Abstract
Among rodents that carry hantaviruses, more males are infected than females. Male rats also have elevated copies of Seoul virus RNA and reduced transcription of immune-related genes in the lungs than females. To further characterize sex differences in antiviral defenses and whether these differences are mediated by gonadal hormones, we examined viral RNA in the lungs, virus shedding in saliva, and antiviral defenses among male and female rats that were intact, gonadectomized neonatally, or gonadectomized in adulthood. Following inoculation with Seoul virus, high amounts viral RNA persisted longer in lungs from intact males than intact females. Removal of the gonads in males reduced the amount of viral RNA to levels comparable with intact females at 40 days post-inoculation (p.i.). Intact males shed more virus in saliva than intact females 15 days p.i.; removal of the gonads during either the neonatal period or in adulthood increased virus shedding in females and decreased virus shedding in males. Induction of pattern recognition receptors (PRRs; TLR7 and RIG-I), expression of antiviral genes (Myd88, VISA, Jun, IRF7, IFNβ, IFNAR1, JAK2, STAT3, and Mx2), and production of Mx protein was elevated in the lungs of intact females compared with intact males. Gonadectomy had more robust effects on the induction of PRRs than on downstream IFNβ or Mx2 expression. Putative androgen and estrogen response elements are present in the promoters of several of these antiviral genes, suggesting the propensity for sex steroids to directly affect dimorphic antiviral responses against Seoul virus infection.
Keywords: biodefense, estrogen, hemorrhagic fever, interferon, Mx, RNA helicase, Seoul virus, sex steroid, testosterone, TLR7, TLR3
Males and females differ in their susceptibility to several RNA and DNA viruses, including HIV, HSV, hantaviruses, measles virus, coxsackievirus, and VSV (Klein, 2000). Because females typically mount higher immune responses than males, susceptibility to viral infections often is reduced among females. Immune responses to viruses can vary with changes in hormone concentrations caused by natural fluctuations over the menstrual or estrous cycle, contraception use, and pregnancy (Brabin, 2002).
Although behavioral factors can influence exposure to viruses, several studies illustrate that hormonal differences between the sexes cause dimorphic responses to infection (Klein, 2000). Elevated immunity among females creates a double-edge sword, being beneficial against infectious diseases, but detrimental in terms of increased development of autoimmune diseases (Wizemann, 2001). Approximately 80% of all cases of autoimmunity in the US are women (Jacobson et al., 1997). Animal models illustrate that elevated detection of self antigens by toll-like receptors (TLRs), including TLR7, and subsequent production of type I interferons (IFNs) leads to development of autoimmune diseases, including systemic lupus erythematosus and insulin-dependent diabetes mellitus (Pisitkun et al., 2006; Theofilopoulos et al., 2005).
Reported human hantavirus infections in the Americas and Europe, as well as field observations of several rodent-virus systems indicate that more males than females are infected with hantaviruses (Family: Bunyaviridae) (Klein and Calisher, 2007). Sex differences in hantavirus infection only become apparent after puberty, suggesting that sex steroid hormones, including testosterone in males and estradiol in females, may be involved (Childs et al., 1988; Douglass et al., 2007). Old World hantaviruses, including Seoul virus which is maintained in Norway rats (Rattus norvegicus), are found throughout the world. Rodents infected with their species-specific hantaviruses remain persistently infected, without developing notable signs of morbidity or mortality, and shed virus intermittently in saliva, urine, and feces. Hantaviruses disseminate to several organs in the adult rodent host, including the lungs, kidneys, spleen, heart, liver, and brown fat; the lungs, however, are consistently a site of elevated hantavirus replication in rodent reservoirs (Botten et al., 2003; Hutchinson et al., 1998; Klein et al., 2002). Transmission of hantaviruses between rodents and from rodents to humans occurs from inhalation of aerosolized virus in excrement or passage of virus in saliva during wounding (Easterbrook et al., 2007; Glass et al., 1988; Hinson et al., 2004; Hjelle and Yates, 2001).
Laboratory studies of Seoul virus infection of Norway rats reveal that when given the same challenge, males shed Seoul virus longer and via more routes (i.e., a combination of saliva, urine, and feces) and have more viral RNA copies present in target organs, such as the lungs, than females (Klein et al., 2000, 2001; Klein et al., 2002). Additionally, the expression of genes that encode for immunological proteins associated with innate antiviral defenses, T cell responsiveness, and antibody production is higher in females than males (Klein et al., 2004a). Mathematical models utilizing ordinary and stochastic differential equations to predict the emergence of hantaviruses reveal that male rodents are more likely to be exposed to hantaviruses and to be infectious following exposure, suggesting that both extrinsic and intrinsic factors are involved (Allen et al., 2006).
Production of IFNα/β and downstream expression of IFN-stimulated genes (ISGs) is critical for restricting virus replication and establishing antiviral immunity. Viral nucleic acids are recognized by host cells via TLRs and RNA helicases in a cell-specific manner (Melchjorsen et al., 2005). Detection of RNA viruses, including hantaviruses and influenza viruses, by TLR3, TLR7/8, and the cytoplasmic RNA helicase, retinoic acid-inducible gene-I (RIG-I), is critical for production of type I IFNs (Kawai and Akira, 2006). Mx proteins are induced by type I IFNs and possess important antiviral properties. Human MxA and rodent Mx2, in particular, confer resistance against hantaviruses, including Seoul virus, Puumala virus, Hantaan virus, and Andes virus, in vitro (Jin et al., 2001; Khaiboullina et al., 2005; Temonen et al., 1995). The expression of Mx2 is suppressed in male rats during Seoul virus infection and may contribute to increased virus shedding and viral RNA levels in lung tissue (Klein et al., 2004a). Whether sex differences in the expression of Mx2 are caused by an upstream dimorphism in virus recognition has not been reported. Further, the effects of sex steroids on innate antiviral responses to hantavirus infection have not been elucidated.
In vertebrates, sex steroids affect sex-specific development at two distinct times during ontogeny. During perinatal (i.e., prenatal and early neonatal) development, sex steroids can permanently affect sexual differentiation of the brain and peripheral organs (called the organizational effects) (Arnold and Breedlove, 1985). After puberty, exposure to sex steroids can transiently activate pre-existing hormonal targets (i.e., activational effects) (Arnold and Breedlove, 1985). Previous data from our laboratory illustrate that manipulation of sex steroids in adulthood has no effect on antibody responses against Seoul virus whereas neonatal hormone manipulation in males reverses the sex difference in antibody production, suggesting that sex steroids may act early in development to affect the functioning of the immune system later in life (Klein et al., 2000; Klein et al., 2002).
The primary goal of this study was to examine whether the expression of genes that mediate innate antiviral responses against hantavirus infection differs between the sexes and is altered by hormone deprivation during different stages of development. The binding of sex steroids to their respective steroid receptors directly influences transcriptional factors (e.g., NF-κB and cJun) that also are induced by signaling along TLR- and RNA helicase-mediated pathways. Ultimately, convergence of steroid receptor and antiviral pathway activity may affect production of cytokines and chemokines by cells of the immune system (30). Thus, the expression and translation of antiviral genes may be altered by removal of sex steroids via gonadectomy.
Material and Methods
Animals
Adult (60–70 days of age) male and female Long Evans rats were purchased from Charles Rivers Laboratories (Raleigh, NC) and housed individually in polypropylene cages covered with polyester filter bonnets. Animals were housed individually to avoid sporadic re-exposure due to transmission of virus in saliva during wounding or inhalation of aerosolized virus in excrement. All animals were housed in a Biosafety Level (BSL) 3 animal facility. The Johns Hopkins Animal Care and Use Committee (RA98H536) and the Johns Hopkins Office of Health, Safety, and Environment (A9902030102) approved all procedures.
Procedure
Adult female Long Evans rats were bred in the JHU animal facility and gestational day 1 was determined based on the presence of a sperm plug. Pregnancy was monitored and the day of birth constituted postnatal day 1. Each experimental group consisted of 60–72 animals. Animals were assigned to one of six groups: 1) intact-sham males (IM); 2) intact-sham females (IF); 3) neonatally gonadectomized (gdx) males (NGM); 4) neonatally gdx females (NGF); 5) adult gdx males (AGM); and 6) adult gdx females (AGF). At postnatal days 2–4, male and female pups assigned to have their gonads removed were anesthetized by brief hypothermia and gonadectomized bilaterally (Klein et al., 2002). Males and females not assigned to have their gonads removed received sham operations. Animals were weaned at 21 days of age and housed with same sex siblings. At 50–55 days of age, females and males assigned to be gonadectomized as adults were gonadectomized under a Ketamine (80 mg/kg)/Xylazine (6 mg/kg) anesthesia (Phoenix Pharmaceutical, St. Joseph, MO) and given several weeks to recover from surgery.
At 80 days of age, all animals were inoculated in the intraperitoneal cavity with 104 pfu of Seoul virus strain SR-11. Ten to twelve animals from each group were killed 1, 3, 15, 30, or 40 days p.i. and immune responses, virus shedding, and virus replication were monitored. Samples also were collected from uninfected rats (designated Day 0 p.i.) in each treatment group and processed at the same time-points as the infected animals. Saliva was collected to determine virus shedding and lung samples were collected to assess viral RNA, gene expression patterns, and protein production as described below.
Real-time RT-PCR for Seoul virus
RNA was isolated from saliva and lung using Trizol LS and the manufacturer’s protocol as described previously (Invitrogen, Carlsbad, CA) (Klein et al., 2000; Klein et al., 2004a; Klein et al., 2002). First-strand synthesis cDNA was prepared in a single reaction for both the negative- and positive-strands of Seoul virus using 0.1 μM gene-specific primers (S-segment coordinate-828 sense primer: TTCAAGCCCTCAGGCAACA; S-segment coordinate-901 antisense primer: CGTGACTATATCAGACAGAGACAAGGT), the Invitrogen Superscript III First Strand Synthesis reagents, and the manufacturer’s protocol. The positive control was Seoul virus RNA isolated from our virus stock (strain SR-11) and the negative control was DEPC-treated water included in the cDNA syntheses and amplification. For amplification of negative-strand genomic Seoul virus RNA, an 81 bp nucleotide sequence of the S segment was amplified in a reaction mixture containing Platinum Quantitative PCR SuperMix-UDG (Invitrogen), Rox reference dye, 4.5 mM MgCl2, 0.1 μM primers (S-segment coordinate-1006 forward: GTCGGAGGGATGGCTGAAT; coordinate-1087 reverse: CCACAGTTTTTGAAGCCATGATT), and 0.2 μM probe (coordinate-1042 ATACTTCAGGATATGAGGAAC-MGB; Applied Biosystems, Foster City, CA). For amplification of the positive-strand of viral mRNA (i.e., replicative RNA) (Botten et al., 2003), a 60 bp nucleotide sequence was amplified in the same reaction mixture as the negative sense S-segment with 0.1 μM primers (S-segment coordinate-875 forward: TGGCTCCATCCCTGCAA; coordinate-815 reverse: GAATCCTGTGAATCGTGACTATATCAG) and 0.2 μM probe (coordinate-857 GCACCTTGTCTCTG-MGB; Applied Biosystems). All reactions were multiplexed in optical 96-well plates using the ABI 7300 Sequence Detection System (Applied Biosystems). A standard curve ranging from 1 × 107 to 1 × 101 copies of negative and positive sense SR-11 S segment in plasmid pWRG7077 was run on each plate.
DNA Microarrays
For RNA extraction, lungs were homogenized in Trizol LS and processed according to the manufacturer’s protocol (Invitrogen). RNA pellets were resuspended in nuclease-free water. Quantitation of RNA was performed using a Beckman DU640 spectrophotometer and RNA quality assessment was determined by RNA Nano LabChip analysis on an Agilent Bioanalyzer 2100. Equivalent aliquots (5 μg) of RNA from two animals/treatment group were pooled and 3 separate pools were processed on separate Affymetrix GeneChips (n = 3 GeneChips/time point/sex).
Processing of RNA for GeneChip Analysis was in accordance with methods described in the Affymetrix GeneChip Expression Analysis Technical Manual, Revision Three, as previously reported (Klein et al., 2004a). Hybridization cocktails were prepared as recommended prior to pipetting into the Affymetrix GeneChips (Rat Genome RAE230A). Hybridization and the signal amplification protocol for washing and staining of eukaryotic targets were performed as previously described (Klein et al., 2004a). The arrays were scanned at an emission wavelength of 570 nm at 2.5 μm resolution in the GCS3000 laser Scanner (Affymetrix) and the intensity of hybridization for each probe pair was computed by GCOS 1.1 software. For more detailed methods, please refer to the website of the Malaria Research Institute Gene Array Core Facility (MRI-GACF) at the Johns Hopkins Bloomberg School of Public Health http://jhmmi.jhsph.edu/.
Real-time RT-PCR for host genes
Custom primer and probe sets were generated for β-actin 3, IFNβ, Mx2, RIG-I, TLR3, TLR7, and VISA using Primer Express 2.0 software (Applied Biosystems). Reverse transcription of 1 μg of RNA per 20 μl reaction from each individual animal sample (n = 8–12/time-point/treatment group) was conducted using oligo (dT)17–20 primers and the manufacturer’s protocol for SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen). Complementary DNA was amplified in a reaction mixture containing TaqMan Universal PCR Master Mix, 0.05 μM primers, and 0.250 μM probe using the manufacturer’s protocol (Applied Biosystems). All reactions were run in optical 96-well plates using the ABI 7300 Sequence Detection System (Applied Biosystems). Serial dilutions of pooled cDNA from randomly selected animals were used to generate a standard curve, which was run on each plate. Gene expression patterns for each target gene were reported relative to the values from same-treatment uninfected control animals.
Immunohistochemistry (IHC)
Lungs were fixed in 10% neutral, buffered formaldehyde, embedded in paraffin, cut into 5 μm sections, and mounted on glass slides. Formaldehyde- induced autofluorescence was photobleached with a 40 W cool white 800 fluorescent lamp at a distance of 6 in for 24 h (Neumann and Gabel, 2002). Sections were deparaffinized with xylene, rehydrated in graded ethanol, and post-fixed with Streck’s Tissue Fixative (STF Streck, Omaha, NE). Antigen retrieval was maximized using Antigen Unmasking Solution (1:100; Vector Labs, Burlingame, CA). Tissue sections were blocked with 10% normal horse serum, followed by blocking with avidin D and biotin to block all endogenous biotin, biotin receptors, and avidin binding sites. Slides were incubated for 60 min at room temperature in a humidified chamber with a monoclonal antibody against the Seoul virus nucleocapsid protein (NP; 1:250; EC01-AB08, gift of Connie Schmaljohn, Fort Detrick, MD) or Mx (1:800; M143 antibody against human MxA that cross-reacts with rat Mx2 and Mx3, obtained from Georg Kochs, University of Freiburg). Sections were incubated with biotinylated, rat-adsorbed, anti-mouse IgG (5 μg/ml; Vector Labs, Burlingame, CA). The signal was revealed using streptavidin conjugated Fluorescein or Texas Red (15 μg/ml or 10 μg/ml, respectively; Vector Labs) as specified. Tissues were counterstained with 4′,6 diamidino-2-phenylindole (DAPI, Vector Labs) and stored in the dark at 4°C until viewed. Fluorescence emission images were acquired and merged using a SPOT charge-coupled device camera and software (Diagnostic Instruments, Sterling Heights, MI). Merged images were counted and analyzed at a magnification of 100x. Semiquantitative scoring was conducted by calculating the average number of cells in 4 randomly selected visual fields as follows: 0 = no cells present; 1 = 1 cell/visual field; 2 = 2–3 cells/visual field; 3 = 4–10 cells/visual field; 4 = 10+ cells/visual field.
Western blot analysis
Lung protein extracts were prepared by homogenizing tissue in lysis buffer containing 50 mM Tris (pH 7.4), 0.5 mM EDTA, 1% Nonidet P40, 0.01 mM PMSF, and 0.1% protease inhibitor cocktail (Sigma Aldrich, St. Louis, MO). Proteins were separated onto a 4%–12% polyacrylamide-sodium dodecyl sulfate gels (Laemmli, 1970) and transferred to nitrocellulose by standard procedures. Membranes were blocked for 2 h in TBS containing 5% nonfat dry milk and 0.5% Tween 20 (TBST). Monoclonal antibody against Mx (1:2000; M143 antibody) diluted in PBS containing 0.1 % Tween 20 (PBST), was incubated with shaking at 4°C for 16 h. Membranes were washed in TBST and incubated for 1 h with HRP-conjugated goat anti-mouse IgG (Cayman Chemical, Ann Arbor, MI) diluted 1:5000 in PBST. Membranes were washed in TBST and incubated for 2 h with shaking in mouse monoclonal β actin-HRP (Abcam, Cambridge, MA) diluted 1:5000 in PBST and detected with 3-aminophthathic hydrazide and 4-iodophenylboronic acid (Haan and Behrmann, 2007). The positive control for Mx protein was protein extracts from lungs collected from uninfected rats (n = 3) 6 h after ip injection of 25,000 units of rat rIFNα (Sigma).
Identification of putative hormone response elements (HREs)
For 16 differentially expressed genes, as well as for several control genes (four negative control and one positive control gene) (see Table 3), we identified putative HREs in promoters. Promoter sequences covering the region [−3000, +100] bp relative to the transcription start site at the 5′ end of the gene were selected using genomic rat sequences obtained from NCBI. To identify HREs, we used eight mammalian matrix models of HRE sites from Transfac Professional version 10.4 (Matys et al., 2006). The selected matrices correspond to the response elements of three sex steroids, estrogen, androgen and progesterone, as well as to the response element of glucocorticoids. The matrices used have the following Transfac IDs: V$ER_Q6, V$AR_Q2, V$AR_01, V$AR_02, V$AR_03, V$PR_01, V$PR_02, and V$GR_01. These HRE sites were mapped to the promoters of the selected genes by the Match program (Kel et al., 2003) of the Transfac package using the optimized ‘minFP’ thresholds for matrix and core scores. The ‘minFP’ thresholds are provided by Transfac and correspond to settings that enable the minimum number of false positive predictions; thus, this approach maximizes the probability that the predicted sites are very similar to the actual binding sites of the respective transcription factors used to build matrix models. Details of the methodology used to determine the ‘minFP’ thresholds are described elsewhere (Kel et al., 2003).
Table 3.
Putative hormone response elements in the promoters of antiviral and housekeeping genes
| Gene | GenBank # | Chromosomal location |
Putative HRE |
HRE sequence | Core Threshold |
Matrix Threshold |
Strand | Starting Position |
Matrix |
|---|---|---|---|---|---|---|---|---|---|
| Genes differentially expressed between the sexes | |||||||||
| Ccl5 | NM_031116 | 10q26 | -- | -- | -- | -- | |||
| Ccl11 | NM_019205 | 10q26 | -- | -- | -- | -- | |||
| Hsp70 | NW_047597 | 20p12 | ARE | acccgcaGAACAccgtgttcgacgcga | 1 | 0.961 | −1 | 1670 | V$AR_02 |
| ARE | acccgcaGAACAccgtgttcgacgcga | 1 | 0.975 | −1 | 1670 | V$AR_03 | |||
| ARE | agaacaccgTGTTCg | 1 | 0.915 | 1 | 1676 | V$AR_01 | |||
| ARE | aGAACAccgtgttcg | 1 | 0.945 | −1 | 1676 | V$AR_01 | |||
| ARE | agaaccggtcGTTCT | 1 | 0.89 | 1 | 1814 | V$AR_Q2 | |||
| ERE | ccagatcgaggTGACCttc | 1 | 0.959 | 1 | 2901 | V$ER_Q6 | |||
| GRE | acccgcaGAACAccgtgttcgacgcga | 1 | 0.981 | −1 | 1670 | V$GR_01 | |||
| Ifnar1 | NW_047354 | 11q11 | ERE | ctagcttaaaaTGACCcga | 1 | 0.955 | 1 | 7 | V$ER_Q6 |
| Ifnβ1 | NW_047715 | 5q31–q33 | GRE | agatcaaagctgcaaTGTTCtcatcca | 1 | 0.922 | 1 | 3054 | V$GR_01 |
| Irf7 | NW_047563 | 1q41 | ARE | AGTACaccttgcact | 0.939 | 0.874 | −1 | 1423 | V$AR_Q2 |
| Jun | NW_047717 | 5q34.1 | ERE | tatGGTCAaaatcaactca | 1 | 0.951 | −1 | 2575 | V$ER_Q6 |
| Mapk3 | NW_047562 | 1q36 | ARE | aGGACAgccagttcc | 0.975 | 0.934 | −1 | 447 | V$AR_01 |
| Mx2 | NW_001084662 | 11q21 | ARE | aGAACTcatggtgcc | 0.905 | 0.897 | −1 | 3076 | V$AR_01 |
| ERE | gaagaaaactaTGACCaga | 1 | 0.947 | 1 | 10 | V$ER_Q6 | |||
| Myd88 | NW_047803 | 8q32 | ERE | gtagcccaggcTGACCtta | 1 | 0.951 | 1 | 2268 | V$ER_Q6 |
| Rbd2 | NW_047475 | 16q12.5 | GRE | gagttaaGAACAataggtaaccatcac | 1 | 0.951 | −1 | 1844 | V$GR_01 |
| RIG-I | NW_047711 | 5q22 | -- | -- | -- | -- | |||
| Stat3 | NW_047339 | 10q32.1 | ERE | gccGGTCAccttgtcttac | 1 | 0.956 | −1 | 1549 | V$ER_Q6 |
| ERE | gatggtgaattTGACCtgg | 1 | 0.95 | 1 | 1819 | V$ER_Q6 | |||
| ERE | ataggtttgcaTGACCaca | 1 | 0.94 | 1 | 2680 | V$ER_Q6 | |||
| Tlr3 | NW_047473 | 16q11 | ERE | tcgggatattcTGACCtta | 1 | 0.946 | 1 | 1281 | V$ER_Q6 |
| Tlr7 | NW_048039 | Xq21 | ERE | gcaagtgtggcTGACCttc | 1 | 0.943 | 1 | 863 | V$ER_Q6 |
| VISA | NW_047658 | 3q36 | -- | -- | -- | -- | |||
| Negative control genes (genes not differentially expressed between the sexes) | |||||||||
| Irf3 | NW_047558 | 1q22 | -- | -- | -- | -- | |||
| α-actin1 | NW_047536 | 19q12 | -- | -- | -- | -- | |||
| β-actin | NW_047369 | 12p11 | -- | -- | -- | -- | |||
| GAPDH | NW_047696 | 4q42 | -- | -- | -- | -- | |||
| Positive control gene for EREs | |||||||||
| PR | NW_047798 | 8q11 | ERE | ggaagtaagaaTGACCggc | 1 | 0.946 | 1 | 1006 | V$ER_Q6 |
| ERE | caaGGTCActgtgagcaat | 1 | 0.941 | −1 | 1284 | V$ER_Q6 | |||
The capitalized portion of each sequence (length of 5 nt) is the ‘core’ portion of the sequence that the Match program first identified in the process of mapping binding sites to the DNA; this is the most conserved portion of the binding site.
Statistical Analyses
Virus prevalence (i.e., the proportion of animals with detectable virus) was compared among groups using chi-square analyses. Quantitative variables, including virus copy number, cell counts, and gene expression values were analyzed using 2-way ANOVAs with 2 between subjects variables (day post-inoculation and treatment group). Data that were not normally distributed (e.g., virus copy data) were log transformed prior to statistical analyses. Significant interactions were further analyzed using planned comparisons or the Tukey method for pairwise multiple comparisons. Mean differences were considered statistically significant if p < 0.05.
For microarray analyses, Affymetrix CEL file data were preprocessed using GC-RMA in GeneSpring 7.2. Gene expression patterns for each gene from infected animals were normalized to the expression levels from same-treatment uninfected control (i.e., Day 0) animals (i.e., per gene normalization) (Klein et al., 2004a). Filtering by probe intensity for raw levels ≥ 200 units in at least 2 of the 8 conditions was conducted prior to statistical analyses, which reduced the initial list of gene probes from 15,923 to 7,118. This filtered list was subsequently used for all statistical analyses. Two-way ANOVAs (variables: day post-inoculation and sex) were used to compare gene expression patterns between intact males and intact females and mean differences were considered statistically significant if p < 0.05. All gene expression data from the Affymetrix output are located on the NCBI Gene Expression Omnibus (GEO accession #: GSE7271 [public access pending] ; www.ncbi.nlm.nih.gov/geo/).
Results
The amount of Seoul virus RNA is higher in the lungs of intact males than intact females; gonadectomy of males reduces Seoul virus RNA loads
Previous data from our laboratory reveal that the lungs are a target organ of Seoul virus replication (Klein et al., 2004b) and host immunological defenses (Klein et al., 2004a) in rats. In the current study, gonadally intact male and female rats differed in the amount of virus present in the lungs (F(4,101) = 2.48, p = 0.049; Figure 1A). Among intact females, the quantity of viral RNA in the lungs peaked at 15 days p.i. and declined thereafter resulting in almost no virus present in lungs by Day 40 p.i. Among intact males, there was a temporal shift in the amount of viral RNA, in which the number of Seoul virus copies peaked at 30 days p.i. and remained high 40 days p.i. Removal of the gonads in males reduced the amount of viral RNA 40 days p.i. (F(8,163) = 2.15, p = 0.034; Figure 1B); conversely, removal of the ovaries did not affect the amount of viral RNA in the lungs of female rats (p > 0.05; Figure 1C).
Figure 1.
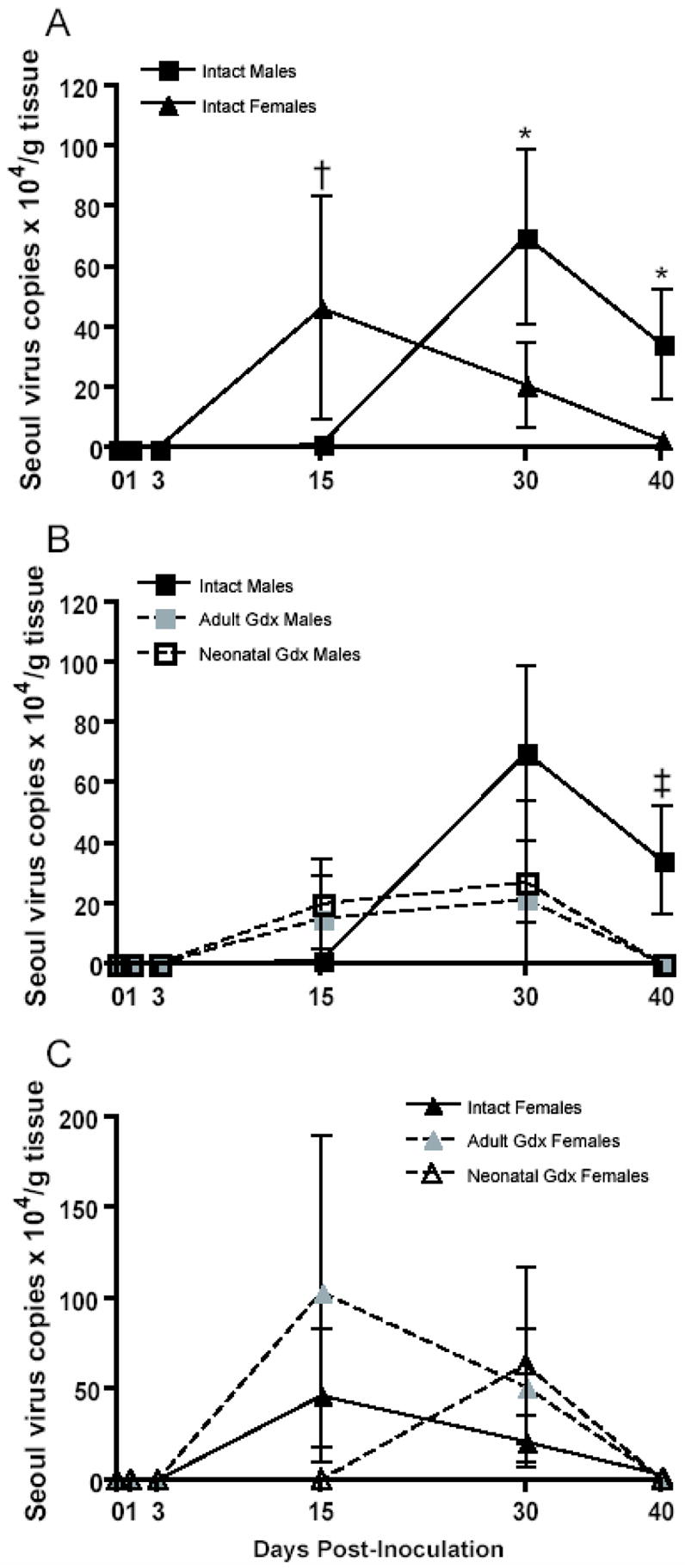
Numbers of genomic Seoul virus RNA copies (± SEM) in lung tissue collected 0, 1, 3, 15, 30, or 40 days after inoculation with Seoul virus from intact males (IM), adult gonadectomized males (AGM), neonatally gonadectomized males (NGM), intact females (IF), adult gonadectomized females (AGF), or neonatally gonadectomized females (NGF). * IM > IF, † IF > IM, ‡ IM > AGM and NGM, p < 0.05, two-way ANOVAs (A–C).
Production of positive-sense hantavirus mRNA is carried out by the viral RNA-dependent RNA polymerase using the negative-sense genomic viral RNA as a template for transcription and replication. Thus, positive-sense viral mRNA can be measured as an indication of hantavirus replication in target organs, including the lungs (Botten et al., 2003). Among animals with detectable genomic Seoul virus RNA, neither sex nor gonadectomy affected the number of viral mRNA copies, which was highly variable among individual rats (p > 0.05 in each case; data not shown). Most animals with detectable genomic Seoul virus RNA had detectable viral mRNA (Table 1). Neither sex nor gonadectomy significantly affected the proportion of animals with detectable genomic Seoul virus RNA or mRNA in the lungs over the course of infection (p > 0.05; Table 1). There were approximately 95 times more copies of genomic viral RNA than viral mRNA copies in the lungs of infected animals (t = 3.99, df = 662, p < 0.001).
Table 1.
Proportion of rats with detectable genomic and replicative Seoul virus RNA in lungs and saliva
| Negative-strand Seoul virus RNA | Positive-strand Seoul virus RNA | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sample and group | Day post-inoculation | Day post-inoculation | ||||||||
| 1 | 3 | 15 | 30 | 40 | 1 | 3 | 15 | 30 | 40 | |
| Lungs | ||||||||||
| Intact Males | 1/11 | 3/11 | 12/15 | 9/12 | 6/11 | 0/1 | 2/3 | 10/12 | 9/9 | 6/6 |
| Adult Gdx Males | 1/12 | 3/12 | 10/11 | 8/11 | 6/12 | 1/1 | 3/3 | 9/10 | 8/8 | 5/6 |
| Neonatal Gdx Males | 1/12 | 1/12 | 10/12 | 3/12 | 8/13 | 0/1 | 0/1 | 10/10 | 3/3 | 5/8 |
| Intact Females | 3/10 | 1/10 | 6/11 | 5/10 | 6/11 | 1/3 | 1/1 | 5/6 | 5/5 | 6/6 |
| Adult Gdx Females | 2/10 | 1/10 | 8/10 | 8/10 | 4/10 | 0/2 | 1/1 | 8/8 | 8/8 | 4/4 |
| Neonatal Gdx Females | 0/9 | 2/11 | 5/10 | 5/11 | 4/10 | 0/0 | 2/2 | 5/5 | 5/5 | 3/4 |
| Saliva | ||||||||||
| Intact Males | 6/10 | 5/11 | 12/15 | 7/12 | 5/12 | |||||
| Adult Gdx Males | 6/10 | 6/12 | 2/11 | 7/11 | 10/12 | |||||
| Neonatal Gdx Males | 7/12 | 9/12 | 6/11 | 8/12 | 8/13 | |||||
| Intact Females | 8/10 | 4/10 | 4/10 | 6/10 | 8/11 | |||||
| Adult Gdx Females | 5/10 | 6/10 | 8/10 | 5/10 | 7/10 | |||||
| Neonatal Gdx Females | 5/9 | 6/11 | 6/10 | 7/11 | 7/10 | |||||
Intact males shed more Seoul virus RNA in saliva than do intact females and gonadectomy of both sexes reverses the sex difference
Rodents infected with species-specific hantaviruses remain persistently infected and shed virus in saliva, urine, and feces for a limited period during the acute phase of infection (Klein and Calisher, 2007). Overall, the greatest amount of virus was shed in saliva 15 days p.i. (F(4, 296) = 14.15, p < 0.001; Figure 2). Intact males shed more viral RNA in saliva than intact females 15 days p.i.; removal of the gonads during either the neonatal period or in adulthood increased virus shedding in females (F(8,135) = 2.43, p = 0.018; Figure 2) and decreased virus shedding in males (F(8,161) = 8.95, p < 0.001; Figure 2). Neither sex nor gonadectomy affected the proportion of animals with detectable viral RNA in saliva (p > 0.05; Table 1).
Figure 2.
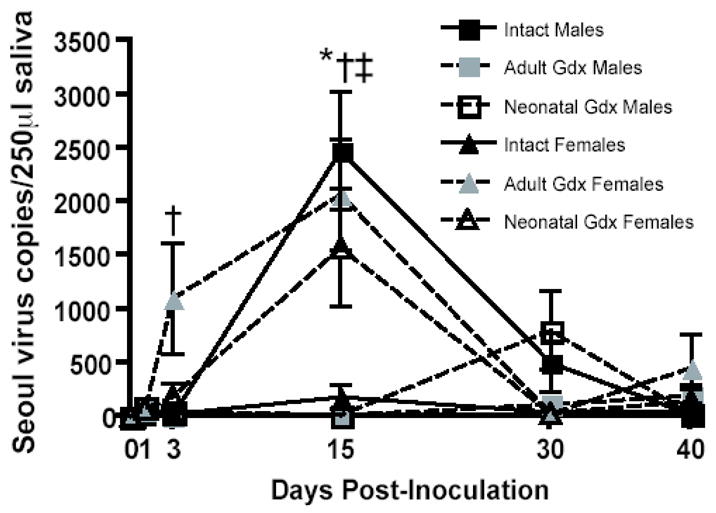
Numbers of genomic Seoul virus RNA copies (± SEM) shed in saliva collected 0, 1, 3, 15, 30, or 40 days after inoculation with Seoul virus from intact males (IM), adult gonadectomized males (AGM), neonatally gonadectomized males (NGM), intact females (IF), adult gonadectomized females (AGF), or neonatally gonadectomized females (NGF). * IM > IF, ‡ IM > AGM and NGM, † IF < AGF and/or NGF, p < 0.05, two-way ANOVAs.
Induction of pattern recognition receptors by Seoul virus differs between the sexes and is altered by gonadectomy
To assess whether the activation of pattern recognition receptors (PRRs) by Seoul virus differs between the sexes, we examined the expression of TLR3, TLR7, RIG-I, and VISA by real-time RT-PCR. The housekeeping gene, β-actin 3 also was assessed and did not differ among the treatment groups over the course of Seoul virus infection (p > 0.05, data not shown). During Seoul virus infection, the expression of TLR3 was approximately 4-fold higher than the expression of TLR7 (t = 20.98, df = 647, p < 0.001), which likely reflects that TLR3 is expressed in multiple cells, whereas TLR7 is primarily expressed in plasmacytoid DCs. TLR7 mRNA expression was 2–4-fold higher among intact females than intact males during Seoul virus infection (F(1, 114) = 35.24, p < 0.001; Figure 3A). Removal of the gonads during adulthood increased TLR7 expression in males (F(2, 189) = 15.69, p < 0.001; Figure 3B). Removal of the gonads, during either neonatal development or in adulthood, reduced the expression of TLR7 in females (F(2, 162) = 24.37, p < 0.001; Figure 3C). The expression of TLR3 was higher in the lungs of infected intact male than intact female rats (F(1, 116) = 65.12, p < 0.01; Figure 3D). Gonadectomy of males reduced (F(10, 188) = 3.67, p < 0.001), whereas gonadectomy of females had no effect on TLR3 expression (p > 0.05; Figures 3E and F).
Figure 3.
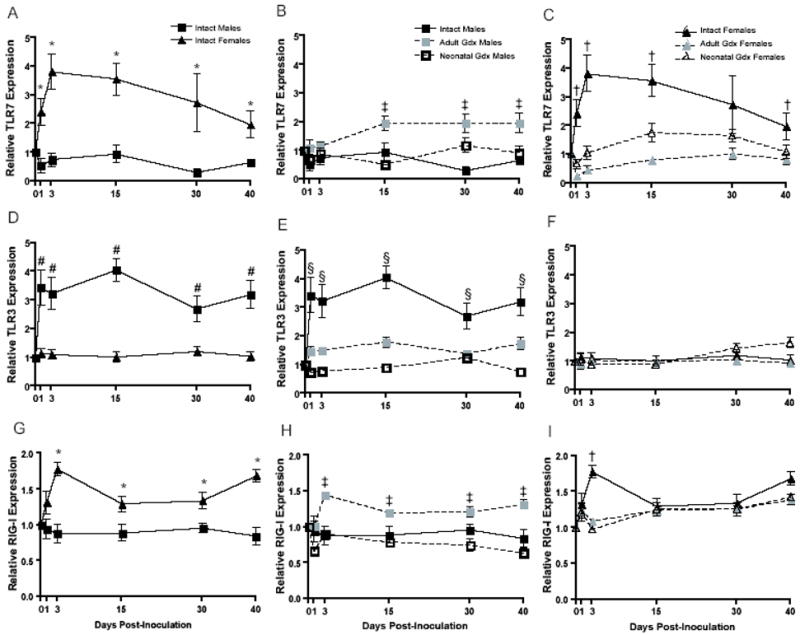
Expression of TLR7 (A–C), TLR3 (D–F), and RIG-I (G–I) mRNA in the lungs of intact males (IM), adult gonadectomized males (AGM), neonatally gonadectomized males (NGM), intact females (IF), adult gonadectomized females (AGF), and neonatally gonadectomized females (NGF) collected 0, 1, 3, 15, 30, or 40 days after inoculation with Seoul virus. * IF > IM, # IM > IF, ‡ IM < AGM, § IM > AGM and NGM, † IF > AGF and NGF, p < 0.05, two-way ANOVAs.
RNA helicases, in particular RIG-I, are critical PRRs for RNA viruses and recent data illustrate that hantaviruses, including NY-1 and Hantaan virus, can suppress RIG-I activity in human endothelial cell lines (Alff et al., 2006). Because Seoul virus persistence and shedding is greater among intact male than intact female rats, we hypothesized that Seoul virus suppression of RIG-I-mediated transcription may be sex specific. Activation of RIG-I and the downstream CARD-containing adaptor protein VISA was higher among intact females than intact males during Seoul virus infection (RIG-I: F(1, 114) = 44.85, p < 0.001; VISA: F(1, 114) = 12.63, p < 0.001; Figure 3G and Table 2). Sex differences in the expression of RIG-I and VISA were most pronounced 72 h following Seoul virus inoculation. Removal of the gonads, in neonates or adults, reduced the expression of RIG-I and VISA in females 72 h after inoculation with Seoul virus (RIG-I: F(2, 162) = 7.16, p < 0.001; VISA: F(2, 162) = 6.59, p = 0.002; Figure 3I and data not shown). Expression of these genes was increased in males gonadectomized as adults only (RIG-I: F(2, 189) = 31.33, p < 0.001; VISA: F(2, 189) = 25.84, p < 0.001; Figure 3H and data not shown).
Table 2.
Genes that are differentially expressed in the lungs of intact female and male Norway rats during Seoul virus infection
| Normalized Intensity
|
|||||||
|---|---|---|---|---|---|---|---|
| Gene name | Unigene ID | P value | Sex | Day 3 | Day 15 | Day 40 | Chromosome location |
| Antiviral/interferon-related genes assessed by microarray analyses | |||||||
| chemokine (C-C motif) ligand 5 (Ccl5) | Rn.8019 | 0.0024 |
F
M |
1.031
0.699 |
1.638
0.861 |
0.94
0.816 |
10q26 |
| chemokine (C-C motif) ligand 11 (Ccl11) | Rn.10632 | 0.0377 |
F
M |
1.995
0.933 |
1.184
0.726 |
1.143
0.863 |
10q26 |
| Mitogen-activated protein kinase 3 (Mapk3) | Rn.2592 | 0.0301 |
F
M |
1.198
1.31 |
1.226
0.488 |
1.232
0.5 |
1q36 |
| Myeloid differentiation primary response gene 88 (Myd88) | Rn.37341 | 0.0132 |
F
M |
1.1
0.839 |
1.026
0.975 |
0.938
0.943 |
8q32 |
| Feline sarcoma viral oncogene homolog (Fgr) | Rn.11309 | 0.0250 |
F
M |
1.119
0.566 |
1.143
1.06 |
0.817
0.944 |
5q36 |
| Heat shock protein 70 (Hsp70) | Rn.1950 | 0.0053 |
F
M |
0.154
0.777 |
0.357
2.382 |
0.272
0.801 |
20p12 |
| Interferon regulatory factor 7 (Irf7) | Rn.101159 | 0.0312 |
F
M |
0.523
0.526 |
1.487
0.501 |
0.812
0.442 |
1q41 |
| Interferon alpha receptor 1 (Ifnar1) | Rn.105738 | 0.0105 |
F
M |
0.656
1.265 |
1.78
0.526 |
1.471
0.527 |
11q11 |
| Janus-activated kinase 2 (Jak2) | Rn.18909 | 0.0132 |
F
M |
1.077
0.583 |
0.926
0.983 |
0.943
1.05 |
1q51–q53 |
| Jun oncogene (Jun) | Rn.93714 | 0.0026 |
F
M |
0.809
1.27 |
1.803
0.477 |
1.571
0.608 |
5q34.1 |
| Myxovirus resistance 2 (Mx2) | Rn.10374 | 0.0401 |
F
M |
1.214
0.841 |
2.229
0.579 |
0.867
0.664 |
11q21 |
| Protein tyrosine phosphatase 16 (Ptp16) | Rn.98260 | 0.0202 |
F
M |
0.39
1.148 |
1.713
0.165 |
2.058
0.197 |
10q12 |
| Protein tyrosine phosphatase receptor F (Ptprf) | Rn.11386 | 0.0035 |
F
M |
1.654
1.073 |
1.437
0.578 |
1.423
0.612 |
1q22 |
| Rat beta defensin 2 (Rbd2) | Rn.2267 | 0.0001 |
F
M |
0.853
0.559 |
2.537
0.621 |
1.943
0.575 |
16q12.5 |
| RNA helicase, DEAD-box protein 6 (Ddx6) | Rn.4270 | 0.0007 |
F
M |
2.419
1.711 |
6.338
0.372 |
9.861
0.358 |
8q22 |
| Signal transducers and activators of transcription 3 (Stat3) | Rn.10247 | 0.0081 |
F
M |
1.023
1.408 |
2.153
0.454 |
2.669
0.488 |
10q32.1 |
| Antiviral/interferon-related genes examined by real-time RT-PCR | |||||||
| Interferon beta 1 (Ifnb1) | Rn.119051 | 0.0010 |
F
M |
1.2
0.5 |
1.62
0.37 |
0.69
0.44 |
5q31–q33 |
| RNA helicase, DEAD-box protein 58 (RIG-I) | Rn.38642 | 0.0010 |
F
M |
1.78
0.878 |
1.295
0.888 |
1.686
0.841 |
5q22 |
| Toll-like receptor 3 (Tlr3) | Rn.15273 | 0.0010 |
F
M |
1.109
3.224 |
1.02
4.05 |
1.038
3.2 |
16q11 |
| Toll-like receptor 7 (Tlr7) | Rn.14819 | 0.0010 |
F
M |
3.82
0.748 |
3.563
0.953 |
1.965
0.665 |
Xq21 |
| Virus-induced signaling adaptor (Visa) | Rn.34996 | 0.0010 |
F
M |
1.59
0.942 |
1.183
0.924 |
1.16
0.905 |
3q36 |
Males have lower type I IFN responses than females during Seoul virus infection and gonadectomy differentially affects these responses
Type I IFNs are critical for restricting virus replication and establishing antiviral immunity, including production of Mx proteins. Interferon β expression was downregulated among intact male rats relative to intact females during Seoul virus infection (F (5,115) = 5.18, p < 0.001; Figure 4A). Removal of the ovaries neonatally reduced expression of IFNβ during the acute phase of infection; conversely, removal of the ovaries in adulthood increased the expression of IFNβ, as compared with intact females, during the persistent phase of infection (F (10,164) = 3.188, p < 0.001; Figure 4C). Seoul virus infection downregulated the expression of IFNβ in all groups of males, but to a lesser extent among males gonadectomized as adults (F (2,188) = 11.19, p < 0.001; Figure 4B).
Figure 4.
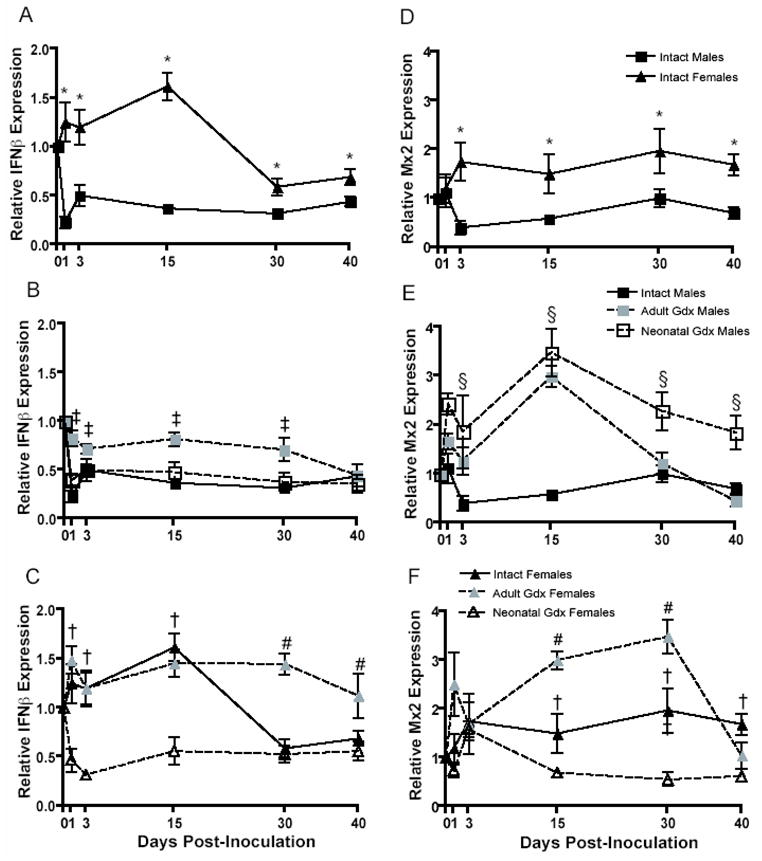
Expression of IFNβ (A–C) and Mx2 (D–F) mRNA (± SEM) in the lungs of intact males (IM), adult gonadectomized males (AGM), neonatally gonadectomized males (NGM), intact females (IF), adult gonadectomized females (AGF), and neonatally gonadectomized females (NGF) collected 0, 1, 3, 15, 30, or 40 days after inoculation with Seoul virus. * IF > IM, ‡ IM < AGM and/or NGM, † IF > AGF and/or NGF, # IF < AGF, p < 0.05, two-way ANOVAs.
Expression of Mx2 was consistently higher among intact female than intact male rats during Seoul virus infection (F (1,108) = 20.08, p < 0.001; Figure 4D). Removal of the testes in males, during either the neonatal period or in adulthood, elevated expression of Mx2 during infection (F (10,186) = 3.28, p < 0.001; Figure 4E). Ovariectomy of females during neonatal development reduced, whereas ovariectomy during adulthood increased, expression of Mx2 among female rats (F (10,159) = 4.21, p < 0.001; Figure 4F).
To examine Mx protein production in relation to Seoul virus NP, fluorescent IHC and western blotting were used on lung samples collected from intact male and intact female rats 0, 3, 15, and 40 days p.i. Seoul virus NP antigen expression was more extensive in lungs collected from intact male than intact female rats 15 and 40 days p.i. (F (3,14) = 1.94, p < 0.001; Figure 5A). Although the number of Mx-producing cells in the lungs increased in both sexes over the course of infection, intact females exhibited higher numbers of Mx-expressing cells than intact males 40 days p.i. (F (3,14) = 2.87, p < 0.001; Figure 5B). During persistent infection (i.e., Day 40 p.i.), intact males had a higher proportion of cells expressing both Seoul virus antigen and Mx than intact females (F (3,14) = 7.21, p < 0.001; Figure 5C). In addition to having more Mx-producing cells in their lungs, intact females had greater amounts of Mx protein in their lungs than intact males 15 and 40 days p.i. (Figure 5D).
Figure 5.

Mx protein production in lung tissues collected 0, 3, 15, or 40 days after inoculation with Seoul virus. Semiquantitative scoring (± SEM) of Seoul virus nucleocapsid protein (SEOV NP; A) and Mx protein (B) examined by IHC. Scores: 0 = no cells present; 1 = 1 cell/visual field; 2 = 2–3 cells/visual field; 3 = 4–10 cells/visual field; 4 = 10+ cells/visual field. Each data point represents the average score from 3 animals/time point and each individual score represents the mean cell count from 4 randomly selected visual fields. Colocalization of SEOV NP and Mx protein in the lungs of intact male (IM) and intact female (IF) rats collected 40 days p.i. (C). Western blot analyses of protein extracts from lung tissues collected 0, 3, 15, or 40 days p.i. or from uninfected rats injected with rIFNα (positive control [+]) incubated with antibody against human MxA, that cross-reacts with rat Mx2 and Mx3 (M143), or β actin (D); results are representative of 3 independent experiments. See Materials and Methods for additional details. * IM > IF, † IF > IM, p < 0.05, two-way ANOVAs (A–B).
The expression of genes along antiviral pathways is upregulated in the lungs of intact females compared with intact males
To further elucidate whether IFN-related responses downstream of PRRs are upregulated in intact females during Seoul virus infection, microarray analyses were conducted using RNA isolated from lung tissue collected 0, 3, 15, and 40 days p.i. Microarray analyses were used on samples from intact males and intact females only. After normalization, approximately 17% of the genes arrayed on the RAE230A Rat GeneChip were differentially expressed between intact males and females during Seoul virus infection, as reported previously (Klein et al., 2004a). Differential expression of genes during Seoul virus infection was observed 3, 15, and 40 days p.i. (Table 2); differences in gene expression levels between the sexes, however, were most pronounced 15 days p.i. (Figure 6).
Figure 6.
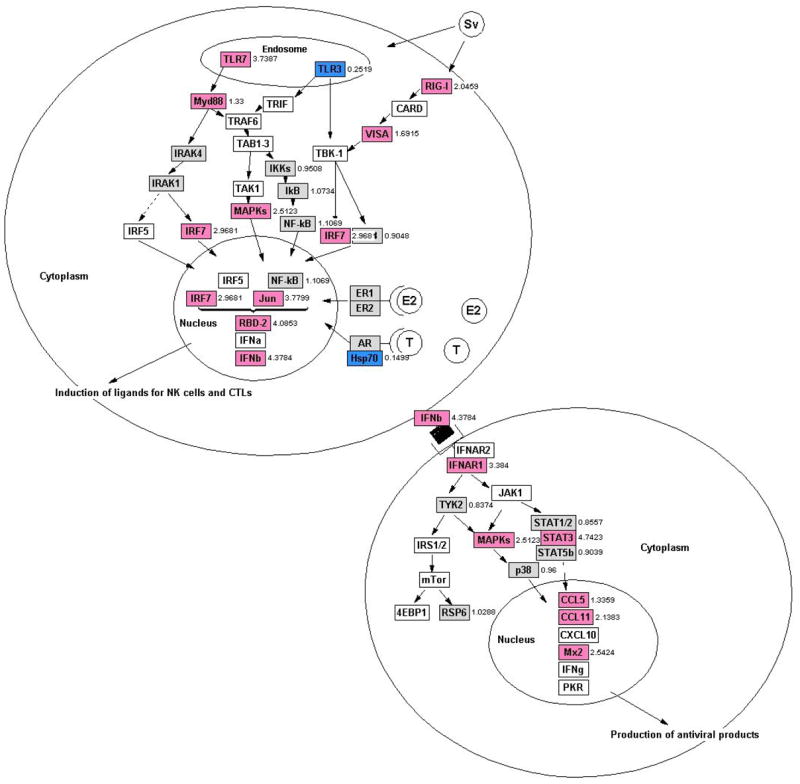
TLR and RIG-I-mediated responses are upregulated in female rats during Seoul virus infection based on integration of microarray and real-time RT-PCR data analyses using GenMAPP 2.1 pathway analysis software, two-way ANOVAs, p < 0.05. Numbers to the right of each gene are fold-change expression in females relative to males during the acute phase of infection (Days 3–15 p.i.). Genes shaded pink are upregulated in females, genes colored blue are upregulated in males, genes shaded gray are not differentially expressed between the sexes, and genes that are white either have raw expression values < 200 units or are not arrayed on the RAE230A GeneChip (Affymetrix; see text for details).
Using GO Biological Processes descriptions, we identified genes associated with innate antiviral signaling pathways that had expression levels that differed significantly between intact males and intact females during infection (p < 0.05 in each case; Table 2 and Figure 6). Neither TLR7, TLR3, nor RIG-I were arrayed on the RAE230A GeneChip; the PRRs on the RAE230A chip, including TLR2, TLR4, and TLR5, were not differentially expressed between the sexes during infection. The expression of several genes associated with TLR3, TLR7, and RIG-I signaling, including Myd88, MAP kinases, Jun, IRF7, and rat β-defensin 2 (RBD-2) was elevated among intact females compared with intact males (p < 0.05 in each case; Table 2 and Figure 6).
Downstream of the TLR signaling pathway, the expression of the type I IFN receptor (IFNAR1), Janus-activated kinase 2 (JAK2), signal transducers and activators of transcription 3 (STAT3), Chemokine ligand (Ccl) 5, Ccl11, and Mx2 was consistenly higher among intact females than intact males (p < 0.05 in each case; Table 2 and Figure 6). As illustrated in Figure 6, several genes along these antiviral signaling pathways were not differentially expressed between males and females (genes shaded gray), had raw expression values < 200 units (genes shaded white; including Ar, Cxcl10, Esr1, Esr2, IFNα, IFNγ, Irak4, Pkr, and 4ebp1), or were not arrayed on the RAE230A GeneChips (genes shaded white; including Irak1, Irf5, Jak1, Mda5, mTor, Tlr9, Trif, Tram, and Traf6).
Putative HREs are present on antiviral genes
Steroid hormones can bind to their respective receptors, that then translocate to the nucleus and bind to specific DNA sequences, called HREs, in the promoter region of hormone-responsive genes, thereby influencing gene transcirption. The extent to which genes associated with antiviral defenses are transcriptionally regulated by sex steroids is not well characterized (but see (Brahmachary et al., 2006; Fox et al., 1991). To test the hypothesis that genes associated with antiviral defenses against hantaviruses are transcriptionally regulated by steroids, we conducted computational analyses to identify HREs (i.e., estrogen response elements (EREs), androgen response elements (AREs), progesterone response elements (PREs), and glucocorticoid response elements (GREs)) in promoters of antiviral genes in rats. We focused our analyses on genes that were differentially expressed between the sexes and/or affected by gonadectomy. For controls, we also included several genes that were not differentially expressed between the sexes, such as the housekeeping genes, α-actin 1, β-actin, and GAPDH, as well as Irf3, while the progesterone receptor we used as a positive control for EREs. Based on our analysis of putative HREs, TLR7, TLR3, Myd88, Jun, Hsp70, IFNAR1, STAT3, and Mx2 contain EREs in their promoters (Table 3). Several genes, including Hsp70, IRF7, Mapk3, and Mx2, contain AREs in their promoters (Table 3). Very few genes (Hsp70, Ifnβ1, Rbd2) along these antiviral pathways contain GREs and none of these genes contain PREs suggesting that these steroids may not be critical for antiviral defenses or they may only affect specific subclasses of genes. No HREs that we analyzed were identified in the promoters of the housekeeping genes or of antiviral genes not differentially expressed between the sexes (e.g., IRF3). We found putative ERE or ARE in 62.5% (10 out of 16) of the differentially expressed genes, but none in the negative control genes. If we also consider GRE with the motivation that glucocorticoids differentially affect immune responses in females and males, then we find 75% (12 out of 16) of the differentially expressed genes contain putative ERE, ARE or GRE, while none of the negative control genes contain these sequences.
Discussion
In the present study, antiviral responses to Seoul virus were higher and the amount of Seoul virus RNA and antigen in the lungs was lower in female than male rats. In particular, the induction of PRRs by Seoul virus differed between the sexes, in which females showed higher expression of TLR7 and RIG-I, but lower expression of TLR3, than males within 72 h p.i. Downstream antiviral responses in the lungs, including the expression of IFNβ and production of Mx2, also were higher in female than male rats during Seoul virus infection. The identification of putative AREs and EREs in the promoters of several of antiviral genes, including TLR3, TLR7, Myd88, IRF7, Jun, Hsp70, IFNAR1, and Mx2, suggests that sex steroids may directly affect dimorphic antiviral responses against infection. Despite the presence of putative HREs on these antiviral genes, the effects of sex hormone deprivation on the expression of IFN-related genes was not uniformly consistent.
Viral nucleic acids are recognized by host cells via TLRs and RNA helicases (Kawai and Akira, 2006). Toll-like receptor 7 mediates production of type I IFNs via Myd88, is critical for initiating antiviral responses against other negative ssRNA viruses, including influenza viruses (Diebold et al., 2004), and is encoded on the X chromosome (Pisitkun et al., 2006). Intact females exhibited higher expression of TLR7 as well as Myd88 in their lungs than did males during Seoul virus infection. Although critical for recognizing dsRNA, a common feature of replicating RNA viruses, the role of TLR3 in regulating IFN responses against RNA viruses is less well characterized than for TLR7 (Edelmann et al., 2004). TLR3 recently has been shown to mediate detrimental inflammation of the bronchoalveolar airspace during influenza infection of male mice (Le Goffic et al., 2006). Intact male rats had higher expression of TLR3 in the lungs than intact females during Seoul virus infection. Whether increased hantavirus replication faciliates higher expression of TLR3 for recognition of dsRNA requires investigation.
Many host cells produce type I IFNs and several transcriptional factors, including IRF3, IRF7, NF-κB, cJun, and AP-1, are involved in the synthesis of type I IFNs (Weber et al., 2004). The elevated expression of IRF7 in the lungs of female rats during infection is of particular interest because IRF7 is necessary for both Myd88-dependent and –independent induction of type I IFNs (Honda et al., 2005). Activation of IFNβ during the acute phase of infection and production of Mx during the persistent phase of infection was enhanced in intact females as compared with intact males. Heightened antiviral responses may ultimately explain why females have reduced numbers of Seoul virus RNA copies (30–40 days p.i.) and expression of Seoul virus antigen (15–40 days p.i.) in their lungs as compared with intact male rats during persistent infection. In addition to modulating expression of ISGs, type I IFNs increase expansion and survival of CD8+ T cells, regulate NK cell and naïve T cell activation, and enhance production of Th1-related cytokines (Kolumam et al., 2005; Thompson et al., 2006; Tough et al., 1996). Recent data from our laboratory illustrate that intact females have more CD8+ T cells in their lungs than intact males during persistent Seoul virus infection (JD Easterbrook and SL Klein, unpublished data).
The amount of Seoul virus RNA in target organs for virus replication as well as shed in saliva differed between the sexes. Males shed more viral RNA than females during the acute phase of infection. In the lungs, there was a temporal relationship between viral RNA loads and sex, such that females had more viral RNA than males during the acute phase of infection, whereas this sex difference was reversed during the persistent phase of infection. Recent data from our laboratory further indicate that male rats have detectable Seoul virus RNA in their lungs for at least 60 days p.i., suggesting that males are not clearing virus at a later time-point p.i. (J.D. Easterbrook and S.L. Klein, unpublished data). Although males had lower viral RNA copies than females 15 days p.i., the number of cells containing viral antigen was higher in males than females at 15 days p.i. The dissociation between viral RNA and antigen loads 15 days p.i. in males may indicate that the activity of the viral RNA-dependent RNA polymerase encoded by the L segment of the virus is more efficient in males as compared with females. Thus, the dynamics of Seoul virus replication may be sexually dimorphic.
Our data provide evidence for and against a role for sex steroids in mediating sex differences in antiviral defenses and Seoul virus persistence and shedding in Norway rats. Gonadectomy of males, during either neonatal development or in adulthood, reduced the amount of virus shed in saliva and detectable in the lungs during persistent infection. Although sex hormone deprivation in females did not affect viral RNA load in tissue, removal of the ovaries, either neonatally or in adulthood, increased the amount of virus shed in saliva during the acute phase of infection. Because manipulation of sex steroids either during sexual differentiation or after puberty resulted in similar effects on the amount of detectable Seoul virus RNA, sex differences in response to Seoul virus may be caused by the effects of sex steroids on antiviral defenses at the time of infection (i.e., activation effects) and not during earlier stages of development (i.e., organizational effects). Additionally, removal of the testes during neonatal development did not affect the expression of several genes associated with type I IFNs, including TLR7, RIG-I, VISA, and IFNβ, as compared with the effects of gonadectomy in adulthood. Thus, compensatory processes during development may have mitigated the effects of neonatal gonadectomy on responses to Seoul virus later in life. These observations are in contrast to our previous report that neonatal, but not adult, gonadectomy of males reversed antibody responses against Seoul virus (Klein et al., 2002). Based on our data, the timing of hormone deprivation may differentially affect innate and adaptive immunity. The hypothesis that the immune system can be directly influenced by early sex hormone exposure to affect responses to infection later in life has not been adequately tested and requires additional investigation (Martin, 2000)
Sex hormone deprivation consistently had more robust effects on the induction of PRRs than on downstream type I IFNs or ISGs. Thus, sex steroids may have their greatest impact on the initial recognition of hantaviruses by host cells. Gonadectomy of females had a greater effect on the expression of TLR7 than did gonadectomy of males, which may reflect the presence of a putative ERE in the promoter of TLR7. Our identification of putative EREs and AREs in the promoters of 62.5% of the differentially expressed antiviral genes, including TLR3, TLR7, Myd88, IRF7, Jun, Hsp70, IFNAR1, and Mx2, but not in the negative control genes, suggests that these antiviral genes can be transcriptionally regulated by sex steroids. Conversely, neither RIG-I nor VISA contain HREs in their promoters, suggesting that sex differences and the effects of gonadectomy on the expression of these genes may be caused by indirect effects of sex steroids. Because hormone replacement was not conducted in the present study, these data leave open the possibility that there may be sex steroid independent effects on antiviral responses in males and females. Seoul virus infection of primary cell lines from rats is being utilized to address sex steroid-specific effects on intracellular signaling and antiviral defenses. Another testable hypothesis is that sex steroids may directly affect Seoul virus replication and shedding, independent of their effects on host immunity, as has been demonstrated for glucocorticoids and Epstein-Barr virus (Cacioppo et al., 2002; Glaser et al., 1995).
Transmission of zoonotic pathogens, including hantaviruses, to humans requires contact with infectious reservoir hosts, their excrement, or other secretions. Hantaviruses are shed in saliva, feces, and urine and one prominent mode of horizontal transmission is through the passage of virus in saliva during aggressive encounters (Glass et al., 1988; Hinson et al., 2004). Our data collectively illustrate that male rodents shed more virus in saliva, are more aggressive, and are more likely to be infected with hantaviruses than females (Easterbrook et al., 2007; Hinson et al., 2004; Klein et al., 2000, 2001). The data from the present study further illustrate that induction of PRRs and antiviral responses are down-regulated in male rats during infection. Reduced antiviral defenses, increased virus shedding, and the propensity to engage in heightened aggression may make males better transmitters of hantaviruses than females.
Acknowledgments
We thank Judith Easterbrook and Jenifer Kaplan for laboratory assistance; Greg Glass, Diane Griffin, and Richard Johnson for helpful comments on an earlier draft; Anne Jedlicka and Alan Scott for assistance with the microarray experimentation and analyses, which were provided by the Johns Hopkins Malaria Research Institute. Financial support for this study was provided by NIH R01 AI054995 (SLK). VBB was supported by NRF grants FA2006040900002 and ICD2006071000003. The authors have no conflicting financial interests.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alff PJ, Gavrilovskaya IN, Gorbunova E, Endriss K, Chong Y, Geimonen E, Sen N, Reich NC, Mackow ER. The pathogenic NY-1 hantavirus G1 cytoplasmic tail inhibits RIG-I- and TBK-1-directed interferon responses. J Virol. 2006;80:9676–9686. doi: 10.1128/JVI.00508-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen LJ, McCormack RK, Jonsson CB. Mathematical models for hantavirus infection in rodents. Bull Math Biol. 2006;68:511–524. doi: 10.1007/s11538-005-9034-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold AP, Breedlove SM. Organizational and activational effects of sex steroids on brain and behavior: a reanalysis. Horm Behav. 1985;19:469–498. doi: 10.1016/0018-506x(85)90042-x. [DOI] [PubMed] [Google Scholar]
- Botten J, Mirowsky K, Kusewitt D, Ye C, Gottlieb K, Prescott J, Hjelle B. Persistent Sin Nombre virus infection in the deer mouse (Peromyscus maniculatus) model: sites of replication and strand-specific expression. J Virol. 2003;77:1540–1550. doi: 10.1128/JVI.77.2.1540-1550.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brabin L. Interactions of the female hormonal environment, susceptibility to viral infections, and disease progression. AIDS Patient Care STDS. 2002;16:211–221. doi: 10.1089/10872910252972267. [DOI] [PubMed] [Google Scholar]
- Brahmachary M, Schonbach C, Yang L, Huang E, Tan SL, Chowdhary R, Krishnan SP, Lin CY, Hume DA, Kai C, Kawai J, Carninci P, Hayashizaki Y. [Google Scholar]
- Bajic VB. Computational promoter analysis of mouse, rat and human antimicrobial peptide-coding genes. BMC Bioinformatics. 2006;7(Suppl 5):S8. doi: 10.1186/1471-2105-7-S5-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacioppo JT, Kiecolt-Glaser JK, Malarkey WB, Laskowski BF, Rozlog LA, Poehlmann KM, Burleson MH, Glaser R. Autonomic and glucocorticoid associations with the steady-state expression of latent Epstein-Barr virus. Horm Behav. 2002;42:32–41. doi: 10.1006/hbeh.2002.1801. [DOI] [PubMed] [Google Scholar]
- Childs JE, Glass GE, Korch GW, LeDuc JW. The ecology and epizootiology of hantaviral infections in small mammal communities of Baltimore: A review and synthesis. Bulletin of the Society of Vector Biology. 1988;13:113–122. [Google Scholar]
- Diebold SS, Kaisho T, Hemmi H, Akira S, Reis e Sousa C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 2004;303:1529–1531. doi: 10.1126/science.1093616. [DOI] [PubMed] [Google Scholar]
- Douglass RJ, Calisher CH, Wagoner KD, Mills JN. Sin nombre virus infection of deer mice in montana: characteristics of newly infected mice, incidence, and temporal pattern of infection. J Wildl Dis. 2007;43:12–22. doi: 10.7589/0090-3558-43.1.12. [DOI] [PubMed] [Google Scholar]
- Easterbrook JD, Kaplan JB, Glass GE, Pletnikov MV, Klein SL. Elevated testosterone and reduced 5-HIAA concentrations are associated with wounding and hantavirus infection in male Norway rats. Horm Behav. 2007 doi: 10.1016/yhbeh.2007.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelmann KH, Richardson-Burns S, Alexopoulou L, Tyler KL, Flavell RA, Oldstone MB. Does Toll-like receptor 3 play a biological role in virus infections? Virology. 2004;322:231–238. doi: 10.1016/j.virol.2004.01.033. [DOI] [PubMed] [Google Scholar]
- Fox HS, Bond BL, Parslow TG. Estrogen regulates the IFN-gamma promoter. J Immunol. 1991;146:4362–4367. [PubMed] [Google Scholar]
- Glaser R, Kutz LA, MacCallum RC, Malarkey WB. Hormonal modulation of Epstein-Barr virus replication. Neuroendocrinology. 1995;62:356–361. doi: 10.1159/000127025. [DOI] [PubMed] [Google Scholar]
- Glass GE, Childs JE, Korch GW, LeDuc JW. Association of intraspecific wounding with hantaviral infection in wild rats (Rattus norvegicus) Epidemiol Infect. 1988;101:459–472. doi: 10.1017/s0950268800054418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haan C, Behrmann I. A cost effective non-commercial ECL-solution for Western blot detections yielding strong signals and low background. J Immunol Methods. 2007;318:11–19. doi: 10.1016/j.jim.2006.07.027. [DOI] [PubMed] [Google Scholar]
- Hinson ER, Shone SM, Zink MC, Glass GE, Klein SL. Wounding: the primary mode of Seoul virus transmission among male Norway rats. Am J Trop Med Hyg. 2004;70:310–317. [PubMed] [Google Scholar]
- Hjelle B, Yates T. Modeling hantavirus maintenance and transmission in rodent communities. Curr Top Microbiol Immunol. 2001;256:77–90. doi: 10.1007/978-3-642-56753-7_5. [DOI] [PubMed] [Google Scholar]
- Honda K, Yanai H, Negishi H, Asagiri M, Sato M, Mizutani T, Shimada N, Ohba Y, Takaoka A, Yoshida N, Taniguchi T. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. 2005;434:772–777. doi: 10.1038/nature03464. [DOI] [PubMed] [Google Scholar]
- Hutchinson KL, Rollin PE, Peters CJ. Pathogenesis of a North American hantavirus, Black Creek Canal virus, in experimentally infected Sigmodon hispidus. Am J Trop Med Hyg. 1998;59:58–65. doi: 10.4269/ajtmh.1998.59.58. [DOI] [PubMed] [Google Scholar]
- Jacobson DL, Gange SJ, Rose NR, Graham NM. Epidemiology and estimated population burden of selected autoimmune diseases in the United States. Clin Immunol Immunopathol. 1997;84:223–243. doi: 10.1006/clin.1997.4412. [DOI] [PubMed] [Google Scholar]
- Jin HK, Yoshimatsu K, Takada A, Ogino M, Asano A, Arikawa J, Watanabe T. Mouse Mx2 protein inhibits hantavirus but not influenza virus replication. Arch Virol. 2001;146:41–49. doi: 10.1007/s007050170189. [DOI] [PubMed] [Google Scholar]
- Kawai T, Akira S. Innate immune recognition of viral infection. Nat Immunol. 2006;7:131–137. doi: 10.1038/ni1303. [DOI] [PubMed] [Google Scholar]
- Kel AE, Gossling E, Reuter I, Cheremushkin E, Kel-Margoulis OV, Wingender E. MATCH: A tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res. 2003;31:3576–3579. doi: 10.1093/nar/gkg585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaiboullina SF, Rizvanov AA, Deyde VM, St Jeor SC. Andes virus stimulates interferon-inducible MxA protein expression in endothelial cells. J Med Virol. 2005;75:267–275. doi: 10.1002/jmv.20266. [DOI] [PubMed] [Google Scholar]
- Klein SL. The effects of hormones on sex differences in infection: from genes to behavior. Neurosci Biobehav Rev. 2000;24:627–638. doi: 10.1016/s0149-7634(00)00027-0. [DOI] [PubMed] [Google Scholar]
- Klein SL, Bird BH, Glass GE. Sex differences in Seoul virus infection are not related to adult sex steroid concentrations in Norway rats. J Virol. 2000;74:8213–8217. doi: 10.1128/jvi.74.17.8213-8217.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein SL, Bird BH, Glass GE. Sex differences in immune responses and viral shedding following Seoul virus infection in Norway rats. Am J Trop Med Hyg. 2001;65:57–63. doi: 10.4269/ajtmh.2001.65.57. [DOI] [PubMed] [Google Scholar]
- Klein SL, Calisher CH. Emergence and persistence of hantaviruses. Current Topics in Microbiology and Immunology. 2007;315:217–252. doi: 10.1007/978-3-540-70962-6_10. [DOI] [PubMed] [Google Scholar]
- Klein SL, Cernetich A, Hilmer S, Hoffman EP, Scott AL, Glass GE. Differential expression of immunoregulatory genes in male and female Norway rats following infection with Seoul virus. J Med Virol. 2004a;74:180–190. doi: 10.1002/jmv.20163. [DOI] [PubMed] [Google Scholar]
- Klein SL, Marson AL, Scott AL, Ketner G, Glass GE. Neonatal sex steroids affect responses to Seoul virus infection in male but not female Norway rats. Brain Behav Immun. 2002;16:736–746. doi: 10.1016/s0889-1591(02)00026-0. [DOI] [PubMed] [Google Scholar]
- Klein SL, Zink MC, Glass GE. Seoul virus infection increases aggressive behaviour in male Norway rats. Animal Behaviour. 2004b;67:421–429. [Google Scholar]
- Kolumam GA, Thomas S, Thompson LJ, Sprent J, Murali-Krishna K. Type I interferons act directly on CD8 T cells to allow clonal expansion and memory formation in response to viral infection. J Exp Med. 2005;202:637–650. doi: 10.1084/jem.20050821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Le Goffic R, Balloy V, Lagranderie M, Alexopoulou L, Escriou N, Flavell R, Chignard M, Si-Tahar M. Detrimental contribution of the Toll-like receptor (TLR)3 to influenza A virus-induced acute pneumonia. PLoS Pathog. 2006;2:e53. doi: 10.1371/journal.ppat.0020053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin JT. Sexual dimorphism in immune function: the role of prenatal exposure to androgens and estrogens. Eur J Pharmacol. 2000;405:251–261. doi: 10.1016/s0014-2999(00)00557-4. [DOI] [PubMed] [Google Scholar]
- Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, Voss N, Stegmaier P, Lewicki-Potapov B, Saxel H, Kel AE, Wingender E. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchjorsen J, Jensen SB, Malmgaard L, Rasmussen SB, Weber F, Bowie AG, Matikainen S, Paludan SR. Activation of innate defense against a paramyxovirus is mediated by RIG-I and TLR7 and TLR8 in a cell-type-specific manner. J Virol. 2005;79:12944–12951. doi: 10.1128/JVI.79.20.12944-12951.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann M, Gabel D. Simple method for reduction of autofluorescence in fluorescence microscopy. J Histochem Cytochem. 2002;50:437–439. doi: 10.1177/002215540205000315. [DOI] [PubMed] [Google Scholar]
- Pisitkun P, Deane JA, Difilippantonio MJ, Tarasenko T, Satterthwaite AB, Bolland S. Autoreactive B cell responses to RNA-related antigens due to TLR7 gene duplication. Science. 2006;312:1669–1672. doi: 10.1126/science.1124978. [DOI] [PubMed] [Google Scholar]
- Temonen M, Lankinen H, Vapalahti O, Ronni T, Julkunen I, Vaheri A. Effect of interferon-alpha and cell differentiation on Puumala virus infection in human monocyte/macrophages. Virology. 1995;206:8–15. doi: 10.1016/s0042-6822(95)80014-x. [DOI] [PubMed] [Google Scholar]
- Theofilopoulos AN, Baccala R, Beutler B, Kono DH. Type I interferons (alpha/beta) in immunity and autoimmunity. Annu Rev Immunol. 2005;23:307–336. doi: 10.1146/annurev.immunol.23.021704.115843. [DOI] [PubMed] [Google Scholar]
- Thompson LJ, Kolumam GA, Thomas S, Murali-Krishna K. Innate inflammatory signals induced by various pathogens differentially dictate the IFN-I dependence of CD8 T cells for clonal expansion and memory formation. J Immunol. 2006;177:1746–1754. doi: 10.4049/jimmunol.177.3.1746. [DOI] [PubMed] [Google Scholar]
- Tough DF, Borrow P, Sprent J. Induction of bystander T cell proliferation by viruses and type I interferon in vivo. Science. 1996;272:1947–1950. doi: 10.1126/science.272.5270.1947. [DOI] [PubMed] [Google Scholar]
- Weber F, Kochs G, Haller O. Inverse interference: how viruses fight the interferon system. Viral Immunol. 2004;17:498–515. doi: 10.1089/vim.2004.17.498. [DOI] [PubMed] [Google Scholar]
- Wizemann TM, Pardue M, editors. Exploring the biological contributions to human health: does sex matter? National Academy Press; Washington DC: 2001. [PubMed] [Google Scholar]


