Abstract
Drosophila melanogaster MTF-1 (dMTF-1) is a copper-responsive transcriptional activator that mediates resistance to Cu, as well as Zn and Cd. Here, we characterize a novel cysteine-rich domain which is crucial for sensing excess intracellular copper by dMTF-1. Transgenic flies expressing mutant dMTF-1 containing alanine substitutions of two, four or six cysteine residues within the sequence 547CNCTNCKCDQTKSCHGGDC565 are significantly or completely impaired in their ability to protect flies from copper toxicity and fail to up-regulate MtnA (metallothionein) expression in response to excess Cu. In contrast, these flies exhibit wild-type survival in response to copper deprivation thus revealing that the cysteine cluster domain is required only for sensing Cu load by dMTF-1. Parallel studies show that the isolated cysteine cluster domain is required to protect a copper-sensitive S. cerevisiae ace1Δ strain from copper toxicity. Cu(I) ligation by a Cys-rich domain peptide fragment drives the cooperative assembly of a polydentate [Cu4-S6] cage structure, characterized by a core of trigonally S3 coordinated Cu(I) ions bound by bridging thiolate ligands. While reminiscent of Cu4-L6 (L = ligand) tetranuclear clusters in copper regulatory transcription factors of yeast, the absence of significant sequence homology is consistent with convergent evolution of a sensing strategy particularly well suited for Cu(I).
INTRODUCTION
Metal ions play myriad essential roles in all of biology. As a result, all cell types have evolved the ability to extract specific metal ions from their environment and ultimately maintain the intracellular concentrations of each in a range compatible with cellular needs (1). This is critical for the survival of the organism since even essential transition metal ions, e.g. Fe, Cu and Zn are toxic in excess (2). The same is true for Ni (3) and Mn (4), although acquisition of these ions ensures that specialized microorganisms are capable of surviving in a strongly acidic or potently oxidizing environment, respectively. Cu and Fe are particularly toxic since their reduced forms, Cu(I) and Fe(II), when weakly chelated in an aerobic environment, will catalyze the production of damaging hydroxyl radicals via redox cycling; as a result, the ‘free’ or bioavailable concentrations of these ions, as well as Zn, may likely be vanishingly small (5,6). The control of metal homeostasis is mediated by the balancing of uptake, efflux and intracellular sequestration or compartmentalization of essential metal ions, and is largely regulated transcriptionally by gene regulatory proteins, collectively coined metal sensor proteins (2). Metal sensor proteins directly bind a particular metal ion, or groups of metal ions that form similar coordination complexes, to the exclusion of all others (7); this, in turn, allows an organism to turn on or turn off the expression of specific genes in order to mount a metabolic response to either deprivation or excess of a particular metal ion in the cell.
Metal-responsive transcription factor-1 (MTF-1) is a heavy metal sensing transcriptional activator that up-regulates the expression of genes that allow an organism to mitigate zinc, cadmium and copper toxicity (8) (for reviews, see (9–13)). MTF-1 has been identified and at least partially characterized from human, mouse (14), pufferfish Fugu rubripes (15), zebrafish Danio rerio (16,17) and Drosophila (18). Human and mouse MTF-1 as well as the Drosophila homolog, termed dMTF-1, contain multiple functional domains, including a highly conserved zinc-finger domain that recognizes the cognate DNA sequence termed metal response element (MRE) (8). MTF-1 also harbors multiple domains for transcriptional activation (19), and short sequences that mediate intracellular trafficking into and out of the nucleus (20).
How a particular metal ion mediates MTF-1-dependent metalloregulation of gene expression is the subject of current debate (13,21); however, multiple levels of regulation clearly exist (20,22,23). Zn(II) binding to the zinc-finger domains clearly stabilizes an MRE-MTF-1 complex (8), particularly in chromatin (24). Biochemical studies of the finger domain fragment (25–27) have revealed that at least part of the zinc-sensing mechanism is mediated by the zinc-finger domain itself (21). However, MTF-1 also senses other cell stress conditions including Cd(II) (28), oxidative stress (29), hypoxia (30), and the synergistic influence of heavy metal load and heat shock (31). It seems unlikely that such inducers would act directly on the finger domain. For example, it is known that Cd(II) does not bind to the finger domain in a way that preserves the canonical ββα-structure for DNA binding (26). However, substantial data support an indirect sensing model, in which MTF-1 senses Zn(II) that is mobilized by other inducers from intracellular stores of cytoplasmic Zn(II) (29).
Previous functional studies of mammalian MTF-1 reveal that a 13-amino acid domain containing four conserved cysteines just C-terminal to a transcriptional activation domain is required for Zn(II)/Cd(II)-induced transcriptional activation in transiently transfected mouse MTF-1−/− cells (23). The mechanistic role of this domain in metalloregulation is not yet clear. However, it functions downstream of nuclear translocation and MRE-binding, perhaps activating transcription via a metal-dependent protein-protein interaction at the promoter. Indeed, when Drosophila S2 cells are stimulated with exogenous copper salts, dMTF-1 recruits TFIID to the MtnA (metallothionein A) promoter (32).
Drosophila MTF-1 differs from mammalian MTF-1 in two crucial respects. First, MRE- and MTF-1-dependent expression of metallothionein genes (mtnA-D) is strongly induced by Cu and Cd, relative to Zn, whereas Zn and Cd are the most potent inducers of mammalian metallothioneins (18). Second, disruption of the MTF-1 gene by targeted insertional mutagenesis (MTF-1 KO flies) results in a strong sensitivity to not only Cu, Cd and Zn toxicity but also to Cu depletion (33,34). The requirement for dMTF-1 to mitigate the effects of Cu deprivation is unique to dMTF-1, and originates with the ability of dMTF-1 to activate expression of a high affinity Cu importer Ctr1B under normal or low-Cu growth conditions. As a result, dMTF-1 plays a central role in copper homeostasis in the fly by regulating both import and sequestration of this essential yet toxic metal (34,35).
We reasoned that some aspect of copper-dependent metalloregulation of dMTF-1 requires the direct binding of Cu(I), analogous to the direct binding of Zn(II) to the zinc fingers of hMTF-1. Such a Cu(I)-sensing mechanism is however unlikely to function through the zinc finger domain itself, which is predicted to have a low affinity for Cu(I); thus, some other Cu(I)-binding domain would have to be present in dMTF-1. Inspection of the amino acid sequence reveals two candidate cysteine-rich Cu(I)-binding domains, both located in the C-terminal one-third of the protein (9). Here, we present evidence that the six cysteine residues from residues 547-565 are necessary for dMTF-1 to sense copper load. When challenged with copper stress, flies harboring Cys-to-Ala substitutions are unable to up-regulate the transcription of metallothionein MtnA, the major effector of copper-resistance (36,37). We also show that a peptide harboring this Cys-rich domain protects a Cu-sensitive S. cerevisiae strain (38) from Cu-toxicity, presumably by mediating intracellular storage/chelation of the metal. Binding studies show that the Cu-sensing domain of dMTF-1 binds four Cu(I) ions tightly and highly cooperatively to form a Cu4-Cys6 polynuclear cluster. This cluster is reminiscent of known Cu-sensing domains of S. cerevisiae Mac1 and Ace1 and paralogs in other organisms (39–41). The mechanistic implications of these findings are discussed.
MATERIALS AND METHODS
Plasmids and fly transformation
Cys-to-Ala mutations were generated using pUAST-dMTF-1 as a template by a quick change mutagenesis technique. pUAST-dMTF-14C-4A, pUAST-dMTF-12C-2A and pUAST-dMTF-16C-6A constructs were used to generate transgenic flies with P-element mediated transformation as described earlier (37).
Fly food, fly stocks and genetics
One liter of standard fly food was composed of 55 g corn, 10 g wheat, 100 g yeast, 75 g glucose, 8 g agar and 15 ml anti-fungal agent nipagin (15% in ethanol). For toxicity experiments, food was supplemented with CuSO4 or CdCl2 or bathocuproinedisulfonate (BCS) disodium salt hydrate (Sigma-Aldrich No. 14,662-5) to the indicated concentrations. BCS is a specific copper chelator used to deplete copper in the food. Flies were raised at 25°C and 65% humidity. UAS-dMTF-1, UAS-dMTF-14CA, UAS-dMTF-12CA, and UAS-dMTF-16CA transgenes were crossed into dMTF-1140-1R (dMTF-1 null allele) background respectively. The expression of the transgenes was induced by a ubiquitous Gal4 transactivator (actin-Gal4).
Drosophila toxicity experiments
The flies that were homozygous for dMTF-1140-1R and UAS-dMTF-1 (or its derivatives) were crossed with y w;; dMTF-1140-1R, actin-Gal4/TM6B,y+ flies on standard food or food containing copper or BCS. From the cross, two types of progeny could be obtained: Progeny (A) flies that were expressing the transgene and were dMTF-1 null mutant. Progeny (B) flies that were not expressing the transgene and contained endogenous dMTF-1. The survival index (Is) was calculated as follows: Is = 2A/(A + B).
Quantitation of MtnA and dMTF-1 transcripts in transgenic flies
To determine the level of MtnA transcripts, larvae were raised on either standard food or food containing 100 µM CuSO4. Only third instar stage larvae were collected for analysis. Total RNA was extracted using the TRIzol reagent (Life Technologies) and nuclease S1 mapping of transcripts (100 μg of total RNA) was performed as described previously (42). The gels were developed using FLA-7000 system and bands were quantified using ImageGauge software (Fuji Film). The transcripts of the endogenous actin5c gene were measured and used for normalization of MtnA transcript levels. To monitor dMTF-1 expression levels, the gut tissue was dissected from the third instar stage larvae raised on standard food. Total RNA was extracted using TRizol and first-strand cDNA synthesis was performed with 5 µg total RNA using reverse transcriptase (RT). mRNA levels were measured by quantitative (q) PCR using a SybrGreen Q-PCR reagent kit (Sigma) in combination with the MX3000P light cycler (Stratagene, Amsterdam, The Netherlands). Initial template concentrations of each sample were calculated by comparison with serial dilutions of a calibrated standard. To verify RNA integrity and equal input levels, actin mRNA was used as a reference.
Cu(I) binding experiments by absorption and luminescence spectroscopies
All Cu(I) titration samples of C-dMTF_81 were prepared anaerobically in a glovebox ([O2] < 2 ppm) with deoxygenated buffers and solvents in 10 mM MES, 0.1 M NaCl, pH 6.3, 25°C. The samples were kept in sealed containers, including during transfer from the glovebox for characterization. Then, 500 µM Cu(I) was titrated into 800 µL of 20 µM apo-protein in anaerobic environment and the absorption was monitored over the wavelength range 200–500 nm on a Hewlett-Packard model 8452A spectrophotometer. In magfura-2 competition experiments, Zn(II) was titrated into the mixture of 15.8 µM C-dMTF_131 and 16.3 µM magfura-2 (43). For the competition experiments with BCS, 282 µM C-dMTF_81 was titrated into a mixture of 100 µM BCS and 30 µM Cu(I) and the absorption spectra recorded from 250 to 600 nm. Luminescence spectra were recorded on an ISS PC1 Photon Counting spectrofluorometer. Also, 1.0 mM Cu(I) was titrated into 1700 µL of 20 µM apo-C-dMTF_81 and the full emission spectra were collected from 400 to 800 nm with excitation at 300 nm essentially as described (44).
RESULTS
Domain structure of Drosophila MTF-1
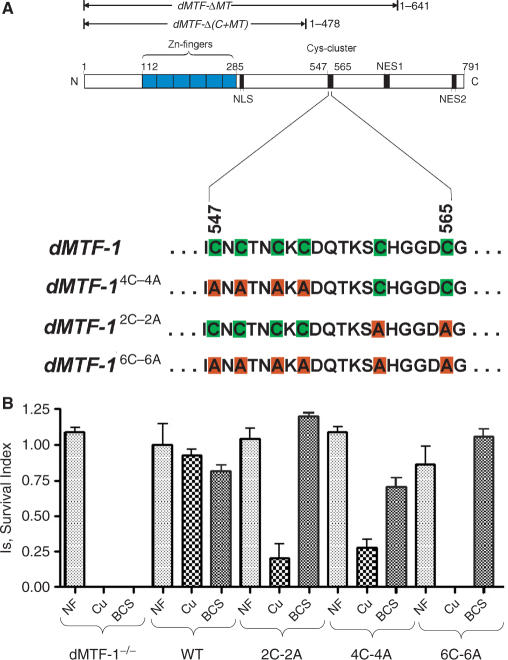
The domain structure of D. melanogaster MTF-1 (dMTF-1) is shown in Figure 1 (18). The functional domains of dMTF-1 have not yet been extensively mapped and the amino acid sequence has diverged considerably from mammalian MTF-1 outside of the DNA-binding zinc finger domain (18). However, dMTF-1 contains a cluster of six cysteines within 19 consecutive amino acids (residues 547-565) that bears some resemblance to the Cys4 cluster that has been functionally characterized in hMTF-1 (23). This cluster is followed by a Thr/Ser-rich domain (13 Thr/Ser in 19 residues), which is connected via a seryl-glycyl linker to a C-terminal metallothionein (MT)-like domain, which also contains several cysteines (residues 641-791). This C-terminal MT-domain bears strong resemblance to domain IV of S. pombe Pccs, a copper chaperone for copper-zinc superoxide dismutase (SOD1) which have been shown to protect S. pombe and a S. cerevisiae ace1Δ mutant strain from copper toxicity (45). To probe the copper-binding ability of the cysteine cluster (residues 547-565), we performed a functional analysis of this region of dMTF-1 both in Drosophila and in S. cerevisiae.
Figure 1.
The cysteine-rich region plays a critical role in protecting Drosophila from copper toxicity. (A) Domain structure of Drosophila melanogaster (dMTF-1) highlighting the short Cys-cluster region (residues 547-565) and dMTF-1 Cys-to-Ala substitution alleles characterized in transgenic flies. In addition to the zinc-finger DNA-binding domain, a putative nuclear localization signal (NLS) and two nuclear export signals (NES1 and NES2) are also indicated (V. Günther and W.S., unpublished). 131- (C-dMTF_131), 81- (C-dMTF_81) and 51- (C-dMTF_51) residue constructs of dMTF-1 characterized here correspond to amino acid residues 499-629, 499-579 and 529-579, respectively. Two C-terminal domain deletion mutants of dMTF-1, ΔMT and Δ(C + MT) characterized in S. cerevisiae (see Figure 3) are also shown. MT, metallothionein-like segment (residues 642-791); C, Cys-rich domain (residues 479-641). (B) Survival of dMTF-1 null flies and flies expressing dMTF-12C-2A, dMTF-14C-4A or dMTF-16C-6A on a standard food source (NF), or on food supplemented with 400 µM CuSO4 (Cu) or 160 µM bathocuprione disulfonate (BCS).
Transgenic flies expressing wild-type and mutant dMTF-1 genes
To investigate whether the Cys-cluster in Drosophila MTF-1 plays any role in copper homeostasis, we generated transgenic flies with constructs in which subsets of cysteines, or all six of them, are substituted by alanines (Figure 1A). As mentioned, dMTF-1 mutant flies are sensitive not only to excess copper but also to copper depletion (34). This is due to the fact that dMTF-1 activates two sets of genes that are working in opposing conditions, namely, metallothioneins at high copper, and the copper importer Ctr1B at times of copper deprivation (35). The sensitivity of dMTF-1 mutants can be rescued by co-expression of wild-type dMTF-1 transgene. To examine the role of the cysteine-rich domain, we introduced the mutant constructs dMTF-12C-2A, dMTF-14C-4A or dMTF-16C-6A encoding double (C560A/C565A), quadruple (C547A/C549A/C552A/C554A) or complete (C547A/C549A/C552A/C554A/C560A/C565A) alanine substitutions (Figure 1A), into dMTF-1 mutant flies lacking endogenous dMTF-1 and tested whether these constructs could rescue the sensitivity to either copper supplementation or copper depletion. All of the three dMTF-1 derivatives are able to rescue the sensitivity to copper starvation as well as the wild-type dMTF-1 transgene (Figure 1B). This result demonstrates that the wild-type and mutant forms of dMTF-1 are expressed to similar levels since a functional dMTF-1 is required for this.
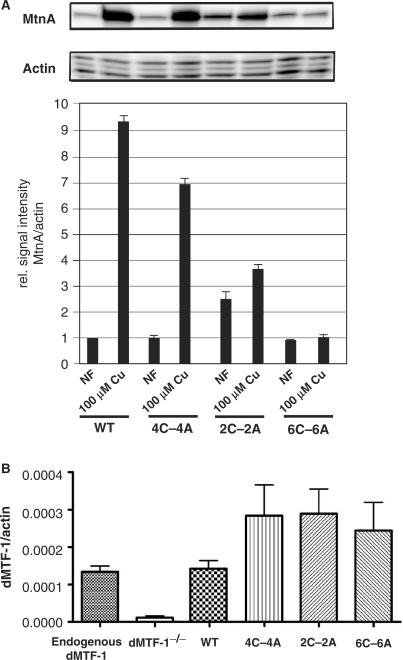
In contrast, dMTF-12C-2A and dMTF-14C-4A could only partially rescue the sensitivity to copper while dMTF-1 mutant flies expressing dMTF-16C-6A failed to survive to adulthood in copper supplemented food (Figure 1B). To further understand the molecular mechanism of the copper sensitivity phenotype, we examined the expression of metallothionein A (MtnA) in flies expressing either the wild-type or mutant alleles of dMTF-1 (Figure 2A). MtnA transcript abundance was measured by quantitative S1 nuclease mapping experiments. These data show that under copper stress (100 µM), the wild-type dMTF-1 transgene strongly activates the transcription of MtnA while dMTF-16C-6A transgene is completely unable to induce MtnA transcription. Interestingly, dMTF-14C-4A and dMTF-12C-2A transgenes mediate some Cu(I)-induced MtnA expression, but to a lesser extent than wild-type dMTF-1. Control experiments reveal that the wild-type and mutant dMTF-1 transgenes are expressed to similar levels in the larval gut, with the expression of the mutant dMTF-1 alleles perhaps even slightly (≈2-fold) higher; this and the fact that all transgenes equally confer resistance to copper starvation (see Figure 1B) render unlikely the possibility that the observed phenotypes could be due to insufficient expression of mutant alleles (Figure 2B). Taken together, these data show that the cysteine-rich domain of dMTF-1 is critical for copper-induced transcriptional activation but is clearly dispensable for sensing copper scarcity.
Figure 2.
The Cys-rich domain of dMTF-1 is required to activate MtnA expression in transgenic flies. (A) Total RNA was isolated from transgenic Drosophila at the third instar larval stage expressing either a wild-type dMTF-1, dMTF-14A-4A, dMTF-12C-2A or dMTF-16C-6A allele raised on normal food (NF) or on 100 µM CuSO4 (Cu). MtnA and actin5c-specific transcripts were measured by S1 nuclease mapping and are shown as a ratio of transcript abundance. (B) Drosophila with indicated genotypes was allowed to develop on standard food until third instar larval stage. Total RNA was isolated from larval gut and analyzed by quantitative RT-PCR to quantify transcripts of dMTF-1. Actin-5c transcripts served as a normalization reference.
The cysteine-rich region protects S. cerevisiae against copper toxicity
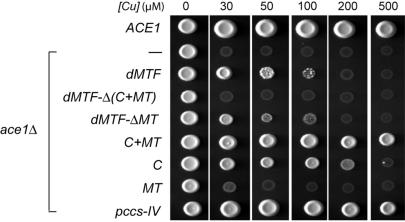
The above results indicate that the cysteine-rich domain plays an essential role in protecting Drosophila from copper toxicity by mediating up-regulation of metallothionein genes. In order to assess the importance of this domain relative to other domains in dMTF-1, we have carried out a parallel experiment in a Cu-sensitive S. cerevisiae strain, Δace1, which lacks the gene for the Cu-dependent activator of CUP1, the Cu-binding yeast metallothionein (Figure 3) (38). This strain exhibits severely attenuated survival on Cu-supplemented growth media (first two rows, Figure 3). Expression of dMTF-1 reverses some of this sensitivity in a manner that absolutely requires the Cys-rich domain (rows 3–5, Figure 3). Interestingly, expression of the entire C-terminal domain of dMTF-1 (C + MT) induces resistance to Cu-toxicity equivalent to that of the MT-like domain of the Cu-chaperone pccS of S. pombe (45), with most of the protection mediated by the Cys-rich domain itself (rows 6–8, Figure 3).
Figure 3.
The Cys-rich domain of dMTF-1 protects a Cu-sensitive strain of baker's yeast from the effects of Cu toxicity. S. cerevisiae strain DTY59 (ace1Δ) was transformed with a plasmid expressing either intact dMTF-1 (dMTF) or the indicated domain fragments of dMTF-1 and spotted onto agar plates in a defined medium containing the indicated concentration of CuSO4. A fragment encoding domain IV of the copper chaperone for SOD1 in S. pombe Pccs (labeled pccs-IV) is a positive control for this experiment. –, empty vector control; ACE1, isogenic wild-type strain DTY7 transformed with empty vector.
The cysteine-rich region of dMTF-1 binds four mol•equiv of Cu(I)
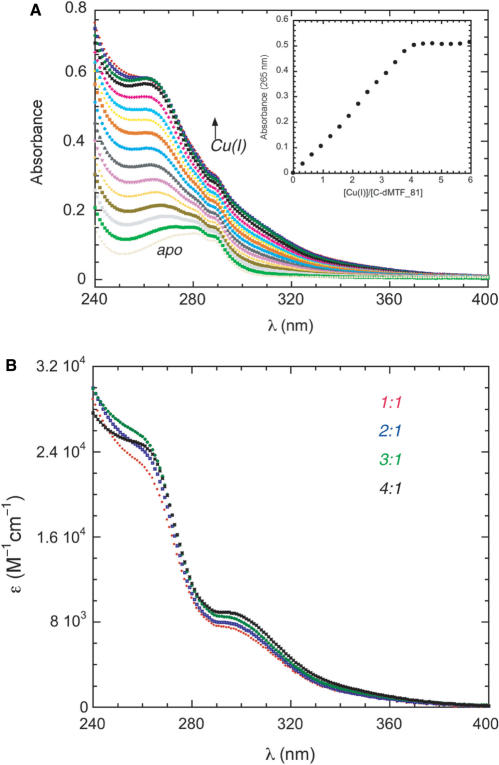
We hypothesized that the direct binding of Cu(I) by the sequence encompassing residues 547-565 in dMTF-1 is the basis for copper sensing in cells. To test this, we purified three recombinant dMTF-1 fragments of 131, 81 and 51 amino acids each of which contains the Cys-rich motif, encompassing residues 499-629 (denoted C-dMTF_131), 499-579 (C-dMTF_81) and 529-579 (C-dMTF_51). C-dMTF_81 was chosen for detailed study. Cu(I) titration of C-dMTF_81 (carried out at pH 6.0, 22°C) exhibits intense metal-to-ligand charge transfer absorption (Figure 4A), indicative of coordination to Cys thiolates (44). Similar spectra were obtained for C-dMTF_131 and C-dMTF_51 as well (data not shown). The absorption spectra for C-dMTF_81 saturate at 4 mol•equiv of Cu(I) and the binding is stoichiometric (tight) under these conditions (Figure 4A). Further examination of the absorption spectra at subsaturating amounts of Cu(I) added are consistent with the formation of a single molecular species throughout the course of the titration since molar (per bound Cu(I)) absorptivity spectra of the species formed at 1:1, 2:1, 3:1 and 4:1 Cu(I):C-dMTF_81 molar ratios are identical (Figure 4B) (vide infra). These spectra are virtually identical to previously published spectra of Cu4-Ace1 (46), and are consistent with highly cooperative assembly of Cu(I)4 polynuclear cluster in C-dMTF_81.
Figure 4.
Representative anaerobic titration of C-dMTF_81 with Cu(I). (A) Full absorption spectra are shown corrected for dilution, with the apoprotein contribution (gray curve) not subtracted. Inset, apoprotein-subtracted absorbance at 265 nm from the main body of the figure plotted as a function of Cu(I)/C-d-MTF-1 ratio. (B) Apoprotein-subtracted corrected molar absorptivity spectra of Cu(I):C-dMTF_81 mixtures at 1 : 1 (red) 2 : 1 (blue), 3 : 1 (green) and 4 : 1 (black) molar ratios. Conditions: 20 µM apo C-dMTF_81, with Cu(I) concentrations ranging from 0.3 to 6.0 molar equivalents, pH 6.0, 22°C.
C-dMTF_81 also binds Zn(II) (KZn⩾1010 M−1) and Cd(II) (KCd≈3 × 106 M−1) to form saturating 1:1 complexes under the same solution conditions (Supplementary Figure S1). However, preincubation of C-dMTF_81 with 4 mol•equiv of Zn(II) has virtually no influence on the Cu(I) binding titration; i.e., Cu(I) still binds stoichiometrically (Supplementary Figure S2A). This suggests that the Cu4 complex is far more thermodynamically stable than other metallated complexes of C-dMTF_81. Consistent with this, 30 µM C-dMTF_81 is capable of stripping ⩾80% of the Cu(I) from 30 µM Cu(I)-(BCS)2, the latter of which forms with an affinity constant KCu≈1019 M−1(Supplementary Figure S2B). This suggests that the affinity constants for BCS and C-dMTF_81 may be comparable.
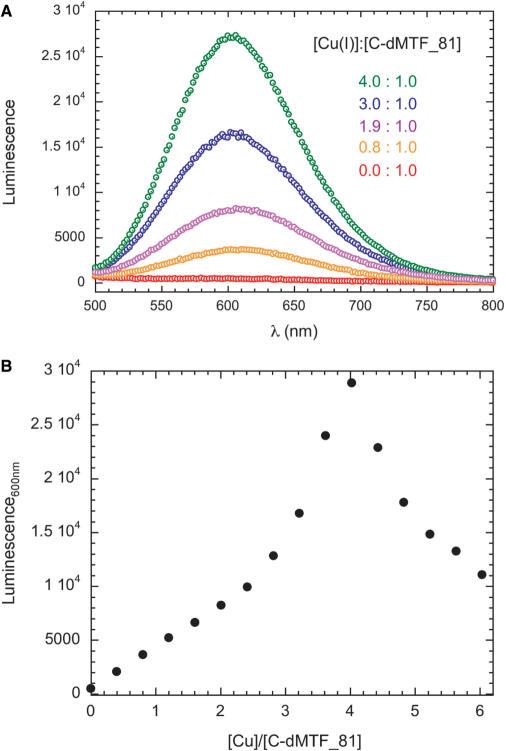
Anaerobic titrations like those shown in Figure 4 were also acquired using luminescence spectroscopy (λex = 300 nm). The results of a representative titration are shown in Figure 5, with full luminescence emission spectra (Figure 5A) and a plot of the λem, 600 vs. Cu(I):C-dMTF_81 molar ratio (Figure 5B) shown. These spectra reveal an intensely luminescent species that shows maximum intensity at a molar ratio of 4:1, after which point the intensity sharply decreases. These data reveal that the Cu(I) ions in the Cu4 polynuclear cluster are significantly shielded from solvent, as has been previously observed for other polynuclear metalloregulatory clusters in S. cerevisiae Mac1 and Ace1 (47). Further titration beyond four mol•equiv of Cu(I) results in significant bleaching of the luminescence intensity, which is not observed in an anaerobic optical titration (Figure 2). This suggests that Cu(I) ions that are added beyond saturation induce significant reorganization in the structure, which leads to a less solvent-shielded average environment for the Cu(I) ions. Addition of greater than 4 mol•equiv of Cu(I) to apo-C-dMTF_81 also leads to significant degradation of the 1H-15N HSQC spectrum (data not shown) consistent with conformational exchange broadening at greater than saturating Cu(I). These complexes may well by oligomeric in nature.
Figure 5.
Representative anaerobic titration of apo C-dMTF_81 with Cu(I) as monitored by luminescence spectroscopy. (A) Full luminescence spectra (λex = 300 nm) acquired as a function of Cu(I):C-dMTF_81 ratio, as indicated. (B) Luminescence emission intensity at 600 nm (from panel A) plotted a s function of Cu(I):C-dMTF_81 ratio. Conditions: pH 6.0, 25°C.
X-ray absorption spectroscopy reveals a Cu4-S6 polynuclear cluster
X-ray absorption spectroscopy was carried out to structurally characterize the copper binding to C-dMTF_81. Figure 6A shows that the Cu K-edge near-edge spectra from Cu(I)-C-dMTF_81 complex prepared with 1.0 and 3.5 mol•equiv of Cu(I) are essentially identical. The peak centered at around 8983 eV, is a 1s→4p transition that is commonly used as a fingerprint for determining the coordination environment of Cu(I) compounds (48). The spectra of the Cu(I)-peptide complexes are very similar to trigonally-coordinated [Cu4(SPh)6]2- and distinct from digonally-coordinated [Cu(SC10H12)2]2- (48,49) (Figure 6A), suggesting the former coordination environment in the peptide.
Figure 6.
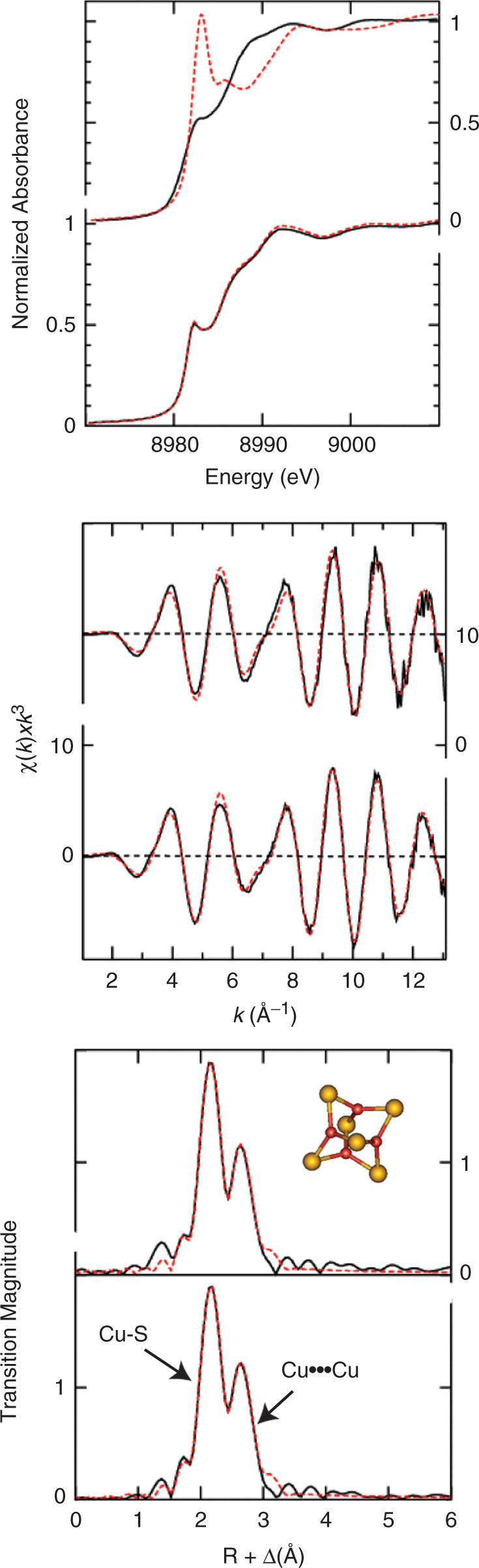
X-ray absorption spectroscopy (XAS) of Cu(I)- C-dMTF_81 complex. (A) Cu K-edge near-edge comparison of Cu(I)- C-dMTF_81 complex with two model Cu(I) thiolate compounds. In the upper panel are the trigonal Cu(I) thiolate model [Cu4(SPh)6]2- (or [Cu4(SR)6]2-, black solid line) forming a four-Cu(I) cluster, and the diagonal Cu(I) thiolate model [Cu(SC10H12)2]2- (or [Cu(SR)2]2-, red dash line) containing a single Cu(I) ion. The lower panel shows Cu(I)- C-dMTF_81 complex with metal stoichiometries of 1.0 (black solid line) and 4.0 (red dash line), respectively. (B) Copper K-edge EXAFS spectra and C) Cu-S phase-corrected EXAFS Fourier Transforms of Cu(I)-C-dMTF_81 complex mixing with 1 mol·equiv. Cu (upper panel) and 4 mol·equiv. Cu (lower panel), respectively. Black solid curves represent the experimental data, while the red dash curves are for best fits with the parameters listed in Supplementary Table S1. The inset shows a structural model representing the proposed metal coordination of the Cu(I)- C-dMTF_81 complex based on the XAS data. The red balls represent copper atoms, while the yellow ones are for sulfur atoms.
More structural detail is available from analysis of the Cu K-edge extended X-ray absorption fine structure (EXAFS) spectra. Figure 6B and C show the EXAFS, and corresponding Fourier transforms of the Cu(I)-C-dMTF_81 complexes with both 1 : 1 and 3.5 : 1 Cu:peptides, together with best fits. EXAFS curve-fitting parameters are listed in Table SI (Supplementary Material). As with the near-edge spectra, the EXAFS of the two stoichiometries are essentially identical, and gave curve fitting analysis (discussed below) that were also very similar. Two major Fourier transform peaks are observed at ≈2.3 and ≈2.7 Å, and are attributable to Cu—S and Cu ··· Cu interactions, respectively. In agreement with the near-edge spectra (Figure 6A), EXAFS curve fitting indicates three Cu—S at 2.26 Å. Inclusion of lighter scatterers such as N or O resulted in unreasonably small Debye-Waller factors for Cu—S, indicating a sulfur-only Cu(SR)3 coordination. The 2.7 Å Fourier transform peak is best fitted by including two different types of Cu ··· Cu interactions, with two short and one long Cu ··· Cu interactions at 2.70 Å and 2.82 Å, respectively, for 1:1 Cu(I):C-dMTF_81; similar fitted parameters characterize 3.5 : 1 Cu(I):C-dMTF_81 sample as well. The overall similarity of the XAS for both Cu(I):C-dMTF_81 stoichiometries suggests the same Cu center structure and provides direct evidence that C-dMTF-1 binds to Cu(I) cooperatively. Based on the XAS results a Cu4S6 polynuclear cluster is proposed to form in Cu(I)-C-dMTF_81, as shown in inset of Figure 6C. MALDI-TOF mass spectroscopy of a 1:1 Cu(I):C-dMTF_81 mixture, i.e. identical to the 1:1 sample probed by XAS, as well as a 2:1 Cu(I):C-dMTF_131 mixture, is consistent with this picture, and further suggests that an intramolecular (monomolecular) polynuclear cluster is the dominant conformer in solution (see Supplementary Figure S3).
C-dMTF_81 binds Cu(I) in an all-or-none manner
We first performed a preliminary NMR analysis of 131, 81 and 51 residue fragments of dMTF-1 encompassing residues 499-629 (C-dMTF_131), 499-579 (C-dMTF_81) and 529-579 (C-dMTF_51) by acquiring 1H-15N HSQC and 1H-{15N}heteronuclear NOE (ssNOE) spectra in the presence and absence of Cu(I). The latter experiment carried out with C-dMTF_81 revealed that only ≈27 crosspeaks were characterized by positive 1H-{15N} ssNOE values and were significantly shifted following the addition of 4.0 mol•equiv of Cu(I). This finding is consistent with the idea that Cu(I) folds the region immediately around the Cys cluster with little additional long-range folding evident in these spectra (Supplementary Figures S4-S5); in the absence of Cu(I), all resolvable crosspeaks have strongly negative ssNOE values revealing little or no stable structure in the absence of Cu(I) (spectra not shown). Further evidence for limited and localized Cu-dependent folding is that amide resonances that shift upon addition of Cu(I) have virtually identical chemical shifts in the context of a fusion protein in which 27-residues of dMTF-1 (542-568) are C-terminally appended to protein G B1 domain (GB1) (spectra not shown) (50).
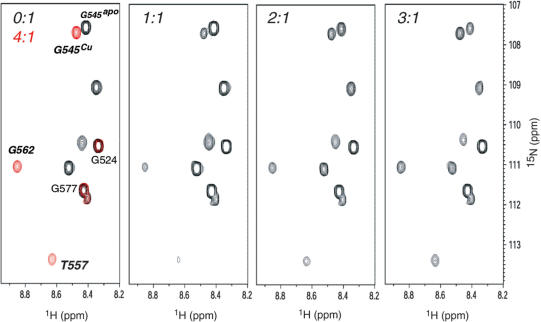
We next used NMR spectroscopy to investigate the cooperativity of Cu4 cluster formation by acquiring 1H-15N HSQC spectra as a function of Cu(I):C-dMTF_81 molar ratio (Figure 7). These spectra reveal that at subsaturating Cu(I), the spectrum corresponds to a superposition of apo- and Cu4 conformers with no evidence of a non-native structural intermediate. Quantitation of the crosspeak intensities of selected resonances (Supplementary Figure S6) as a function of Cu(I) loading is fully compatible with scenario, i.e. the intensity of apo-C-dMTF_81 crosspeaks decrease monotonically as Cu4 crosspeak areas increase. The assembly of the Cu4 cluster is therefore highly cooperative, a result consistent with the findings by XAS and mass spectrometry, which reveal significant Cu4 polynuclear cluster upon addition of sub-stoichiometric Cu(I). Despite the highly cooperative Cu-binding by C_dMTF_81, the peptide is characterized by a high degree of internal dynamics, a characteristic not unprecedented from previous studies of Cu- and Zn/Cd-loaded metallothioneins (51).
Figure 7.
Substoichiometric addition of Cu(I) to apo-C-dMTF_81 results in cooperative assembly of a Cu4 cluster from the apoprotein. A subregion of 1H-15N HSQC spectrum is shown as a function of Cu(I): C-dMTF_81 molar ratio indicated. Select resonances within the Cys cluster domain are italicized, while those outside this region (G524, G577) do not shift upon Cu(I) binding. The three remaining crosspeaks are unassigned apoprotein crosspeaks.
DISCUSSION
MTF-1 of Drosophila is capable of activating the transcription of distinct metallothionein genes (MtnA-D) in response to several metal ions, including Cu(I), Cd(II) and Zn(II). A recent characterization of transgenic flies carrying deletions of one of the four Mtn genes reveals that MtnA (the expression of which is studied here) is primarily responsible for protecting flies against exogenous copper load, while MtnB−/− flies are most sensitive to cadmium toxicity. The biological roles of MtnC and MtnD, which are closely related to MtnB, remain enigmatic since flies harboring a deletion of one or both of these genes exhibit near wild-type resistance against Cu and Cd toxicity (37). Binding studies revealed that MtnA is most strongly stabilized by Cu(I) binding, while MtnB binds Cd(II) preferentially over Zn(II) and Cu(I). These findings are generally consistent with the characteristics of flies harboring a metallothionein gene family knockout; these flies are viable and develop normally on standard food, but are highly sensitive to copper and cadmium toxicity. In particular, these experiments establish that MtnA and its regulator MTF-1 are responsible for the intense orange copper-mediated luminescence (when excited in the ultraviolet; see Figure 5) associated with specialized cells from the intestinal tract, termed midgut ‘copper cells’ (36). These cells likely function as storage depots for excess Cu(I), essentially protecting the organism against the effects of Cu(I)-mediated oxidative stress as well as a source of intracellular copper under conditions of copper deprivation (35). Interestingly, in contrast to mammalian MTF-1, Drosophila metallothioneins appear to play only a minor role against zinc toxicity (36). On the other hand, the expression of the zinc efflux transporter ZnT35C, thought to be analogous to the mammalian zinc exporter ZnT1, is strongly induced by Zn in an MTF-1-dependent manner (52).
How a single transcriptional activator, dMTF-1, is capable of up-regulating the expression of specific genes in response to distinct metal ions is unclear. One plausible scenario is that the metal selectivity of gene expression is dictated by the promoter-specific nature of the protein complex containing MTF-1 that mediates a specific transcriptional response. There is some evidence in support of this idea, since when MREs are excised from their context in the Ctr1B promoter (which is induced by Cu-scarcity) and placed in a non-native, mini-promoter context, they simply function as activating elements in response to copper overload, just like those derived from metallothionein genes (which are activated upon Cu-overload) (34). Along this vein of thought, MTF-1 might function as a promoter-specific adaptor molecule, in which the Zn(II)-bound zinc fingers mediate a direct interaction with the MRE, and another domain of the molecule mediates a Cu- or Cd- or Zn-specific complex with a putative co-activator or co-repressor. The foundational tenet of this hypothesis is that MTF-1 should be capable of forming complexes with Cu or Cd/Zn, with the distinct structures of each coordination complex (Cu vs. Zn/Cd) required to mediate metal-specific protein-protein interactions.
In the work presented here, we show that a C-terminal Cys-cluster of dMTF-1 encompassing six closely spaced cysteines forms a very stable, highly cooperative brightly luminescent Cu4-S6 polynuclear cluster. This cluster is essential for dMTF-1 to drive the expression of its target gene MtnA, because a complete Cys substitution (6C-6A) in dMTF-1 abolishes its activity under copper stress and keeps the MtnA gene uninduced. Such a defect at molecular level results in a copper sensitive phenotype of mutant Drosophila. Partial alanine substitution of two (2C-2A) or four (4C-4A) of the Cu(I)-liganding cysteines also results in a severely attenuated survival index; this suggests that formation of the Cu4-L6 (L = ligand) complex optimally protects flies against copper toxicity by inducing MtnA expression. This short 19-amino acid domain is necessary and sufficient to bind four mol•equiv of Cu tightly and stoichiometrically in vitro and in vivo, the latter measured by examining the viability of Cu-sensitive S. cerevisiae strain on Cu-supplemented media.
Strikingly, the cysteine-rich domain of dMTF-1 is reminiscent of Cu-sensing domains of other Cu-regulators from lower eukaryotes, including Mac1 and Ace 1 from S. cerevisiae, Cuf1 from the fission yeast S. pombe (40), GRISEA from Podospora anserina (39), and Amt1 from Candida glabrata (41), in the complete absence of amino acid sequence homology. Spectroscopic studies of Amt1, Mac and Ace1 reveal that each forms intensely luminescent tetranuclear Cu4•L6 ‘cage-like’ clusters containing trigonally coordinated solvent-shielded Cu(I) ions, with significant Cu•••Cu interactions, that either stimulate (Ace1, Amt1) or inhibit (Mac1) promoter DNA binding and/or transcriptional activation (41,47). A characteristic feature of the Cu complexes formed by Amt1, Ace and Mac1 is a short 2.7 Å Cu-Cu distance, also found here for dMTF-1 (41,47). Extensive molecular genetic studies have been carried out on S. cerevisiae Mac1, and these experiments are consistent with a model in which the Cu-binding domain forms a direct intramolecular protein–protein interaction with the N-terminal DNA-binding and nearby transactivation domain that allosterically blocks Mac1 function at multiple levels (53–55). Since Mac1 regulates the expression of the two high affinity Cu-importers CTR1 and CTR3, Cu-replete cells turn off the transcriptional activity of Mac1 in a Cu-dependent manner. In contrast, Cu-binding to both Ace1 and Amt1 strongly activates binding to the CuREs (copper response elements) positioned upstream of the genes encoding two metallothioneins, CUP1 and CRS5, and superoxide dismutase SOD1. It seems plausible that Cu-binding to the C-terminal domain in dMTF-1 might unmask a critical transcription activation domain that allows the recruitment of TFIID to the promoter (32) or perhaps the chromatin remodeling enzymes, Swi5/Snf and Gcn5, as has been demonstrated for C. glabrata Amt1 (56).
The Cu-regulatory complexes formed by dMTF-1 and yeast transcriptional activators contrast sharply with those found in known copper metalloregulatory proteins in prokaryotes, which form either digonal (57), mononuclear trigonal planar (44), or binuclear Cu2•S4 coordination complexes (2,58). Unlike each of these systems which are highly specific for Cu(I) (and its structural surrogate Ag(I)) (46), the intrinsic metal specificity of the metal sensing domain of dMTF-1 may well be relaxed since dMTF-1 has to bind and metalloregulate gene expression from a variety of promoters in response to a number of different metal ions, including Cd(II) and Zn(II). A direct role of the Cys-cluster in sensing both Cd(II) and Zn(II) would require that the Cys-cluster of dMTF-1 bind these metal ions as well. In fact, the Cys cluster in dMTF-1 forms stoichiometric 1:1 complexes with both Cd(II) and Zn(II), rather than a polynuclear cluster; however, Cu(I) easily outcompetes Zn(II), with Zn(II) binding considerably more tightly than Cd(II) (Supplementary Figure S1). It seems likely then that Cu(I) is the ‘cognate’ metal and others may have to be recruited under specialized intracellular conditions at specific promoters. It will be interesting to determine the degree to which inactivation of the Cys-cluster by mutagenesis influences the metal-selectivity and inducibility at other promoters, in particular those that respond to other metal ions. In any case, under the chelator conditions tested, the cysteine mutants of dMTF-1 were no more sensitive to copper starvation than wild-type flies (Figure 1B). This likely indicates that the regulation of the Ctr1B copper importer gene by dMTF-1 (34) involves protein domain(s) other than the cysteine cluster characterized here.
In conclusion, we have identified a novel Cu-binding domain in dMTF-1 derived from a cluster of six cysteines that is required to regulate metallothionein expression in transgenic flies in response to toxic intracellular levels of Cu(I). The structural features of this Cu-sensing domain while reminiscent of those previously identified in a number of fungal copper regulators, occurs in the absence of significant sequence homology and is therefore consistent with convergent evolution (Figure 8). These findings reveal a functional conservation of Cu homeostasis and detoxification from fungi to flies, with the added twist that just one transcription factor, dMTF-1, which must have evolved independently of the fungal regulators, handles both the uptake and detoxification arms of the Cu homeostasis system in Drosophila (34). Ongoing studies in our laboratories are directed toward understanding the molecular mechanism of differential sensing and regulation performed by MTF-1 in response to a variety of inducers.
Figure 8.

Cysteine clusters of metalloregulatory transcription factors. (A) Conservation of the Cys-rich region in MTF-1 of Drosophilidae and a mosquito. Dm, Drosophila melanogaster; Dps, Drosophila pseudoobscura; Dmo, Drosophila mojavensis; Dgr, Drosophila grimshawi; An, Anopheles gambiae (23). (B) Cys-rich domains of yeasts and a filamentous fungus (39,40,47). Mac1 and Cuf1, copper-regulated transcription factors of baker's yeast (S. cerevisiae) and fission yeast (S. pombe), respectively. Grisea, copper-responsive transcription factor of the fungus Podospora anserina. (C) Tetracysteine cluster of human and mouse MTF-1, required for transcriptional response to zinc and cadmium load (9,23).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Simón Labbé, University of Sherbrooke, for the gift of plasmids p426GPD and p426GPD-pccs-IV and Dennis Thiele, Duke University, for the S. cerevisiae strains used here. We also thank Viola Günther for communicating unpublished data. We thank Jon Karty and Zhen Ma, Indiana University, for their help in acquiring the mass spectrometry data. This work was supported by grants from the US National Institutes of Health (GM042569) and the Robert A. Welch Foundation (A-1295) to D.P.G., and the Swiss National Science Foundation (3100-064139), the Kanton Zurich, and the European Union (GENINTEG project ‘Mechanisms of Gene Integration’ LSHG-CT-2003-503303) to W.S. Work at the University of Saskatchewan was supported by a Canada Research Chair award to G.N.G. Portions of this work were carried out at the Stanford Synchrotron Radiation Laboratory which is funded by the US Department of Energy with additional support from the US National Institutes of Health, the Engineering Research Council (Canada), and the Canadian Institute for Health Research. Open Access charges for this article were waived by Oxford University Press.
Conflict of interest statement. None declared.
REFERENCES
- 1.O’Halloran TV. Transition metals in control of gene expression. Science. 1993;261:715–725. doi: 10.1126/science.8342038. [DOI] [PubMed] [Google Scholar]
- 2.Giedroc DP, Arunkumar AI. Metal sensor proteins: nature's metalloregulated allosteric switches. Dalton Trans. 2007:3107–3120. doi: 10.1039/b706769k. [DOI] [PubMed] [Google Scholar]
- 3.Whitmire JM, Gancz H, Merrell DS. Balancing the double-edged sword: metal ion homeostasis and the ulcer bug. Curr. Med. Chem. 2007;14:469–478. doi: 10.2174/092986707779941069. [DOI] [PubMed] [Google Scholar]
- 4.Johnston JW, Myers LE, Ochs MM, Benjamin W.H., Jr., Briles DE, Hollingshead SK. Lipoprotein PsaA in virulence of Streptococcus pneumoniae: surface accessibility and role in protection from superoxide. Infect. Immun. 2004;72:5858–5867. doi: 10.1128/IAI.72.10.5858-5867.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rae TD, Schmidt PJ, Pufahl RA, Culotta VC, O'Halloran TV. Undetectable intracellular free copper: the requirement of a copper chaperone for superoxide dismutase. Science. 1999;284:805–808. doi: 10.1126/science.284.5415.805. [DOI] [PubMed] [Google Scholar]
- 6.Outten CE, O'Halloran TV. Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science. 2001;292:2488–2492. doi: 10.1126/science.1060331. [DOI] [PubMed] [Google Scholar]
- 7.Pennella MA, Giedroc DP. Structural determinants of metal selectivity in prokaryotic metal-responsive transcriptional regulators. Biometals. 2005;18:413–428. doi: 10.1007/s10534-005-3716-8. [DOI] [PubMed] [Google Scholar]
- 8.Westin G, Schaffner W. A zinc-responsive factor interacts with a metal-regulated enhancer element (MRE) of the mouse metallothionein-I gene. EMBO J. 1988;7:3763–3770. doi: 10.1002/j.1460-2075.1988.tb03260.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giedroc DP, Chen X, Apuy JL. Metal response element (MRE)-binding transcription factor-1 (MTF-1): structure, function, and regulation. Antioxid. Redox. Signal. 2001;3:577–596. doi: 10.1089/15230860152542943. [DOI] [PubMed] [Google Scholar]
- 10.Lichtlen P, Schaffner W. The ‘metal transcription factor' MTF-1: biological facts and medical implications. Swiss Med. Wkly. 2001;131:647–652. doi: 10.4414/smw.2001.09672. [DOI] [PubMed] [Google Scholar]
- 11.Lichtlen P, Schaffner W. Putting its fingers on stressful situations: the heavy metal-regulatory transcription factor MTF-1. Bioessays. 2001;23:1010–1017. doi: 10.1002/bies.1146. [DOI] [PubMed] [Google Scholar]
- 12.Andrews GK. Cellular zinc sensors: MTF-1 regulation of gene expression. Biometals. 2001;14:223–237. doi: 10.1023/a:1012932712483. [DOI] [PubMed] [Google Scholar]
- 13.Laity JH, Andrews GK. Understanding the mechanisms of zinc-sensing by metal-response element binding transcription factor-1 (MTF-1) Arch. Biochem. Biophys. 2007;463:201–210. doi: 10.1016/j.abb.2007.03.019. [DOI] [PubMed] [Google Scholar]
- 14.Brugnera E, Georgiev O, Radtke F, Heuchel R, Baker E, Sutherland GR, Schaffner W. Cloning, chromosomal mapping and characterization of the human metal-regulatory transcription factor MTF-1. Nucleic Acids Res. 1994;22:3167–3173. doi: 10.1093/nar/22.15.3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Auf der Maur A, Belser T, Elgar G, Georgiev O, Schaffner W. Characterization of the transcription factor MTF-1 from the Japanese pufferfish (Fugu rubripes) reveals evolutionary conservation of heavy metal stress response. Biol. Chem. 1999;380:175–185. doi: 10.1515/BC.1999.026. [DOI] [PubMed] [Google Scholar]
- 16.Chen WY, John JA, Lin CH, Chang CY. Molecular cloning and developmental expression of zinc finger transcription factor MTF-1 gene in zebrafish, Danio rerio. Biochem. Biophys. Res. Commun. 2002;291:798–805. doi: 10.1006/bbrc.2002.6517. [DOI] [PubMed] [Google Scholar]
- 17.Chen WY, John JA, Lin CH, Chang CY. Expression pattern of metallothionein, MTF-1 nuclear translocation, and its DNA-binding activity in zebrafish (Danio rerio) induced by zinc and cadmium. Environ. Toxicol. Chem. 2007;26:110–117. doi: 10.1897/06-153r.1. [DOI] [PubMed] [Google Scholar]
- 18.Zhang B, Egli D, Georgiev O, Schaffner W. The Drosophila homolog of mammalian zinc finger factor MTF-1 activates transcription in response to heavy metals. Mol. Cell. Biol. 2001;21:4505–4514. doi: 10.1128/MCB.21.14.4505-4514.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muller HP, Brungnera E, Georgiev O, Badzong M, Muller KH, Schaffner W. Analysis of the heavy metal-responsive transcription factor MTF-1 from human and mouse. Somat. Cell Mol. Genet. 1995;21:289–297. doi: 10.1007/BF02257464. [DOI] [PubMed] [Google Scholar]
- 20.Saydam N, Georgiev O, Nakano MY, Greber UF, Schaffner W. Nucleo-cytoplasmic trafficking of metal-regulatory transcription factor 1 is regulated by diverse stress signals. J. Biol. Chem. 2001;276:25487–25495. doi: 10.1074/jbc.M009154200. [DOI] [PubMed] [Google Scholar]
- 21.Li Y, Kimura T, Laity JH, Andrews GK. The zinc-sensing mechanism of mouse MTF-1 involves linker peptides between the zinc fingers. Mol. Cell. Biol. 2006;26:5580–5587. doi: 10.1128/MCB.00471-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saydam N, Adams TK, Steiner F, Schaffner W, Freedman JH. Regulation of metallothionein transcription by the metal-responsive transcription factor MTF-1: identification of signal transduction cascades that control metal-inducible transcription. J. Biol. Chem. 2002;277:20438–20445. doi: 10.1074/jbc.M110631200. [DOI] [PubMed] [Google Scholar]
- 23.Chen X, Zhang B, Harmon PM, Schaffner W, Peterson DO, Giedroc DP. A novel cysteine cluster in human metal-responsive transcription factor 1 is required for heavy metal-induced transcriptional activation in vivo. J. Biol. Chem. 2004;279:4515–4522. doi: 10.1074/jbc.M308924200. [DOI] [PubMed] [Google Scholar]
- 24.Jiang H, Daniels PJ, Andrews GK. Putative zinc-sensing zinc-fingers of metal response element-binding transcription factor-1 stabilize a metal-dependent chromatin complex on the endogenous metallothionein-I promoter. J. Biol. Chem. 2003;278:30394–30402. doi: 10.1074/jbc.M303598200. [DOI] [PubMed] [Google Scholar]
- 25.Chen X, Chu M, Giedroc DP. MRE-Binding transcription factor-1: weak zinc-binding finger domains 5 and 6 modulate the structure, affinity, and specificity of the metal-response element complex. Biochemistry. 1999;38:12915–12925. doi: 10.1021/bi9913000. [DOI] [PubMed] [Google Scholar]
- 26.Giedroc DP, Chen X, Pennella MA, LiWang AC. Conformational heterogeneity in the C-terminal zinc fingers of human MTF-1: an NMR and zinc-binding study. J. Biol. Chem. 2001;276:42322–42332. doi: 10.1074/jbc.M106517200. [DOI] [PubMed] [Google Scholar]
- 27.Apuy JL, Chen X, Russell DH, Baldwin TO, Giedroc DP. Ratiometric pulsed alkylation/mass spectrometry of the cysteine pairs in individual zinc fingers of MRE-binding transcription factor-1 (MTF-1) as a probe of zinc chelate stability. Biochemistry. 2001;40:15164–15175. doi: 10.1021/bi0112208. [DOI] [PubMed] [Google Scholar]
- 28.Smirnova IV, Bittel DC, Ravindra R, Jiang H, Andrews GK. Zinc and cadmium can promote rapid nuclear translocation of metal response element-binding transcription factor-1. J. Biol. Chem. 2000;275:9377–9384. doi: 10.1074/jbc.275.13.9377. [DOI] [PubMed] [Google Scholar]
- 29.Zhang B, Georgiev O, Hagmann M, Gunes C, Cramer M, Faller P, Vasak M, Schaffner W. Activity of metal-responsive transcription factor 1 by toxic heavy metals and H2O2 in vitro is modulated by metallothionein. Mol. Cell. Biol. 2003;23:8471–8485. doi: 10.1128/MCB.23.23.8471-8485.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murphy BJ, Andrews GK, Bittel D, Discher DJ, McCue J, Green CJ, Yanovsky M, Giaccia A, Sutherland RM, Laderoute KR, et al. Activation of metallothionein gene expression by hypoxia involves metal response elements and metal transcription factor-1. Cancer Res. 1999;59:1315–1322. [PubMed] [Google Scholar]
- 31.Saydam N, Steiner F, Georgiev O, Schaffner W. Heat and heavy metal stress synergize to mediate transcriptional hyperactivation by metal-responsive transcription factor MTF-1. J. Biol. Chem. 2003;278:31879–31883. doi: 10.1074/jbc.M302138200. [DOI] [PubMed] [Google Scholar]
- 32.Marr MT, 2nd, Isogai Y, Wright KJ, Tjian R. Coactivator cross-talk specifies transcriptional output. Genes Dev. 2006;20:1458–1469. doi: 10.1101/gad.1418806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Egli D, Selvaraj A, Yepiskoposyan H, Zhang B, Hafen E, Georgiev O, Schaffner W. Knockout of ‘metal-responsive transcription factor' MTF-1 in Drosophila by homologous recombination reveals its central role in heavy metal homeostasis. EMBO J. 2003;22:100–108. doi: 10.1093/emboj/cdg012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Selvaraj A, Balamurugan K, Yepiskoposyan H, Zhou H, Egli D, Georgiev O, Thiele DJ, Schaffner W. Metal-responsive transcription factor (MTF-1) handles both extremes, copper load and copper starvation, by activating different genes. Genes Dev. 2005;19:891–896. doi: 10.1101/gad.1301805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Balamurugan K, Egli D, Hua H, Rajaram R, Seisenbacher G, Georgiev O, Schaffner W. Copper homeostasis in Drosophila by complex interplay of import, storage and behavioral avoidance. EMBO J. 2007;26:1035–1044. doi: 10.1038/sj.emboj.7601543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Egli D, Yepiskoposyan H, Selvaraj A, Balamurugan K, Rajaram R, Simons A, Multhaup G, Mettler S, Vardanyan A, Georgiev O, et al. A family knockout of all four Drosophila metallothioneins reveals a central role in copper homeostasis and detoxification. Mol. Cell. Biol. 2006;26:2286–2296. doi: 10.1128/MCB.26.6.2286-2296.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Egli D, Domenech J, Selvaraj A, Balamurugan K, Hua H, Capdevila M, Georgiev O, Schaffner W, Atrian S. The four members of the Drosophila metallothionein family exhibit distinct yet overlapping roles in heavy metal homeostasis and detoxification. Genes Cells. 2006;11:647–658. doi: 10.1111/j.1365-2443.2006.00971.x. [DOI] [PubMed] [Google Scholar]
- 38.Gralla EB, Thiele DJ, Silar P, Valentine JS. ACE1, a copper-dependent transcription factor, activates expression of the yeast copper, zinc superoxide dismutase gene. Proc. Natl. Acad. Sci. U. S. A. 1991;88:8558–8562. doi: 10.1073/pnas.88.19.8558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Borghouts C, Osiewacz HD. GRISEA, a copper-modulated transcription factor from Podospora anserina involved in senescence and morphogenesis, is an ortholog of MAC1 in Saccharomyces cerevisiae. Mol Gen Genet. 1998;260:492–502. doi: 10.1007/s004380050922. [DOI] [PubMed] [Google Scholar]
- 40.Beaudoin J, Mercier A, Langlois R, Labbe S. The Schizosaccharomyces pombe Cuf1 is composed of functional modules from two distinct classes of copper metalloregulatory transcription factors. J. Biol. Chem. 2003;278:14565–14577. doi: 10.1074/jbc.M300861200. [DOI] [PubMed] [Google Scholar]
- 41.Graden JA, Posewitz MC, Simon JR, George GN, Pickering IJ, Winge DR. Presence of a copper(I)-thiolate regulatory domain in the copper-activated transcription factor Amt1. Biochemistry. 1996;35:14583–14589. doi: 10.1021/bi961642v. [DOI] [PubMed] [Google Scholar]
- 42.Weaver RF, Weissmann C. Mapping of RNA by a modification of the Berk-Sharp procedure: the 5' termini of 15 S beta-globin mRNA precursor and mature 10 s beta-globin mRNA have identical map coordinates. Nucleic Acids Res. 1979;7:1175–1193. doi: 10.1093/nar/7.5.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu T, Reyes-Caballero H, Li C, Scott RA, Giedroc DP. Multiple metal binding domains enhance the Zn(II) selectivity of the divalent metal ion transporter AztA. Biochemistry. 2007;46:11057–11068. doi: 10.1021/bi7006367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu T, Ramesh A, Ma Z, Ward SK, Zhang L, George GN, Talaat AM, Sacchettini JC, Giedroc DP. CsoR is a novel Mycobacterium tuberculosis copper-sensing transcriptional regulator. Nat. Chem. Biol. 2007;3:60–68. doi: 10.1038/nchembio844. [DOI] [PubMed] [Google Scholar]
- 45.Laliberte J, Whitson LJ, Beaudoin J, Holloway SP, Hart PJ, Labbe S. The Schizosaccharomyces pombe Pccs protein functions in both copper trafficking and metal detoxification pathways. J. Biol. Chem. 2004;279:28744–28755. doi: 10.1074/jbc.M403426200. [DOI] [PubMed] [Google Scholar]
- 46.Casas-Finet JR, Hu S, Hamer D, Karpel RL. Characterization of the copper- and silver-thiolate clusters in N-terminal fragments of the yeast ACE1 transcription factor capable of binding to its specific DNA recognition sequence. Biochemistry. 1992;31:6617–6626. doi: 10.1021/bi00143a036. [DOI] [PubMed] [Google Scholar]
- 47.Brown KR, Keller GL, Pickering IJ, Harris HH, George GN, Winge DR. Structures of the cuprous-thiolate clusters of the Mac1 and Ace1 transcriptional activators. Biochemistry. 2002;41:6469–6476. doi: 10.1021/bi0160664. [DOI] [PubMed] [Google Scholar]
- 48.Kau LS, Spira-Solomon DJ, Penner-Hahn JE, Hodgson KO, Solomon EI. X-ray absorption-edge determination of the oxidation-state and coordination-number of copper – application to the type-3 site in rhus-vernicifera laccase and its reaction with oxygen. J. Am. Chem. Soc. 1987;109:6433–6442. [Google Scholar]
- 49.Dance I. The structural chemistry of metal thiolate complexes. Polyhedron. 1986;5:1037–1104. [Google Scholar]
- 50.Gronenborn AM, Clore GM. Rapid screening for structural integrity of expressed proteins by heteronuclear NMR spectroscopy. Protein Sci. 1996;5:174–177. doi: 10.1002/pro.5560050123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vasak M, Hasler DW. Metallothioneins: new functional and structural insights. Curr. Opin. Chem. Biol. 2000;4:177–183. doi: 10.1016/s1367-5931(00)00082-x. [DOI] [PubMed] [Google Scholar]
- 52.Yepiskoposyan H, Egli D, Fergestad T, Selvaraj A, Treiber C, Multhaup G, Georgiev O, Schaffner W. Transcriptome response to heavy metal stress in Drosophila reveals a new zinc transporter that confers resistance to zinc. Nucleic Acids Res. 2006;34:4866–4877. doi: 10.1093/nar/gkl606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Graden JA, Winge DR. Copper-mediated repression of the activation domain in the yeast Mac1p transcription factor. Proc. Natl. Acad. Sci. U. S. A. 1997;94:5550–5555. doi: 10.1073/pnas.94.11.5550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jensen LT, Winge DR. Identification of a copper-induced intramolecular interaction in the transcription factor Mac1 from Saccharomyces cerevisiae. EMBO J. 1998;17:5400–5408. doi: 10.1093/emboj/17.18.5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Keller G, Gross C, Kelleher M, Winge DR. Functional independence of the two cysteine-rich activation domains in the yeast Mac1 transcription factor. J. Biol. Chem. 2000;275:29193–29199. doi: 10.1074/jbc.M001552200. [DOI] [PubMed] [Google Scholar]
- 56.Koch KA, Allard S, Santoro N, Cote J, Thiele DJ. The Candida glabrata Amt1 copper-sensing transcription factor requires Swi/Snf and Gcn5 at a critical step in copper detoxification. Mol. Microbiol. 2001;40:1165–1174. doi: 10.1046/j.1365-2958.2001.02458.x. [DOI] [PubMed] [Google Scholar]
- 57.Changela A, Chen K, Xue Y, Holschen J, Outten CE, O'Halloran TV, Mondragon A. Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR. Science. 2003;301:1383–1387. doi: 10.1126/science.1085950. [DOI] [PubMed] [Google Scholar]
- 58.Cobine PA, George GN, Jones CE, Wickramasinghe WA, Solioz M, Dameron CT. Copper transfer from the Cu(I) chaperone, CopZ, to the repressor, Zn(II)CopY: metal coordination environments and protein interactions. Biochemistry. 2002;41:5822–5829. doi: 10.1021/bi025515c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.