Abstract
To investigate the binding of 5′–CpG–3′ sequences by small molecules, two pyrrole (Py)–imidazole (Im) hairpin polyamides, PyImPyIm–γ–PyImPyIm–β–Dp (1) and PyIm–β–Im–γ–PyIm–β–Im–β–Dp (2), which recognize the sequence 5′–CGCG–3′, were synthesized. The binding affinities of the 5′–CGCG–3′ sequence to the Py–Im hairpin polyamides were measured by surface plasmon resonance (SPR) analysis. SPR data revealed that dissociation equilibrium constants (Kd) of polyamides 1 and 2 were 1.1 (± 0.3) × 10–6 M and 1.7 (± 0.4) × 10–8 M, respectively. Polyamide 2 possesses great binding affinity for this sequence, 65-fold higher than polyamide 1. Moreover, when all cytosines in 5′–CpGpCpG–3′ were replaced with 5-methylcytosines (mCs), the Kd value of polyamide 2 increased to 5.8 (± 0.7) × 10–9 (M), which indicated about 3-fold higher binding than the unmethylated 5′–CGCG–3′ sequence. These results suggest that polyamide 2 would be suitable to target CpG-rich sequences in the genome.
INTRODUCTION
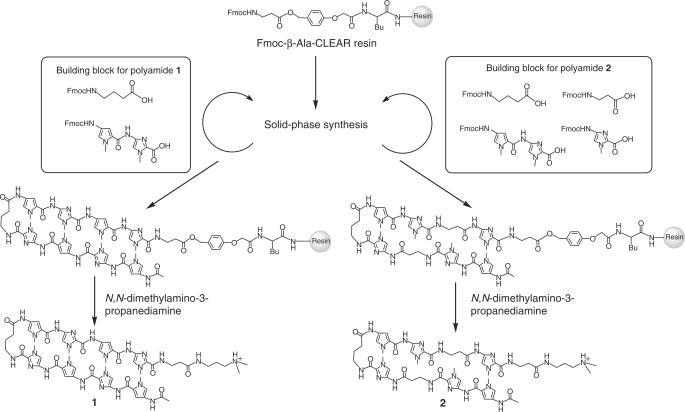
N-Methylpyrrole (Py)–N-methylimidazole (Im) polyamides can recognize pre-determined DNA sequences in the minor groove of B-DNA (1,2). All four Watson–Crick base pairs can be recognized using different pairings of three aromatic amino acids oriented in the aminocarboxyl (N–C) direction with respect to the 5′–3′ direction of the DNA helix, i.e. with a pairing of Im/Py or Py/Im specifying G·C or C·G base pairs, respectively, a pairing of 3-hydroxypyrrole (Hp)/Py or Py/Hp specifying T·A or A·T base pairs, respectively, and a Py/Py pairing being able to degenerately recognize A·T or T·A base pairs (1,2). Aliphatic β-alanine can be replaced with Py. Py/β and β/Py pairings are found to recognize A·T/T·A pairs relative to G·C/C·G pairs. The Im/β pairing is found to target G·C relative to C·G, A·T, and T·A, while the β/Im pair targets C·G relative to G·C, A·T, and T·A. (3,4). As Py–Im polyamides bind to DNA with affinity and sequence-specificity comparable to DNA-binding proteins, gene expression can be regulated by competitive displacement of transcription factor from DNA target sequences (5). Several approaches have been reported for targeting DNA sequence at transcription factor binding sites in a promoter region (6–9). This region is highly GC rich and possesses high relative densities of 5′–CpG–3′ sequences, called CpG islands (10–12). In addition, cytosines in this 5′–CpG–3′ sequences can be methylated to 5-methylcytosines (5-meCs) in post-replication processes. Methylation of DNA is closely associated with transcriptional regulation (13,14). Methylated DNA binding proteins (MDBs) recruit histone deacetylases (HDACs) and form heterochromatin structures to inactivate gene expression. Aberrant methylation of CpG islands in promoter regions of tumor suppressor genes (such as Rb, p16 and other genes) causes cancer proliferation (15–20). Although targeting 5′–CpG–3′ sequences has great potential for controlling biological activities, there are few examples of hairpin polyamides recognizing regions including 5′–CpG–3′ sequences (1–3). In this article, two Py–Im hairpin polyamides that recognize 5′–CGCG–3′ sequences 1 and 2 were synthesized. Polyamide 2 possesses two β-alanine units, replacing the Py units of 1 (Figure 1a). The binding affinities of these polyamides to unmethylated and methylated 5′–CGCG–3′ sequences were investigated using surface plasmon resonance (SPR) analysis.
Figure 1.
Synthetic scheme of Py–Im polyamides 1 and 2 recognizing 5′–CGCG–3′ sequence.
MATERIALS AND METHODS
Synthesis of Fmoc–Py–Im–COOH dimer unit
Py–Im hairpin polyamides that recognize 5′–CpG–3′ sequences are limited because of the difficulty of coupling Py after Im on solid phase (21). To solve this problem, the Fmoc–Py–Im–COOH dimer unit (4-[9-fluorenylmethoxycarbonyl-{(4-amino-1-methylpyrrole-2-yl)–carbonylamino}]-1-methylimidazole–2-carboxylic acid) was prepared from methyl-{4-(4-nitro-1-methylpyrrole-2-yl)–carbonylamino}]-1-methylimidazole–2-carboxylate (22). To a solution of starting material in methanol/dichloromethane (v/v: 2/1), Pd/C was added and stirred overnight at room temperature under H2 gas pressure (3.0 atm). After the completion of the reaction, as determined by TLC, Pd/C was filtered, and the filtrate was concentrated under reduced pressure. To the resultant red oil product, aqueous NaOH solution was added and stirred overnight at 45°C under an argon atmosphere. After the completion of hydrolysis, as determined by TLC, the solution was neutralized with dilute hydrochloric acid and concentrated under reduced pressure. The resultant yellow solid was dissolved in saturated aqueous sodium hydrogen carbonate and 1,2-dimethoxyethane. The 9-fluorenylmethyl-succimidyl carbonate was added and stirred overnight at room temperature under an argon atmosphere. After the completion of the reaction, as determined by TLC, the solution was concentrated, and dilute hydrochloric acid was added. The orange precipitation was filtered to obtain 4.0 g (69% yield) of product. The compound was identified by ESI–MS using an API165E (Applied Biosystems) and 1H-NMR using a JEOL JNM–FX 400 nuclear magnetic resonance spectrometer (JEOL). 1H NMR (400 MHz, DMSO-d6) δ 10.61 (s, 1H; NH), 9.47 (s, 1H; NH), 7.89 (d, J = 6.8 Hz, 2H; Ph-H), 7.73 (d, J = 6.8 Hz, 2H; Ph-H), 7.59 (s, 1H; Im-H), 7.42 (t, J = 7.2 Hz, 2H; Ph-H), 7.34 (t, J = 7.2 Hz, 2H; Ph-H), 7.00 (s, H; Py-H), 6.94 (s, H; Py-H), 4.40 (d, J = 5.6 Hz, 2H; CH2), 4.27 (t, J = 6.0 Hz, 1H; CH), 3.91 (s, 3H; NCH3), 3.90 (s, 3H; NCH3). ESI–MS m/z calcd for C25H22N6O5 [M + H] + 486.17, found 486.2.
Synthesis of PyImPyIm–γ–PyImPyIm–β–Dp (1)
Py–Im polyamide 1 was synthesized by Fmoc-based solid-phase methods on Fmoc-β-Alanine–CLEAR-Acid resin (Peptides International), as described previously (22,23). After completion of the synthesis, polyamides were cleaved from the resin using N,N-dimethylamino-3-propaneamine to afford 1. A product was purified by reverse-phase HPLC, and 1 was obtained as 26 mg (22% yield) of yellow powder. The structures of polyamides 1 and 2 were identified by 1H-NMR using a JEOL JNM–FX 400 nuclear magnetic resonance spectrometer (JEOL) and ESI–TOFMS using a BioTOF II (Bruker Daltonics) mass spectrometer. UV-visible spectra were measured with a NanoDrop spectrophotometer (NanoDrop Technologies) and the concentrations of the polyamides were determined. 1H NMR (400 MHz, DMSO-d6) δ 10.44 (s, 1H; NH), 10.36 (s, 1H; NH), 10.30 (s, 2H; NH), 9.80 (s, 1H; NH), 9.79 (s, 1H; NH), 7.92 (br, 5H; NH), 7.48 (s, 2H; Im-H), 7.39 (s, 2H; Im-H), 7.24 (s, 1H; Py-H), 7.23 (s, 1H; Py-H), 6.89 (s, 2H; Py-H), 3.93 (s, 6H; NCH3), 3.90 (s, 6H; NCH3), 3.80 (s, 6H; NCH3), 3.50 (br, 4H; CH2), 3.41 (q, J = 6.0 Hz, 2H; CH2), 3.04 (q, J = 6.0 Hz, 2H; CH2), 2.58 (q, J = 6.0 Hz, 4H; CH2), 2.32 (t, J = 6.0 Hz, 2H; CH2), 2.26 (br, 4H; CH2), 2.14 (s, 6H; NCH3), 1.95 (s, 3H; COCH3), 1.78 (qui, J = 7.2 Hz, 2H; CH2), 1.51 (qui, J = 7.2 Hz, 2H; CH2); ESI–TOFMS m/z calcd for C57H71N25O11 [M + H]+ 1179.57, found 1179.55. UV (DMSO): 310 (78,400).
PyIm–β–Im–γ–PyIm–β–Im–β–Dp (2)
Py–Im polyamide 2 was synthesized according to a procedure similar to that for polyamide 1. A product was purified by reverse-phase HPLC, and 1 was obtained as 6.0 mg (4.7% yield) of yellow powder. 1H NMR (400 MHz, DMSO-d6) δ 10.33 (s, 1H; NH), 10.27 (s, 1H; NH), 10.26 (s, 2H; NH), 9.88 (s, 1H; NH), 9.85 (s, 1H; NH), 9.84 (s, 1H; NH), 9.81 (s, 1H; NH), 8.01 (t, J = 6.0 Hz, 1H; NH), 7.93 (br, 2H; NH), 7.54 (s, 2H; Im-H), 7.50 (s, 1H; Im-H), 7.49 (s, 1H; Im-H), 7.38 (s, 1H; Py-H), 7.37 (s, 1H; Py-H), 7.26 (s, 1H; Py-H), 7.23 (s, 1H; Py-H), 7.14 (s, 2H; Py-H), 6.92 (s, 2H; Py-H), 3.97 (s, 6H; NCH3), 3.93 (s, 6H; NCH3), 3.86 (s, 3H; NCH3), 3.85 (s, 3H; NCH3), 3.83 (s, 6H; NCH3), 3.44 (q, J = 6.4 Hz, 2H; CH2), 3.37 (q, J = 6.4 Hz, 2H; CH2), 3.07 (q, J = 6.4 Hz, 2H; CH2), 2.34 (t, J = 6.4 Hz, 2H; CH2), 2.26 (br, 4H; CH2), 1.96 (s, 6H; NCH3), 1.90 (s, 3H; COCH3), 1.81 (t, J = 7.2 Hz, 2H; CH2), 1.56 (t, J = 7.2 Hz, 2H; CH2); ESI–TOFMS m/z calcd for C58H73N24O11 [M + H]+ 1281.59, found 1281.96. UV (DMSO): 295 (58 800).
Measurement of binding constants by SPR
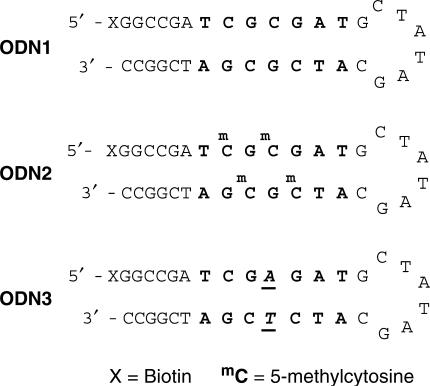
SPR experiments were performed using a BIACORE X instrument (GE Healthcare), as described in previous studies (24,25). Biotinylated hairpin DNA ODN1 (5′–Biotin–GGCCGATCGCGATGCTATAGCATCGCGATCGGCC–3′) and ODN2 (5′–Biotin–GGCCGATmCGmCGATGCTATAGCATmCGmCGATCGGCC–3′) and ODN3 (5′–Biotin–GGCCGATCGAGATGCTATAGCATCTCGATCGGCC–3′), including one base mismatch base pair for recognition by polyamide 2, were purchased from Sigma–Genosys (Figure 2). ODNs were immobilized to streptavidin-coated sensor chip SA at a flow rate of 5 μl/min to obtain the wanted immobilization level (up to approximately 1400 RU rise). Experiments were accomplished using HBS–EP (10 mM HEPES, 150 mM NaCl, 3 mM EDTA, and 0.005% Surfactant P20) buffer with 0.1% DMSO at 25°C, pH 7.4. A series of sample solutions with various concentrations were prepared in HBS–EP buffer with 0.1% DMSO and injected at a flow rate of 20 μl/min. In order to measure dissociation equilibrium constants (Kd), data processing was performed with an appropriate fitting model using the BIAevaluation 4.1 program (26). The sensorgrams of polyamides 1 (Figure 3) and 2 (Figure 4) for ODN1 and ODN2 were fitted by using a 1:1 Langmuir binding model with mass transfer effect. The sensorgrams of polyamide 2 for ODN3 (Figure 5) were fitted using a general fitting mode because of non-kinetic curves resulting from the rapid dissociation (26). The values of Kd for ODN1, ODN2 and ODN3 are summarized in Table 1.
Figure 2.
Sequences of 5′-biotinylated hairpin DNA (ODN1- 3). X represents biotin, and mC represents 5-methylcytosine. The binding sequences of the polyamides are shown in bold. The mismatch-recognition bases are denoted in italics and underlined.
Figure 3.
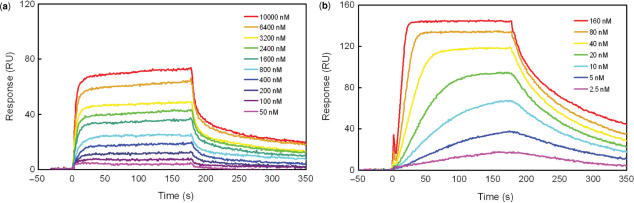
SPR sensorgrams for the interaction of polyamides with ODN1 in HBS–EP buffer (pH 7.4) with 0.1% DMSO at 25°C. (a) Sensorgrams of 1 at concentrations from 50 to 10000 nM. (b) Sensorgrams of 2 at concentrations from 2.5 to 160 nM.
Figure 4.
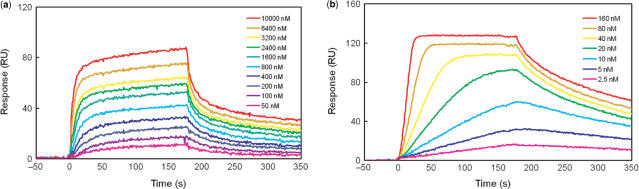
SPR sensorgrams for the interaction of polyamides with ODN2 in HBS-EP buffer (pH 7.4) with 0.1% DMSO at 25°C. (a) Sensorgrams of 1 at concentration from 50 to 10000 nM. (b) Sensorgrams of 2 at concentration from 2.5 to 160 nM.
Figure 5.
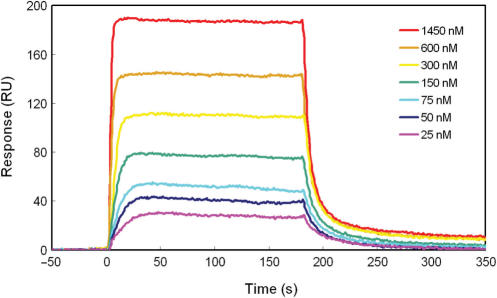
(a) SPR sensorgrams for the interaction of polyamides 2 with ODN3. Sensorgrams of polyamide 2 at concentration from 25 to 1450 nM.
Table 1.
Dissociation equilibrium constants (M) for polyamides 1–2
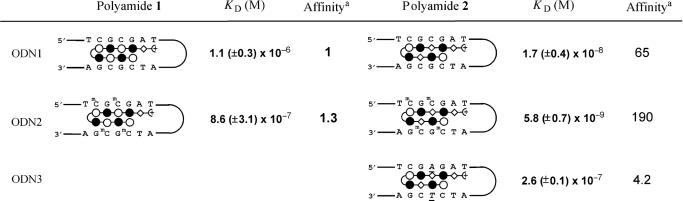 |
KD values are shown as the average and standard derivative from two experiments for each sample.
aAffinity indicates the relative binding affinity to polyamide 1 for ODN1. White circles represent pyrroles, black circles imidazoles, diamondes β-alanine, and the terminal moiety is N,N-dimethylamino-3-propanediamine.
RESULTS AND DISCUSSION
Design and synthesis of Py–Im polyamides
Py–Im polyamides 1 and 2, which bind to a target 5′–CGCG–3′ sequence, are shown in Figure 1. Replacement of Py with a β-alanine can increase binding affinity (3). β-alanine was introduced to polyamide 2 to provide flexibility in the polyamide structure. Both polyamides were prepared by Fmoc-based solid-phase synthesis. Because of the difficulty of coupling Py after Im (21), Fmoc–Py–Im–COOH dimer units were prepared and used for the solid-phase synthesis of 1. After completion of the synthesis, the polyamides were cleaved from the resin using N,N-dimethylamino-3-propaneamine to afford 1 and 2. Products were purified by reverse-phase HPLC, and the structures of polyamides 1 and 2 were identified by 1H-NMR and ESI–TOFMS.
Binding to the unmethylated 5′–CGCG–3′ sequence (ODN1) by polyamides 1 and 2
For the measurement of binding by polyamides 1 and 2, SPR experiments were performed. 5′-Biotinylated hairpin DNA (ODN1) containing a single match site (5′–TCGCGAT–3′) was prepared. DNA was immobilized to a streptavidin-coated sensor chip, and the polyamide solutions were injected. The SPR sensorgrams obtained are shown in Figure 3a and b. In the case of polyamide 1, modest response curves were observed, even at micromolar concentrations (Figure 3a). The Kd was determined at 1.1 (±0.3) × 10−6 M. Compared with 1, the response curve was observed from 2.5 nM (Figure 3b). Polyamide 2 has an affinity high enough to bind DNA at nanomolar concentrations. The Kd was determined at 1.7 (±0.4) × 10−8 M, 65-fold lower than for 1. These results showed that polyamide 2 exhibited higher binding affinity relative to polyamide 1 for this sequence. The increased binding affinity of 2 can be explained as in the case of binding to 5′–GCGC–3′ sequence by hairpin polyamides. Dervan and coworkers demonstrated that the polyamide Im–β–ImPy–γ–Im–β–ImPy–β–Dp has a 100-fold greater binding affinity over the polyamide ImPyImPy–γ–ImPyImPy–β–Dp for the sequence 5′–GCGC–3′ (3). The aliphatic β-alanine unit gives flexibility to the polyamide structure and optimizes the positioning of the imidazole amino acids on binding to the 5′–CGCG–3′ sequence. Therefore, polyamide 2 binds the 5′–CGCG–3′ sequence with good affinity.
Binding to methylated 5′–CGCG–3′ sequence (ODN2)
Mammalian promoter regions contain highly methylated 5′–CpG–3′ sequences. Therefore, we investigated whether cytosine methylation may influence DNA binding by Py–Im hairpin polyamides. We measured the binding affinities of 1 and 2 by using ODN2, which contains four mCs in place of cytosines in the binding site of ODN1. The obtained SPR sensorgrams are shown in Figure 4a–b. From the SPR data, the Kd of polyamides 1 and 2 were determined at 8.6 (±3.1) × 10−7 M and 5.8 (±0.7) × 10−9 M, respectively. As with the results with ODN1, the binding affinity of polyamide 2, with β-alanine introduced, was much higher (over 100-fold, Table 1) than polyamide 1. Interestingly, the binding affinity of polyamide 2 to ODN2 increased 2.9-fold over ODN1. Conversely, polyamide 1 binding to ODN2 is comparable to ODN1. These results suggest that cytosine methylation does not disrupt the binding of Py–Im polyamides and would positively contribute to the binding of polyamide 2. This result was somewhat surprising because methylation of cytosines is assumed to cause little effect on the global structure of the B form of DNA, and the methyl groups of the mCs protrude into the major groove of DNA, which is located opposite to the polyamide binding site in the minor groove. However, it is known that the reactivities of some DNA-interacting drugs are affected by cytosine methylation. For example, benzo[a]pyrene and mitomycin C show increased covalent adduct formation (27–30), while bleomycin gives a decreased cleavage reaction (31), although these drugs act in the DNA minor groove. The precise reason for the stronger binding of polyamide to the methylated DNA is not clear, however, hydrophobic and electronic factors would contribute to this phenomenon.
Binding to a mismatch sequence (ODN3)
To confirm the sequence-specificity of polyamide 2, we measured the binding affinities of 2 using ODN3, which contains a single mismatch in the binding site of ODN1. The average Kd of polyamide 2 to ODN3 was determined to be 2.6 (±0.1) × 10−7 M from the steady-state binding levels of the fitting curves, because it was inaccurate to evaluate the Kd value from kinetic parameters as a result of the rapid dissociation. This Kd value indicated that the binding of polyamide 2 is 15 times weaker to a single mismatch sequence ODN3 than to match sequence ODN1, and polyamide 2 binds almost 45 times more strongly to methylated ODN2 than to a single mismatch ODN3. This result suggests that polyamide 2 has a great affinity for the methylated 5′–CGCG–3′ sequence.
CONCLUSION
We synthesized and evaluated Py–Im hairpin polyamides that bind the sequence 5′–CGCG–3′. Polyamide 2 has a high affinity to this sequence at nanomolar concentrations. Methylation of cytosines in the binding site affected the binding of polyamide 2, with about a 3-fold increase. The influence of binding by Py–Im polyamides on subsequent cytosine methylation was not investigated. These results suggest that proteins that bind to unmethylated and methylated CpG sequences may be inhibited by Py–Im polyamides such as 2. Methylation of CpG islands and binding to methylated 5′–CpG–3′ sequences by methylated DNA-specific binding proteins play significant roles in transcriptional regulation and maintenance of genomic stability (32–36). Novel biological phenomena may be observed by regulating binding in this region using Py–Im polyamides.
ACKNOWLEDGEMENTS
Mr M. Minoshima was supported by a research fellowship of the Global COE (Center of Excellence) program, International Center for Integrated Research and Advanced Education in Material Science, Kyoto University, Japan. We would like to thank Gentier Biosystems for synthesis of polyamide 1. Funding to pay the Open Access publication charges for this article was provided by a Grant-in-Aid for Priority Research from the Ministry of Education, Culture, Sports, Science, and Technology, Japan.
Conflict of interest statement. None declared.
REFERENCES
- 1.Dervan PB, Edelson BS. Recognition of the DNA minor groove by pyrrole-imidazole polyamides. Curr. Opin. Struct. Biol. 2003;13:284–299. doi: 10.1016/s0959-440x(03)00081-2. [DOI] [PubMed] [Google Scholar]
- 2.Dervan PB, Poulin-Kerstein AT, Fecher EJ, Edelson BS. Regulation of gene expression by synthetic DNA-binding ligands. Top. Curr. Chem. 2005;253:1–31. [Google Scholar]
- 3.Turner JM, Swalley SE, Baird EE, Dervan PB. Aliphatic/aromatic amino acid pairings for polyamide recognition in the minor groove of DNA. J. Am. Chem. Soc. 1998;120:6219–6226. [Google Scholar]
- 4.Wang CCC, Ellervik U, Dervan PB. Expanding the recognition the minor groove of DNA by incorporation of beta-alanine in hairpin polyamides. Bioorg. Med. Chem. 2001;9:653–657. doi: 10.1016/s0968-0896(00)00282-0. [DOI] [PubMed] [Google Scholar]
- 5.Gottesfeld JM, Neely L, Trauger JW, Baird EE, Dervan PB. Regulation of gene expression by small molecules. Nature. 1997;387:202–205. doi: 10.1038/387202a0. [DOI] [PubMed] [Google Scholar]
- 6.Olenyuk BZ, Zhang GJ, Klco JM, Nickols NG, Kaelin W.G., Jr., Dervan PB. Inhibition of vascular endothelial growth factor with a sequence-specific hypoxia response element antagonist. Proc. Natl Acad. Sci. USA. 2004;101:16768–16773. doi: 10.1073/pnas.0407617101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nickols NG, Dervan PB. Suppression of androgen receptor-mediated gene expression by a sequence-specific DNA-binding polyamide. Proc. Natl Acad. Sci. USA. 2007;104:10418–10423. doi: 10.1073/pnas.0704217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matsuda H, Fukuda N, Ueno T, Tahira Y, Ayame H, Zhang W, Bando T, Sugiyama H, Saito S, Matsumoto K, et al. Development of gene silencing pyrrole-imidazole polyamide targeting the TGF-beta1 promoter for treatment of progressive renal diseases. J. Am. Soc. Nephrol. 2006;17:422–432. doi: 10.1681/ASN.2005060650. [DOI] [PubMed] [Google Scholar]
- 9.Kageyama Y, Sugiyama H, Ayame H, Iwai A, Fujii Y, Huang LE, Kizaka-Kondoh S, Hiraoka M, Kihara K. Suppression of VEGF transcription in renal cell carcinoma cells by pyrrole-imidazole hairpin polyamides targeting the hypoxia responsive element. Acta. Oncologica. 2006;45:317–324. doi: 10.1080/02841860500486648. [DOI] [PubMed] [Google Scholar]
- 10.Bird AP. CpG islands as gene markers in the vertebrate nucleus. Trends Genet. 1987;3:342–347. [Google Scholar]
- 11.Antequera F, Bird AP. Number of CpG islands and genes in human and mouse. Proc. Natl Acad. Sci. USA. 1993;90:11995–11999. doi: 10.1073/pnas.90.24.11995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saxonov S, Berg P, Brutlag DL. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc. Natl Acad. Sci. USA. 2006;103:1412–1417. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bird AP. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 14.Bird AP, Wolffe AP. Methylation-induced repression-belts, braces, and chromatin. Cell. 1999;99:451–454. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- 15.Jones PA, Laird PW. Cancer epigenetics comes of age. Nat. Genet. 1999;21:163–167. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- 16.Esteller M, Corn PG, Baylin SB, Herman JG. A gene hypermethylation profile of human cancer. Cancer Res. 2001;61:3225–3229. [PubMed] [Google Scholar]
- 17.Herman JG, Baylin SB. Gene silencing in cancer association with promoter hypermethylation. N. Engl. J. Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 18.Herman JG, Merlo A, Mao L, Lapidus RG, Issa JP, Davidson NE, Baylin SB. Inactivation of the CDKN2/p16/MTS1 gene is frequently associated with aberrant DNA methylation in all common human cancers. Cancer Res. 1995;55:4525–4530. [PubMed] [Google Scholar]
- 19.Merlo A, Herman JG, Mao L, Lee DJ, Gabrielson E, Burger PC, Baylin SB, Sidransky D. 5′ CpG island methylation is associated with transcriptional silencing of the tumour suppressor p16/CDKN2/MTS1 in human cancers. Nat. Med. 1995;1:686–692. doi: 10.1038/nm0795-686. [DOI] [PubMed] [Google Scholar]
- 20.Robertson KD, Ait-Si-Ali S, Yokochi T, Wade PA, Jones PL, Wolffe AP. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat. Genet. 2000;25:338–342. doi: 10.1038/77124. [DOI] [PubMed] [Google Scholar]
- 21.Swalley SE, Baird EE, Dervan PB. Discrimination of 5′-GGGG-3′, 5′-GCGC-3′, and 5′-GGCC-3′ sequences in the minor groove of DNA by eight-ring hairpin polyamides. J. Am. Chem. Soc. 1997;119:6953–6961. [Google Scholar]
- 22.Baird EE, Dervan PB. Solid phase synthesis of polyamides containing imidazole and pyrrole amino acids. J. Am. Chem. Soc. 1996;118:6141–6146. [Google Scholar]
- 23.Wurtz NR, Turner JM, Baird EE, Dervan PB. Fmoc solid phase synthesis of polyamides containing pyrrole and imidazole amino acids. Org. Lett. 2001;3:1201–1203. doi: 10.1021/ol0156796. [DOI] [PubMed] [Google Scholar]
- 24.Zhang W, Bando T, Sugiyama H. Discrimination of hairpin polyamides with an alpha-substituted-gamma-aminobutyric acid as a 5′-TG-3′ reader in DNA minor groove. J. Am. Chem. Soc. 2006;128:8766–8776. doi: 10.1021/ja0580587. [DOI] [PubMed] [Google Scholar]
- 25.Nguyen B, Tanious FA, Wilson WD. Biosensor-surface plasmon resonance: Quantitative analysis of small molecule–nucleic acid interactions. Methods. 2007;42:150–161. doi: 10.1016/j.ymeth.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 26.Rapsgatan, Sweden: Biacore AB; 1999. BIAevaluation Software Handbook. Version 3. [Google Scholar]
- 27.Denissenko MF, Pao A, Tang M, Pfeifer GP. Preferential formation of benzo[a]pyrene adducts at lung cancer mutational hotspots in p53. Science. 1996;274:430–432. doi: 10.1126/science.274.5286.430. [DOI] [PubMed] [Google Scholar]
- 28.Zhang N, Lin C, Huang X, Kolbanovskiy A, Hingerty BE, Amin S, Broyde S, Geacintov NE, Patel DJ. Methylation of cytosine at C5 in a CpG sequence context causes a conformational switch of a benzo[a]pyrene diol epoxide-N2-guanine adduct in DNA from a minor groove alignment to intercalation with base displacement. J. Mol. Biol. 2005;346:951–965. doi: 10.1016/j.jmb.2004.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson WS, He Q-Y, Tomasz M. Selective recognition of the m5CpG dinucleotide sequence in DNA by mitomycin C for alkylation and cross-linking. Bioorg. Med. Chem. 1995;3:851–860. doi: 10.1016/0968-0896(95)00067-q. [DOI] [PubMed] [Google Scholar]
- 30.Li V-S, Reed M, Zheng Y, Kohn H, Tang M-S. C5 cytosine methylation in CpG sites enhances sequence selectivity of mitomycin C-DNA bonding. Biochemistry. 2000;39:2612–2618. doi: 10.1021/bi991307h. [DOI] [PubMed] [Google Scholar]
- 31.Hertzberg RP, Caranfa MJ, Hecht SM. DNA methylation diminishes bleomycin-mediated strand scission. Biochemistry. 1985;24:5285–5289. doi: 10.1021/bi00341a001. [DOI] [PubMed] [Google Scholar]
- 32.Meehan RR, Lewis JD, McKay S, Kleiner EL, Bird AP. Identification of a mammalian protein that binds specifically to DNA containing methylated CpGs. Cell. 1989;58:499–507. doi: 10.1016/0092-8674(89)90430-3. [DOI] [PubMed] [Google Scholar]
- 33.Hendrich B, Bird AP. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol. Cell. Biol. 1998;18:6538–6547. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fujita N, Shimotake N, Ohki I, Chiba T, Saya H, Shirakawa M, Nakao M. Mechanism of transcriptional regulation by methyl-CpG binding protein MBD1. Mol. Cell. Biol. 2000;20:5107–5118. doi: 10.1128/mcb.20.14.5107-5118.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ballestar E, Wolffe AP. Methyl-CpG-binding proteins; targeting specific gene repression. Eur. J. Biochem. 2001;268:1–6. doi: 10.1046/j.1432-1327.2001.01869.x. [DOI] [PubMed] [Google Scholar]
- 36.Prokhortchouk A, Hendrich B, Jorgensen H, Ruzov A, Wilm M, Georgiev G, Bird AP, Prokhortchouk E. The p120 catenin partner Kaiso is a DNA methylation-dependent transcriptional repressor. Genes Dev. 2001;15:1613–1618. doi: 10.1101/gad.198501. [DOI] [PMC free article] [PubMed] [Google Scholar]