Abstract
Although RNA interference as a tool for gene knockdown is a great promise for future applications, the specificity of small interfering RNA (siRNA)-mediated gene silencing needs to be thoroughly investigated. Most research regarding siRNA specificity has involved analysis of affected off-target genes instead of exploring the specificity of the siRNA itself. In this study we have developed an efficient method for generating a siRNA target library by combining a siRNA target validation vector with a nucleotide oligomix. We have used this library to perform an analysis of the silencing effects of a functional siRNA towards its target site with double-nucleotide mismatches. The results indicated that not only the positions of the mismatched base pair have an impact on silencing efficiency but also the identity of the mismatched nucleotide. Our data strengthen earlier observations of widespread siRNA off-target effects and shows that ∼35% of the double-mutated target sites still causes knockdown efficiency of >50%. We also provide evidence that there may be substantial differences in knockdown efficiency depending on whether the mutations are positioned within the siRNA itself or in the corresponding target site.
INTRODUCTION
Since the finding of siRNAs as mediators of gene expression knockdown, first discovered in Caenorhabditis elegans (1), great expectations have been laid on the mechanism of RNA interference (RNAi) and the use of siRNA as a tool for functional gene studies. Numerous studies have also explored RNAi as a potential therapeutic mediator for cancer treatment and viral diseases like HIV as well as for drug target discovery (2–4). However, what first seemed as a straightforward and highly effective tool for posttranscriptional gene silencing turned out to involve more obstacles than first anticipated. Nonspecific side effects, such as antiviral responses caused by in vitro-transcribed siRNAs and off-target effects resulting in unspecific knockdown of nontargeted genes, are issues that need to be resolved before siRNA can be used as a reliable tool for gene knockdown in various diseases (5–9).
As a result, a great deal of RNAi research has been performed with the aim to define the distinct functions of individual siRNA molecules and what features are crucial for its effectiveness and specificity. The majority of these analyses have been conducted by altering the siRNA sequence itself and monitoring the resulting knockdown pattern (10). This type of data generates a well-defined analysis of each mutated siRNA molecule but does not give a complete and true picture of all possible targets of a single siRNA molecule. Alteration of the siRNA sequence changes its sequence-specific characteristics and affects the thermal stability, which determines the siRNA's ability to incorporate into the RNA-induced silencing complex (RISC) (11). Low internal stability at the 3′ sense strand facilitates strand selection and RISC incorporation, whereas very low or very high internal stability tend to cause reduced target affinity and ineffective unwinding, respectively (12). These sequential effects all have impact on siRNA functionality and therefore limit the conclusions you can draw from analysis of siRNA specificity and the possible off-target effects caused by using sequence-modified siRNAs.
Other studies have used global gene expression analyses to evaluate the effects of siRNAs other than the expected knockdown of the target gene expression (7). In this study, we use an alternative method to investigate siRNA specificity. By altering the target site of a functional siRNA, instead of altering the siRNA sequence itself, we overcome the possible conformational changes in siRNA uptake into RISC. In a previous study, Du et al. (13) analyzed single-base mutations of the 19-base target of a functional siRNA. Here, we analyze multiple combinations of double-nucleotide mismatches of the same target. We have prepared and optimized a straightforward and cost effective method to generate a siRNA target library by combining the siQuant luciferase expression vector and a single-tube chip-synthesized oligomix, containing all possible combinations of double mutations in the 19-nt long target sequence of the functional siCD46 (14). This method avoids the cumbersome procedure of multiple transformation reactions of all unique constructs and provides a relatively fast generation of a large siRNA target library to utilize for siRNA specificity studies. In this study we provide further insight in the field of siRNA off-target effects by showing that double mutations in the siRNA target site still give rise to a substantial amount of remaining knockdown activity by its corresponding siRNA. We also show that nucleotide alterations in the antisense strand of an siRNA molecule creates a different knockdown pattern when compared to the knockdown efficiency of a wild type siRNA's ability to suppress a target site with corresponding mutations.
MATERIALS AND METHODS
Oligo design and plasmid insert preparation
The target of siCD46 (XM_036622, nucleotides 604–622) was used as a template to create our 19-nt double-mutated siRNA targets. We designed a total of 1539 target sites, each containing double-nucleotide mismatches in all possible combinations (Table 1). The oligos consists of a BglII restriction site at the 5′ region followed by the siCD46 target site and a 3′ restriction site for ApaI. The sequences were ordered in a single-tube oligomix (Atactic Technologies Inc). The oligomix was PCR amplified using the flanking primers CD46-F3T7 (5′-TAATACGACTCACTATAGGGGGTACAGCGGAGAT-3′) and CD46-R3T7 (5′-TAATACGACTCACTATAGGGAGCCTGTACTGGG-3′), giving a product of 85 bp in length (Figure 1). Several PCR reactions were pooled and ethanol precipitated to obtain a high concentration. The purified PCR product was cleaved with restriction enzymes FastDigest ApaI and BglII (Fermentas GmbH, St. Leon-Rot, Germany) followed by analysis on a 2% MetaPhor agarose gel for verification of correct size (33 bp). The forward and reverse strands of the wild type target of siCD46 were ordered separately and annealed to form a ready-to-ligate target sequence. Sense strand: 5′-GATCACACTTATTGGAGAGAGCACGAGGCGGCC-3′ and the antisense strand: 5′-GCCTCGTGCTCTCTCCAATAAGTGT-3′. All nucleotides except the oligomix were purchased from MWG Biotech, Ebersberg, Germany.
Table 1.
The 19-mer target sequences were designed to represent every possible double-nucleotide mismatch, a total of 1539 variations
| Sequence name | siQuant CD46 target oligo |
|---|---|
| CD6 wt-target | CTTATTGGAGAGAGCACGA |
| BglII ApaI | |
| siQ46-lA_2A | CAGCGG AGATCT CA AATATTGGAGAGAGCACGA TA GGGCCC AGTACA |
| siQ46-lA_2C | CAGCGG AGATCT CA ACTATTGGAGAGAGCACGA TA GGGCCC AGTACA |
| siQ46-lA_2G | CAGCGG AGATCT CA AGTATTGGAGAGAGCACGA TA GGGCCC AGTACA |
| siQ46-lA_3A | CAGCGG AGATCT CA ATAATTGGAGAGAGCACGA TA GGGCCC AGTACA |
| siQ46-lA_3C | CAGCGG AGATCT CA ATCATTGGAGAGAGCACGA TA GGGCCC AGTACA |
| siQ46-lA_3G | CAGCGG AGATCT CA ATGATTGGAGAGAGCACGA TA GGGCCC AGTACA |
| siQ46-lA_4C | CAGCGG AGATCT CA ATTCTTGGAGAGAGCACGA TA GGGCCC AGTACA |
| siQ46-lA_4G | CAGCGG AGATCT CA ATTGTTGGAGAGAGCACGA TA GGGCCC AGTACA |
| siQ46-lA_4T | CAGCGG AGATCT CA ATTTTTGGAGAGAGCACGA TA GGGCCC AGTACA |
| siQ46-lA_5A | CAGCGG AGATCT CA ATTAATGGAGAGAGCACGA TA GGGCCC AGTACA |
| siQ46-lA_5C | CAGCGG AGATCT CA ATTACTGGAGAGAGCACGA TA GGGCCC AGTACA |
| siQ46-lA_5G | CAGCGG AGATCT CA ATTAGTGGAGAGAGCACGA TA GGGCCC AGTACA |
| etc. |
The mutations are marked in bold letters. Target sites for restriction enzymes BglII and ApaI were introduced in the 5′ and 3′ of the target site respectively to enable subsequent ligation to the siQuant vector (underlined sequences). In this table only the first 15 combinations are listed.
Figure 1.
The oligomix containing all 1539 sequence variations of the siCD46 target site were PCR amplified with flanking primers resulting in a 85-bp long DNA molecule. The target sequence in this figure represents the wild-type version.
Construction of siQuant-CD46 vector
In this study, we used the previously described siRNA validation vector siQuant to analyze siRNA specificity (14). To generate circular constructs of the siQuant vector and the siCD46-target restriction products, we performed a ligation reaction in 4°C overnight with 100 ng siQuant vector, ∼25 ng of CD46-insert restriction product, 1U T4 DNA Ligase (Invitrogen Ltd, Paisley, PA, UK) at a final volume of 20 µl. 5 µl of the ligation reaction was then analyzed on 0.6% agarose gel and the remaining 15 µl were diluted 1:4 in 45 µl TE. 5 µl of this dilution was used for transformation of 50 µl Max-efficiency DH5α cells (Invitrogen) according to the manufacturer's protocol. 200 µl of the transformation reaction was then spread on prewarmed LB Agar plates containing 50 mg/l ampicillin and incubated in 37°C overnight. All positive clones were then picked and inoculated in 1.4 ml LB media with ampicillin (50 mg/l) for 16 h at 37°C.
siQ-CD46 target identification
To identify which sequence that had been inserted in the vector for each positive clone, a small amount of each overnight culture was used for subsequent pyrosequencing analysis on a PSQTM96 instrument (Biotage AB, Uppsala, SwedenBiotage). The prepyrosequencing PCR was carried out with the primers pyro-F: 5′-ATCCGCACCATGGGCTGT-3′ and the biotinylated pyro-R-biot: 5′-CTCTCCAGCGGTTCCATCTT-3′. The reaction was carried out in 95°C for 3 min; 35 cycles of 95°C for 30 s, 65°C for 30 s, 72°C for 40 s; 72°C for 5 min. 5 µl of the PCR product was analyzed on 2% agarose gel for visualization of band of correct size: 132 bp. The pyrosequencing procedure was carried out using the sequencing primer pyro-S: 5′-CATGGGCTGTGAGATC-3′ and according to manufacturer's protocol. The resulting pyrograms were transferred to Word documents and the sequences analyzed manually. Positive clones of siQuant vectors with correct and unique inserts were purified with Qiagens's QIAprep 96 Turbo Miniprep kit. Concentrations were measured on a ND-1000 Spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA).
Cell transfections
Human embryonic kidney cells (HEK293A) were cultured with GIBCO Dulbecco's modified Eagle's medium (Invitrogen) containing 10% fetal bovine serum and 1 × Penicillin–Streptomycin–Glutamine (Invitrogen). For large-scale transfection experiments, the cells were seeded in 96-well plates and grown for 24 h. The confluence was about 50% for the time of transfection. The cells were co-transfected with 0.08 µg of the different siQ-CD46 mutants together with 0.008 µg of pRL-TK and 13 nM of siCD46, using 0.2% Lipofectamine2000 (Invitrogen), GIBCO Opti-Mem media (Invitrogen) in a final volume of 100 μl. Transfections were carried out for 24 h with a media change after 4 h. For the comparative study of knockdown efficiency between mutations in siRNA strand versus mutations in its corresponding targets site, transfections were carried out in 24-well plates and plasmid concentrations were 0.17 and 0.017 µg for fusion constructs and pRL-TK, respectively. All samples were analyzed in triplicates. The siCD46 sequences are as follows: wt-sense: 5′-CUUAUUGGAGAGAGCACGA-3′, and wt-antisense: 5′-UCGUGCUCUCUCCAAUAAG-3′; si46-9T_17A-sense: 5′-CUUAUUGGUGAGAGCAAGA-3′, si46-9T_17A-antisense: 5′-UCUUGCUCUCACCAAUAAG-3′; si46-3A_14C-sense: 5′-CUAAUUGGAGAGACCACGA-3′, si46-3A_14C-antisense: 5′-UCGUGGUCUCUCCAAUUAG-3′; si46-2C_8A-sense: 5′-CCUAUUGAAGAGAGCACGA-3′, si46-2C_8A-antisense: 5′- UCGUGCUCUCUUCAAUAGG-3’. The names of the sequence-altered siRNAs correspond to the sequence substitutions in the DNA sequence of the corresponding siQuant target sites.
Fluorometer analysis
At 24 h posttransfection, the media was removed and the cells washed with a sufficient amount of phosphate buffered saline and treated with 20 μl passive lysis buffer (PLB 1X, Promega Corporation, Madison, WI, USA) for each well of the 96-well plates and 120 μl for each well in the 24-well plates. The samples were incubated on rocking tables for 15 min in room temperature. To measure siRNA silencing efficiency of our mutated target sites, the Dual-Luciferase Reporter Assay System (Promega) was used to detect luciferase expression. This was feasible since the CD46-targets were fused with the reporter gene, hence giving a luciferase expression proportional to the siRNA silencing efficiency. The Dual-Luciferase System was performed according to the standard protocol from Promega Corporation. Samples from the transfection in the 96-well were analyzed on a fluorometer (NOVOstar, BMG Labtechnologies GmbH, Germany). The firefly luciferase signal was normalized to the renilla luciferase signal for each sample. All siQuant constructs were transfected with or without siCD46 to obtain the relative silencing efficiency for each unique clone.
RESULTS
Generation of siQuant CD46-target library
By using a nucleotide oligomix we were able to generate a large set of double-mutated target sequences for the functional siRNA siCD46. Correct inserts were verified by Pyrosequencing. The sequences surrounding the two restriction sites were also analyzed for consistency and only plasmids with the exactly correct sequence were selected for future transfection. This is of importance because even a single nucleotide substitution, insertion or deletion can change the reading frame and subsequent transcription. About 50% of all positive colonies were verified as correct siQuant-CD46-target constructs. Following transfection assays showed that the majority of the siQuant-CD46-target constructs obtained by this method were functional and a total amount of 709 out of 1539 possible constructs were analyzed in this study (Supplementary Table 1 a–c).
Silencing effects of siCD46 on target sites with double-nucleotide mismatches
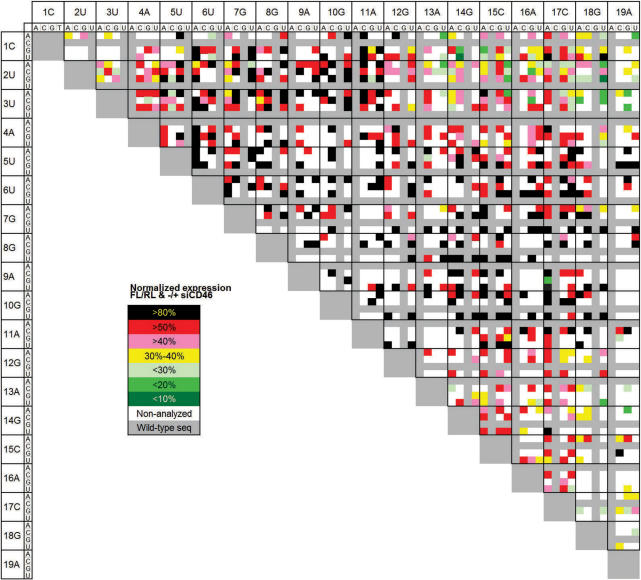
The fusion luciferase reporter plasmids were used for analysis of siRNA target specificity in a cell-based assay system and validated by Dual-Luciferase Reporter Assay System. The generated firefly luciferase activity is proportional to the siCD46-target expression, which makes it possible to validate the siRNA specificity by its ability to repress luciferase activity. Each siQuant reporter plasmid was co-transfected with the renilla luciferase expressing pRL-TK control plasmid. To compensate for possible variations in expression efficiency, the luciferase activity values were normalized to the expression levels of renilla luciferase and each reporter plasmid was also transfected both in absence or presence of siCD46 to verify that the knockdown efficiency corresponds to every reporter plasmids own ability to express firefly luciferase. The resulting data shows remaining expression levels of firefly luciferase (Supplementary Table 1 a–c). To facilitate the comprehensibility of the large data set, the expression levels were categorized into different subgroups and visualized in a colour-coded chart ranging from green—symbolizing low expression levels (efficient knockdown ability)—to red/black to symbolize high expression levels, i.e. reduced knockdown ability (Figure 2).
Figure 2.
Color-coded expression chart visualizing positional and regional trends for siCD46 knockdown efficiency and tolerance towards double-mutations in its specific target site. The vertical axis represents the position of the first mutation and the horizontal axis represents the position of the second mutation. The cross-section of each pair illustrates the remaining luciferase expression of each clone. Seven hundred and nine out of 1539 different combinations were identified and analyzed. These clones are each represented in the chart with a color corresponding to the remaining expression activity after targeting with siCD46. Dark green represents <10% remaining expression activity, lime green: <20%, light green: <30%, yellow: 30–40%, pink: >40%, red: >50% and black: >80% remaining activity. Light gray represents wild type target sequence and white nonidentified clones.
The knockdown efficiency of the siCD46 when targeting a wild type reporter is ∼92% (13). Out of our target library of 709 reporter plasmids, 18 (3%) double-mutated constructs gave a knockdown efficiency of >80%, 123 (17%) of the constructs resulted in 60–80% knockdown and 318 (45%) of them reduced the knockdown ability to 20–60%. The remaining 250 (35%) of the constructs gave <20% knockdown which is considered to be nonsignificant down regulation when it comes to siRNA efficiency. It is remarkable that 227 (35%) of analyzed construct causes a silencing efficiency of more than 50%.
Different tolerance regions for mismatch insertions
Looking at the colour-coded expression chart (Figure 2), we can see a clear pattern of positional significance for the siRNAs ability to knockdown targets with different double-nucleotide mismatches. In accordance to earlier studies (13), we can confirm that position 5–11 seems to be highly sensitive to nucleotide alterations and most of the constructs with at least one mutation in this area causes reduced knockdown efficiency, which results in remaining luciferase expression levels of more than 50%. We can see this pattern in both the vertical and horizontal levels. Mutations in position 1–2 and 18–19 are the most tolerable alterations but of course depending on were the second mutation is positioned. Furthermore, double mutations both situated in the 5′-region of the siRNA target site tend to cause greater knockdown inhibition than double mutations in the 3′-region of the same target. The least changes in knockdown efficiency results when one mutation occurs at position 1 or 2 and the other one is positioned at the far 3′-end. It is notable that double mutations in region 1–3 or 17–19 seems to have greater impact than when one of the mutations occurs in region 1–3 and the other in position 17–19. Another remarkable observation is how well mutations in region 13–17 (medium tolerance region) are tolerated, especially when the second mutation lies not only in region 1–3 but also when the second mutation is positioned in region 13–17. As reported previously (13), variations of knockdown efficiency in the low tolerance regions may occur depending on which nucleotide is substituted and in what position. In this study we can, for instance, see that the substitution of guanosine to adenosine in position 8 for siQ46-3C_8A and alteration of adenosine to guanosine in position 11 for siQ46-2G_11G are examples of highly tolerated mismatches in a low tolerance region. These occurrences are, however, not as frequent in this study as compared with analyses of siRNA targets with single-nucleotide mismatches, most probably due to the additional nucleotide mismatch, which further affects the siRNA's ability to recognize its target.
If we compare our results with earlier studies we can see that the siRNA knockdown efficiency of our double-mutated siRNA target constructs, follows the pattern of previously identified tolerance regions (13). Mismatches in the high tolerance regions affect the specificity less than mismatches in the low tolerance regions, which are critical and often lead to full protein expression. Mutations in the medium tolerance region differ a lot in their ability to affect siRNA specificity depending on the identity of the substituting nucleotide. We were, however, surprised to see that a large part of the double-mutated target site still causes little or no alterations in knockdown efficiency.
Comparison of site-specific alterations in siRNA or target sequence
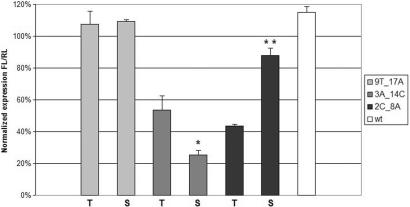
As discussed above, one of the benefits of using a reporter-based vector system for validation of siRNA target specificity is the avoidance of altering the siRNA uptake into RISC. Thermal stability of the siRNA duplex is considered to be crucial for RISC assembly and uptake of the correct antisense strand. By altering the siRNA sequence, when performing studies of siRNA specificity and investigations of possible off-target effects, you might also change its ability to enter RISC and hence affect the silencing efficiency negatively in an availability—rather than specificity—based manner. To analyze this further, we selected three different siQuant constructs with varying effect on siCD46 knockdown efficiency: siQ46-9T_17A gives total abolition of siRNA knockdown; siQ46-3A_14C and siQ46-2C_8A are both silenced by ∼50%. Three siRNAs corresponding to these double-mutated target sites were designed to compare the knockdown efficiency towards reporter constructs with wild type targets. The results of this analysis indicate that in some of the cases, there are indeed differences in knockdown efficiency depending on whether the mutations is positioned on the siRNA target or the siRNA itself (Figure 3). The si-3A_14C gives a knockdown efficiency of 75% on its wild type target as compared to the remaining 46% knockdown efficiency when targeting siQ46-3A_14C with the functional siCD46. To the opposite, si-2C_8A is only able to knockdown the luciferase expression with 12%, as compared to siQ46-2C_8A which results in 57% knockdown. The si-9T_17A does not seem to alter any preferences for knockdown efficiency and has no effect on the expression levels as is the case also for the corresponding siQ46-9T_17A. The results from these experiments indicate that the sequence alterations in si-3A_14C gives a positive effect in terms of knockdown efficiency and the corresponding changes in si-2C_8A gives a negative impact on its silencing ability.
Figure 3.
Comparative study of mutations occurring in the siRNA antisense strand or in the corresponding positions of its target site. Light gray bars: siQ46-9T_17A and si-9T_17A, medium gray bars: siQ46-3A_14C and si-3A-14C, dark gray bars: siQ46-2C_8A and si-2C_8A, white bar: siQ46-wt co-transfected with unrelated siRNA. T—mutation of siRNA target, S—mutation of siRNA antisense strand. The bars represent remaining expression levels of firefly luciferase activity normalized to renilla luciferase. All values are also normalized to each reporter construct's own ability to express luciferase. *P < 0.05 and **P < 0.01, respectively are significant differences of luciferase expression between cells transfected with either mutated siQuant target constructs or mutated siRNAs.
DISCUSSION
Earlier studies about siRNA specificity show that the location of the mutation is of great importance (10). The 3′ and central regions of the siRNA antisense strand are considered highly sensitive for sequence alterations and to cause great impact on knockdown efficiency (15,16). When using siRNA as a tool for functional genomics studies, molecules should at least give more than 70% knockdown of its target expression to be considered as an effective siRNA. However, when evaluating the possible off-target effects of a single siRNA, >50% knockdown of various off-targeted gene is a fairly large knockdown effect and might cause major alterations in cell function. In this study we only examine the mismatch tolerance of a single siRNA, which might not be a representative of all siRNAs. Even so, the fact that we from our present study could see that ∼35% of nonspecific targets with double-nucleotide mismatches were down regulated with more than 50% efficiency, demonstrates the need for careful investigation of possible unwanted side effects caused by siRNAs. This is a particularly important matter to consider before any efforts of using siRNA-mediated gene silencing as a therapeutic tool can be made.
By using the siRNA-target library construction method described above, combining the siQuant reporter vector with a single-tube oligomix, we were able to produce a large set of reporter constructs in a high-throughput and cost-effective manner. 709 double-mismatched targets were derived out of 1539 possible combinations. In theory, all 1539 permutations should be included in our target library, but the time and effort for getting additional clones eventually reached a steady-state level. As we started to examine the 709 targets, we found that they could already give a very clear trend.
Subsequent validation experiments have given us further insight in siRNA off-target effects. There seems to be two types of off-target effects from siRNA: (i) off-target effects mediated by Ago2 on targets that are highly similar to the wild type targets of the siRNA. This type of off-target effects seems to be very potent although the relative abundance of such ‘off-targets’ is relatively low. (ii) off-target effects mediated by Ago1 (or maybe partially by Ago2 or other Ago's) on targets that contain the ‘seed region’ (8,17). Clearly, not all sequences that contain a seed region will be affected by a siRNA and currently there are no clear indications about what are the additional determinants that would render a mRNA an ‘off-target’ of this type. In addition, off-target effects of this type are normally pretty weak. Also there is no knowledge about when the type-1 off-target effects will fade and the type-2 off-target effects take over. The current article is a useful exploration about the scope of type-1 off-target effects. Results of this article cannot be deduced from the study of type-2 off-target effects.
We have also shown that nucleotide mismatches in the antisense strand of the siRNA molecule can give a different knockdown pattern when comparing it to a wild type siRNAs ability to suppress expression from target sites with corresponding mutations. These data indicates that the usage of mutated siRNA molecules is a less reliable tool for evaluating siRNA specificity and further strengthens the advantages of using the siQuant siRNA validation vectors for these purposes.
Numerous studies exploring the functions of siRNA-mediated RNAi have resulted in a large amount of factors that all need to be taken under consideration when evaluating siRNA specificity. In an indirect approach, comprehensive mutations were done on siRNAs and the mutated versions of each siRNA were analyzed for their silencing efficiency on native targets of the siRNA (17). Of 171 mutated siRNAs, silencing efficiency of about 60% of the siRNAs (102 siRNAs) were above 50%, which suggested that many mutated siRNAs have significant ‘off-target’ effects on native target transcripts. These observations largely confirm the results from our system in terms of the widespread off-target effects of siRNAs and significance of positions as well as nature of mismatches in determining the level of off-target effects. However, it should be stressed that the results from a siRNA mutating approach should be viewed as the off-target effects of many siRNAs on a single target whereas our study provides a comprehensive view of the off-target effects of a single siRNA. In assessment of the specificity of future siRNA drugs, approaches employed in the current study could potentially be more applicable.
Except from sequence similarities between siRNAs and nontargeted mRNAs as discussed in this article, there are also other parameters like strand length, 3′ overhangs, internal stability of siRNA strands and siRNA seed region complementarity to 3′ UTR of mRNAs to take under consideration when designing an siRNA of high specificity and efficiency (17–21). It has also been suggested that surrounding sequences and mRNA secondary structure may have an impact on siRNA targeting and that mRNA-binding proteins might interfere with the accessibility of the siRNA target site. There are probably several other features of RNAi action to be determined and in order be able to predict siRNA specificity and possible off-target effects, computational methods combining all of these parameters need to be established (22,23).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
The authors would like to thank Selim Sengul for helpful advice in Pyrosequencing. This research was supported by the Swedish Research Council (Vetenskapsrådet grant 2005-5397), Pfizer Inc. and KI Travel fund. Funding to pay the Open Access publication charges for this article was provided by the Swedish Research Council.
Conflict of interest statement. None declared.
REFERENCES
- 1.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Scherer L, Rossi JJ, Weinberg MS. Progress and prospects: RNA-based therapies for treatment of HIV infection. Gene Therapy. 2007;14:1057–1064. doi: 10.1038/sj.gt.3302977. [DOI] [PubMed] [Google Scholar]
- 3.Lu PY, Xie FY, Woodle MC. Modulation of angiogenesis with siRNA inhibitors for novel therapeutics. Trends Mol. Med. 2005;11:104–113. doi: 10.1016/j.molmed.2005.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aigner A. Applications of RNA interference: current state and prospects for siRNA-based strategies in vivo. Appl. Microbiol. Biotechnol. 2007;76:9–21. doi: 10.1007/s00253-007-0984-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dahlgren C, Wahlestedt C, Thonberg H. No induction of anti-viral responses in human cell lines HeLa and MCF-7 when transfecting with siRNA or siLNA. Biochem. Biophys. Res. Commun. 2006;341:1211–1217. doi: 10.1016/j.bbrc.2006.01.085. [DOI] [PubMed] [Google Scholar]
- 6.Kim D-H, Longo M, Han Y, Lundberg P, Cantin E, Rossi JJ. Interferon induction by siRNAs and ssRNAs synthesized by phage polymerase. Nat. Biotechnol. 2004;22:321–325. doi: 10.1038/nbt940. [DOI] [PubMed] [Google Scholar]
- 7.Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, Li B, Cavet G, Linsley PS. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- 8.Jackson AL, Burchard J, Schelter J, Chau BN, Cleary M, Lim L, Linsley PS. Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity. RNA. 2006;12:1179–1187. doi: 10.1261/rna.25706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Persengiev SP, Zhu X, Green M. Nonspecific, concentration-dependent stimulation and repression of mammalian gene expression by small interfering RNAs (siRNAs) RNA. 2004;10:12–18. doi: 10.1261/rna5160904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amarzguioui M, Holen T, Babaie E, Prydz H. Tolerance for mutations and chemical modifications in a siRNA. Nucleic Acids Res. 2003;31:589–595. doi: 10.1093/nar/gkg147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwarz DS, Hutvágner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 12.Reynolds A, Leake D, Boese Q, Scaringe S, Marshall W, Khvorova A. Rational siRNA design for RNA interference. Nat. Biotechnol. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- 13.Du Q, Thonberg H, Wang J, Wahlestedt C, Liang Z. A systematic analysis of the silencing effects of an active siRNA at all single-nucleotide mismatched target sites. Nucleic Acids Res. 2005;33:1671–1677. doi: 10.1093/nar/gki312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Du Q, Thonberg H, Zhang H.-YZ, Wahlestedt C, Liang Z. Validating siRNA using a reporter made from synthetic DNA oligonucleotides. Biochem. Biophys. Res. Commun. 2004;325:243–249. doi: 10.1016/j.bbrc.2004.09.222. [DOI] [PubMed] [Google Scholar]
- 15.Elbashir SM, Matrinez J, Patkaniowska A, Lendeckel W, Tuschl T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 2001;20:6877–6888. doi: 10.1093/emboj/20.23.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwarz DS, Ding H, Kennington L, Moore JT, Schelter J, Burchard J, Linsley PS, Aronin N, Xu Z, Zamore PD. Designing siRNA that distinguish between genes that differ by a single nucleotide. PLoS Genet. 2006;2:1308–1318. doi: 10.1371/journal.pgen.0020140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Birmingham A, Anderson EM, Reynolds A, Ilsley-Tyree D, Leake D, Fedorov Y, Baskerville S, Maksimova E, Robinson K, Karpilow J, et al. 3′ UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat. Methods. 2006;3:199–204. doi: 10.1038/nmeth854. [DOI] [PubMed] [Google Scholar]
- 18.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 19.Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W. Single processing center models for human Dicer and bacterial RNase III. Cell. 2004;118:56–68. doi: 10.1016/j.cell.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 20.Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin SE, Caplen NJ. Mismatched siRNAs downregulate mRNAs as a function of target site location. FEBS Lett. 2006;580:3694–3698. doi: 10.1016/j.febslet.2006.05.056. [DOI] [PubMed] [Google Scholar]
- 22.Naito Y, Yamada T, Matsumiya T, Ui-Tei K, Saigo K, Morishita S. dsCheck: highly sensitive off-target search software for double-stranded RNA-mediated RNA interference. Nucleic Acids Res. 2005;33:W589–W591. doi: 10.1093/nar/gki419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qiu S, Adema CM, Lane T. A computational study of off-target effects of RNA interference. Nucleic Acids Res. 2005;33:1834–1847. doi: 10.1093/nar/gki324. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.