Abstract
Ionizing radiation induces various clustered DNA lesions, including double-strand breaks (DSBs) accompanied by nearby oxidative base damage. Previous work showed that, in HeLa nuclear extracts, DSBs with partially complementary 3′ overhangs and a one-base gap in each strand are accurately rejoined, with the gaps being filled by DNA polymerase λ. To determine the possible effect of oxidative base damage on this process, plasmid substrates were constructed containing overhangs with 8-oxoguanine or thymine glycol in base-pairing positions of 3-base (-ACG or -GTA) 3′ overhangs. In this context, 8-oxoguanine was well tolerated by the end-joining machinery when present at one end of the break, but not when present at both ends. Thymine glycol was less well tolerated than 8-oxoguanine, reducing gap filling and accurate rejoining by at least 10-fold. The results suggest that complex DSBs can be accurately rejoined despite the presence of accompanying base damage, but that nonplanar bases constitute a major barrier to this process and promote error-prone joining. A chimeric DNA polymerase, in which the catalytic domain of polymerase λ was replaced with that of polymerase β, could not substitute for polymerase λ in these assays, suggesting that this domain is specifically adapted for gap filling on aligned DSB ends.
INTRODUCTION
Radiation-induced DSBs are typically formed by fragmentation of deoxyribose (1), and thus they often bear partially complementary overhangs, the annealing of which will produce a structure with staggered one-base gaps in opposite strands. Numerous in vivo and in vitro data suggest that the major nonhomologous end-joining pathway, whose core components are DNA-PK and the XRCC4/DNA ligase IV (X4L4) complex, can accurately restore the original sequence at such breaks, by a mechanism involving annealing of the residual complementarities in the overhangs, replacement of the fragmented nucleotides by gap filling on the aligned ends and ligation (2–8). The gap filling step is carried out primarily by DNA polymerase lambda (polλ) and to a lesser extent polymerase μ (polμ), both of which form complexes with DNA-PK and X4L4 on DNA ends, as detected by electrophoretic mobility shift assays (9–12). Gap filling on aligned ends of partially complementary 3′ overhangs also requires the core end-joining proteins Ku, XRCC4 and DNA ligase IV, while resection-based end-joining not involving gap filling can occur in the absence of Ku, at least in extracts (6,13,14).
Although few direct measurements have been performed, track structure calculations predict that a significant fraction of radiation-induced DSBs will be accompanied by nearby oxidative base damage (15). If such damage occurs in base-pairing positions of aligned overhangs, gap filling may be significantly compromised, potentially reducing both the efficiency and the accuracy of repair. However, the demonstrated ability of polλ to catalyze polymerization on substrates with mismatched bases near the 3′ end of the primer (16) raises the possibility that it might tolerate damaged bases in annealed overhangs of DSBs as well. To test this hypothesis, end-joining of DSB substrates containing oxidatively damaged bases in base-pairing positions of partially complementary overhangs was examined. The results indicate a tolerance for some but not all damaged bases in this context by both polλ and polμ.
MATERIALS AND METHODS
Substrates
An oligomer with sequence 5′-CGAGGAACGCGACG and containing 8-oxoguanine at the 3′-terminal G was obtained from Midland Certified Reagents (Midland, TX, USA). It was HPLC-purified and subjected to electrospray mass spectrometry, which yielded the expected molecular mass of 4338. It was 5′-phosphorylated with T4 polynucleotide kinase and [γ-32P]ATP, and then purified by polyacrylamide gel electrophoresis followed by reverse-phase HPLC, as described (17).
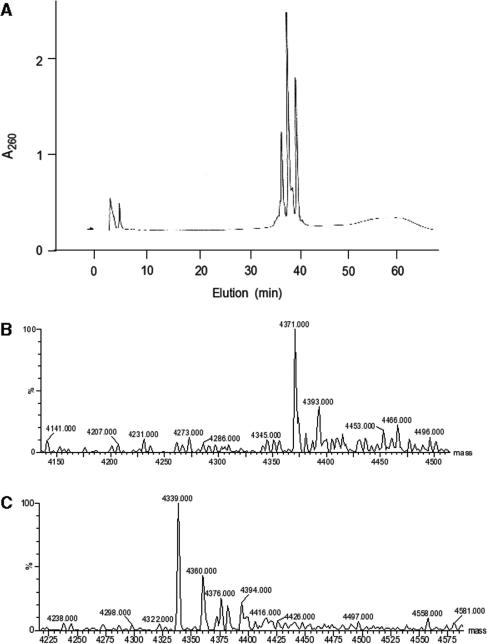
To oxidize the single T residue in the oligomer 5′-CGAGGAACGCGGTA to thymine glycol, 40 nmol of oligomer were incubated at 22°C for 40 h in an unbuffered 2% aqueous solution of OsO4 (Sigma, St. Louis, MO). Reaction mixtures were extracted three times with chloroform to remove excess OsO4 and then evaporated to dryness and dissolved in 250 μl of water (18). Ten microliters of 0.3 mM NaHSO4 were added and the mixture was stirred at room temperature for 16 h. The samples were evaporated, dissolved in 0.1 M triethylammonium acetate pH 7.0 and subjected to reverse-phase HPLC with elution at 1 ml/min for 10 min with 5% acetonitrile, followed by a 30-min gradient of 5–27.5% acetonitrile. Three major products were recovered, and an aliquot of each was end-labeled and analyzed on a sequencing gel. The earliest eluting peak from HPLC showed slightly slower electrophoretic mobility than the corresponding unmodified oligomer, and mass spectrometry yielded a deconvoluted Mr of 4371, consistent with a single thymine glycol substitution. By analogy to previous work (18), this species likely contained the (5S,6R) stereoisomer of thymine glycol. The second peak consisted exclusively of unmodified oligomer (Mr = 4339) and the third peak was predominantly thymine glycol-containing oligomer, presumably the (5R,6S) isomer. However, by mass spectrometry, the third peak contained some unmodified oligomer as well. Therefore, the earliest eluting oligomer was used for preparation of DSB substrates. It was 5′-32P-end-labeled and again purified by gel electrophoresis and HPLC.
For construction of internally labeled DSB substrates, the plasmid pRZ56 (19) was cut with MluI and subjected to controlled 3′→5′ resection with T4 polymerase to generate 10- and 11-base 5′ overhangs. An unmodified unlabeled 13-mer, and a 5′-32P-labeled modified or unmodified 14-mer, were sequentially ligated into these overhangs to yield site-specifically labeled substrates with partially complementary 3-base 3′ overhangs (-ACG/-ACG, -GTA/-GTA or -ACA/-ATG; see Figure 2A), as described previously (6,17). A ‘mismatched’ substrate bearing overhangs with only a single potential base pair (-ATG/-ACG; see Figure 3A) was similarly constructed. Because of apparent instability of the 8-oxoguanine-containing oligomer, it was stored at −80°C and was ligated onto resected plasmid for 5 min at 25°C using a Quick-Ligase kit (New England Biolabs, Beverly, MA). Control experiments showed that both unmodified and modified oligomers were ligated onto at least 90% of the overhangs (data not shown). Each labeled plasmid was purified by agarose gel electrophoresis, electroeluted, concentrated (Amicon Centricon 100, Millipore, Bedford MA) and precipitated.
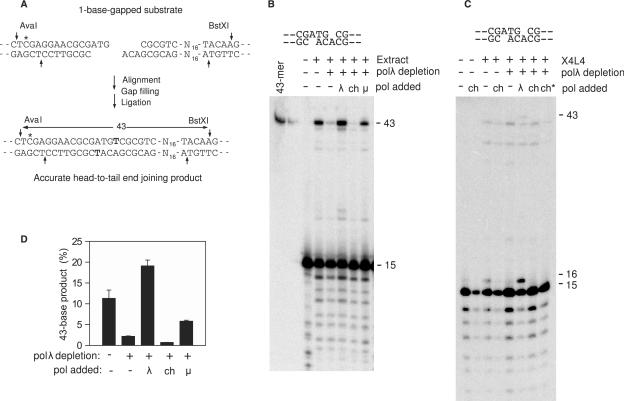
Figure 2.
Gap filling and accurate end joining of a DSB substrate bearing partially complementary overhangs. (A) Schematic of the site-specifically labeled (asterisk) DSB substrate showing accurate end joining by gap filling and ligation, resulting in a 43-base BstXI/AvaI fragment. All sequences read 5′→3′ in the top strand and 3′→5′ in the bottom strand. Bolded T indicates the filled-in base. (B) The substrate shown, bearing -ACA and -ATG 3′ overhangs, was incubated for 6 h with X4L4-supplemented HeLa extracts that had been immunodepleted of polλ or mock depleted. Polλ, polμ or pol-ch (70 ng) was also added as indicated. After incubation, samples were cut with BstXI and AvaI, and analyzed on denaturing gels. (C) Same as (B), except the reaction was performed in the presence of ddTTP instead of dTTP, and only some samples contained X4L4, as indicated. ‘ch*’ indicates 280 ng of pol-ch was added. (D) Quantitation of data from (B) and from a replicate experiment; error bars show the range of values obtained in the two experiments.
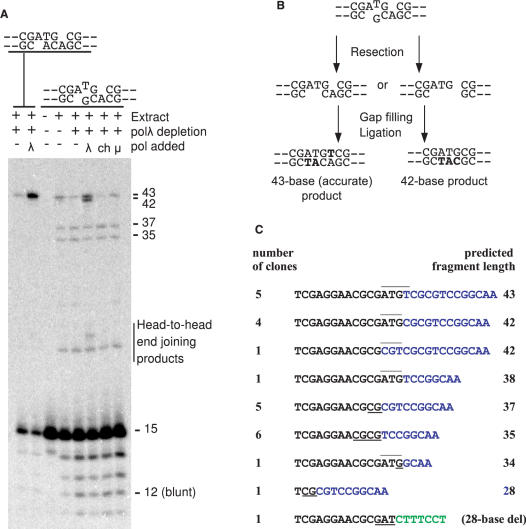
Figure 3.
Effect of a single-base mismatch on end joining. (A) The 3-base 3′ overhang substrates shown were incubated in polλ-depleted extracts supplemented with various polymerases as in Figure 2, cut with BstXI and AvaI and analyzed on sequencing gels. (B) Proposed mechanisms of formation of 43- and 42-base products from the mismatched substrate. (C) Sequencing of repair joints in recircularized products. Sequences are expressed in terms of the radiolabeled (top) strand. Black and blue letters indicate sequences originating from the left and right ends of the break, respectively. Underlined nucleotides could have originated from either end and may have resulted from microhomology annealing and splicing. Overlines indicate apparent preservation of 3′ overhangs. The green letters in the last sequence begin 28 bases from the 5′ terminus of the right end. The first two sequences correspond to the two pathways shown in (B).
Mass spectrometry
Electrospray mass spectra were obtained on a Waters/Micromass Qtof-2 (Manchester, UK) instrument equipped with a custom-built nano-spray source operated in negative ion mode. Samples at a concentration of 5–10 μM in a 50:50 mixture of methanol/water containing 25 mM ammonium acetate were introduced into the inlet at 1.0 μl/min with a capillary voltage of −1.9 kV and a cone voltage of 36 V. Source temperature was maintained at 110°C. Full-scan mass spectra were acquired over a mass range of 100–2000 m/z. Data were collected and processed using the Mass Lynx 4.0 software. Deconvolution to molecular mass scale was performed using the Max Ent software supplied with the instrument.
Proteins
To generate a chimeric DNA polymerase, sequences encoding the BRCT and serine/proline-rich domains of polλ (residues 1–253), and most of polβ (residues 12–335), were amplified through PCR with overlapping oligonucleotides. A full-length λ/β product was generated through linked PCR and cloned in the NdeI/NcoI sites of pET22(+). The resulting chimera was overexpressed in E. coli and purified as described earlier (20). Normal full-length polλ and polμ were similarly prepared.
End joining and gap filling in nuclear extracts
Reaction mixtures contained HeLa nuclear extract (27 μg protein, in vitro transcription grade, Promega, Madison, WI), 0.1 μg of XRCC4/LigIV (Trevigen, Gaithersburg, MD), 50 mM triethanolamine–NaOH, 10 mM Tris–HCl pH 7.9, 1 mM Mg(OAc)2, 40 mM KOAc, 0.5 mM dithiothreitol, 1 mM ATP, 50 μM of each dNTP or dideoxynucleoside 5′-triphosphate (ddNTP), 50 μg/ml BSA and 16 ng DNA substrate in a total volume of 16 μl. Following substrate addition, samples were incubated for 6 h at 37°C (2 h for 8-oxoguanine-containing substrates and the corresponding controls), and then deproteinized by treatment with proteinase K followed by phenol/chloroform extraction. DNA was digested with BstXI and AvaI, and then precipitated (6). To minimize degradation of 8-oxoguanine, the proteinase K digestion was shortened to 1 h at 65°C, and the restriction enzyme digestions to 2 h at 37°C. Following digestion, some samples were dissolved in 100 μl 30% NH4OH, bubbled with O2, and heated at 60°C for 16 h to cleave 8-oxoguanine residues (21), and then evaporated to 10 μl. All samples were then denatured in formamide and analyzed on 20% denaturing polyacrylamide gels (22). Intensities of bands corresponding to repair products and intermediates were quantitated by phosphorimaging, and are expressed as a fraction of the total phosphorimage intensity in the lane, excluding any material that remained in the well. For sequencing, deprotenized end-joining samples (1–2 μl) were transfected into DH5α competent cells (Invitrogen). Clones were sequenced by the fluorescent dideoxy method at the Massey Cancer Center Nucleic Acids Shared Resource, using an ABI sequencer (GMI, Ramsey, MN) and the primer GCAAACAATGGCCTGAGTGTG. Assays for polymerase activity were performed at 37°C in 10 μl of end-joining buffer containing 30 fmol of the 5′-end-labeled 15-mer *pGATCACAGTGAGTAC annealed to a 1.5-fold excess of the 33-mer ACTGGCCGTCGTTCTAATGTACTCACTGTGATC.
Immunodepletion
Generation of polyclonal antibodies to polλ has been described (10). Antiserum (10 μl) or affinity-purified antibodies (2.7 μg) were bound to 5 μl (bead volume) of Protein A agarose beads (Sigma) by mixing at 900 r.p.m. in antibody-binding buffer (0.1 M KOAc, 20% glycerol, 0.2 mM EDTA, 0.5 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, 50 μg/ml BSA) with total volume of 250 μl at 4°C for 16 h in an Eppendorf Thermomixer. Antibody-bound beads were washed three times with 400 μl of antibody-washing buffer (10 mM Tris–HCl pH 7.9, 65 mM KOAc, 0.25 mM EDTA, 0.5 mM dithiothreitol, 1 mM Mg(OAc)2) in order to remove unbound antibodies. HeLa nuclear extract (15 μl) was directly added to the beads and mixed at 900 r.p.m. for 1.5 h at 4°C. Beads were briefly pelleted in a microfuge, 10 μl of supernatant was removed and 5 μl was used for each reaction. Samples were incubated and processed as described earlier. For reconstitution, 70 ng of recombinant polλ or polμ was added to each reaction mixture.
Electrophoretic mobility shift
Purified recombinant Ku (Trevigen), X4L4 (Trevigen), and/or DNA polymerase were allowed to bind to a 5′-32P-labeled 60-base duplex for 30 min at 4°C in 10 μl of 25 mM Tris–HCl pH 8, 100 mM KCl, 0.1 mM EDTA, 2 mM dithiothreitol, 0.05% Triton X-100, 50 μg/ml BSA (11). Complexes were resolved on a 3.5% nondenaturing polyacrylamide gel at 12 V/cm at 4°C, and detected by phosphorimaging.
RESULTS
A polλ/polβ chimera does not support gap filling on aligned DSB ends
The catalytic domain of polλ possesses significant differences from that of polβ (9,23,24), suggesting that polλ structure may be specifically adapted for gap filling on aligned DSB ends. Previous work has shown that polβ cannot substitute for polλ in gap filling on aligned DSB ends (10). However, inasmuch as the BRCT domain of polλ is essential for its recruitment to DSB ends by DNA-PK and X4L4 (12), and polβ lacks a BRCT domain, the failure of polβ to support gap filling may reflect simply a lack of recruitment to the aligned DSB ends.
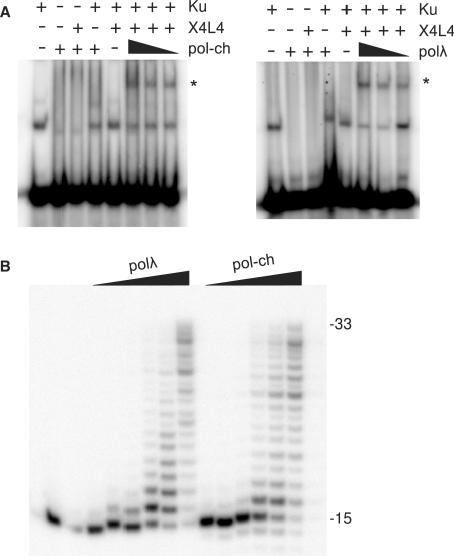
To address this question, a chimeric polymerase was prepared, in which the catalytic domain of polλ was replaced with that of polβ. In an electrophoretic mobility shift assay, the chimeric polymerase formed an apparent complex with Ku and X4L4 on a short DNA duplex, with an efficiency similar to that of normal polλ (Figure 1A). Unexpectedly, normal full-length polβ lacking a BRCT domain did form a trace of a similar complex, but about 10-fold less efficiently than the chimera (Supplementary Figure S1). Using a labeled 15-mer primer annealed to a 33-mer template as a substrate, the ability of the chimera to extend a recessed 3′ end was equal to that of normal recombinant polλ (Figure 1B). Thus, the chimeric polymerase has the same degree of intrinsic polymerase activity as polλ, and is effectively recruited to the end-joining repair complex.
Figure 1.
Catalytic activity of a chimeric DNA polymerase and interaction with core end joining proteins. (A) A 5′-32P-end-labeled 60-base duplex (90 nM) was incubated with Ku (5 nM), X4L4 (25 nM) and 25, 50 or 100 polλ or a chimeric DNA polymerase (pol-ch) wherein the catalytic core of polλ was replaced with that of polβ (‘+’ = 100 nM). Complexes were resolved on a nondenaturing gel. The uppermost band (asterisk) is formed only when all three proteins are present, and presumably represents a stable complex of the three on DNA (11). This complex forms equally well with either polλ or pol-ch. (B) A partial duplex consisting of a 15-base primer annealed to a 33-base template (3 nM) was treated with 3.5, 7, 14, 35, 70 or 140 nM polλ or pol-ch. Following incubation for 1 h at 37°C, samples were analyzed on a sequencing gel.
However, whereas addition of polλ to polλ-depleted X4L4-supplemented nuclear extracts dramatically stimulated accurate repair of a DSB substrate with partially complementary (–ATG and –ACA) 3′ overhangs, the chimeric polymerase did not (Figure 2B). Indeed, addition of the chimera to polλ-depleted extract decreased the extent of accurate end joining below that seen with polλ-depleted extract alone (Figure 2D), suggesting that its binding to the repair complex may have displaced other polymerases (polμ or residual polλ) capable of gap filling. When the final ligation step was blocked by replacement of dTTP with ddTTP in the reaction, polλ efficiently catalyzed single-base extension of the aligned end, whereas the chimeric polymerase did not (Figure 2C).
Thus, even though it was effectively recruited to the repair complex, the chimeric polymerase did not support gap filling on aligned DSB ends. These results are consistent with the view that the catalytic domain of polλ is specifically adapted for this function.
A single-base mismatch substantially impairs joining of a gapped substrate
Previous work with Xenopus egg extracts, as well as with purified enzymes, has suggested that gaps in aligned DSB ends can be filled and the ends ligated despite base mismatches in annealed overhangs (12,25). As a prelude to studies with oxidatively modified bases, end joining of a DSB substrate bearing 3-base 3′ overhangs with one paired and one unpaired base (–ATG and –ACG) was examined in the same immunodepleted and supplemented extracts, and compared to overhangs with a two-base complementarity (–ACA/–ATG) (Figure 3A). Analysis of ligated products indicated that the mismatched base was a significant barrier, reducing overall end joining in the polλ-supplemented extract 4-fold (from 45% to 11%, total of 35- to 43-base products), and reducing the yield of the 43-base product 10-fold (from 45% to 4.7%). Nevertheless, there was detectable 43-base product formed even in the unsupplemented extract. Sequencing of clones recovered from a replicate experiment indicated the presence of –CGATGTCG- but not –CGACGTCG- joints (Figure 3C), consistent with a model wherein the terminal mismatch was not extended, but rather the terminal G in the bottom strand was removed and replaced with A at some point in the repair process (Figure 3B). Sequencing also confirmed the presence of -CGATGTCG- joints derived from the matched (–ATG and –ACA) substrate (3/16 sequences, Supplementary Table S1), as well as the presence of -CGATGCG- joints derived from the mismatched substrate (Figure 3C), consistent with complete removal of the bottom strand overhang, and then alignment-based gap filling using the overhang in the top strand as template (Figure 3B). This mechanism would yield a 42-mer, and presumably accounts for the product migrating just below the 43-mer band. No 42-mer product was formed when polμ was added to immunodepleted extract, consistent with a previous finding that polμ is very inefficient in filling in even a two-base gap on aligned DSB ends (10). The mismatched substrate also yielded substantial polymerase-independent 35- and 37-base products (Figure 3A). Sequencing (Figure 3C) suggests that these products correspond to annealing of and splicing at the 5′-terminal CG- and CGCG- sequences, respectively, presumably preceded by 3′ resection, as depicted in Figure 6A.
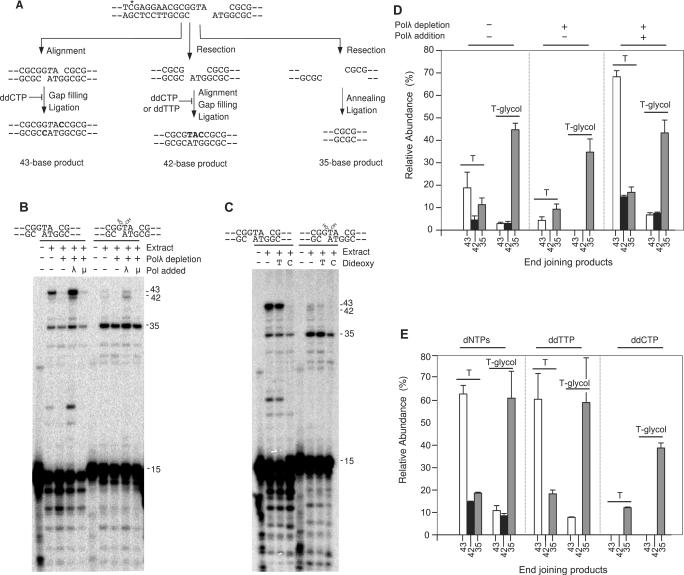
Figure 6.
End joining of a substrate containing a thymine glycol base. (A) Schematic of the internally labeled (asterisk) substrate, showing proposed mechanisms of formation of products giving 43-, 42- and 35-base BstXI/AvaI fragments. The italicized T is the site of thymine glycol substitution. Bolded nucleotides indicate gap filling. The 35-base product is shown as arising by 3′→5′ resection, but it could in principle be generated by 5′→3′ resection as well. (B) Unmodified or thymine glycol-substituted substrate was incubated for 6 h with X4L4-supplemented HeLa nuclear extracts that had been immunodepleted of polλ or mock depleted. Polλ or polμ (70 ng) was added as indicated. (C) Same as (B), except extracts were not immunodepleted, polλ was added to all samples and samples contained either ddCTP or ddTTP in place of normal dNTPs as indicated. (D) Quantitative data from (B) and similar experiments; the abundance of each product is normalized to the total of all head-to-tail end-joining products in the sample with the unmodified substrate (‘T’) and added polλ; ‘T-glycol’ = thymine glycol-substituted substrate. Error bars show mean and SE of 3–4 experiments. (E) Similar quantitation of data from (C) and a replicate experiment. Error bars show the range of values obtained in the two experiments.
8-oxoguanine at one end of a DSB does not prevent accurate joining of a gapped substrate
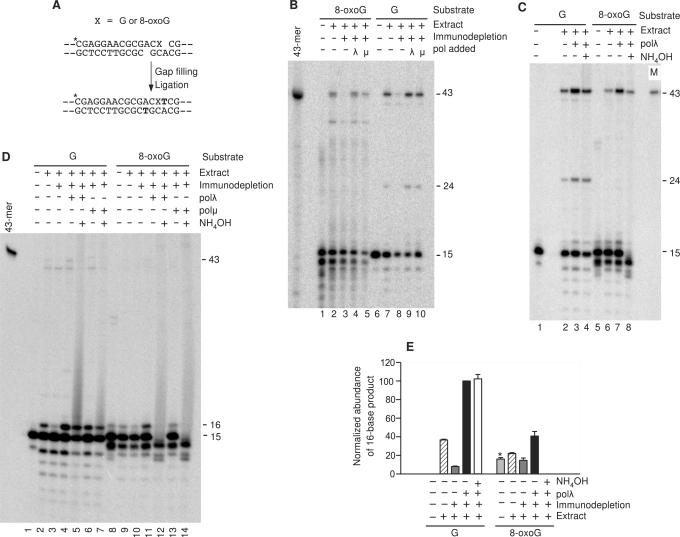
8-oxoguanine is a common and highly mutagenic oxidized DNA base that can pair with either cytosine or, by flipping into a syn conformation, with adenine. This mispairing results in misincorporation, the extent of which varies greatly among various DNA polymerases (26). To determine the possible effect of 8-oxoguanine on DSB alignment and gap filling, a substrate was prepared bearing 3-base -ACG overhangs on each end, with 8-oxoguanine replacing the terminal G at the labeled end of the break (Figure 4A).
Figure 4.
End joining of a substrate containing an 8-oxoguanine base. (A) Schematic of the substrate and mechanism of formation of the accurate end-joining product (X = guanine or 8-oxoguanine). Bolded T indicates the filled-in base. (B) The substrates were incubated in polλ-depleted or mock-depleted nuclear extracts supplemented with polλ or polμ as indicated, cut with BstXI and AvaI, and analyzed on denaturing gels. The 43-base product corresponds to gap filling and accurate head-to-tail joining of the plasmid, as shown in (A). The 24-base product (formed only with the unmodified substrate) corresponds to accurate head-to-head joining of two plasmid molecules. The band migrating as a ∼14-mer is an apparent spontaneous degradation product of 8-oxoguanine (see text). (C) Same as (B), except that extracts were not immunodepleted, and some samples were treated with NH4OH to cleave 8-oxoguanine residues. The lane marked ‘M’ contains a labeled 43-base marker of the expected sequence. (D) Same as (C), except that the end-joining buffer contained ddTTP instead of dTTP, in order to trap the filled-in but unligated intermediate (16-mer band), and some extracts were immunodepleted as indicated. (E) Quantitative gap-filling data from (D) and a replicate experiment. The abundance of the 16-mer band was calculated for each sample and normalized to the value obtained for the unmodified substrate in the polλ-supplemented extract. The value shown for the 8-oxoguanine substrate (asterisk) includes an apparent degradation product of the initial substrate that comigrates with the 16-mer.
Incubation of this substrate in extracts supplemented with X4L4 yielded predominantly a 43-base end-joining product, suggesting that alignment-based gap filling on the annealed ends resulted in accurate repair of the break (Figure 4B, lane 2 and Figure 4C, lane 6). Previous sequencing of repaired plasmid clones derived from repair of a similar vector (6) confirmed the predominance of the predicted accurately repaired product shown in Figure 4A. Formation of this product was dramatically reduced by immunodepletion of polλ from the extract (Figure 4B, lane 3), but was stimulated by addition of recombinant polλ to either immunodepleted or undepleted extract (Figure 4B, lane 4 and Figure 4C, lane 7). With polλ added, the extent of accurate repair was about half that seen with the corresponding unmodified substrates. Polμ also supported by gap filling and accurate end joining of the 8-oxoguanine-containing substrate (Figure 4B, lane 5). For both polymerases, a 24-base product was only seen with the unmodified and not with the 8-oxoguanine-containing substrate. Previous work showed that generation of this fragment did not require BstXI cleavage, suggesting that it corresponds to accurate head-to-head joining of two substrate molecules (6). Moreover, it migrates at precisely the expected mobility (19), it is only generated from substrates with self-complementary overhangs (compare Figure 2B with Figure 4C), and sequencing indicates that there are no major head-to-tail recircularization products consistent with this fragment size. Thus, the absence of this product with the modified substrate suggests that when 8-oxoguanine is present on overhangs at both ends of a DSB, accurate end joining is precluded, presumably because the two cytosine•8-oxoguanine base pairs cannot be properly aligned and annealed with sufficient stability.
Although generation of the accurate 43-base product is consistent with direct gap filling and ligation of the 8-oxoguanine-containing substrate, the same product could arise from exonucleolytic removal of the terminal 8-oxoguanine base (or its degradation product), and annealing of the remaining single G•C base pair, followed by two-base gap filling and ligation. Therefore, to determine whether 8-oxoguanine was present in the 43-base end-joined product, the BstXI/AvaI-cut DNA samples were treated with hot ammonia under 100% oxygen, which efficiently cleaves DNA at 8-oxoguanine sites (21). This treatment resulted in complete loss of the 15-mer band corresponding to unprocessed substrate (Figure 4C, lane 8), thus confirming the efficiency of ammonia-induced 8-oxoguanine cleavage. The principal degradation product, migrating between the 14- and 15-mer bands (and comigrating with an apparent spontaneous degradation product seen in all samples), is presumably a 14-mer with a 3′-terminal deoxyribose, generated by alkaline hydrolysis of 8-oxoguanine base and cleavage of the 5′ phosphodiester bond (21). Ammonia treatment also eliminated most (58 ± 2%, N = 4) of the 43-base product derived from the 8-oxoguanine substrate (lane 8), but had little effect on that derived from the unmodified substrate (<10% reduction in each of four independent experiments) (lane 4). This result strongly suggests that a significant fraction of the accurately repaired products still contained 8-oxoguanine, implying that gap filling can occur despite the presence of 8-oxoguanine in the overhang. The proportion of accurately repaired product that contained 8-oxoguanine may have actually been higher than was measured, as some minor fraction of the modified 43-mer probably degraded spontaneously during experimental manipulations.
When dTTP in the reaction buffer was replaced with ddTTP, end joining was eliminated as expected but a prominent, one-base-elongated, unligated intermediate was generated from both the modified and the unmodified substrates (16-mer in Figure 4D). For the modified substrate, this putative intermediate comigrated with a minor degradation product that was seen even in the absence of extract (Figure 4D, lane 8); nevertheless, the intensity of this band was clearly greater when extract and polλ were added (lane 11; quantitation in Figure 4E). The abundance of intermediate from the unmodified substrate was unaffected by ammonia treatment, as expected (Figure 4E). However, for the 8-oxoguanine-containing substrate, hot ammonia completely eliminated the putative intermediate (Figure 4D, lane 12), implying that it still contained 8-oxoguanine. This result provides further confirmation that alignment-based gap filling occurred without prior 8-oxoguanine removal. Moreover, this gap filling must have occurred without ligation of the complementary strand, as ddTTP incorporation would preclude such ligation.
Thymine glycol inhibits accurate repair of a gapped DSB substrate
Thymine glycol is a frequent product of oxidative DNA base damage, and it constitutes a major block to replicative DNA polymerases due to its nonplanar structure (27). To assess the effect of thymine glycol on polλ-mediated gap filling on aligned DSB ends, an oligomer with a single thymine base was treated with OsO4, resulting in conversion of thymine to thymine glycol. Separation by HPLC revealed three major products (Figure 5A). The earliest eluting peak was subjected to mass spectrometry (Figure 5B), which confirmed the presence of thymine glycol [presumably the (5S,6R) isomer (18)]. This oligomer was used to construct a DSB substrate bearing partially complementary -GTA overhangs at both ends, but with thymine glycol replacing normal thymine in the overhang of the labeled end (Figure 6A). This substrate, as well as an analogous unmodified substrate, was incubated in X4L4-supplemented HeLa nuclear extracts, with or without added purified polλ.
Figure 5.
Generation of a thymine glycol-containing oligomer. (A) HPLC absorbance (260 nm) trace showing elution of products of oxidation of the oligomer CGAGGAACGCGGTA by OsO4. (B) Deconvoluted electrospray mass spectrum of the earliest-eluting peak (36 min), consistent with oxidation of the single T in the sequence to thymine glycol. (C) Mass spectrum of the second-eluting peak (37.5 min), apparently containing unmodified oligomer.
As expected, a substantial fraction of the substrate with unmodified -GTA overhangs was rejoined to yield predominantly a 43-base product (Figure 6B), consistent with gap filling on annealed overhangs prior to ligation (Figure 6A). More than 90% of this product could be cleaved by KpnI, yielding the expected 16-mer and thus verifying the predicted GGTACC sequence of the repair joint (Supplementary Figure S2). Just as with the −ATG and −ACA overhangs described above, formation of the 43-base product was diminished by immunodepletion of polλ, and enhanced by reconstitution with purified polλ (Figure 6B and D). In polλ-supplemented extracts, presence of thymine glycol in the overhang of the labeled strand resulted in a 10-fold decrease in the yield of 43-base product, and an increase in resected products, particularly a 35-base product consistent with annealing of the self-complementary CGCG sequences adjacent to the overhangs at both ends of the break (see Figure 3C). Nevertheless, a detectable level of the 43-base product was consistently formed, and only in the extract supplemented with polλ. In principle, this product could only have been formed by annealing of a two-base complementarity that includes an adenine•(thymine glycol) base pair, and one-base extension by polλ prior to ligation (Figure 6A).
To verify that the 43-base product corresponds to fill-in of a single C residue in the annealed overhangs, the gap filling reaction was carried out in the presence of either ddCTP or ddTTP. As predicted, ddTTP had little effect on formation of the 43-base product generated either from the normal or the thymine glycol-containing substrate, while ddCTP eliminated formation of this product completely (Figure 6C and E). The finding that ddTTP did not block formation of this product excludes the possibility that it was formed by exonucleolytic removal of the thymine glycol nucleotide, followed by its replacement with dTTP using the overhang in the opposite strand as template. Presence of ddTTP did, however, eliminate the 42-base product, consistent with the view that it was formed by resection of the thymine glycol-containing overhang to a blunt end followed by resynthesis of a 3-base -TAC 3′ overhang complementary to the overhang in the opposite strand (Figure 6A). As expected, ddCTP eliminated formation of this 42-base product as well. For the substrate with unmodified -GTA overhangs, polμ promoted formation of the 43-base product, although much less efficiently than polλ. However, unlike polλ, polμ did not promote detectable 43-base product formation from the thymine glycol-substituted substrate. The strikingly lower efficiency of polμ in promoting gap filling on the substrate with unmodified -GTA overhangs, as compared to the substrate with -ACG overhangs (Figure 4B), may be due to the intrinsically lower stability of A•T base pairs.
Because the thymine glycol-containing oligomer was generated by OsO4 treatment of an unmodified oligomer, a question arises as to whether the residual 43-base product might be an artifact resulting from a contamination of the glycol-containing substrate with normal substrate. This possibility appears unlikely, as there is no detectable 4339-Da peak in the mass spectrum of the purified, modified oligomer, and certainly not the 10% contamination that would be required to explain the residual accurate product (Figure 5B). Moreover, the two oligomers are relatively well resolved by both gel electrophoresis and HPLC, and before use the purified modified oligomer was subjected to a second round of purification by gel electrophoresis and HPLC after radiolabeling. However, while contamination with unmodified oligomer is unlikely, the possibility of contamination with some other form of oxidized oligomer that copurifies more closely with the thymine glycol oligomer, cannot be definitively excluded. Thus, strictly speaking, the residual gap filling and accurate end joining seen with the thymine glycol oligomer must be regarded as an upper limit, and it is formally possible that thymine glycol blocks these processes completely.
DISCUSSION
Although nonhomologous end joining is error prone, diverse lines of evidence suggest that this pathway is capable of aligning, annealing and patching partially complementary overhangs (2–8). In principle, such a process could restore the original DNA sequence at sites of staggered DSBs, including free radical-mediated DSBs formed by nucleotide fragmentation. Moreover, gap filling and ligation can occur even when the annealed overhangs include a base mismatch (8,25), suggesting a remarkable tolerance for structural perturbations. Subsequent work has confirmed that polλ and DNA ligase IV, the prime candidate enzymes for carrying out nonhomologous end joining, are both highly tolerant of substrates containing mismatched or misaligned bases (12,16,23).
This demonstrated tolerance for perturbed DNA structures raises the possibility that the end-joining machinery might have similar tolerance for modified bases, and thus might be capable of accurately rejoining DSBs despite accompanying base damage. To assess this possibility, end joining was examined in human nuclear extracts, a system that exhibits dependence on most of the known nonhomologous end-joining proteins, including Ku, DNA-PK and X4L4 (6,13,28). In this system, gap filling on aligned DNA ends is largely dependent on polλ (10), reflecting a dependence recently demonstrated in vivo (8). Thus, it is likely that this system recapitulates most biochemical features of in vivo end joining.
In this extract-based system, 8-oxoguanine was relatively well tolerated in base pairing positions of aligned overhangs, promoting polλ-mediated gap filling and ligation at about half the efficiency of unmodified guanine when present at one end of a DSB. Although it is possible that gap filling and ligation occurred in the nonsubstituted strand prior to gap filling on the substituted strand, the initial gap filling still must have occurred on an end-aligned structure involving a base-paired 8-oxoguanine. Recent kinetic studies have indicated that, in the absence of other proteins, 8-oxoguanine at the 3′ terminus of a template-primer was a more favorable substrate for extension by polλ than normal guanine, also suggesting a remarkable tolerance for this oxidized base (29). Polμ also extended the 8-oxoguanine template at least as efficiently as the normal template (29). These results raise the possibility that these polymerases may have evolved a specific tolerance for 8-oxoguanine due to their participation in repair of radiation-induced complex DSBs.
As might be expected, thymine glycol, the most common pyrimidine oxidation product, was much less well tolerated in a base-pairing position than 8-oxoguanine, reducing the efficiency of gap filling on aligned ends by at least a factor of 10. Although it is capable of normal Watson–Crick hydrogen bonding, thymine glycol is nonplanar, and will necessarily disrupt normal DNA structure much more than 8-oxoguanine. In may be noted, however, that the position of thymine glycol in this substrate (penultimate base in the overhang) was not precisely equivalent to that of 8-oxoguanine (terminal base); this difference as well as the presumed difference in stability of the adjacent unmodified base pair (A•T versus G•C) may have contributed to the different gap filling efficiencies for the two substrates. The apparent decrease in gap filling resulting from thymine glycol (Figure 6) was comparable to that for a mismatched base (Figure 3). However, unlike a mismatched base, thymine glycol appeared to actively promote resection-based end joining at a microhomology near the two DNA ends (Figure 6).
The structural basis of alignment-based gap filling during nonhomologous end joining for the most part remains to be determined, but it presumably reflects a combination of a multiprotein scaffold that holds the two ends in proximity and a polymerase that can accept a minimally base-paired template-primer. It is presently unclear what structural features would be required of such a polymerase, as in principle it is possible that any gap-filling polymerase would suffice provided the ends are properly and stably aligned. We previously showed that polβ did not support alignment-based gap filling (10). However, while the catalytic domain of polβ differs from that of polλ, most notably in having a more negatively charged template-binding channel (23,24), polβ also lacks a BRCT domain, which is essential for alignment-based gap filling by polλ (10). Thus, failure of polβ to support gap filling could be due to lack of recruitment to the DSB repair complex. To test this possibility, a chimeric polymerase was constructed by replacing the catalytic domain of polλ with that of polβ. Despite efficiently binding to and stabilizing a complex of Ku and X4L4 on a DNA duplex, the chimera did not support gap filling during end joining at all (Figure 2). This result suggests that the polλ catalytic domain contains specific structural features that promote formation of an aligned substrate by juxtaposed DNA ends.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank John B. Mangrum for performing mass spectrometry, and Samuel Wilson and Rajendra Prasad for the polβ clone. This work was supported by Grant CA40615 from the National Cancer Institute, DHHS to L.F.P., and by the Intramural Research Program of the National Institutes of Health, National Institute of Environmental Health Sciences to T.A.K. Funding to pay the Open Access publication charges for this article was provided by CA40615.
Conflict of interest statement. None declared.
REFERENCES
- 1.Ward JF. DNA damage produced by ionizing radiation in mammalian cells: identities, mechanisms of formation, and reparability. Prog. Nucleic Acid. Res. Mol. Biol. 1988;35:95–125. doi: 10.1016/s0079-6603(08)60611-x. [DOI] [PubMed] [Google Scholar]
- 2.Roth DB, Wilson JH. Nonhomologous recombination in mammalian cells: role for short sequence homologies in the joining reaction. Mol. Cell. Biol. 1986;6:4295–4304. doi: 10.1128/mcb.6.12.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guirouilh-Barbat J, Huck S, Bertrand P, Pirzio L, Desmaze C, Sabatier L, Lopez BS. Impact of the KU80 pathway on NHEJ-induced genome rearrangements in mammalian cells. Mol. Cell. 2004;14:611–623. doi: 10.1016/j.molcel.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 4.Pfeiffer P, Thode S, Hancke J, Vielmetter W. Mechanisms of overlap formation in nonhomologous DNA end joining. Mol. Cell. Biol. 1994;14:888–895. doi: 10.1128/mcb.14.2.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daza P, Reichenberger S, Göttlich B, Hagmann M, Feldmann E, Pfeiffer P. Mechanisms of nonhomologous DNA end-joining in frogs, mice and men. Biol. Chem. 1996;377:775–786. doi: 10.1515/bchm3.1996.377.12.775. [DOI] [PubMed] [Google Scholar]
- 6.Chen S, Inamdar KV, Pfeiffer P, Feldmann E, Hannah MF, Yu Y, Lee JW, Zhou T, Lees-Miller SP, Povirk LF. Accurate in vitro end-joining of a DNA double-strand break with partially cohesive 3′-overhangs and 3′-phosphoglycolate termini: effect of Ku on repair fidelity. J. Biol. Chem. 2001;276:24323–24330. doi: 10.1074/jbc.M010544200. [DOI] [PubMed] [Google Scholar]
- 7.Povirk LF. Biochemical mechanisms of chromosomal translocations resulting from DNA double-strand breaks. DNA Repair. 2006;5:1199–1212. doi: 10.1016/j.dnarep.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 8.Capp JP, Boudsocq F, Bertrand P, Laroche-Clary A, Pourquier P, Lopez BS, Cazaux C, Hoffmann JS, Canitrot Y. The DNA polymerase lambda is required for the repair of non-compatible DNA double strand breaks by NHEJ in mammalian cells. Nucleic Acids Res. 2006;34:2998–3007. doi: 10.1093/nar/gkl380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nick McElhinny SA, Havener JM, Garcia-Diaz M, Juarez R, Bebenek K, Kee BL, Blanco L, Kunkel TA, Ramsden DA. A gradient of template dependence defines distinct biological roles for family X polymerases in nonhomologous end joining. Mol. Cell. 2005;19:357–366. doi: 10.1016/j.molcel.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 10.Lee JW, Blanco L, Zhou T, Bebenek K, Garcia-Diaz M, Kunkel TA, Wang Z, Povirk LF. Implication of DNA polymerase lambda in alignment-based gap filling for nonhomologous DNA end joining in human nuclear extracts. J. Biol. Chem. 2004;279:805–811. doi: 10.1074/jbc.M307913200. [DOI] [PubMed] [Google Scholar]
- 11.Mahajan KN, Nick McElhinny SA, Mitchell BS, Ramsden DA. Association of DNA polymerase mu (polμ) with Ku and ligase IV: role for pol μ in end-joining double-strand break repair. Mol. Cell. Biol. 2002;22:5194–5202. doi: 10.1128/MCB.22.14.5194-5202.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma Y, Lu H, Tippin B, Goodman MF, Shimazaki N, Koiwai O, Hsieh CL, Schwarz K, Lieber MR. A biochemically defined system for mammalian nonhomologous DNA end joining. Mol. Cell. 2004;16:701–713. doi: 10.1016/j.molcel.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 13.Lee JW, Yannone SM, Chen DJ, Povirk LF. Requirement for XRCC4 and DNA ligase IV in alignment-based gap filling for nonhomologous DNA end joining in vitro. Cancer Res. 2003;63:22–24. [PubMed] [Google Scholar]
- 14.Feldmann E, Schmiemann V, Goedecke W, Reichenberger S, Pfeiffer P. DNA double-strand break repair in cell-free extracts from Ku80-deficient cells: implications for Ku serving as an alignment factor in non-homologous DNA end joining. Nucleic Acids Res. 2000;28:2585–2596. doi: 10.1093/nar/28.13.2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nikjoo H, O’Neill P, Goodhead DT, Terrissol M. Computational modelling of low-energy electron-induced DNA damage by early physical and chemical events. Int. J. Radiat. Biol. 1997;71:467–483. doi: 10.1080/095530097143798. [DOI] [PubMed] [Google Scholar]
- 16.Picher AJ, Garcia-Diaz M, Bebenek K, Pedersen LC, Kunkel TA, Blanco L. Promiscuous mismatch extension by human DNA polymerase lambda. Nucleic Acids Res. 2006;34:3259–3266. doi: 10.1093/nar/gkl377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bennett RAO, Gu X-Y, Povirk LF. Construction of a vector containing a site-specific DNA double-strand break with 3′-phosphoglycolate termini and analysis of the products of end-joining in CV-1 cells. Intl. J. Radiat. Biol. 1996;70:623–636. doi: 10.1080/095530096144509. [DOI] [PubMed] [Google Scholar]
- 18.Miller H, Fernandes AS, Zaika E, McTigue MM, Torres MC, Wente M, Iden CR, Grollman AP. Stereoselective excision of thymine glycol from oxidatively damaged DNA. Nucleic Acids Res. 2004;32:338–345. doi: 10.1093/nar/gkh190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Povirk LF, Zhou RZ, Ramsden DA, Lees-Miller SP, Valerie K. Phosphorylation in the serine/threonine 2609–2647 cluster promotes but is not essential for DNA-dependent protein kinase-mediated nonhomologous end joining in human whole-cell extracts. Nucleic Acids Res. 2007;5:3869–3878. doi: 10.1093/nar/gkm339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garcia-Diaz M, Dominguez O, Lopez-Fernandez LA, de Lera LT, Saniger ML, Ruiz JF, Parraga M, Garcia-Ortiz MJ, Kirchhoff T, del Mazo J, et al. DNA polymerase lambda (Pol lambda), a novel eukaryotic DNA polymerase with a potential role in meiosis. J. Mol. Biol. 2000;301:851–867. doi: 10.1006/jmbi.2000.4005. [DOI] [PubMed] [Google Scholar]
- 21.Meyer C, Meyer D, Bickle TA, Giese B. Chemical restriction: strand cleavage by ammonia treatment at 8-oxoguanine yields biologically active DNA. Chembiochem. 2003;4:610–614. doi: 10.1002/cbic.200300587. [DOI] [PubMed] [Google Scholar]
- 22.Gu XY, Weinfeld M, Povirk LF. Implication of DNA-dependent protein kinase in an early, essential, local phosphorylation event during end-joining of DNA double-strand breaks in vitro. Biochemistry. 1998;37:9827–9835. doi: 10.1021/bi980198o. [DOI] [PubMed] [Google Scholar]
- 23.Garcia-Diaz M, Bebenek K, Gao G, Pedersen LC, London RE, Kunkel TA. Structure-function studies of DNA polymerase lambda. DNA Repair. 2005;4:1358–1367. doi: 10.1016/j.dnarep.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 24.Garcia-Diaz M, Bebenek K, Krahn JM, Blanco L, Kunkel TA, Pedersen LC. A structural solution for the DNA polymerase lambda-dependent repair of DNA gaps with minimal homology. Mol. Cell. 2004;13:561–572. doi: 10.1016/s1097-2765(04)00061-9. [DOI] [PubMed] [Google Scholar]
- 25.Pfeiffer P, Thode S, Hancke J, Keohavong P, Thilly WG. Resolution and conservation of mismatches in DNA end joining. Mutagenesis. 1994;9:527–535. doi: 10.1093/mutage/9.6.527. [DOI] [PubMed] [Google Scholar]
- 26.Grollman AP, Moriya M. Mutagenesis by 8-oxoguanine: an enemy within. Trends Genet. 1993;9:246–249. doi: 10.1016/0168-9525(93)90089-z. [DOI] [PubMed] [Google Scholar]
- 27.Aller P, Rould MA, Hogg M, Wallace SS, Doublie S. A structural rationale for stalling of a replicative DNA polymerase at the most common oxidative thymine lesion, thymine glycol. Proc. Natl Acad. Sci. USA. 2007;104:814–818. doi: 10.1073/pnas.0606648104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baumann P, West SC. DNA end-joining catalyzed by human cell-free extracts. Proc. Natl Acad. Sci. USA. 1998;95:14066–14070. doi: 10.1073/pnas.95.24.14066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Picher AJ, Blanco L. DNA polymerase lambda is a proficient extender of primer ends paired to 7,8-dihydro-8-oxoguanine. DNA Repair. 2007;6:1749–1756. doi: 10.1016/j.dnarep.2007.06.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.