC motifs in the viral DNA. ‘RefSeq’ number represent GeneBank accession number. ‘#’ no RefSeq FPV sequence available.
DNA VIRUSES AND CYTOSINE METHYLATION
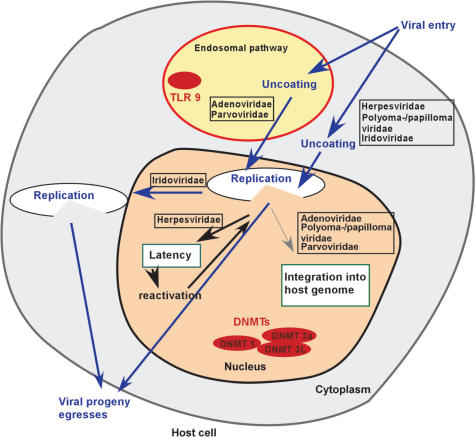
To understand the impact of cytosine methylation on the viral life cycle and the evolution of base composition, the particularities of each virus will need to be considered. Differences will inevitably exist between actively replicating viral DNA and that which is integrated into the host genome. The type of viral persistence will also be of importance. The integration of adeno- or polyomavirus DNA into the host genome is usually a terminal process since the viruses cannot liberate their genomes and are therefore no longer infectious. The evolutionary roles of methylation in these cases will likely differ from those seen in other viruses, such as Herpesviruses, which can liberate their genome after periods of latency. But differences may also exist between large and small viruses—with many larger viruses encoding their own replication machinery and additional proteins which modify host cell processes and immune responses. The susceptibility of the viral genome to methylation and immune recognition will also be affected by other factors, such as the location of replication within the cell and the specific intracellular trafficking route (Figure 1).
Figure 1.
Comparison of DNA virus infection pathways. Major intracellular trafficking routes and characteristics of replication are shown for the DNA virus families discussed. The importance of methylation and immune recognition is indicated. Simplifications and generalizations were made for the purpose of clarity.
Viruses that integrate into the host genome and the effects of methylation on their life cycles have long been of particular interest. Many studies have focused on adenoviruses, but comprehensive knowledge of the methylation status of other viruses, such as polyomaviruses and herpesviruses, has also been obtained (Table 3).
Table 3.
Overview of GC content, CpG frequency and methylation status of small and large DNA viruses
| Virus | Genome sizea (kb) | GC frequencya | CpG contenta (ρCPG) | Methylation status during active replication | Methylation status during latency | Host species | Effect on host methylation | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Replicating | Referenceb | Integrated | Episomal | Referenceb | ||||||
| Large dsDNA viruses | ||||||||||
| Adenoviridae | 28–45 | 0.3–0.65 | 0.5–1.13 | Un/hypomethylated | (121) | Methylated | – | (121) | Mammal, bird | DNMT upregulation |
| Alpha-herpesvirinae | 130–150 | 0.4–0.71 | 0.9–1.17 | Un/hypomethylated | (67) | – | Un/hypo- mehtylated | (89) | Mammal, bird | DNMT upregulation |
| Beta-herpesvirinae | 140–240 | 0.4–0.67 | 1.0–1.25 | Unknown | N/A | – | Unknown | N/A | Mammal | Unknown |
| Gamma-herpesvirinae | 110–185 | 0.3–0.61 | 0.3–0.66 | Un/hypomethylated | (66) | – | Methylated | (65) | Mammal | DNMT upregulation |
| (122) | ||||||||||
| Ranid herpesvirusc | 220–230 | 0.5–0.55 | 0.8–0.95 | Methylated | (70) | – | Unknown | N/A | Amphibian | Viral 5-cytosine methyltransferases? |
| Iridoviridae | 140–383 | 0.2–0.56 | 0.5–0.84 | Methylated | (74) | – | – | – | Amphibian, fish | Viral 5-cytosine methyltransferases? |
| Poxviridae | 130–375 | 0.2–0.64 | 0.8–1.23 | Unknown | N/A | – | – | – | Mammal bird invertebrate | Unknown |
| Small dsDNA viruses | ||||||||||
| Papilloma-viridae | 7–8 | 0.4–0.54 | 0.1–0.57 | Partially methylated | (60) | Methylated | – | (123) | Mammal | DNMT upregulation |
| Polyoma-viridae | 5 | 0.4–0.48 | 0.05–0.78 | Un/hypomethylated | (51) | Methylated | – | (51) | Mammal, bird | DNMT upregulation |
| Small ssDNA viruses | ||||||||||
| Autonomous Parvoviridae | 4–6 | 0.3–0.5 | 0.3–0.71 | Unknown | N/A | – | – | – | Mammal | Unknown |
| Dependo-virinae | 4–6 | 0.4–0.58 | 0.6–1.03 | Unknown | N/A | – | – | – | Mammal | Unknown |
| Circoviridae | 2 | 0.5–0.57 | 0.4–0.87 | Unknown | N/A | – | – | – | Mammal, bird | Unknown |
| Anellovirus | 4 | 0.5 | 0.67 | Unknown | N/A | – | – | – | Human | Unknown |
The GC content, CpG content and, where known, methylation status is shown for viral families/subfamilies. Where applicable, a distinction is made between active replication and latency. If latent, the state of the genome (i.e. integrated or episomal) is specified. Preferred host species are indicated along with any known effect the virus has on host cell methylation. Where relevant, representative references are provided. Dash indicates this form is not known to occur for this virus.
aGC and CpG contents as determined by authors [here or in (83)] according to methods described therein); GC content represents the relative frequency of G and C in the sequence. CpG content represents the observed divided by the expected frequency of the dinucleotide CpG in the sequence.
bReference refers to a representative study, which focuses on one member of the virus family.
cUnpublished results, analysis done by authors as described above (83).
Early studies of viral methylation focused on DNA from polyomavirus-infected cells (51). The early methods (e.g. quantification of methyl-H3 incorporation into the genome) used to measure methylation were limited in their sensitivity and did not show the genomic distribution of methylated sites. Technical advances later made it possible to study cytosine methylation in a more detailed and site-specific manner. However, the various roles of methylation during persistent viral infections, in the silencing of viral genomes, and in immune evasion and tumor formation are still being uncovered.
The impact of methylation during active viral replication is generally incompletely understood, and for many small DNA viruses it is currently not known whether the viral genome is methylated during active replication.
Adenovirus DNA methylation: summarizing technical advances in viral methylation studies
Early studies of adenovirus methylation relied on chromatographic and radioactive techniques to distinguish methylated from unmethylated bases. The techniques were complicated by limited sensitivity, the difficulty of isolating pure viral fractions and the inability to discriminate between host and integrated viral DNA. The investigators nevertheless carefully compared actively replicating and integrated viral DNA and their conclusions have stood the test of time. Genomic DNA from adenovirus type 2 and 12 infected cells was heavily methylated, more than cellular DNA from uninfected control cells (52). Whether the integration of viral DNA altered methylation patterns in the host genome or whether the observed differences were due exclusively to the integrated viral DNA was not determined. However, actively replicating adenovirus type 2 and 12 DNA appeared to have few or no methylated bases (52).
More sensitive analysis was allowed by pairs of methylation-sensitive and insensitive restriction endonucleases. In particular, the isoschizomers HpaII and MspI have been used frequently in studies of DNA methylation. Both enzymes recognize the motif CCGG, but the catalytic activity of HpaII is inhibited by CpG methylation while the activity of MspI is not affected. The new technique verified the previous results with actively replicating adenovirus type 2 and 12 DNA showing no detectable methylated CCGG sites and the restriction patterns were consistent among viral DNAs from different cells (53). Inference from restriction enzyme analyses, however, remained limited to those sites recognizable by restriction enzyme pairs. Several subsequent studies focused on adenovirus DNA and the absence of methylated sites could later be conclusively established through bisulfate sequencing (51–53). Bisulfate converts unmethylated cytosines to thymines and thus allows the sensitive detection of specific methylated sites by sequencing the genome prior to and after treatment. The absence of methylated sites in the replicating adenovirus 2 genome has been shown using this technique (54–56).
Many studies of adenovirus DNA have focused on integrated viral genomes or genomic segments, largely because of their oncogenic potential. Integrated adenoviral DNA is known to be hypermethylated, but the regulatory events which determine the methylation status of replicating or integrated sequences are not well understood. A recombinant between adenovirus 12 and host cell DNA (referred to as SYREC) is unmethylated over the whole genome during active replication including the host-derived sequence, which is methylated in the chromosome. This led to speculations about the existence of adenovirus-encoded de-methylation proteins, or a role for the virus-encoded replication machinery in methylation avoidance (57). However, no specific mechanisms have been defined, and the speed of viral replication or compartmentalization of this process within the host cell has also been proposed as factors contributing to the lack of methylation.
Methylation status of polyoma- and papillomaviruses
Studies of papovavirus DNA methylation also focused on integrated sequences, mostly of human papillomavirus strains and SV40 polyomavirus. Polyomavirus and papillomavirus genomes are CpG-depleted (Table 3) and any CpG motifs present are clustered within certain regions of the viral genome (58), complicating the analysis of methylation patterns. The early studies, however, concluded that there was little methylation in replicating polyomaviruses (51), with the possible exception of a CpG site near the early promoter (59). Several other studies concurred and found several human papillomavirus strains to be hypomethylated during active replication, again with methylated sites clustering in specific regions of the viral genome. The integrated viral DNA, on the contrary, appeared heavily methylated. However, distinguishing between actively replicating and integrated DNA appears difficult in some cases (60–62).
Methylation status of herpesvirus genomes
Herpesviruses establish latent infections without integrating into the host genome, and reactivate to re-establish active replication. During latency, the circularized viral genome remains quiescent in an episomal state, and can be replicated by the host cell replication machinery. Active (e.g. lytic) viral replication differs in several respects. Production of viral progeny inevitably results in cell lysis, utilizes the viral-encoded replication machinery and involves a separate origin of replication [e.g. in the case of Epstein–Barr Virus (EBV) oriLyt functions as the origin of replication in the lytic cycle while oriP is responsible for replication of episomal EBV DNA (63)]. The methylation status during lytic DNA replication is likely different from that during latency, but most studies have focused on latent infections and the differences have rarely been studied explicitly.
Differences might also exist among herpesvirus subfamilies, as CpG motifs are highly underrepresented in gammaherpesvirus genomes, but not in alpha- or betaherpesvirus genomes (64). The reasons for these differences remain elusive. Gammaherpesviruses cause predominantly persistent, lymphoproliferative diseases. Studies of some gammaherpesviruses, such as EBV and Kaposi's Sarcoma-associated herpesvirus (KSHV), indicate that methylation serves an intricate regulatory role during the viral life cycle (see below). The EBV genome appears to be hypomethylated or unmethylated during lytic infection, but highly methylated during latency, and appears to become demethylated during reactivation (65). There is some evidence for methylation during active replication of another gammaherpesvirus, herpesvirus saimiri, since Kaschka-Dierich et al. (66) found both the linear and circular viral DNA to be heavily methylated. The alphaherpesvirus herpes simplex virus (HSV) appears unmethylated during active replication and latency (67–69) while two frog herpesviruses (ranid herpesvirus 1 and 2) appear to be methylated during replication. The ranid herpesvirus genomes also contain putative DNA cytosine-5 methyltransferases sequences located in ORFs 86 and 120 (70).
Methylation plays a pivotal role in the life cycle of EBV (71,72), where methylation of specific genes appears to be involved in the transition from lytic to latent infection. Most EBV nuclear antigens (EBNAs) are expressed only for short periods during the lytic infection, after which their transcription is silenced through hypermethylation of the viral promoters Cp and Wp. Production of the indispensable EBNA1 is then achieved by expression from an alternative ‘TATA-less’ promoter Qp. Reinitiation of lytic infection can occur spontaneously or may be triggered by events such as immunoglobulin crosslinking of the host cell and cell proliferation. Reactivation is mediated through the expression of the immediate early genes Zta and Rta, which are regulated by the promoters Zp and Rp, the latter of which is hypermethylated during latency. While Rp hypermethylation is a major regulator of latent infection, Zp hypermethylation is dispensable. EBV therefore seems to use methylation-induced gene silencing as an immune evasion strategy. While a role for methylation in the EBV lifecycle is widely accepted, its importance for other herpesviruses is still disputed.
Methylation status of iridiviruses and ascoviruses
The genomes of iridoviruses appear to be heavily methylated during active replication and methylation might again have a regulatory role. Iridoviruses are large DNA viruses that infect fish, amphibians and reptiles. Initial stages of replication occur in the nucleus, after which the nascent viral genomes are transported into the cytoplasm where the second stage of replication occurs (73). Willis and Granoff (74) analyzed the iridovirus frog virus 3 (FV3) by radioactive labeling/restriction enzyme digestion and found the viral DNA to be heavily methylated, with an estimated 20% of cytosines methylated. Time-course experiments later indicated changing methylation patterns over the course of infection. Willis et al. (75) analyzed nuclear and cytoplasmic extracts collected at various times postinfection (p.i.) and found methylated viral DNA after ∼6–7 h p.i. This DNA appeared to be located in the cytoplasm, while nuclear DNA, collected at earlier time points, appeared to be hypomethylated. The authors also provided evidence against demethylation of parental viral genomes upon host cell infection. Further, Schetter et al. (76) showed that nascent FV3 DNA in the nucleus is unmethylated or hypomethylated at early times p.i, but later becomes hypermethylated (after ∼6 h p.i. in their assay). Cytoplasmic viral DNA, on the other hand, appears to be methylated at all times. The authors reported DNA methyltransferase activity in nuclear extracts of FV3 infected cells, beginning at a time p.i. which coincided with the appearance of methylated viral genomes. This activity was absent from cytoplasmic extracts, indicating FV3 might regulate methylation of its genome. It was later discovered that the FV3 genome encodes a putative DNA cytosine-5 methyltransferase sequence, which bears similarities to other known methyltransferases (77). The putative methyltransferase, however, is more homologous to prokaryotic enzymes than to eukaryotic methyltransferases. While transcription (from an early promoter) and translation of this FV3 protein have been verified, direct evidence for its methylation activity is still limited.
Another iridovirus, fish lymphocystis disease virus (FLDV), was also found to have a highly methylated genome, even though its GC content is considerably lower than that of FV3 (78,79). FLDV also contains a putative 5-cytosine methyltransferase (53.3% identical to that of FV3) (80) but evidence for its function is, thus far, scarce. Besides the frog herpesviruses and iridoviruses, ascoviruses, which infect insects and are closely related to the iridoviruses, are also heavily methylated during replication (81).
Methylation status of small DNA viruses
In contrast to most large DNA viruses, CpG is underrepresented in the majority of small DNA viruses (64,82,83). It is possible that cytosine methylation is partly responsible for this underrepresentation. Small DNA viruses encode only a minimal number of proteins and use the host cell replication machinery for replication. Low CpG frequencies may have been selected to avoid methylation by host methyltransferases, to maximize translation efficiencies, or as a means of reducing CpG mediated immune responses. However, apart from the small polyomaviruses and papillomaviruses described above, there appears to have been few close examinations of the methylation status of small DNA viruses (particularly those that do not integrate), such as autonomous parvoviruses, circoviruses and anelloviruses. The roles and effects of methylation might be different from those seen in integrating and/or large DNA viruses, but further research is needed to understand the specific effects of methylation on small, nonintegrating DNA viruses.
Adeno-associated virus (AAV) integrates into the host genome, and has been examined primarily in the context of guanine methylation. Productive infection with an AAV normally requires co-infection with a helper virus (usually an adenovirus or, less frequently, a herpesvirus). In the absence of a helper virus, AAVs may remain in the cell as persistent dsDNA, or they may be stably integrated into the host genome. Upon infection by a helpervirus, the integrated AAV DNA can be rescued and undergo active replication. The CpG contents of AAVs more closely resemble those of their helper viruses than those of the closely related autonomous parvoviruses, which show strong CpG suppression. The mechanisms of integration have been extensively studied in AAV serotype 2 (AAV2), which predominantly integrates in a specific region of chromosome 19 (locus 19q13.3t). Integration is directed by AAV-encoded Rep proteins, which likely recognize guanine residues within GCTC repeating motifs (located in the AAV terminal hairpin). In vitro methylation of these guanine residues has been shown to effectively inhibit DNA–protein interactions and likely interferes with integration in vivo (84). Similar results were obtained when examining the integration of AAV4 (which utilizes a slightly different recognition motif) into African green monkey cells (85). Nevertheless, it is not known what role, if any, cytosine-5 methylation plays in the natural AAV life cycle, if AAV methylation influences integration, or if guanines or cytosines are methylated during active replication.
METHYLATION AND INTEGRATED OR LATENT VIRAL DNA
Methylation of integrated viral DNA has been studied extensively and the topic is extensively covered in the scientific literature. Areas of interest have included changes in methylation patterns during integration, the effects of viral methylation on host cells and the correlation between virally induced methylation and tumor formation. Excellent reviews have been published on these topics and the reader is referred to these articles, indicated in the following discussion, for additional detail. Here we briefly summarize what is known about methylation during integration and latency and the regulatory elements involved, contrasting the level of understanding to what is known about methylation during active replication.
As already mentioned above, most DNA viruses such as adenoviruses (57,59,86–88) and polyomaviruses (83) appear specifically methylated during latency or when in an integrated form. In contrast, the alphaherpesvirus HSV does not appear to be methylated during latency (89), but the reasons for this difference are unclear. The processes by which integrating viruses are de novo methylated are only partly understood, but it is clear that the extent and pattern of de novo methylation depend upon the time after integration and site in the host genome (90), the integrated sequences (90–94), and possibly the cell type (57,90). De novo methylation tends to be initiated within confined genomic regions and then spreads throughout the viral genome (90,93,95,87). For example, in a study examining the methylation pattern of adenovirus 12 over the course of integration into hamster cells, methylation began in the center of the viral DNA (between map units 30 and 75) and subsequently spread outward in both directions. Some regions of the viral genome remained unmethylated throughout the study, namely the right terminal repeat and regions near the left terminus (95). It is not known which factors initiate methylation of the genome, nor the determinants of the sequential methylation pattern.
Methylation patterns appear to be site specific. In many viruses there seems to be an inverse correlation between viral gene expression and degree of methylation, with late viral genes being generally more susceptible to methylation effects than early genes (53,91). Promoter regions of integrated viruses are frequently hypomethylated. For example, promoter regions in adenovirus 12 are hypomethylated at CCGG sites while the viral genome is otherwise heavily methylated (57,59,96). Methylation of viral genes, particularly at promoter sequences, appears to be a reversible event (96) but the mechanism by which viral genomes are demethylated remains to be determined. Demethylation has also been shown to occur after in vitro methylation of several viral DNAs. For example, SV40 DNA recovered 20 h after microinjection into cells retained the artificial methylation pattern under nonpermissive culture conditions, while allowing early gene transcription. The DNA, however, became demethylated after viral replication (97). Loss of methylation has also been observed in adenoviruses, in the case of the SYRECs mentioned above (86,98) which are methylated when covalently linked to the host genome, but unmethylated during active replication. They require the presence of fully functional adenovirus helper for efficient lytic replication, which led some to postulate a role for the viral transcription machinery in the loss of methylation. Unfortunately, experimental data in support of this hypothesis are so far scarce.
Infection with integrating (and even some nonintegrating) viruses can induce alterations in the methylation patterns of host DNA, a potential factor in the development of malignant tumors. For example, herpesviruses KSHV and EBV, polyomaviruses BK and SV40, papillomavirus HPV, several adenoviruses and hepatitis B virus encode proteins that activate/upregulate DNMT1, 3a, or 3b. This results in the hypermethylation and, therefore, the downregulation of a number of cellular genes. One of the cellular genes frequently hypermethylated in these infections is the cell cycle regulating tumor suppressor gene p16INK4a, which is commonly hypermethylated in many types of cancer (99). Differentiation of host cell tumors and the level of genome methylation of integrated viruses generally appear to be positively correlated, as seen for various papillomaviruses such as Shope papillomavirus (100) and human papillomavirus (HPV) (101,102), the herpesvirus EBV (72,103) and various adenoviruses (96).
METHYLATION AND VIRAL GENE EXPRESSION
For iridoviruses, gene expression at functional levels can be maintained despite genome hypermethylation. Methylated sites, however, are not distributed randomly across the genome and methylation levels differ among CpG motifs. FLDV, for example, has a considerably higher level of methylation at CCGG than CGCG motifs (79). The physiological significance of this, however, is still disputed. In vitro studies of the related FV3 showed that specific methylation (using HpaII methyltransferase) of CCGG motifs in a late promoter, L1140, abolished its function. However, the methylation of all CpG sites by the indiscriminate SssI methyltransferase or the specific methylation of GCGC motifs by HhaI methyltransferase had no dramatic effect. This appears to be a rare example where, counterintuitively, complete methylation of a late promoter does not abolish its function (methylation frequently abolishes the function of late viral promoters, while early promoters tend to be less susceptible to the repressive effect of cytosine methylation). The underlying mechanism has not been elucidated, but steric hindrance in asymmetrically methylated DNA might play a role (104). It seems possible that methylation of CCGG motifs results in complex secondary structures in the promoter region, whereas indiscriminant CpG methylation or methylation of GCGC motifs balances these secondary structures. A similar phenomenon was seen in adenovirus type 2, where methylation of CCGG sites within the (late) E2a promoter inhibited the transcription of viral DNA, while methylation of GCGC motifs had no repressive effect (105,106). The transcription activity of the polyomavirus SV40 was found not drastically decreased when in vitro methylated with indiscriminant DNMTs from rat liver (107,108). In contrast, upon specific methylation of HpaII sites (CCGG-specific), transcription activity was reduced markedly (59). However, the interpretation of these latter results is complicated by the possibility of incomplete methylation by the rat liver extract, as rat liver DNA is a competitive inhibitor of adenovirus methylation (55) and low-level contaminants might likewise have inhibited complete methylation of SV40.
In vitro methylation studies involving human papillomavirus type 16 (HPV16) showed that indiscriminate SssI methylation significantly decreased the transcriptional activity of the long control region (LCR), which is devoid of ORFs but contains several cis-acting regulatory elements. Enhancers located in the LCR are responsive to both cellular and viral factors, such as the viral E2 gene products. Transcriptional silencing was measured using an LCR-containing reporter plasmid and inhibition was again believed to be steric, specifically due to interference with the binding of transcription regulator E2. Notably, hypomethylation of HPV16 E2-binding sites has been observed in highly differentiated, but not in undifferentiated, tumor cells (101). However, direct comparison of the HPV16 and SV40 studies is difficult. The effects of indiscriminate methylation on early and late viral gene products in the genomic context are hard to infer from this study.
The time of transcription appears to impact the effects of methylation in several viruses. For example, the in vitro HpaII (CCGG-specific) methylation of SV40 early genes (i.e. those coding for the large T-antigen) does not lead to transcriptional repression, and the in vitro methylated sites are subsequently lost during active replication (108). Late viral gene expression, however, can be efficiently inhibited by in vitro methylation of HpaII sites within the 5′ part of this region (109). Site-specific methylation might be involved in the transition to latency in some cases (as in the case of EBV). On the other hand, methylation does not seem to be involved in the switch from early to late gene expression in HSV (89). Likewise, the major late promoter of adenovirus 2, which induces the switch from early to late gene expression, is unmethylated in nonintegrated viruses (56), suggesting methylation does not play a major role in the regulation of early versus late gene expression for this virus.
CONCLUSIONS
A number of studies show that CpG methylation can greatly affect the life cycles of DNA viruses, but its exact role in natural infections remains unclear. Those effects likely differ between different viruses and are dependent upon many factors, such as the stage in the viral life cycle, the host species and infected tissue, the flanking nucleotide motifs, the genes in question and the genomic location. Where it has been studied, major differences have been seen for some viruses between the actively replicating and the latent or integrated viral DNA; the former is often unmethylated or hypomethylated, while the later often shows specific and regulated methylation. Differences may also exist between large and small DNA viruses, potentially because of their rate and site of replication and differences in their ability to manipulate the host responses to unmethylated DNA. Thus, it is necessary to not only examine individual viruses, but to examine their dynamic methylation status and the effects of any viral methylation on both the virus and the host cell. The various ways in which mutational bias, codon usage and host selections (e.g. through TLR9-dependent responses) influence CpG frequency and methylation status need to be defined. It will be especially important to determine the unique roles and regulators of methylation, and their divergent effects on the genome evolution of small and large DNA viruses during active or latent infection, and in packaged, integrated or episomal DNA. A deeper knowledge of the relationships among the virus, the host cell and methylation (and de-methylation) machinery will provide essential insights into determinants of methylation status, gene expression, replicative behavior, as well as activation of pattern recognition receptors and immune responses.
ACKNOWLEDGEMENTS
Funding to pay the Open Access publication charges for this article was provided by National Institutes of Health Grant R01 GM080533 to ECH.
REFERENCES
- 1.Bird AP. DNA methylation and the frequency of CpG in animal DNA. Nucleic Acids Res. 1980;8:1499–1504. doi: 10.1093/nar/8.7.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schorderet DF, Gartler SM. Analysis of CpG suppression in methylated and nonmethylated species. Proc. Natl Acad. Sci. USA. 1992;89:957–961. doi: 10.1073/pnas.89.3.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Swartz MN, Trautner TA, Kornberg A. Enzymatic synthesis of deoxyribonucleic acid. XI. Further studies on nearest neighbor base sequences in deoxyribonucleic acids. J. Biol. Chem. 1962;237:1961–1967. [PubMed] [Google Scholar]
- 4.Burge C, Campbell AM, Karlin S. Over- and under-representation of short oligonucleotides in DNA sequences. Proc. Natl Acad. Sci. USA. 1992;89:1358–1362. doi: 10.1073/pnas.89.4.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sponer J, Gabb HA, Leszczynski J, Hobza P. Base-base and deoxyribose-base stacking interactions in B-DNA and Z-DNA: a quantum-chemical study. Biophys. J. 1997;73:76–87. doi: 10.1016/S0006-3495(97)78049-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andersson SG, Kurland CG. Codon preferences in free-living microorganisms. Microbiol. Rev. 1990;54:198–210. doi: 10.1128/mr.54.2.198-210.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krieg AM. A role for toll in autoimmunity. Nat. Immunol. 2002;3:423–424. doi: 10.1038/ni0502-423. [DOI] [PubMed] [Google Scholar]
- 8.Bachmann MF, Kopf M. On the role of the innate immunity in autoimmune disease. J. Exp. Med. 2001;193:F47–F50. doi: 10.1084/jem.193.12.f47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Janeway CA, Jr, Medzhitov R. Innate immune recognition. Ann. Rev. Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 10.Coulondre C, Miller JH, Farabaugh PJ, Gilbert W. Molecular basis of base substitution hotspots in Escherichia coli. Nature. 1978;274:775–780. doi: 10.1038/274775a0. [DOI] [PubMed] [Google Scholar]
- 11.Sved J, Bird A. The expected equilibrium of the CpG dinucleotide in vertebrate genomes under a mutation model. Proc. Natl Acad. Sci. USA. 1990;87:4692–4696. doi: 10.1073/pnas.87.12.4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fryxell KJ, Zuckerkandl E. Cytosine deamination plays a primary role in the evolution of mammalian isochores. Mol. Biol. Evol. 2000;17:1371–1383. doi: 10.1093/oxfordjournals.molbev.a026420. [DOI] [PubMed] [Google Scholar]
- 13.Kass SU, Pruss D, Wolffe AP. How does DNA methylation repress transcription? Trends Genet. 1997;13:444–449. doi: 10.1016/s0168-9525(97)01268-7. [DOI] [PubMed] [Google Scholar]
- 14.Ng HH, Bird A. DNA methylation and chromatin modification. Curr. Opin. Genet. Dev. 1999;9:158–163. doi: 10.1016/s0959-437x(99)80024-0. [DOI] [PubMed] [Google Scholar]
- 15.Antequera F, Bird A. Number of CpG islands and genes in human and mouse. Proc. Natl Acad. Sci. USA. 1993;90:11995–11999. doi: 10.1073/pnas.90.24.11995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McClelland M, Ivarie R. Asymmetrical distribution of CpG in an ‘average’ mammalian gene. Nucleic Acids Res. 1982;10:7865–7877. doi: 10.1093/nar/10.23.7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takai D, Jones PA. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc. Natl Acad. Sci. USA. 2002;99:3740–3745. doi: 10.1073/pnas.052410099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caiafa P, Zampieri M. DNA methylation and chromatin structure: the puzzling CpG islands. J. Cell. Biochem. 2005;94:257–265. doi: 10.1002/jcb.20325. [DOI] [PubMed] [Google Scholar]
- 19.Pradhan S, Esteve PO. Mammalian DNA (cytosine-5) methyltransferases and their expression. Clin. Immunol. 2003;109:6–16. doi: 10.1016/s1521-6616(03)00204-3. [DOI] [PubMed] [Google Scholar]
- 20.Hermann A, Gowher H, Jeltsch A. Biochemistry and biology of mammalian DNA methyltransferases. Cell. Mol. Life Sci. 2004;61:2571–2587. doi: 10.1007/s00018-004-4201-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bird AP, Wolffe AP. Methylation-induced repression–belts, braces, and chromatin. Cell. 1999;99:451–454. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- 22.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 23.Robertson KD, Uzvolgyi E, Liang G, Talmadge C, Sumegi J, Gonzales FA, Jones PA. The human DNA methyltransferases (DNMTs) 1, 3a and 3b: coordinate mRNA expression in normal tissues and overexpression in tumors. Nucleic Acids Res. 1999;27:2291–2298. doi: 10.1093/nar/27.11.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Golding MC, Westhusin ME. Analysis of DNA (cytosine 5) methyltransferase mRNA sequence and expression in bovine preimplantation embryos, fetal and adult tissues. Gene Expression Patterns. 2003;3:551–558. doi: 10.1016/s1567-133x(03)00121-2. [DOI] [PubMed] [Google Scholar]
- 25.Mizuno S, Chijiwa T, Okamura T, Akashi K, Fukumaki Y, Niho Y, Sasaki H. Expression of DNA methyltransferases DNMT1, 3A, and 3B in normal hematopoiesis and in acute and chronic myelogenous leukemia. Blood. 2001;97:1172–1179. doi: 10.1182/blood.v97.5.1172. [DOI] [PubMed] [Google Scholar]
- 26.Salozhin SV, Prokhorchuk EB, Georgiev GP. Methylation of DNA–one of the major epigenetic markers. Biochemistry. 2005;70:525–532. doi: 10.1007/s10541-005-0146-8. [DOI] [PubMed] [Google Scholar]
- 27.Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- 28.Fulka H, Mrazek M, Tepla O, Fulka J., Jr. DNA methylation pattern in human zygotes and developing embryos. Reproduction. 2004;128:703–708. doi: 10.1530/rep.1.00217. [DOI] [PubMed] [Google Scholar]
- 29.Aranyi T, Paldi A. The constant variation: DNA methylation changes during preimplantation development. FEBS Lett. 2006;580:6521–6526. doi: 10.1016/j.febslet.2006.11.018. [DOI] [PubMed] [Google Scholar]
- 30.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 2003;33(Suppl.):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 31.Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Ann. Rev. Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 32.Attwood JT, Yung RL, Richardson BC. DNA methylation and the regulation of gene transcription. Cell. Mol. Life Sci. 2002;59:241–257. doi: 10.1007/s00018-002-8420-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singal R, Ginder GD. DNA methylation. Blood. 1999;93:4059–4070. [PubMed] [Google Scholar]
- 34.Hendrich B, Tweedie S. The methyl-CpG binding domain and the evolving role of DNA methylation in animals. Trends Genet. 2003;19:269–277. doi: 10.1016/S0168-9525(03)00080-5. [DOI] [PubMed] [Google Scholar]
- 35.Slotkin RK, Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 2007;8:272–285. doi: 10.1038/nrg2072. [DOI] [PubMed] [Google Scholar]
- 36.Robertson KD, Wolffe AP. DNA methylation in health and disease. Nat. Rev. Genet. 2000;1:11–19. doi: 10.1038/35049533. [DOI] [PubMed] [Google Scholar]
- 37.Rothenfusser S, Hornung V, Krug A, Towarowski A, Krieg AM, Endres S, Hartmann G. Distinct CpG oligonucleotide sequences activate human gamma delta T cells via interferon-alpha/-beta. Eur. J. Immunol. 2001;31:3525–3534. doi: 10.1002/1521-4141(200112)31:12<3525::aid-immu3525>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 38.Kojima Y, Xin K.-Q, Ooki T, Hamajima K, Oikawa T, Shinoda K, Ozaki T, Hoshino Y, Jounai N, Nakazawa M, et al. Adjuvant effect of multi-CpG motifs on an HIV-1 DNA vaccine. Vaccine. 2002;20:2857–2865. doi: 10.1016/s0264-410x(02)00238-4. [DOI] [PubMed] [Google Scholar]
- 39.Krieg AM, Yi AK, Matson S, Waldschmidt TJ, Bishop GA, Teasdale R, Koretzky GA, Klinman DM. CpG motifs in bacterial DNA trigger direct B-cell activation. Nature. 1995;374:546–549. doi: 10.1038/374546a0. [DOI] [PubMed] [Google Scholar]
- 40.Krug A, Towarowski A, Britsch S, Rothenfusser S, Hornung V, Bals R, Giese T, Engelmann H, Endres S, Krieg AM, et al. Toll-like receptor expression reveals CpG DNA as a unique microbial stimulus for plasmacytoid dendritic cells which synergizes with CD40 ligand to induce high amounts of IL-12. Eur. J. Immunol. 2001;31:3026–3037. doi: 10.1002/1521-4141(2001010)31:10<3026::aid-immu3026>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 41.Krieg AM. CpG motifs in bacterial DNA and their immune effects. Ann. Rev. Immunol. 2002;20:709–760. doi: 10.1146/annurev.immunol.20.100301.064842. [DOI] [PubMed] [Google Scholar]
- 42.Hartmann G, Krieg AM. Mechanism and function of a newly identified CpG DNA motif in human primary B cells. J. Immunol. 2000;164:944–953. doi: 10.4049/jimmunol.164.2.944. [DOI] [PubMed] [Google Scholar]
- 43.Mutwiri G, Pontarollo R, Babiuk S, Griebel P, van Drunen Littel-van den Hurk S, Mena A, Tsang C, Alcon V, Nichani A, Ioannou X, et al. Biological activity of immunostimulatory CpG DNA motifs in domestic animals. Vet. Immunol. Immunopathol. 2003;91:89–103. doi: 10.1016/s0165-2427(02)00246-5. [DOI] [PubMed] [Google Scholar]
- 44.Iho S, Yamamoto T, Takahashi T, Yamamoto S. Oligodeoxynucleotides containing palindrome sequences with internal 5′-CpG-3′ act directly on human NK and activated T cells to induce IFN-gamma production in vitro. J. Immunol. 1999;163:3642–3652. [PubMed] [Google Scholar]
- 45.Kanellos TS, Sylvester ID, Butler VL, Ambali AG, Partidos CD, Hamblin AS, Russell PH. Mammalian granulocyte-macrophage colony-stimulating factor and some CpG motifs have an effect on the immunogenicity of DNA and subunit vaccines in fish. Immunology. 1999;96:507–510. doi: 10.1046/j.1365-2567.1999.00771.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Han J, Hsu C, Zhu Z, Longshore JW, Finley WH. Over-representation of the disease associated (CAG) and (CGG) repeats in the human genome. Nucleic Acids Res. 1994;22:1735–1740. doi: 10.1093/nar/22.9.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lenert P, Stunz L, Yi AK, Krieg AM, Ashman RF. CpG stimulation of primary mouse B cells is blocked by inhibitory oligodeoxyribonucleotides at a site proximal to NF-kappaB activation. Antisense Nucleic Acid Drug Dev. 2001;11:247–256. doi: 10.1089/108729001317022241. [DOI] [PubMed] [Google Scholar]
- 48.Krieg AM, Wu T, Weeratna R, Efler SM, Love-Homan L, Yang L, Yi AK, Short D, Davis HL. Sequence motifs in adenoviral DNA block immune activation by stimulatory CpG motifs. Proc. Natl Acad. Sci. USA. 1998;95:12631–12636. doi: 10.1073/pnas.95.21.12631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krug A, Luker GD, Barchet W, Leib DA, Akira S, Colonna M. Herpes simplex virus type 1 activates murine natural interferon-producing cells through toll-like receptor 9. Blood. 2004;103:1433–1437. doi: 10.1182/blood-2003-08-2674. [DOI] [PubMed] [Google Scholar]
- 50.Roach JC, Glusman G, Rowen L, Kaur A, Purcell MK, Smith KD, Hood LE, Aderem A. The evolution of vertebrate Toll-like receptors. Proc. Natl Acad. Sci. USA. 2005;102:9577–9582. doi: 10.1073/pnas.0502272102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Winocour E, Kaye AM, Stollar V. Synthesis and transmethylation of DNA in polyoma-infected cultures. Virology. 1965;27:156–169. doi: 10.1016/0042-6822(65)90155-8. [DOI] [PubMed] [Google Scholar]
- 52.Gunthert U, Schweiger M, Stupp M, Doerfler W. DNA methylation in adenovirus, adenovirus-transformed cells, and host cells. Proc. Natl Acad. Sci. USA. 1976;73:3923–3927. doi: 10.1073/pnas.73.11.3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vardimon L, Neumann R, Kuhlmann I, Sutter D, Doerfler W. DNA methylation and viral gene expression in adenovirus-transformed and -infected cells. Nucleic Acids Res. 1980;8:2461–2473. doi: 10.1093/nar/8.11.2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wienhues U, Doerfler W. Lack of evidence for methylation of parental and newly synthesized adenovirus type 2 DNA in productive infections. J. Virol. 1985;56:320–324. doi: 10.1128/jvi.56.1.320-324.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.von Acken U, Simon D, Grunert F, Doring HP, Kroger H. Methylation of viral DNA in vivo and in vitro. Virology. 1979;99:152–157. doi: 10.1016/0042-6822(79)90046-1. [DOI] [PubMed] [Google Scholar]
- 56.Kammer C, Doerfler W. Genomic sequencing reveals absence of DNA methylation in the major late promoter of adenovirus type 2 DNA in the virion and in productively infected cells. FEBS Lett. 1995;362:301–305. doi: 10.1016/0014-5793(95)00248-8. [DOI] [PubMed] [Google Scholar]
- 57.Doerfler W. On the biological significance of DNA methylation. Biochemistry. 2005;70:505–524. doi: 10.1007/s10541-005-0145-9. [DOI] [PubMed] [Google Scholar]
- 58.Soeda E, Arrand JR, Smolar N, Griffin BE. Sequence from early region of polyoma virus DNA containing viral replication origin and encoding small, middle and (part of) large T antigens. Cell. 1979;17:357–370. doi: 10.1016/0092-8674(79)90162-4. [DOI] [PubMed] [Google Scholar]
- 59.Doerfler W. DNA methylation–a regulatory signal in eukaryotic gene expression. J. Gen. Virol. 1981;57:1–20. doi: 10.1099/0022-1317-57-1-1. [DOI] [PubMed] [Google Scholar]
- 60.Danos O, Katinka M, Yaniv M. Molecular cloning, refined physical map and heterogeneity of methylation sites of papilloma virus type 1a DNA. Eur. J. Biochem./FEBS. 1980;109:457–461. doi: 10.1111/j.1432-1033.1980.tb04815.x. [DOI] [PubMed] [Google Scholar]
- 61.Danos O, Katinka M, Yaniv M. Human papillomavirus 1a complete DNA sequence: a novel type of genome organization among papovaviridae. EMBO J. 1982;1:231–236. doi: 10.1002/j.1460-2075.1982.tb01152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Burnett TS, Sleeman JP. Uneven distribution of methylation sites within the human papillomavirus la genome: possible relevance to viral gene expression. Nucleic Acids Res. 1984;12:8847–8860. doi: 10.1093/nar/12.23.8847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hammerschmidt W, Sugden B. Identification and characterization of oriLyt, a lytic origin of DNA replication of Epstein–Barr virus. Cell. 1988;55:427–433. doi: 10.1016/0092-8674(88)90028-1. [DOI] [PubMed] [Google Scholar]
- 64.Takacs M, Segesdi J, Balog K, Mezei M, Toth G, Minarovits J. Relative deficiency in CpG dinucleotides is a widespread but not unique feature of Gammaherpesvirinae genomes. Acta Microbiol, Immunol. Hungarica. 2001;48:349–357. doi: 10.1556/amicr.48.2001.3-4.7. [DOI] [PubMed] [Google Scholar]
- 65.Szyf M, Eliasson L, Mann V, Klein G, Razin A. Cellular and viral DNA hypomethylation associated with induction of Epstein–Barr virus lytic cycle. Proc. Natl Acad. Sci. USA. 1985;82:8090–8094. doi: 10.1073/pnas.82.23.8090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kaschka-Dierich C, Bauer I, Fleckenstein B, Desrosiers RC. Episomal and nonepisomal herpesvirus DNA in lymphoid tumor cell lines. Haematol. Blood Transfusion. 1981;26:197–203. doi: 10.1007/978-3-642-67984-1_32. [DOI] [PubMed] [Google Scholar]
- 67.Low M, Hay J, Keir HM. DNA of herpes simplex virus is not a substrate for methylation in vivo. J. Mol. Biol. 1969;46:205–207. doi: 10.1016/0022-2836(69)90068-0. [DOI] [PubMed] [Google Scholar]
- 68.Lundberg P, Welander P, Han X, Cantin E. Herpes simplex virus type 1 DNA is immunostimulatory in vitro and in vivo. J. Virol. 2003;77:11158–11169. doi: 10.1128/JVI.77.20.11158-11169.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kubat NJ, Tran R K, McAnay P, Bloom D C. Specific histone tail modification and NOT DNA methylation is a determinant of herpes simplex virus type 1 latent gene expression. J. Virol. 2004;78:1139–1149. doi: 10.1128/JVI.78.3.1139-1149.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Davison AJ, Cunningham C, Sauerbier W, McKinnell RG. Genome sequences of two frog herpesviruses. J. Gen. Virol. 2006;87:3509–3514. doi: 10.1099/vir.0.82291-0. [DOI] [PubMed] [Google Scholar]
- 71.Ambinder RF, Robertson KD, Tao Q. DNA methylation and the Epstein–Barr virus. Semin. Cancer Biol. 1999;9:369–375. doi: 10.1006/scbi.1999.0137. [DOI] [PubMed] [Google Scholar]
- 72.Tao Q, Robertson KD. Stealth technology: how Epstein–Barr virus utilizes DNA methylation to cloak itself from immune detection. Clin. Immunol. 2003;109:53–63. doi: 10.1016/s1521-6616(03)00198-0. [DOI] [PubMed] [Google Scholar]
- 73.Chinchar VG. Ranaviruses (family Iridoviridae): emerging cold-blooded killers. Arch. Virol. 2002;147:447–470. doi: 10.1007/s007050200000. [DOI] [PubMed] [Google Scholar]
- 74.Willis DB, Granoff A. Frog virus 3 DNA is heavily methylated at CpG sequences. Virology. 1980;107:250–257. doi: 10.1016/0042-6822(80)90290-1. [DOI] [PubMed] [Google Scholar]
- 75.Willis DB, Goorha R, Granoff A. DNA methyltransferase induced by frog virus 3. J. Virol. 1984;49:86–91. doi: 10.1128/jvi.49.1.86-91.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schetter C, Grunemann B, Holker I, Doerfler W. Patterns of frog virus 3 DNA methylation and DNA methyltransferase activity in nuclei of infected cells. J. Virol. 1993;67:6973–6978. doi: 10.1128/jvi.67.12.6973-6978.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kaur K, Rohozinski J, Goorha R. Identification and characterization of the frog virus 3 DNA methyltransferase gene. J. Gen. Virol. 1995;76(Pt 8):1937–1943. doi: 10.1099/0022-1317-76-8-1937. [DOI] [PubMed] [Google Scholar]
- 78.Darai G, Anders K, Koch HG, Delius H, Gelderblom H, Samalecos C, Flugel RM. Analysis of the genome of fish lymphocystis disease virus isolated directly from epidermal tumours of pleuronectes. Virology. 1983;126:466–479. doi: 10.1016/s0042-6822(83)80005-1. [DOI] [PubMed] [Google Scholar]
- 79.Wagner H, Simon D, Werner E, Gelderblom H, Darai C, Flugel RM. Methylation pattern of fish lymphocystis disease virus DNA. J. Virol. 1985;53:1005–1007. doi: 10.1128/jvi.53.3.1005-1007.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tidona CA, Schnitzler P, Kehm R, Darai G. Identification of the gene encoding the DNA (cytosine-5) methyltransferase of lymphocystis disease virus. Virus Genes. 1996;12:219–229. doi: 10.1007/BF00284642. [DOI] [PubMed] [Google Scholar]
- 81.Bigot Y, Stasiak K, Rouleux-Bonnin F, Federici BA. Characterization of repetitive DNA regions and methylated DNA in ascovirus genomes. J. Gen. Virol. 2000;81:3073–3082. doi: 10.1099/0022-1317-81-12-3073. [DOI] [PubMed] [Google Scholar]
- 82.Karlin S, Ladunga I, Blaisdell BE. Heterogeneity of genomes: measures and values. Proc. Natl Acad. Sci. USA. 1994;91:12837–12841. doi: 10.1073/pnas.91.26.12837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shackelton LA, Parrish CR, Holmes EC. Evolutionary basis of codon usage and nucleotide composition bias in vertebrate DNA viruses. J. Mol. Evol. 2006;62:551–563. doi: 10.1007/s00239-005-0221-1. [DOI] [PubMed] [Google Scholar]
- 84.Weitzman MD, Kyostio SR, Kotin RM, Owens RA. Adeno-associated virus (AAV) Rep proteins mediate complex formation between AAV DNA and its integration site in human DNA. Proc. Natl Acad. Sci. USA. 1994;91:5808–5812. doi: 10.1073/pnas.91.13.5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Amiss TJ, McCarty DM, Skulimowski A, Samulski RJ. Identification and characterization of an adeno-associated virus integration site in CV-1 cells from the African green monkey. J. Virol. 2003;77:1904–1915. doi: 10.1128/JVI.77.3.1904-1915.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Doerfler W, Kruczek I, Eick D, Vardimon L, Kron B. DNA methylation and gene activity: the adenovirus system as a model. Cold Spring Harbor Symp. Quant. Biol. 1983;47(Pt 2):593–603. doi: 10.1101/sqb.1983.047.01.070. [DOI] [PubMed] [Google Scholar]
- 87.Doerfler W. Adenoviral DNA integration and changes in DNA methylation patterns: a different view of insertional mutagenesis. Prog. in Nucleic Acid Res. Mol. Biol. 1993;46:1–36. doi: 10.1016/s0079-6603(08)61016-8. [DOI] [PubMed] [Google Scholar]
- 88.Doerfler W. A new concept in (adenoviral) oncogenesis: integration of foreign DNA and its consequences. Biochim. Biophys. Acta. 1996;1288:F79–F99. doi: 10.1016/0304-419x(96)00024-8. [DOI] [PubMed] [Google Scholar]
- 89.Kubat NJ, Tran RK, McAnany P, Bloom DC. Specific histone tail modification and not DNA methylation is a determinant of herpes simplex virus type 1 latent gene expression. J. Virol. 2004;78:1139–1149. doi: 10.1128/JVI.78.3.1139-1149.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Orend G, Knoblauch M, Kammer C, Tjia ST, Schmitz B, Linkwitz A, Meyer G, Maas J, Doerfler W. The initiation of de novo methylation of foreign DNA integrated into a mammalian genome is not exclusively targeted by nucleotide sequence. J. Virol. 1995;69:1226–1242. doi: 10.1128/jvi.69.2.1226-1242.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sutter D, Doerfler W. Methylation of integrated adenovirus type 12 DNA sequences in transformed cells is inversely correlated with viral gene expression. Proc. Natl Acad. Sci. USA. 1980;77:253–256. doi: 10.1073/pnas.77.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kruczek I, Doerfler W. The unmethylated state of the promoter/leader and 5′-regions of integrated adenovirus genes correlates with gene expression. EMBO J. 1982;1:409–414. doi: 10.1002/j.1460-2075.1982.tb01183.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kuhlmann I, Doerfler W. Loss of viral genomes from hamster tumor cells and nonrandom alterations in patterns of methylation of integrated adenovirus type 12 DNA. J. Virol. 1983;47:631–636. doi: 10.1128/jvi.47.3.631-636.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hertz JM, Schell G, Doerfler W. Factors affecting de novo methylation of foreign DNA in mouse embryonic stem cells. J. Biol. Chem. 1999;274:24232–24240. doi: 10.1074/jbc.274.34.24232. [DOI] [PubMed] [Google Scholar]
- 95.Orend G, Kuhlmann I, Doerfler W. Spreading of DNA methylation across integrated foreign (adenovirus type 12) genomes in mammalian cells. J. Virol. 1991;65:4301–4308. doi: 10.1128/jvi.65.8.4301-4308.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Doerfler W. Patterns of DNA methylation–evolutionary vestiges of foreign DNA inactivation as a host defense mechanism. A proposal. Biol. Chem. Hoppe-Seyler. 1991;372:557–564. [PubMed] [Google Scholar]
- 97.Graessmann M, Graessmann A, Wagner H, Werner E, Simon D. Complete DNA methylation does not prevent polyoma and simian virus 40 virus early gene expression. Proc. Natl Acad. Sci. USA. 1983;80:6470–6474. doi: 10.1073/pnas.80.21.6470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Deuring R, Klotz G, Doerfler W. An unusual symmetric recombinant between adenovirus type 12 DNA and human cell DNA. Proc. Natl Acad. Sci. USA. 1981;78:3142–3146. doi: 10.1073/pnas.78.5.3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Flanagan JM. Host epigenetic modifications by oncogenic viruses. Br. J. Cancer. 2007;96:183–188. doi: 10.1038/sj.bjc.6603516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sugawara K, Fujinaga K, Yamashita T, Ito Y. Integration and methylation of shope papilloma virus DNA in the transplantable Vx2 and Vx7 rabbit carcinomas. Virology. 1983;131:88–99. doi: 10.1016/0042-6822(83)90536-6. [DOI] [PubMed] [Google Scholar]
- 101.Kim K, Garner-Hamrick PA, Fisher C, Lee D, Lambert PF. Methylation patterns of papillomavirus DNA, its influence on E2 function, and implications in viral infection. J. Virol. 2003;77:12450–12459. doi: 10.1128/JVI.77.23.12450-12459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Badal S, Badal V, Calleja-Macias IE, Kalantari M, Chuang LS, Li BF, Bernard HU. The human papillomavirus-18 genome is efficiently targeted by cellular DNA methylation. Virology. 2004;324:483–492. doi: 10.1016/j.virol.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 103.Verma M. Viral genes and methylation. Ann. NY Acad. Sci. 2003;983:170–180. doi: 10.1111/j.1749-6632.2003.tb05972.x. [DOI] [PubMed] [Google Scholar]
- 104.Munnes M, Schetter C, Holker I, Doerfler W. A fully 5′-CG-3′ but not a 5′-CCGG-3′ methylated late frog virus 3 promoter retains activity. J. Virol. 1995;69:2240–2247. doi: 10.1128/jvi.69.4.2240-2247.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Vardimon L, Kuhlmann I, Doerfler W, Cedar H. Methylation of adenovirus genes in transformed cells and in vitro: influence on the regulation of gene expression? Eur. J. Cell Biol. 1981;25:13–15. [PubMed] [Google Scholar]
- 106.Vardimon L, Gunthert U, Doerfler W. In vitro methylation of the BsuRI (5′-GGCC-3′) sites in the E2a region of adenovirus type 2 DNA does not affect expression in Xenopus laevis oocytes. Mol. Cell. Biol. 1982;2:1574–1580. doi: 10.1128/mcb.2.12.1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Simon D, Grunert F, von Acken U, Doring HP, Kroger H. DNA-methylase from regenerating rat liver: purification and characterisation. Nucleic Acids Res. 1978;5:2153–2167. doi: 10.1093/nar/5.6.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Subramanian KN. Effect of in vitro methylation at CpG sites on gene expression in a genome functioning autonomously in a vertebrate host. Nucleic Acids Res. 1982;10:3475–3486. doi: 10.1093/nar/10.11.3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Fradin A, Manley JL, Prives CL. Methylation of simian virus 40 Hpa II site affects late, but not early, viral gene expression. Proc. Natl Acad. Sci. USA. 1982;79:5142–5146. doi: 10.1073/pnas.79.17.5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 111.Oka M, Rodic N, Graddy J, Chang L.-J, Terada N. CpG sites preferentially methylated by Dnmt3a in vivo. J. Biol. Chem. 2006;281:9901–9908. doi: 10.1074/jbc.M511100200. [DOI] [PubMed] [Google Scholar]
- 112.Yaman R, Grandjean V. Timing of entry of meiosis depends on a mark generated by DNA methyltransferase 3a in testis. Mol. Reprod. Dev. 2006;73:390–397. doi: 10.1002/mrd.20430. [DOI] [PubMed] [Google Scholar]
- 113.Xie S, Wang Z, Okano M, Nogami M, Li Y, He WW, Okumura K, Li E. Cloning, expression and chromosome locations of the human DNMT3 gene family. Gene. 1999;236:87–95. doi: 10.1016/s0378-1119(99)00252-8. [DOI] [PubMed] [Google Scholar]
- 114.Bourc'his D, Xu GL, Lin CS, Bollman B, Bestor TH. Dnmt3L and the establishment of maternal genomic imprints. Science. 2001;294:2536–2539. doi: 10.1126/science.1065848. [DOI] [PubMed] [Google Scholar]
- 115.Bestor TH, Ingram VM. Two DNA methyltransferases from murine erythroleukemia cells: purification, sequence specificity, and mode of interaction with DNA. Proc. Natl Acad. Sci. USA. 1983;80:5559–5563. doi: 10.1073/pnas.80.18.5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Li E, Beard C, Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993;366:362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- 117.Yoder JA, Walsh CP, Bestor TH. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13:335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- 118.Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N, Strouboulis J, Wolffe AP. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat. Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 119.Kunert N, Marhold J, Stanke J, Stach D, Lyko F. A Dnmt2-like protein mediates DNA methylation in Drosophila. Development. 2003;130:5083–5090. doi: 10.1242/dev.00716. [DOI] [PubMed] [Google Scholar]
- 120.Rankin R, Pontarollo R, Ioannou X, Krieg AM, Hecker R, Babiuk LA, van Drunen Littel-van den Hurk S. CpG motif identification for veterinary and laboratory species demonstrates that sequence recognition is highly conserved. Antisense Nucleic Acid Drug Development. 2001;11:333–340. doi: 10.1089/108729001753231713. [DOI] [PubMed] [Google Scholar]
- 121.Gunthert U, Schweiger M, Stupp M, Doerfler W. DNA methylation in adenovirus, adenovirus-transformed cells, and host cells. Proc. Natl Acad. Sci. USA. 1976;73:3923–3927. doi: 10.1073/pnas.73.11.3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Perlmann C, Saemundsen AK, Klein G. A fraction of Epstein–Barr virus virion DNA is methylated in and around the EcoRI-J fragment. Virology. 1982;123:217–221. doi: 10.1016/0042-6822(82)90308-7. [DOI] [PubMed] [Google Scholar]
- 123.Badal V, Chuang LS, Tan EH, Badal S, Villa LL, Wheeler CM, Li BF, Bernard HU. CpG methylation of human papillomavirus type 16 DNA in cervical cancer cell lines and in clinical specimens: genomic hypomethylation correlates with carcinogenic progression. J. Virol. 2003;77:6227–6234. doi: 10.1128/JVI.77.11.6227-6234.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]