Abstract
Introduction
Developmental vitamin D (DVD) deficiency is a candidate risk factor for schizophrenia. Animal models have confirmed that DVD deficiency is associated with a range of altered genomic, proteomic, structural and behavioural outcomes in the rat. Because the nucleus accumbens has been implicated in neuropsychiatric disorders, in the current study we examined protein expression in this region in adult rats exposed to DVD deficiency
Methods
Female Sprague Dawley rats were maintained on a vitamin D deficient diet for 6 weeks, mated and allowed to give birth, after which a diet containing vitamin D was reintroduced. Male adult offspring (n = 8) were compared to control male (n = 8). 2-D gel electrophoresis-based proteomics and mass spectroscopy were used to investigate differential protein expression.
Results
There were 35 spots, mapped to 33 unique proteins, which were significantly different between the two groups. Of these, 22 were down-regulated and 13 up-regulated. The fold changes were uniformly small, with the largest FC being −1.67. Within the significantly different spots, three calcium binding proteins (calbindin1, calbindin2 and hippocalcin) were altered. Other proteins associated with DVD deficiency related to mitochondrial function, and the dynamin-like proteins.
Conclusions
Developmental vitamin D deficiency was associated with subtle changes in protein expression in the nucleus accumbens. Disruptions in pathways related to calcium-binding proteins and mitochondrial function may underlie some of the behavioural features associated with animal models of developmental vitamin D deficiency
Introduction
Based on clues from epidemiology, we have proposed that low prenatal vitamin D may be a risk factor for later development of schizophrenia [1]. Many studies have shown that those born in winter and spring have a significantly increased risk of developing schizophrenia [2], and that those born at higher latitudes are also at increased risk of schizophrenia [3]. Given that vitamin D levels in the population fluctuate across the seasons and decrease across higher latitude [4], low prenatal vitamin D ‘fits’ these key environmental features and is therefore a plausible candidate risk factor for this disease. In order to explore the biological plausibility of this candidate, we have a rat model that we call the Developmental Vitamin D (DVD) model.
Rats exposed to low prenatal vitamin D have a broad range of neurobiological outcomes that are informative for schizophrenia research. Briefly, DVD-deficient neonates had larger lateral ventricles, increased cellular proliferation and reduced apoptosis, altered neurogenesis, reduced density of neurotrophin receptor (p75NTR), and reduced levels of nerve growth factor (NGF) and glial cell line-derived growth factor (GDNF) compared to controls [5]–[7]. As adults these animals had larger lateral ventricles and reduced NGF expression compared to controls [8]. Behaviourally, adult DVD-deficient rats were more active than controls (i.e. showing “hyperlocomotion”) [9], [10]. 5) DVD-deficient rats also have altered attentional processing indicated by impaired latent inhibition [11]. Some of the most robust and consistent findings in the DVD model have emerged in behavioural pharmacological studies. For example, we have shown that DVD-deficient rats have enhanced locomotion in response to the psychomimetic agents such as NMDA antagonist MK-801 [10], [12]. The DVD adult rat is also more sensitive to haloperidol, a dopamine (DA) D2 receptor antagonist [10], that is a widely used antipsychotic medication used to treat schizophrenia. Dysfunction of DA signalling has been strongly implicated in the pathogenesis of schizophrenia [13]. Dopamine projections involve a range of cortical and subcortical regions, however its role in the nucleus accumbens has been of particular interest with respect to neuropsychiatric disorders [14]–[16]. In the nucleus accumbens, dopamine influences the integration of inputs from the ventral hippocampus, the amygdala, and the prefrontal cortex. Grace and colleagues have suggested that dopamine may modulate a range of limbic and cortical functions relevant to the pathophysiology of schizophrenia via the nucleus accumbens [17].
Previously we explored the genomic and proteomic characteristics of frontal cortex and the hippocampus in the adult DVD rat [18], [19]. In particular, a proteomic study based on two cortical regions of DVD-deficient rats (frontal cortex and hippocampus), identified 36 dysregulated proteins. These proteins are associated with several biological pathways including oxidative phosphorylation, cytoskeleton maintenance, calcium homeostasis, chaperoning, synaptic plasticity and neurotransmission. A computational analysis of these data revealed that many of the proteins dysregulated in the DVD model have also been shown to be altered in schizophrenia post-mortem brain studies [18].
In order to further explore the impact of DVD on brain function, we undertook a proteomic study of the nucleus accumbens.
Methods
To obtain vitamin D3 depletion, female Sprague-Dawley rats (Herston Animal Facility, Queensland, Australia) were kept on a vitamin D deficient diet (Dyets Inc., PA, USA). Animals were housed on a 12-h light/dark cycle (lights on at 06:00 h) using incandescent lighting, to avoid ultraviolet radiation within the vitamin D3 action spectrum. These conditions were maintained for six weeks prior to mating and throughout gestation. Control animals were kept under similar conditions except they received a vitamin D replete diet (Dyets, PA, USA and Specialty Foods, WA, Australia) and were housed under standard lighting conditions. Dams (and corresponding litters) were placed under control conditions for the remainder of the experiment. The male pups were weaned on postnatal day 21 and housed in groups of 3–6. Female rats were not used in these experiments, because the estrous cycle can introduce variability in protein expression in the region of interest [20], [21]. All procedures were performed with approval from the Queensland University Animal Ethics Committee, under the guidelines of the National Health and Medical Research Council of Australia.
Sample Preparation
Tissue was sampled from 8 control and 8 DVD deficient male adult offspring. These animals were from four separate litters per experimental group. The nucleus accumbens (NAc) were sampled from all animals according to boundaries determined from a standard rat brain atlas [22]. Sample preparation and two dimensional gel electrophoresis (2DE) methodology was performed according to Alexander-Kaufman et al [23]. Briefly, 0.04 g–0.07 g of crude fresh frozen NAc tissue was placed in Buffer 1 (7M Urea, 2M Thiourea, 1% C7bZO, 40 mM Tris). Sample suspensions were sonicated 3×10 sec at 40% intensity and centrifuged at 14,000× g for 20 min at 15°C. The supernatant was reduced and alkylated in 5 mM tributylphosphine (TBP) and 1M acrylamide monomer at room temperature for 2 hr. The reaction was quenched by adding 10 mM DTT followed by 20 mg citric acid to adjust pH to approximately pH 6.0. Samples were precipitated using 5 volumes of room temperature acetone for 10 min and centrifuged at 3,500× g for 15 min at 15°C. The pellet was air dried for 5 min and resuspended in Buffer 2 (7M urea, 2M thiourea, 1% C7bZO).
Two Dimensional Gel Electrophoresis
Protein concentration was determined by the Bradford method [24]. The 8 DVD deficient and 8 control sample tissues were used to perform duplicate 2DE analyses, providing a total of 16 gels for each group. Pre-cast immobilised pH gradient strips (IPG, 11 cm, pH 3–10, Proteome Systems, North Ryde, Australia) were passively rehydrated in 200 µg sample protein extract for 6 hr at room temperature. In the first dimension, rehydrated strips were focused using an ElectrophoretIQ3 isoelectric focusing system (Proteome Systems) for a total of 120 kVh. IPG strips were reduced, alkylated and detergent exchanged using SDS equilibration buffer (Proteome Systems, 20 min) and loaded onto pre-cast SDS-PAGE gels (GelChip™ 2D, 6–15%, 10×15 cm; Proteome Systems) for second dimension molecular mass separation using the ElectrophoreticIQ3 system (50mA/gel, 15°C for 90 min).
Image acquisition and analysis
Gels were fixed in solution containing 25% (v/v) methanol and 10% (v/v) acetic acid for 1 hr and stained using colloidal Coomassie Blue for spot visualization. Gels were scanned using a transmissive, flatbed scanner (UMAX) and analyzed using Phoretix 2D Expression software (Nonlinear Dynamics, Newcastle-upon-Tyne, UK). Following background subtraction and volume normalization of all gels, average gels were created for each group to assist comparison and reduce within group variations. Averaging parameters were set at 70%, therefore for a protein spot to appear in the averaged gel it must be present in 70% of all gels within a group. One-way Analysis of Variance (ANOVA) statistical tests were used to reveal statistically significant protein expression differences between the two groups (p<0.05). Protein spots that were significantly altered were excised for identification by mass spectrometry.
Mass Spectrometry
Excised spots were washed in 50 mM ammonium bicarbonate/acetonitrile (60∶40 solution) for 1 hr at room temperature. Spots were dried in a Vacuum Concentrator (Eppendorf, Hamburg Germany) for 25 min and rehydrated at 4°C in tryptic digest solution (10 ng/µl porcine sequencing grade trypsin (Promega) in 50 mM NH4HCO3) for 1 h. Remaining tryptic digest solution was removed and gel pieces suspended overnight at 37°C in 50 mM NH4HCO3.
For protein identification, approximately 0.8 µl of the peptide mixture was spotted onto a target plate and covered with the same volume of matrix solution (α-cyano-4-hydroxy cinnamic acid (Sigma), 8 mg/ml in 70% (v/v) acetonitrile/1% (v/v) formic acid) and allowed to air dry. In several cases, peptides were concentrated and desalted using C18 Perfect Pure Tips (Eppendorf). Tips were activated with acetonitrile and washed with 5×10 µl of 1% (v/v) formic acid. The peptide mixture was then bound and aspirated 5 times through the column and bound peptides washed with 5×10 µl of 1% formic acid. Peptides were eluted in 0.8 µl of matrix solution directly onto a MALDI-TOF target plate. Peptide mass maps of tryptic peptides were generated by matrix assisted laser desorption/ionisation time-of-flight mass spectrometry (MALDI-TOF MS) using an Applied Biosystems Q-STAR Pulsar with MALDI source (APAF, University of Sydney). Mass calibration was performed using trypsin autolysis peaks, 2211.11 Da and 842.51 Da as internal standards.
Database searching and secondary analyses
Data generated from peptide mass mapping (PMM) of each spot were used to perform searches of the SWISS-PROT, NCBI and TrEMBL databases using the programs Aldente (www.expasy.ch) and MASCOT (www.matrixscience.com). Identifications were based on the observed pI and Mr (kDa) of the matched protein, the number of matching peptide masses and the total percentage of the amino acid sequence that those peptides covered, in comparison to other database entries. Generally, a peptide match with at least 30% total sequence coverage was required for confidence in identification, but very low and high mass protein, and those resulting from protein fragmentation may not always meet the criterion. For searches performed using MASCOT, E-value and Score, as well as matching peptides and sequence coverage, were used to determine matches.
Although many proteins were found to be significantly altered in DVD-deficient adults, the fold-change level was insufficient for confirmation analysis by western blot. Bioinformatics was employed as secondary analyses. Depending of the size of the fold changes, we planned to use western blot and/or bioinformatics as secondary analyses. Significantly dysregulated proteins were examined in bioinformatic pathways analysis (Ingenuity Pathway Analysis [IPA]; Ingenuity Systems, Mountain View, CA). This manually-curated database builds hypothetical networks based on the candidate proteins and other potentially associated proteins in the database. For each pathway, scores are calculated as the negative base-10 logarithm of the P value, indicating the likelihood that the dysregulated proteins would be found in a given network by chance. Finally, proteomic analyses are prone to Type I errors. While we did not adjust the p value for the number of comparisons undertaken, we report the false discovery rate, which provides an estimate of the proportion of significantly different proteins (p<0.05) that may be truly null [25], [26].
Results
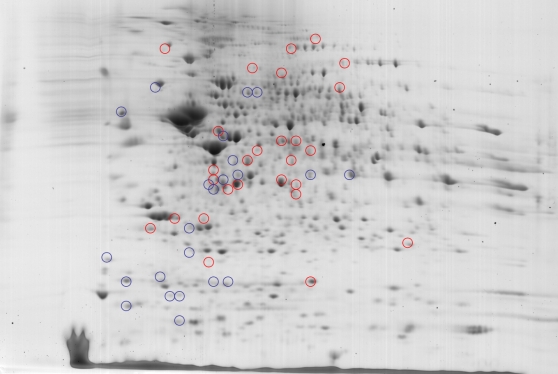
We identified 637 spots in DVD deplete samples and 655 spots in Control samples (see Figure 1 for a representative gel). These spots were matched, normalized and quantified. For each animal, two gels were averaged. There were 35 spots that were significantly altered between the two groups, 22 were down-regulated and 13 up-regulated (Table 1). These spots were mapped to 33 unique proteins. The fold changes were uniformly small, with the largest FC being −1.67 (MEPD). Over half (27 of 49) of the significantly dysregulated spots that were identified as a known protein had a fold change of less than 1.3. Based on a False Discovery Rate of 28%, we predict that nine of these spots would be false positives.
Figure 1. Altered proteins in the nucleus accumbens in adult DVD-deficient and control male rats.
Red circles indicate increased spots, blue circles indicate spots reduced in the average gel in DVD animals.
Table 1. Summary of differentially expressed protein spots in nucleus accumbens tissue from adult DVD-deficient and control rats.
| Fold change | p value | Protein | Uniprot Accession Number | Mowse score | Molecular weight | pH |
| Calcium Binding Proteins | ||||||
| 1.14 | 0.031 | Calreticulin CALR | P18418 | 114 | 47966 | 4.33 |
| -1.25 | 0.050 | Calbindin (calretinin) CALB2 | P47728 | 156 | 31384 | 4.94 |
| -1.18 | 0.045 | Calbindin CALB1 | P07171 | 69 | 29975 | 4.71 |
| 1.17 | 0.032 | Hippocalcin HPCA | P84076 | 79 | 22413 | 4.87 |
| General cellular metabolism proteins | ||||||
| 1.47 | 0.026 | L-lactate dehydrogenase B chain LDHB | P42123 | 119 | 36589 | 5.70 |
| 1.23 | 0.004 | Aldose reductase ALDR | P07943 | 154 | 35774 | 6.26 |
| -1.11 | 0.014 | crystallin, mu CRYM | Q9QYU4 | 96 | 33533 | 5.34 |
| -1.09 | 0.030 | Malate dehydrogenase, cytoplasmic MDHC | O88989 | 69 | 36460 | 6.16 |
| -1.35 | 0.030 | Glycerol-3-phosphate dehydrogenase [NAD+], cytoplasmic GPDA | O35077 | 141 | 37428 | 6.16 |
| -1.17 | 0.043 | Glutathione S-transferase P GSTP1 | P04906 | 78 | 23424 | 6.89 |
| Mitochondria Proteins | ||||||
| -1.54 | 0.041 | Hexokinase 1 HXK1 | P05708 | 76 | 102342 | 6.29 |
| -1.35 | 0.048 | Dynamin 1-like protein DNM1L | O35303 | 106 | 83856 | 6.64 |
| -1.12 | 0.046 | Ubiquinol-cytochrome-c reductase complex core protein 1, UQCR1 | Q68FY0 | 145 | 52815 | 5.57 |
| -1.14 | 0.033 | Isocitrate dehydrogenase (NAD) subunit alpha, mitochondrial IDH3A | Q99NA5 | 113 | 39588 | 6.47 |
| 1.16 | 0.037 | IDH3A | Q99NA5 | 117 | . | . |
| -1.37 | 0.011 | NADH dehydrogenase 1 alpha subcomplex subunit 10, NDUAA | Q561S0 | 219 | 40468 | 7.64 |
| -1.10 | 0.039 | Pyruvate dehydrogenase E1 component subunit beta, ODPB | P49432 | 95 | 38957 | 6.20 |
| -1.18 | 0.033 | Voltage-dependent anion-selective channel protein 2 VDAC2 | P81155 | 59 | 31726 | 7.44 |
| Signal transduction and MAP-related proteins | ||||||
| -1.28 | 0.007 | Mitogen-activated protein kinase 1 (ERK-2; MAPK 2) MK01 | P63086 | 57 | 41249 | 6.50 |
| -1.24 | 0.009 | Serine/threonine-protein phosphatase 2A catalytic subunit beta isoform PP2AB | P62716 | 107 | 35552 | 5.21 |
| 1.23 | 0.036 | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta 1 GBB1 | P54311 | 64 | 37353 | 5.60 |
| -1.15 | 0.031 | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta 2 GBB2 | P54313 | 74 | 37307 | 5.60 |
| -1.24 | 0.029 | Dynamin 1 DYN1 | P21575 | 152 | 95867 | 6.32 |
| -1.67 | 0.047 | Thimet oligopeptidase MEPD | P24155 | 112 | 78264 | 5.54 |
| -1.22 | 0.006 | Dihydropyrimidinase-related protein 2 (DRP-2; CRMP-2) DPYL2 | P47942 | 101 | 62239 | 5.95 |
| 1.17 | 0.038 | DPYL2 | P47942 | 53 | . | . |
| 1.24 | 0.007 | DPYL2 | P47942 | 85 | . | . |
| -1.21 | 0.024 | Syntaxin-binding protein 1 STXB1 | P61765 | 91 | 67526 | 6.49 |
| -1.25 | 0.036 | Adenosylhomocysteinase SAHH | P10760 | 93 | 47507 | 6.07 |
| 1.31 | 0.013 | Ras-related protein Rab-3C RAB3C | P62824 | 64 | 25856 | 5.10 |
| 1.14 | 0.003 | Beta-synuclein SYUB | Q63754 | 56 | 14495 | 4.48 |
| 1.17 | 0.013 | Myosin light polypeptide 6 MYL6 | Q64119 | 54 | 16964 | 4.46 |
| Proteins not otherwise classified | ||||||
| 1.48 | 0.011 | Eukaryotic initiation factor 4A-II IF4A2 | Q5RKI1 | 90 | 46373 | 5.33 |
| 1.19 | 0.001 | Glyceraldehyde-3-phosphate dehydrogenase G3P | P04797 | 78 | 35805 | 8.14 |
| -1.15 | 0.047 | ADP-ribosylation factor 1 ARF1 | P84079 | 128 | 20684 | 6.32 |
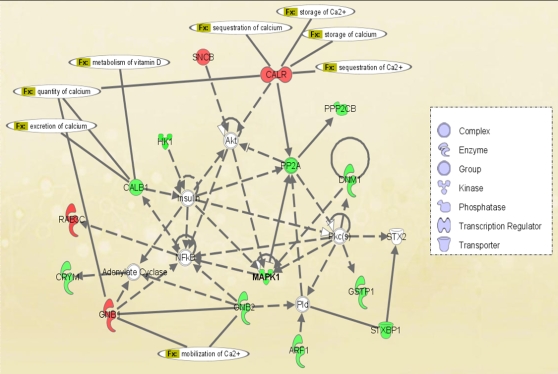
Ingenuity Pathway Analysis identified two major networks; (a) Cellular Movement, cellular assembly and organization, cell signaling; and (b) Protein synthsis, RNA Post-transcriptional Modification, Cancer. These pathways had scores of 32 and 29 respectively (both highly significant). Figure 2 shows the first pathway, annotated for functions related to calcium buffering. Several of the components of the network converge on MAPK1, a MAP kinase that serves to integrate multiple biochemical signals (previously known as ERK2), that was found to be significantly down-regulated in the proteomics study.
Figure 2. Network analysis of the proteins assembled by Ingenuity Pathway Analysis.
Proteins symbols in red were up-regulated, while green were down-regulated. Hub proteins not significantly dysregulated in the study are shown as clear. The proteins with calcium-related functions are annotated (Fx).
Discussion
Developmental vitamin D deficiency is associated with a subtle alteration in the expression of protein involved in functions related to calcium binding proteins, and mitochondrial functioning.
Calcium binding proteins have been of interest to schizophrenia research for some time [27], in particular with respect to the expression in cortical GABAergic interneurons [28]. This study found that four calcium binding proteins were significantly altered in the nucleus accumbens of the adult DVD-deficient rat (calbindin, calbindin2, hippocalcin and calreticulin). Calcium binding proteins are central to a wide range of cellular functions, of which calcium sequestration and buffering are particularly important for neurons. Amongst other functions, it is thought that this family of diffuse cytoplasmic proteins provides a ‘sink’ allowing cells to rapidly buffer intracellular calcium after actions potentials, thus allowing the cell to rapidly repolarise for further firing. Calbindin is strongly induced by vitamin D [29], and thus it is feasible that the reduction in this protein may be a consequence of the early life reduction in vitamin D. The potential links between vitamin D and neuronal calcium binding proteins has been noted in a recent review article [30]
Two members of the dynamin family (dynamin 1 and dynamin 1-like proteins) were also significantly down-regulated. These proteins are essential for clathrin-mediated endocytosis, a role of particular importance in neurons for neurotransmitter release [31]. Apart from this function, these proteins are essential for the insertion of dopamine receptor 2 (DRD2) into the nonsynaptic membrane of dopaminergic neurons [32]. Syntaxin-binding protein 1 was also significantly down-regulated in the DVD-deficient rats. These findings suggest that proteins involved in SNAP and SNARE mediated vesicle release may be disrupted in the DVD model. In addition, six mitochondrial proteins were down-regulated in DVD-deficient rats (NDUAA, UQCR1, ODPB, IDH3A, HKK1, VDAC2). These findings suggest that cellular energics may be altered in this brain region in the DVD-deficient rat. Curiously, it has recently been shown that dynamin 1-like protein is also important for mitochondrial fission and general morphology [33].
It is known that down-regulation of calcium binding proteins shift essential calcium buffering requirements to the mitochondrial compartment, which, in turn, leads to compromised mitochondrial energy production [34]. With respect to the nucleus accumbens, calcium-binding proteins such as calbindin are often used to demarcate discrete neuroanatomical boundaries, inferring that these proteins confer selective functional properties to these cells [35]. Compromised calcium buffering in the nucleus accumbens could disrupt adaptive and goal-directed behaviors. In the core region of the nucleus accumbens, neurons have dense spines, which may reflect the degree of synaptic plasticity required for the integrative function of cells in this region [36]. Many of the dopaminergic cells in this region also express neuropeptides such as bradykinan, neurotensin and substance P. The protein with the greatest fold change in this study was thimet oligopeptidase (down-regulated 1.67), which is involved in the degradation of these small proteins [37]. Neuropeptides such as neurotensin can indirectly influence dopaminergic transmission in the nucleus accumbens via glutamate and GABA-ergic mediated processes [38]. Thus, these neuropeptides are of interest as potential targets for novel antipsychotic agents [39], [40]. Curiously, it has been shown that calcium concentration is an important modulator of thimet oligopeptidase activity [41], thus the disruption of this protein may also be down-stream consequence of altered calcium buffering.
With respect to schizophrenia, several of proteins identified in this study have also been reported to be disrupted in post-mortem brain tissue from patients with schizophrenia. A range of studies (proteomics, genomics, gene association studies) have linked schizophrenia with alterations in malate dehydrogenase cytoplasmic (MDHC, now known as malic enzyme 2) [42]–[47]. Similarly, mitogen-activated protein kinase 1, significantly down-regulated in this study, has been found to be down-regulated at both the mRNA and protein levels in post-mortem schizophrenia brain tissue (thalamus) [48]
Like the previous proteomic study [18], we found no alterations in proteins directly associated with DA signaling. This suggests that baseline DA signaling may be normal in this model and abnormalities only become unmasked in the presence of drugs that alter DA/glutamate balance in the brain. It is conceivable that a slight reduction in calcium buffering proteins may affect the ability of neurons within the nucleus accumbens to repolarise in response to psychomimetic agents. Alternately, any reduction in cellular energetics within this region may delay the integration of cortical and/or sub-cortical inputs.
Disappointingly, we found no overlap with the proteins identified in the current study versus those in the previous publication [18]. However, the previous study was based on cortical and hippocampal tissue from adult female animals, whereas the current study was based on tissue from the nucleus accumbens in adult male animals. Interpretation of the current study is also limited because of the lack of immunoblot confirmation of the differentially expressed spots. The small fold changes found in the study, while statistically significant, were too low to be reliably confirmed via immunoblot [49]. Based on the behavioural findings in DVD-deficient rats, there is a case to explore proteomic dysregulation in rats after exposure to drugs known to disrupt dopaminergic and glutaminergic pathways. For example, we have shown that while habituated DVD rats have normal locomotion activity in the open field at baseline, they have pronounced hyperlocomotion after exposure to MK-801 [10], [12]. We plan to explore these issues in future experiments.
In conclusion, developmental vitamin D deficiency is associated with subtle changes in a range of proteins in the nucleus accumbens. These findings suggest that pathways involved in calcium binding and mitochondrial function may underpin the behavioural features associated with this particular animal model of schizophrenia. Combined with other experimental findings, the current study lends further credibility to the notion that developmental vitamin D deficiency impacts adversely on normal brain development [30].
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This project was supported by the National Health and Medical Research Council and the Schizophrenia Research Institute. Neither agency had any further role in study design, analysis and interpretation of data, writing of the report or decision to submit the manuscript for publication.
References
- 1.McGrath J. Hypothesis: is low prenatal vitamin D a risk-modifying factor for schizophrenia? Schizophr Res. 1999;40:173–177. doi: 10.1016/s0920-9964(99)00052-3. [DOI] [PubMed] [Google Scholar]
- 2.Davies G, Welham J, Chant DC, Torrey EF, McGrath J. Season of birth effect and latitude: a systematic review and meta-analysis of Northern Hemisphere studies. Schizophr Bull. 2003;29:587–593. doi: 10.1093/oxfordjournals.schbul.a007030. [DOI] [PubMed] [Google Scholar]
- 3.Saha S, Chant DC, Welham JL, McGrath JJ. The incidence and prevalence of schizophrenia varies with latitude. Acta Psychiatrica Scandinavica. 2006;114:36–39. doi: 10.1111/j.1600-0447.2005.00742.x. [DOI] [PubMed] [Google Scholar]
- 4.Webb AR, Kline L, Holick MF. Influence of season and latitude on the cutaneous synthesis of vitamin D3: exposure to winter sunlight in Boston and Edmonton will not promote vitamin D3 synthesis in human skin. J Clin Endocrinol Metab. 1988;67:373–378. doi: 10.1210/jcem-67-2-373. [DOI] [PubMed] [Google Scholar]
- 5.Eyles D, Brown J, Mackay-Sim A, McGrath J, Feron F. Vitamin D3 and brain development. Neuroscience. 2003;118:641–653. doi: 10.1016/s0306-4522(03)00040-x. [DOI] [PubMed] [Google Scholar]
- 6.Ko P, Burkert R, McGrath J, Eyles D. Maternal vitamin D(3) deprivation and the regulation of apoptosis and cell cycle during rat brain development. Brain Res Dev Brain Res. 2004;153:61–68. doi: 10.1016/j.devbrainres.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 7.Cui X, McGrath JJ, Burne TH, Mackay-Sim A, Eyles DW. Maternal vitamin D depletion alters neurogenesis in the developing rat brain. Int J Dev Neurosci. 2007;25:227–232. doi: 10.1016/j.ijdevneu.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 8.Feron F, Burne TH, Brown J, Smith E, McGrath JJ, et al. Developmental Vitamin D3 deficiency alters the adult rat brain. Brain Res Bull. 2005;65:141–148. doi: 10.1016/j.brainresbull.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 9.Burne TH, Becker A, Brown J, Eyles DW, Mackay-Sim A, et al. Transient prenatal Vitamin D deficiency is associated with hyperlocomotion in adult rats. Behav Brain Res. 2004;154:549–555. doi: 10.1016/j.bbr.2004.03.023. [DOI] [PubMed] [Google Scholar]
- 10.Kesby JP, Burne TH, McGrath JJ, Eyles DW. Developmental vitamin D deficiency alters MK 801-induced hyperlocomotion in the adult rat: An animal model of schizophrenia. Biol Psychiatry. 2006;60:591–596. doi: 10.1016/j.biopsych.2006.02.033. [DOI] [PubMed] [Google Scholar]
- 11.Becker A, Eyles DW, McGrath JJ, Grecksch G. Transient prenatal vitamin D deficiency is associated with subtle alterations in learning and memory functions in adult rats. Behav Brain Res. 2005;161:306–312. doi: 10.1016/j.bbr.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 12.O'Loan J, Eyles DW, Kesby J, Ko P, McGrath JJ, et al. Vitamin D deficiency during various stages of pregnancy in the rat; its impact on development and behaviour in adult offspring. Psychoneuroendocrinology. 2007;32:227–234. doi: 10.1016/j.psyneuen.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 13.Kapur S, Mamo D. Half a century of antipsychotics and still a central role for dopamine D2 receptors. Prog Neuropsychopharmacol Biol Psychiatry. 2003;27:1081–1090. doi: 10.1016/j.pnpbp.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 14.Schlaepfer TE, Cohen MX, Frick C, Kosel M, Brodesser D, et al. Deep brain stimulation to reward circuitry alleviates anhedonia in refractory major depression. Neuropsychopharmacology. 2008;33:368–377. doi: 10.1038/sj.npp.1301408. [DOI] [PubMed] [Google Scholar]
- 15.Joseph MH, Datla K, Young AM. The interpretation of the measurement of nucleus accumbens dopamine by in vivo dialysis: the kick, the craving or the cognition? Neurosci Biobehav Rev. 2003;27:527–541. doi: 10.1016/j.neubiorev.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 16.Lauer M, Senitz D, Beckmann H. Increased volume of the nucleus accumbens in schizophrenia. J Neural Transm. 2001;108:645–660. doi: 10.1007/s007020170042. [DOI] [PubMed] [Google Scholar]
- 17.Grace AA, Floresco SB, Goto Y, Lodge DJ. Regulation of firing of dopaminergic neurons and control of goal-directed behaviors. Trends Neurosci. 2007;30:220–227. doi: 10.1016/j.tins.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 18.Almeras L, Eyles D, Benech P, Laffite D, Villard C, et al. Developmental vitamin D deficiency alters brain protein expression in the adult rat: implications for neuropsychiatric disorders. Proteomics. 2007;7:769–780. doi: 10.1002/pmic.200600392. [DOI] [PubMed] [Google Scholar]
- 19.Eyles D, Almeras L, Benech P, Patatian A, Mackay-Sim A, et al. Developmental vitamin D deficiency alters the expression of genes encoding mitochondrial, cytoskeletal and synaptic proteins in the adult rat brain. J Steroid Biochem Mol Biol. 2007;103:538–545. doi: 10.1016/j.jsbmb.2006.12.096. [DOI] [PubMed] [Google Scholar]
- 20.Thompson TL, Moss RL. Modulation of mesolimbic dopaminergic activity over the rat estrous cycle. Neurosci Lett. 1997;229:145–148. doi: 10.1016/s0304-3940(97)00450-3. [DOI] [PubMed] [Google Scholar]
- 21.Zhou W, Cunningham KA, Thomas ML. Estrogen regulation of gene expression in the brain: a possible mechanism altering the response to psychostimulants in female rats. Brain Res Mol Brain Res. 2002;100:75–83. doi: 10.1016/s0169-328x(02)00134-1. [DOI] [PubMed] [Google Scholar]
- 22.Paxinos G, Watson C. New York: Academic Press; 2004. The Rat Brain in Stereotaxic Coordinates - the New Coronal Set. [Google Scholar]
- 23.Alexander-Kaufman A, G J, Sheedy D, Harper C, Matsumoto I. Differential protein expression in the prefrontaol white matter of human alcoholics: a proteomics study. 2005 doi: 10.1038/sj.mp.4001741. [DOI] [PubMed] [Google Scholar]
- 24.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 25.Benjamini Y, Hochberg Y. Controlling the false discovery rate, a practical and powerful approach to multiple testing. J Roy Stat Soc, Series B. 1995;57:289–300. [Google Scholar]
- 26.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eyles DW, McGrath JJ, Reynolds GP. Neuronal calcium-binding proteins and schizophrenia. Schizophr Res. 2002;57:27–34. doi: 10.1016/s0920-9964(01)00299-7. [DOI] [PubMed] [Google Scholar]
- 28.Lewis DA, Hashimoto T. Deciphering the disease process of schizophrenia: the contribution of cortical gaba neurons. Int Rev Neurobiol. 2007;78:109–131. doi: 10.1016/S0074-7742(06)78004-7. [DOI] [PubMed] [Google Scholar]
- 29.Christakos S, Dhawan P, Peng X, Obukhov AG, Nowycky MC, et al. New insights into the function and regulation of vitamin D target proteins. J Steroid Biochem Mol Biol. 2007;103:405–410. doi: 10.1016/j.jsbmb.2006.12.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCann JC, Ames BN. Is there convincing biological or behavioral evidence linking vitamin D deficiency to brain dysfunction? FASEB J. 2008;22:982–1001. doi: 10.1096/fj.07-9326rev. [DOI] [PubMed] [Google Scholar]
- 31.Royle SJ, Lagnado L. Endocytosis at the synaptic terminal. J Physiol. 2003;553:345–355. doi: 10.1113/jphysiol.2003.049221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paspalas CD, Rakic P, Goldman-Rakic PS. Internalization of D2 dopamine receptors is clathrin-dependent and select to dendro-axonic appositions in primate prefrontal cortex. Eur J Neurosci. 2006;24:1395–1403. doi: 10.1111/j.1460-9568.2006.05023.x. [DOI] [PubMed] [Google Scholar]
- 33.Scorrano L. Multiple functions of mitochondria-shaping proteins. Novartis Foundation Symposium. 2007;287:47–55. doi: 10.1002/9780470725207.ch4. discussion 55-49. [DOI] [PubMed] [Google Scholar]
- 34.Chen G, Racay P, Bichet S, Celio MR, Eggli P, et al. Deficiency in parvalbumin, but not in calbindin D-28k upregulates mitochondrial volume and decreases smooth endoplasmic reticulum surface selectively in a peripheral, subplasmalemmal region in the soma of Purkinje cells. Neuroscience. 2006;142:97–105. doi: 10.1016/j.neuroscience.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 35.Prensa L, Richard S, Parent A. Chemical anatomy of the human ventral striatum and adjacent basal forebrain structures. J Comp Neurol. 2003;460:345–367. doi: 10.1002/cne.10627. [DOI] [PubMed] [Google Scholar]
- 36.Meredith GE, Baldo BA, Andrezjewski ME, Kelley AE. The structural basis for mapping behavior onto the ventral striatum and its subdivisions. Brain Structure and Function. 2008 doi: 10.1007/s00429-008-0175-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saric T, Graef CI, Goldberg AL. Pathway for degradation of peptides generated by proteasomes: a key role for thimet oligopeptidase and other metallopeptidases. J Biol Chem. 2004;279:46723–46732. doi: 10.1074/jbc.M406537200. [DOI] [PubMed] [Google Scholar]
- 38.Ferraro L, Tomasini MC, Fuxe K, Agnati LF, Mazza R, et al. Mesolimbic dopamine and cortico-accumbens glutamate afferents as major targets for the regulation of the ventral striato-pallidal GABA pathways by neurotensin peptides. Brain Res Rev. 2007;55:144–154. doi: 10.1016/j.brainresrev.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 39.Caceda R, Kinkead B, Nemeroff CB. Neurotensin: role in psychiatric and neurological diseases. Peptides. 2006;27:2385–2404. doi: 10.1016/j.peptides.2006.04.024. [DOI] [PubMed] [Google Scholar]
- 40.Boules M, Shaw A, Fredrickson P, Richelson E. Neurotensin agonists: potential in the treatment of schizophrenia. CNS Drugs. 2007;21:13–23. doi: 10.2165/00023210-200721010-00002. [DOI] [PubMed] [Google Scholar]
- 41.Oliveira V, Garrido PA, Rodrigues CC, Colquhoun A, Castro LM, et al. Calcium modulates endopeptidase 24.15 (EC 3.4.24.15) membrane association, secondary structure and substrate specificity. Febs J. 2005;272:2978–2992. doi: 10.1111/j.1742-4658.2005.04692.x. [DOI] [PubMed] [Google Scholar]
- 42.Beasley CL, Pennington K, Behan A, Wait R, Dunn MJ, et al. Proteomic analysis of the anterior cingulate cortex in the major psychiatric disorders: Evidence for disease-associated changes. Proteomics. 2006;6:3414–3425. doi: 10.1002/pmic.200500069. [DOI] [PubMed] [Google Scholar]
- 43.Escamilla M. Variation in the malic enzyme 2 gene: implications for the pharmacogenomics of psychotic disorders. Pharmacogenomics. 2007;8:691–695. doi: 10.2217/14622416.8.7.691. [DOI] [PubMed] [Google Scholar]
- 44.Lee BD, Walss-Bass C, Thompson PM, Dassori A, Montero PA, et al. Malic enzyme 2 and susceptibility to psychosis and mania. Psychiatry Res. 2007;150:1–11. doi: 10.1016/j.psychres.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 45.Middleton FA, Mirnics K, Pierri JN, Lewis DA, Levitt P. Gene expression profiling reveals alterations of specific metabolic pathways in schizophrenia. J Neurosci. 2002;22:2718–2729. doi: 10.1523/JNEUROSCI.22-07-02718.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vawter MP, Shannon Weickert C, Ferran E, Matsumoto M, Overman K, et al. Gene expression of metabolic enzymes and a protease inhibitor in the prefrontal cortex are decreased in schizophrenia. Neurochem Res. 2004;29:1245–1255. doi: 10.1023/b:nere.0000023611.99452.47. [DOI] [PubMed] [Google Scholar]
- 47.Vawter MP, Ferran E, Galke B, Cooper K, Bunney WE, et al. Microarray screening of lymphocyte gene expression differences in a multiplex schizophrenia pedigree. Schizophr Res. 2004;67:41–52. doi: 10.1016/s0920-9964(03)00151-8. [DOI] [PubMed] [Google Scholar]
- 48.Kyosseva SV. Differential expression of mitogen-activated protein kinases and immediate early genes fos and jun in thalamus in schizophrenia. Prog Neuropsychopharmacol Biol Psychiatry. 2004;28:997–1006. doi: 10.1016/j.pnpbp.2004.05.017. [DOI] [PubMed] [Google Scholar]
- 49.Andrade EC, Krueger DD, Nairn AC. Recent advances in neuroproteomics. Current Opinion in Molecular Therapeutics. 2007;9:270–281. [PMC free article] [PubMed] [Google Scholar]