Abstract
The UL13 protein kinase is conserved among many herpesviruses but HSV-2 UL13 specificity is not known. Here, we found that HSV-2 UL13 is a phosphoprotein that autophosphorylates, and that serines within ERK and Cdc2 motifs were important for autophosphorylation but not for UL13 phosphorylation of exogenous substrates. HSV-2 UL13 phosphorylated a peptide also recognized by ERK and Cdc2. However, mutation of substrate residues critical for Cdc2 or Erk phosphorylation did not alter HSV-2 UL13 phosphorylation of the peptide, and HSV-2 UL13 did not phosphorylate standard Cdc2 or Erk peptide substrates. Mutation of prolines surrounding the peptide phosphoacceptor site reduced phosphorylation by HSV-2 UL13, and a peptide containing serine-proline amid alanines and glycines was phosphorylated. Thus, HSV-2 UL13 does not mimic ERK or Cdc2 substrate recognition and its minimal recognition motif can be serine-proline. This motif's simplicity indicates that distal sequence or protein structure contributes to HSV-2 UL13 substrate specificity.
Keywords: kinase, phosphorylation, motif, UL13, HSV
Introduction
The Herpesvirus family is divided into alpha, beta, and gamma-subfamilies. Herpes simplex viruses (HSV) 1 and 2 are large double-stranded DNA alphaherpesviruses that are co-linear and 83% identical at the nucleotide level (Dolan et al., 1998). HSV virions consist of a host-derived lipid envelope studded with glycoproteins and an icosahedral capsid surrounding the genome. In between the capsid and envelope lies the tegument, consisting of a layer of viral proteins. One of these tegument proteins is the serine/threonine protein kinase, UL13 (Coulter et al., 1993;Overton et al., 1992). Tegument proteins are the first to gain access to the host cell's interior upon infection (Morrison et al., 1998;Roizman & Sears, 1996) Thus, UL13 can immediately exert its effects on both viral and cellular targets. UL13 has homologues among the human alpha-, beta-, and gamma-herpesviruses (Kawaguchi and Kato 2003) and is designated UL13 in the alphaherpesviruses, HSV-1 and HSV-2, UL97 in the betaherpesvirus, Human cytomegalovirus (HCMV), and BGLF4 in the gammaherpesvirus, Epstein Barr virus (EBV) (Chee et al., 1989;McGeoch and Davison, 1986;Smith and Smith, 1989). HSV-1 and HSV-2 UL13 homologues have the closest amino acid sequence identity at 85% (Daikoku et al., 1997). The extensive conservation of UL13 suggests important roles for UL13 in the HSV replication cycle.
Two functions have been proposed for UL13. First, UL13 activity may promote dissociation of the viral tegument by phosphorylating other tegument proteins during entry into newly infected cells (Morrison et al., 1998). Second, UL13 kinase activity may regulate viral gene expression. Growth of HSV-1 UL13-deletion mutants is attenuated in certain cell lines (Purves and Roizman, 1992), possibly due to UL13-mediated effects on expression of the immediate-early protein ICP0 and a subset of late proteins (Purves et al., 1993;Tanaka et al., 2005).
UL13 contains the eleven classical conserved protein kinase subdomains, including the lysine in subdomain II that interacts directly with ATP and is required for high protein kinase catalytic activity (Carrera et al., 1993;Hanks et al., 1988; Iyer et al., 2005;Smith and Smith, 1989). Mutation of this lysine markedly reduces protein kinase activity (Iyer et al 2005;Hanks et al., 1988;He et al., 1997;Kawaguchi et al., 2003;Kenyon et al., 2001). Phosphotransferase activity of a kinase and its binding to other proteins are often regulated by autophosphorylation or phosphorylation by another kinase (Wang and Wu 2002). HSV-1 UL13 and HCMV UL97 autophosphorylate in vitro (Baek et al., 2002a;Cunningham et al., 1992;Ogle et al., 1997). One group of investigators found that the capacity of HCMV UL97 to phosphorylate exogenous substrates is dependent on autophosphorylation (Michel et al., 1999) while another found no relationship between HCMV UL97 autophosphorylation and the phosphorylation of exogenous substrates (Baek et al., 2002a). Whether HSV-2 UL13 autophosphorylates, and whether autophosphorylation is a requirement for HSV-1 or HSV-2 UL13 protein kinase activity on exogenous substrates has not been investigated.
Potential UL13 substrates have been identified by comparing changes in protein phosphorylation in cells infected with wild-type and UL13-deficient HSV-1 or by examining UL13 phosphorylation of proteins in vitro. Certain isoforms of ICP22, ICP0, and the cellular proteins EF-1δ and RNA polII from cells infected with HSV-1 show slower electrophoretic mobility than those observed from cells infected with UL13- HSV-1 (Purves and Roizman, 1992; Ogle et al., 1997, Ng et al., 1998; Long et al., 1999; Kawaguchi et al., 1998). HSV-2 UL13 also may recognize EF-1δ as a substrate (Kawaguchi et al., 1999), however, an HSV-2 UL13-deficient virus has not been generated for use as a negative control. Phosphorylation of gE, gI and ICP0 by HSV-1 UL13 has also been implied because these proteins are more heavily labeled with 32P in cells infected with wild-type virus than UL13-deficient virus (Ogle et al., 1997, Ng et al., 1998). UL13 itself was labeled in wild-type virus-infected cells. These observations have been taken to suggest substrate recognition by UL13, but another kinase activated by UL13 could alternatively have been responsible.
In vitro kinase assays have more definitively identified proteins that can be substrates of UL13. The HSV tegument contains UL13, and isolated HSV-1 tegument preparations release VP22 in a phosphorylation-dependent manner when incubated with 32P-ATP and Mg2+ (Morrison et al., 1998). Phosphorylation of released VP22 occurs on serine residue(s). VP22 in tegument preparations derived from UL13-deficient virus is less labile in the presence of ATP and Mg2+, implying that VP22 is a substrate of UL13, but whether VP22 is phosphorylated in UL13-preparations was not determined. More direct of UL13 substrate specificity derives from immunoprecipitated enzyme mixed with isolated substrate. UL13 immunoprecipitated from HSV-1-infected cells co-immunoprecipitates with gE and gI. Addition of 32P to these complexes reveals phosphorylation of gE and gI (Ng et al., 1998), but not when complexes are derived by anti-gE immunoprecipitation from UL13- virus-infected cells. ICP0 and EF-1δ also can be substrates of UL13 because they are 32P-labeled in an in vitro kinase assay which combines immunoprecipitates of UL13 and ICP0 from infected cell lysates or EF-1δ from uninfected cells (Ng et al., 1998; Kawaguchi et al., 1998). Low level phosphorylation of ICP0 also occurs in immunoprecipitates from cells infected with UL13- virus, but a phosphorylated, slower mobility isoform typically seen in wild-type virus-infected cells is not generated.
To address the lingering possibility that the wild-type UL13 activates or binds another viral or cellular kinase that then phosphorylates the apparent UL13 substrate, recombinant proteins isolated on affinity columns, and enzymatically inactive UL13-K176M have been used. Kawaguchi et al. thus confirmed, using baculovirus-expressed UL13 and EF-1δ fusion proteins, that EF-1δ can be a substrate of HSV-1 UL13 (Kawaguchi et al., 1999). EF-1δ S133A is not phosphorylated in either uninfected or virus-infected cells (Kawaguchi et al., 2003). EF-1δ S133 lies within the sequence SPMR corresponding to the Cdc2 phosphorylation recognition motif S/TPX[R/K/H] (Kawaguchi and Kato, 2003;Kawaguchi et al., 2003;Marin et al., 1992;Shah et al., 2003). In addition, HSV-1 UL13 and EBV-BGLF4 can phosphorylate CKII-β at a serine within the Cdc2 motif, SPVK (Kawaguchi et al., 2003). These data have led investigators to suggest that HSV-1 UL13 and its homologues may recognize the same motif as the cellular kinase Cdc2 (Kawaguchi and Kato, 2003;Kawaguchi et al., 2003). Recently, the same in vitro kinase assay was used to demonstrate that HSV-1 UL13 can directly phosphorylate a recombinant protein containing a domain of the Us3 protein kinase (Kato et al, 2006).
UL13 and cellular kinases mediate phosphorylation of the HSV tegument protein VP22 in co-transfected cells; however, the pattern of VP22 phosphorylation differs between HSV-1 and HSV-2 (Elliott et al., 1999;Geiss et al., 2004). HSV-2 UL13-dependent phosphorylation sites on VP22 have been mapped to serines 28 and 34. Only one of these serines is conserved in HSV-1 VP22, and phosphorylation induced by HSV-1 UL13 at this site has not been demonstrated. HSV-2 VP22 serines 28 and 34 lie within the amino acid sequence PVS28PADPES34PRD containing overlapping ERK and Cdc2 recognition motifs (Geiss et al., 2004). The ERK recognition motif has been defined as PXS/TP (Alvarez et al., 1991;Clark-Lewis et al., 1991;Gonzalez et al., 1991), and Cdc2 can recognize broader versions of its consensus motif including S/TPXD or S/TPXX[R/K/H/D] (Nigg, 1991;Shah et al., 2003;Srinivasan et al., 1995). HSV-2 UL13-mediated phosphorylation of VP22 at serines within these motifs suggests two possibilities: HSV-2 UL13 may recognize a broader version of the Cdc2 motif putatively recognized by HSV-1 UL13, or HSV-2 UL13 may recognize a motif different than that predicted for HSV-1 UL13, either the ERK motif or a novel motif. Defining the HSV-2 UL13 recognition motif would permit identification of sites at which known UL13 protein substrates are phosphorylated and would aid identification of novel UL13 substrates. Therefore, we investigated whether HSV-2 UL13 mimics ERK or Cdc2 substrate recognition by using peptide substrates in in vitro kinase assays.
Results
HSV-2 UL13 is a phosphoprotein
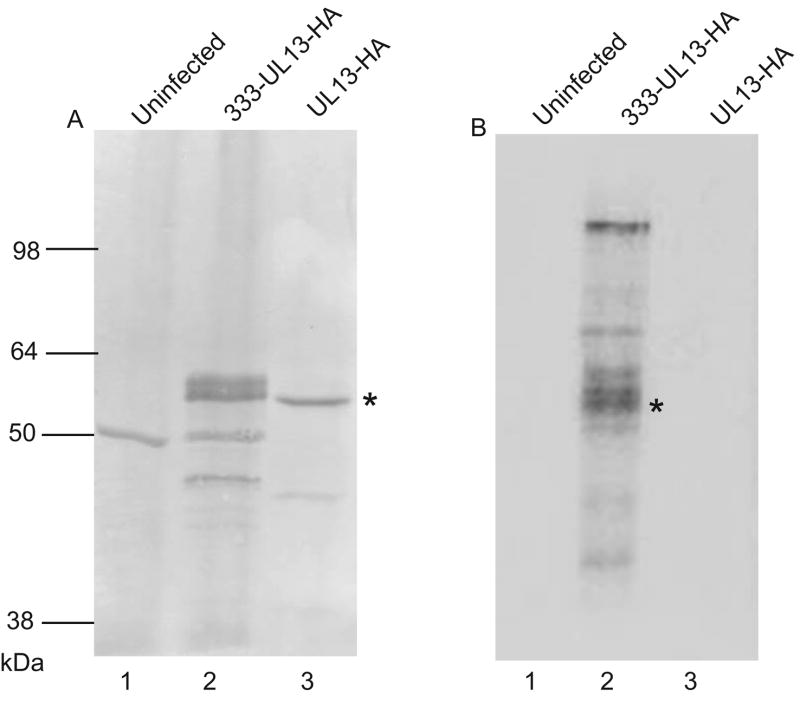
Phosphorylation of UL13 occurs in HSV-1-infected cells (Overton et al. 1992, Ng et al., 1998). To determine whether HSV-2 UL13 is phosphorylated during infection, Vero cells were infected with HSV-2 333-UL13-HA and incubated with 32P-orthophosphate from 4 to 10 hours post-infection. Cells were lysed and immunoprecipitated with the anti-HA monoclonal antibody, 3F-10. The complexes were resolved by SDS-PAGE and subjected to western blot and phosphorimager analysis (Fig 1). Western blot with 3F10 revealed presence of UL13-HA in the immunoprecipitate from cells infected with 333-UL13-HA and in an in vitro translated sample of UL13-HA (Fig 1A, lanes 2 and 3). Slower mobility isoforms of UL13 were apparent in the immunoprecipitate from 333-UL13-HA infected cells (Fig 1A, lane 2). Phosphorimager analysis of the western blot revealed a 32P-labeled band in the 333-UL13-HA sample that corresponded to the isoforms of UL13-HA on the western blot (Fig 1B, lane 2) indicating that HSV-2 UL13 is phosphorylated in infected cells.
Figure 1. HSV-2 UL13 is a phosphoprotein.
Vero cells were uninfected or infected with the HSV-2 recombinant 333-UL13-HA at an MOI of 20, then labeled with 32P-orthophosphate from 4 to 10 hr post-infection. Lysates were immunoprecipitated with an HA-specific mAb, resolved by SDS-PAGE, and subjected to (A) western blot using the 3F-10 MAb and (B) phosphorimager analysis. A UL13-HA in vitro translation product served as a control for detection by western blot. Asterisks denote the position of UL13-HA on the western blot and phosphorimage.
HSV-2 UL13 autophosphorylates
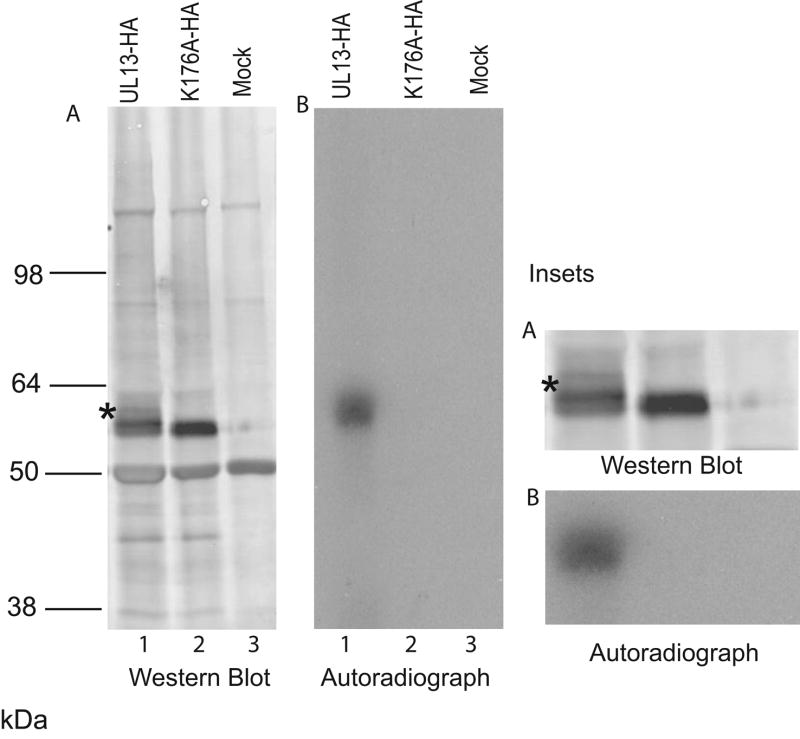
HSV-1 UL13 can autophosphorylate in vitro (Cunningham et al., 1992; Ogle et al., 1997), but HSV-2 UL13 autophosphorylation has not been demonstrated. To determine whether HSV-2 UL13 autophosphorylates, UL13-HA and the enzymatically inactive UL13-K176A-HA were expressed by coupled in vitro transcription/translation, immunoprecipitated, washed extensively, and subjected to an in vitro kinase assay. The products were resolved by SDS-PAGE and detected by both autoradiography and by western blot using an anti-HA antibody (Fig. 2). Autoradiography revealed a band labeled with 32P (Fig. 2B, lane 1) corresponding with the position on the western blot of a small proportion of UL13-HA that migrated with slightly reduced mobility (asterisk, Fig. 2A, lane 1). In comparison, K176A-HA migrated as a tighter band (Fig. 2A, lane 2) and produced only a faint 32P-labeled band on autoradiography (Fig. 2B, lane 2), possibly from background phosphorylation of K176A-HA by a kinase in the translation lysate. Phosphorimager analysis indicated that the 32P signal from UL13-HA was 38-fold greater than from K176A-HA. 32P-labeled product could be re-immunoprecipitated from the UL13-HA reaction with the anti-HA antibody but not from the K176A-HA reaction (data not shown), confirming that the phosphorylated band in lane 3 is HA-tagged UL13. These results indicate that HSV-2 UL13 can autophosphorylate in vitro.
Figure 2. HSV-2 UL13 autophosphorylates.
Empty vector pSG5 (Mock), UL13-HA, and K176A-HA were translated in vitro, isolated by immunoprecipitation, and subjected to an in vitro kinase assay. Proteins were resolved by SDS-PAGE and (A) HA-tagged UL13 and K176A were detected by western blot using HA-specific MAb 3F-10 and inset and (B) 32P-labeled protein was detected by autoradiography of the western blot and inset. Asterisk denotes slower mobility isoform of UL13.
Serines within ERK and Cdc2 recognition motifs are important for HSV-2 UL13 autophosphorylation but are not essential for phosphorylation of exogenous substrates
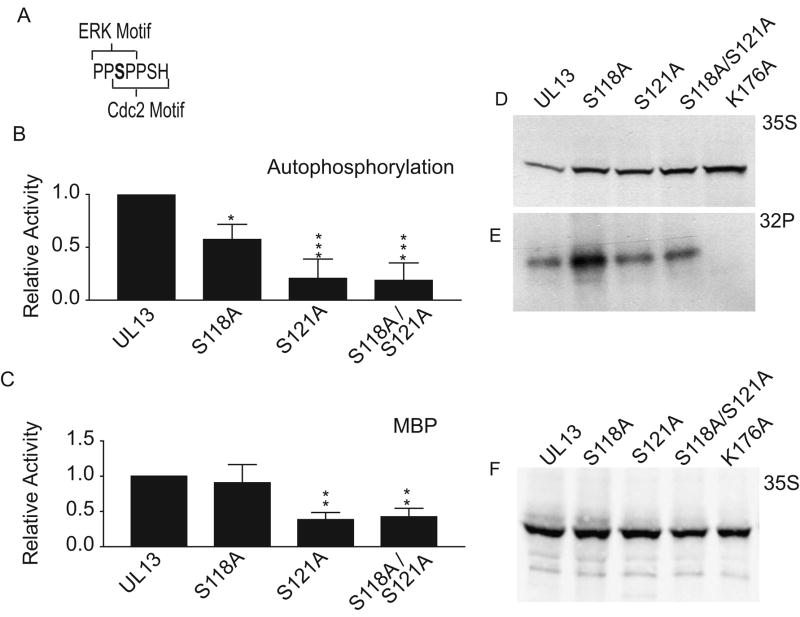
Our previous data suggested that HSV-2 UL13 could phosphorylate a serine residue within Cdc2 and/or ERK recognition motifs (Geiss et al., 2004), and that HSV-2 UL13 could be a substrate for itself. Therefore, we analyzed the HSV-2 UL13 sequence and found that serine 118 lies in the phosphoacceptor sites of overlapping ERK and Cdc2 motifs (Fig. 3A). To investigate whether HSV-2 UL13 can autophosphorylate S118, we made an alanine mutation at S118, translated UL13 and UL13-S118A in vitro, and performed an in vitro kinase assay. The S118A mutation reduced autophosphorylation of UL13 by approximately 40% (Fig. 3B). A serine at position 121 (S121) also lies within the Cdc2 phosphorylation motif but not at the phosphoacceptor site. To determine whether S121 contributes to HSV-2 UL13 autophosphorylation, an alanine mutation was made at S121 within pSG5-UL13-HA (UL13-S121A). The S121A mutation decreased UL13 autophosphorylation by approximately 80% (Fig. 3B). Interestingly, an S118A/S121A double mutant did not decrease UL13 autophosphorylation further than the S121A single mutant (Fig. 3B). Together, these data indicate that S118 and S121 are phosphoacceptor sites, or that they play another important role in UL13 autophosphorylation.
Figure 3. The effect of serine-to-alanine mutations on HSV-2 UL13 autophosphorylation and phosphorylation of exogenous substrates.
(A) Overlapping ERK and Cdc2 phosphorylation recognition motifs of HSV-2 UL13. S118 (bold) is the phosphoacceptor for both motifs. HA-tagged UL13 and UL13 mutants were subjected to in vitro kinase assays (B) in the absence of an exogenous substrate (*P = < 0.03 to *** = < 0.001 relative to WT UL13), or (C) in which the exogenous substrate myelin basic protein was added (**P = < 0.01 relative to WT UL13; see Methods for statistical analyses). Data are the mean ± SD from three experiments. Representative phosphorimages are shown for experiments denoted in 3B (D and E) and 3C (F).
Autophosphorylation often regulates the capacity of a kinase to phosphorylate exogenous substrates. Therefore, we examined the effect of the serine-to-alanine mutations on the capacity of UL13 to phosphorylate an exogenous substrate, myelin basic protein (MBP). UL13-S118A phosphorylated MBP to wild-type levels and UL13-S121A and UL13–S118A/S121A were able to phosphorylate MBP to 25% of wild-type levels. Therefore, autophosphorylation of HSV-2 UL13 is not essential for its activity on exogenous substrates (Fig. 3C).
HSV-2 UL13 can phosphorylate a HSV-2 UL13 peptide
The mutational analysis of HSV-2 UL13 suggested that HSV-2 UL13 phosphorylates a serine within an ERK or Cdc2 recognition motif. To test whether HSV-2 UL13 could phosphorylate a peptide substrate containing these motifs, we generated a peptide based on the wild-type HSV-2 UL13 sequence, APPSPPSHILGGRRR (amino acids 116-124 in bold), containing overlapping ERK and Cdc2 motifs. In vitro translated UL13-HA and K176A-HA were immunoprecipitated and subjected to an in vitro kinase assay to which increasing concentrations of the UL13-WT peptide were added (Fig. 4A). Phosphorylation of UL13-WT peptide by UL13 was readily detectable after 30 min at 0.5mM and became saturated at approximately 3.5mM. A time course experiment showed an increase in phosphorylation of the UL13-WT peptide over a span of 180 min (Fig. 4B) ensuring that the in vitro kinase assay carried out at 30 min is within the linear range. Therefore, the UL13-WT peptide can serve as a substrate for HSV-2 UL13 in vitro.
Figure 4. HSV-2 UL13 can phosphorylate a HSV-2 UL13 peptide.
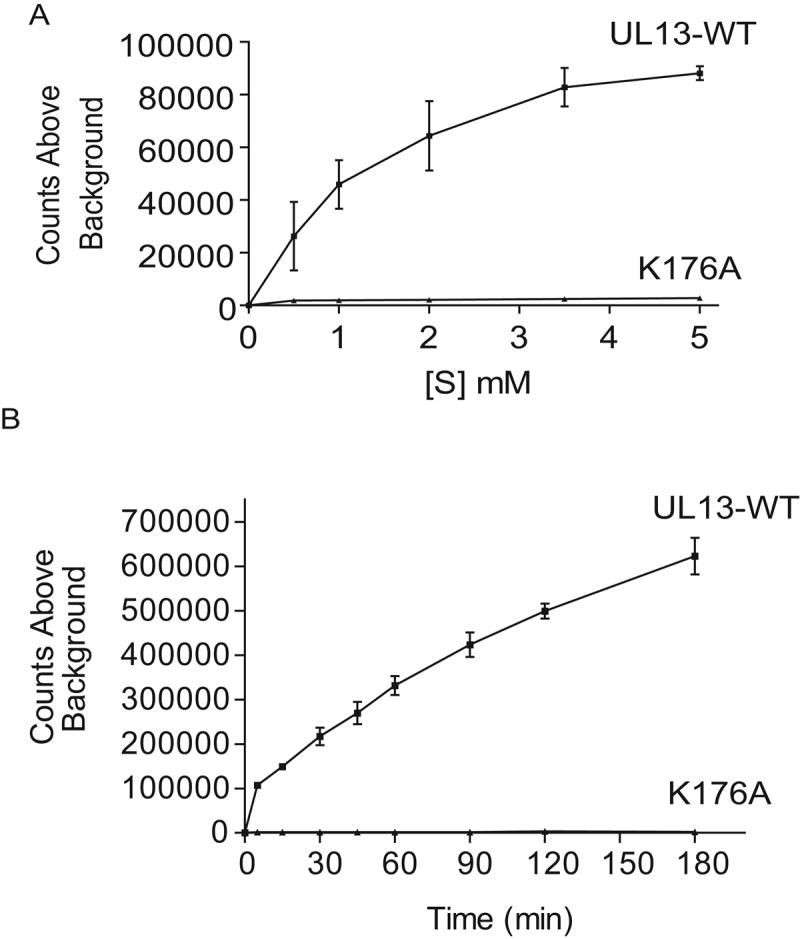
UL13-HA and K176A-HA were translated in vitro, immunoprecipitated, and subjected to an in vitro kinase assay using the UL13-WT peptide as a substrate (A) at increasing concentrations at a single time point of 30 min or (B) over time at a concentration of 1mM. Data are the mean ± SD from three experiments.
HSV-2 UL13 can phosphorylate S118 and S121 in the UL13-WT peptide
The UL13-WT peptide contains S118 at the phosphoacceptor position for both ERK and Cdc2, and S121 within the Cdc2 recognition motif but not at the phosphoacceptor site. Because alanine mutations at both S118 and S121 within HSV-2 UL13 decreased autophosphorylation, we synthesized peptides with alanine substitutions at S118 and S121 in the context of the UL13-WT peptide to determine which serine was the phosphoacceptor. S118A decreased phosphorylation of the UL13 peptide by approximately 60%, S121A decreased phosphorylation by approximately 80%, and the double mutant ablated phosphorylation of the peptide (Fig. 5). Therefore, HSV-2 UL13 can phosphorylate S118 and S121 in the UL13 peptide. Preliminary mass spectrometry data are consistent with the possibility that both residues are phosphorylated simultaneously on the same molecule (not shown).
Figure 5. Serine 118 and serine 121 can be phosphoacceptors for HSV-2 UL13.
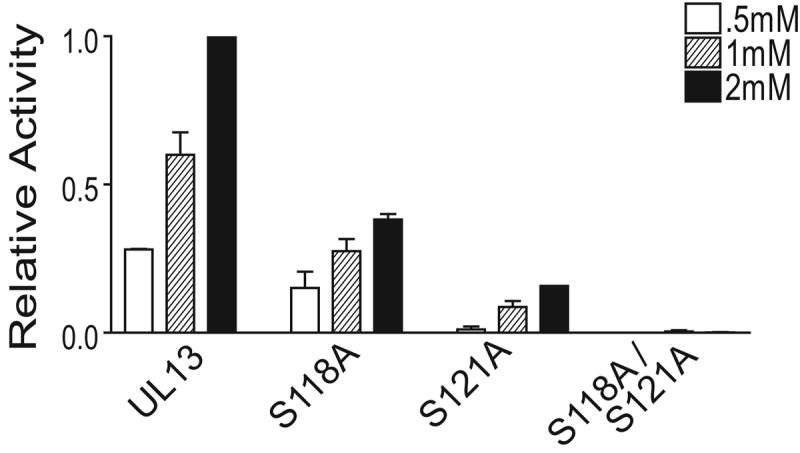
UL13-HA was expressed by in vitro transcription/translation, immunoprecipitated, and subjected to an in vitro kinase assay for 30 min using serine-to-alanine mutant peptides and the UL13-WT peptide as substrates at the indicated concentrations. Values were normalized to UL13 phosphorylation of the UL13-WT peptide at 2mM. The data are the average of three experiments ± SD.
HSV-2 UL13 does not phosphorylate standard ERK and Cdc2 peptide substrates
To verify that HSV-2 UL13 does not mimic ERK or Cdc2 substrate recognition, standard ERK and Cdc2 peptide substrates were tested for phosphorylation by HSV-2 UL13. A myelin basic protein (MBP) peptide corresponding to residues 95-98 of MBP contains the ERK recognition motif PRTP and is used as an ERK substrate in in vitro kinase assays (Clark-Lewis et al., 1991). A CKII-β peptide is commonly used as a Cdc2 substrate in in vitro kinase assays because it contains the Cdc2 recognition sequence SPVK corresponding to residues 209-212 of CKII-β (Litchfield et al., 1995). In addition, this sequence within CKII-ß can be recognized by HSV-1 UL13 and EBV BGLF4 (Kawaguchi et al., 2003). The MBP and CKII-β peptides were phosphorylated by ERK and Cdc2, respectively (Figs. 6A and B). However, phosphorylation of the MBP peptide was barely detectable by HSV-2 UL13, and 32P-incorporation into the CKII-β peptide was 90% less than UL13-WT (Fig. 6C). Thus, peptide substrates containing optimal ERK or Cdc2 recognition motifs are not readily phosphorylated by HSV-2 UL13. Taken together, these data indicate that HSV-2 UL13 does not mimic either ERK or Cdc2 substrate recognition.
Figure 6. HSV-2 UL13 does not phosphorylate ERK or Cdc2 peptide substrates.
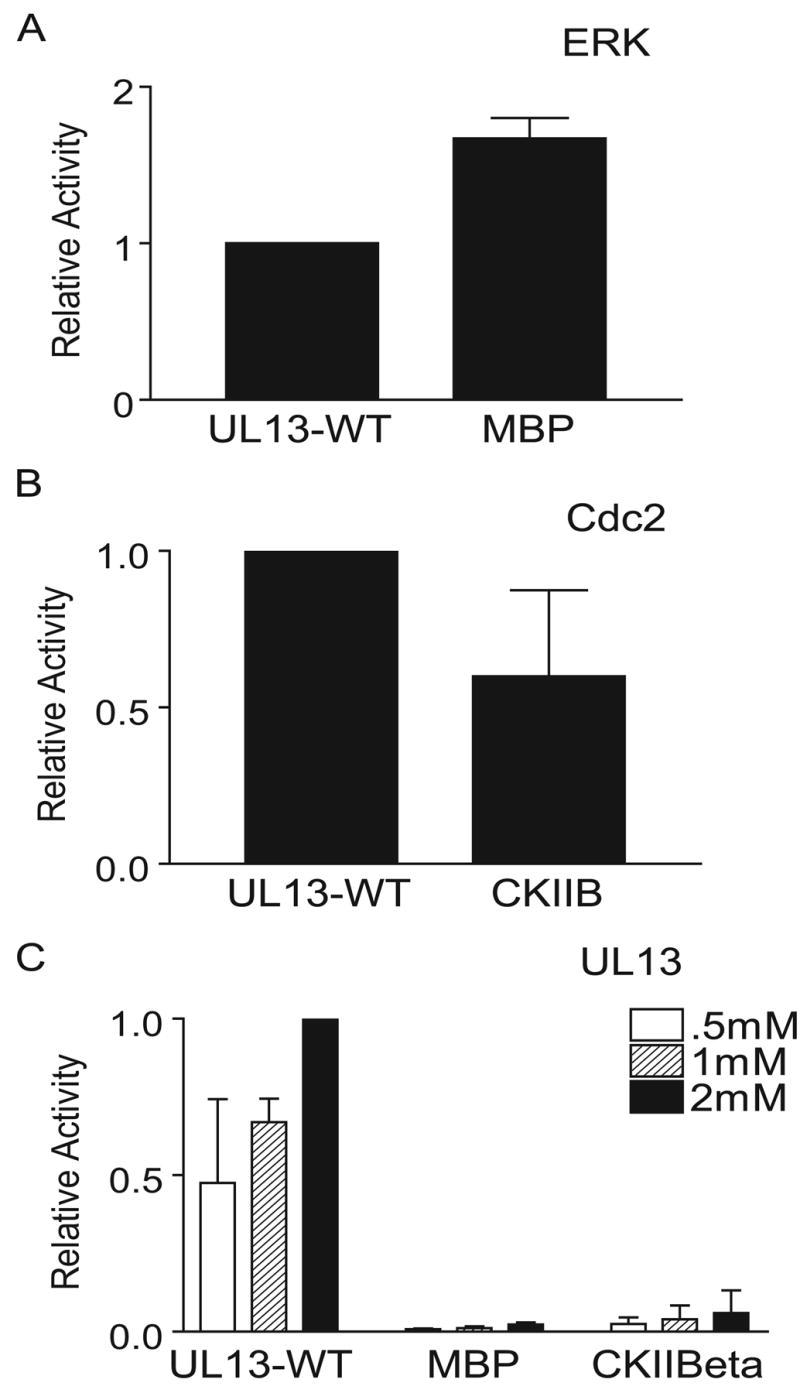
(A) Phosphorylation of MBP and UL13-WT peptides by ERK. (B) Phosphorylation of CKII-β and UL13-WT peptides by Cdc2. (C) UL13-HA was translated in vitro, immunoprecipitated, and subjected to an in vitro kinase assay for 30 min using standard ERK and Cdc2 substrate peptides. Values were normalized to UL13 phosphorylation of the UL13-WT peptide at 2mM. The data are the average of three experiments ± SD.
HSV-2 UL13 is sensitive to mutations in prolines surrounding the phosphoacceptor
To identify residues crucial for HSV-2 UL13 substrate specificity, peptides were synthesized containing substitutions in the context of the UL13-WT peptide (Table 1). A proline at position +1 (P119) relative to the phosphoacceptor S118 is crucial for ERK and Cdc2 substrate recognition; hence peptide P119A was synthesized. A charged residue at position +4 is important for Cdc2 substrate recognition, so peptide H122A contained an alanine substitution at position +4 relative to S118 in the Cdc2 recognition motif. These peptides were used as substrates for HSV-2 UL13, ERK, and Cdc2 in in vitro kinase assays. As expected, P119A ablated recognition of the UL13 peptide by ERK and Cdc2, and H122A greatly decreased Cdc2 recognition of the UL13 peptide (Figs. 7A and B). In contrast, HSV-2 UL13 phosphorylated P119A and H122A at nearly wild-type levels. Surprisingly, ERK phosphorylation of the H122A peptide was also reduced (Fig. 7A), even though a histidine at position +4 is not known to be necessary for ERK recognition (Songyang et al., 1996). To determine whether amino acids in other positions are important for substrate recognition by HSV-2 UL13, alanine substitutions were made at prolines across the UL13-WT peptide. Alanine substitutions at prolines 116, 117, and 120 decreased phosphorylation by HSV-2 UL13 more than 50%. A decrease in phosphorylation by ERK and Cdc2 was also observed when alanine mutations were made at prolines 116, 117, and 120. These results indicate that UL13 kinase activity is sensitive to alanine substitutions at multiple residues flanking the phosphoacceptor sites, and that proline residues in the vicinity of the phosphoacceptor site are important for HSV-2 UL13 phosphorylation.
Table 1. UL13 Peptides.
UL13-WT peptide and UL13 mutant peptides used in in vitro kinase assays. The putative phosphoacceptors are in bold.
| Peptide | Sequence |
|---|---|
| UL13-WT | APPSPPSHGGRRR |
| P116A | AAPSPPSHGGRRR |
| P117A | APASPPSHGGRRR |
| P119A | APPSAPSHGGRRR |
| P120A | APPSPASHGGRRR |
| H122A | APPSPPSAGGRRR |
Figure 7. HSV-2 UL13 is a proline-directed kinase.
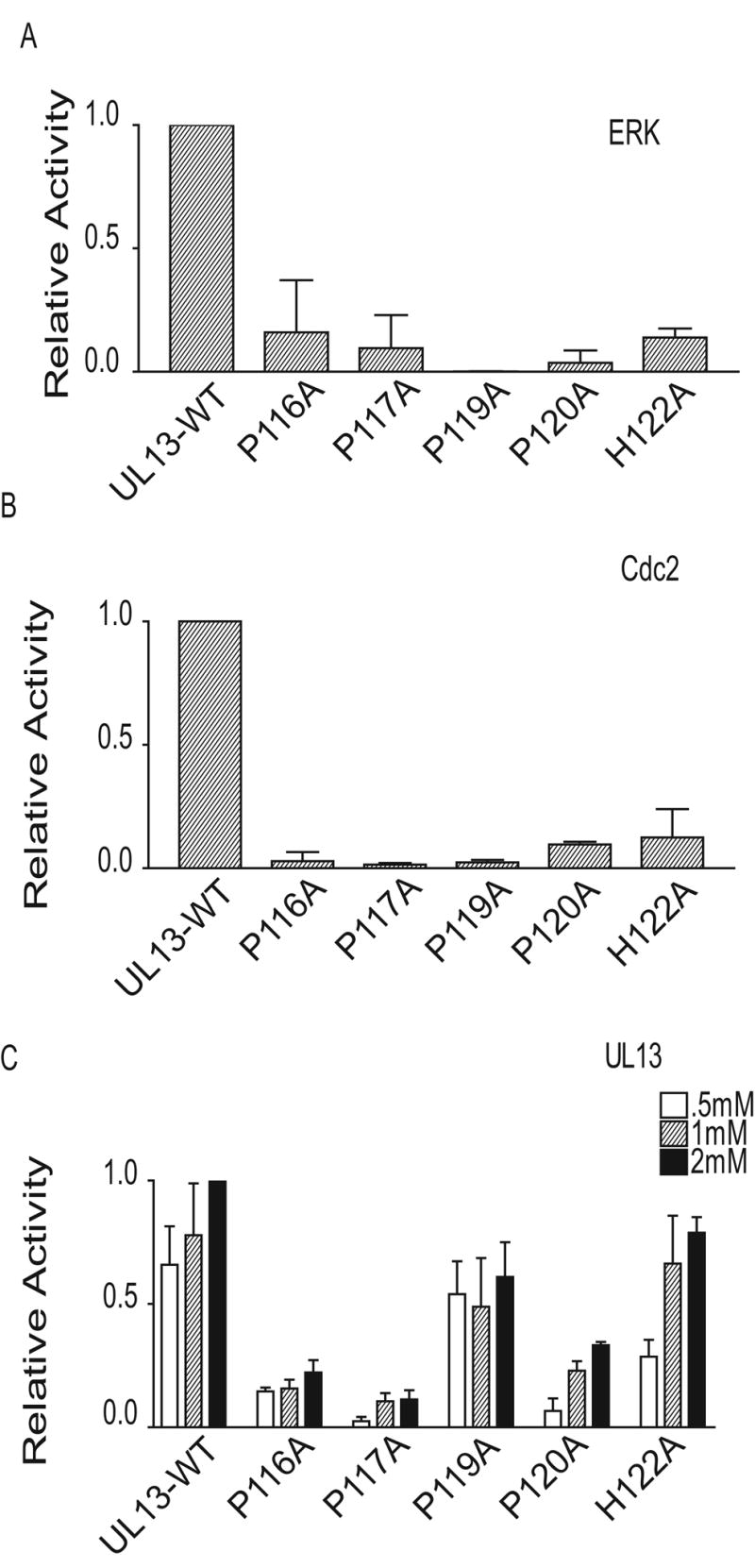
(A) The UL13-WT peptide and mutant peptides were used at 1mM as substrates for ERK in an in vitro kinase assay. Values are the average of 2 experiments. (B) The UL13-WT peptide or mutant peptides were used at 1mM as substrates for Cdc2 in an in vitro kinase assay. Values are the average of 2 experiments ± the range. (C) UL13-HA was expressed by in vitro transcription/translation, immunoprecipitated, and subjected to an in vitro kinase assay for 30 min using the UL13-WT peptide or UL13-WT mutant peptides as substrates. Values were normalized to UL13 phosphorylation of the UL13-WT peptide at 2mM. The data are the average of three experiments ± SD.
HSV-2 UL13 can phosphorylate a simple serine and proline motif
Phosphorylation of the UL13-WT peptide by UL13 was reduced more than 50% only when proline-to-alanine substitutions were made one or two residues before a serine, and to a lesser extent after a serine (Fig. 7C). Thus, the peptide data indicate that HSV-2 UL13 may recognize a motif consisting of a serine adjacent to proline residues (Fig. 7C). To test this prediction, we synthesized peptides containing a serine preceded or followed by a proline. The SP or PS sequences were in this case flanked with alternating alanines and glycines to determine the minimal recognition motif for HSV-2 UL13. The PS and SP peptides were used as substrates for HSV-2 UL13 in an in vitro kinase assay and phosphorylation was compared to that of the UL13-WT peptide. HSV-2 UL13 phosphorylated the peptide containing a serine followed by a proline and to a much lesser extent a proline followed by a serine (Fig. 8). Therefore, SP flanked by alanine and glycine residues can serve as a minimal phosphorylation recognition motif for HSV-2 UL13).
Figure 8. HSV-2 UL13 can phosphorylate a peptide containing a serine followed by a proline.
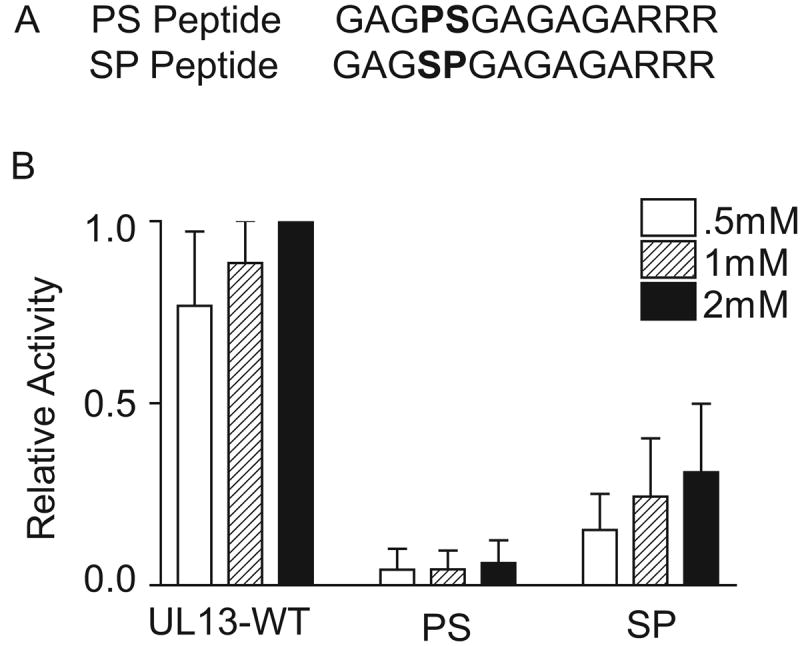
UL13-HA was translated in vitro, immunoprecipitated, and subjected to an in vitro kinase assay for 30 min using the UL13-WT, PS or SP peptides as a substrate. Values were normalized to UL13 phosphorylation of the UL13-WT peptide at 2mM. The data are the average of three experiments ± SD.
Discussion
We have 1) demonstrated that HSV-2 UL13 is phosphorylated in infected cells, 2) demonstrated that HSV-2 UL13 can autophosphorylate, 3) identified potential phosphoacceptor sites for HSV-2 UL13 autophosphorylation, 4) demonstrated that autophosphorylation is not required for HSV-2 UL13 to phosphorylate exogenous substrates, 5) demonstrated that the HSV-2 UL13 recognition motif differs from that of ERK and Cdc2, and 6) determined that SP flanked by alanines and glycines can be a minimal recognition motif for HSV-2 UL13.
The inactivity of UL13-K176A strongly suggests that lysine 176 in subdomain II is required for catalytic activity and autophosphorylation, as has been seen when the lysine in subdomain II is mutated in UL13 homologues (He et al., 1997;Kenyon et al., 2001). A significant decrease in HSV-2 UL13 autophosphorylation occurred when alanine mutations were made at either S118 or S121. This result suggests that S118 and S121 are sites of HSV-2 UL13 autophosphorylation or form important components of the recognition motif. We cannot rule out the possibility that these mutations altered UL13 folding, resulting in loss of activity or decreased phosphorylation at another phosphoacceptor site, or reducing the affinity of UL13 for its substrate. However, S118 and S121 do not lie within catalytic subdomains of Ser/Thr protein kinases but instead are found 39 and 36 residues, respectively, upstream of subdomain I. In addition, neither mutation observably altered protein accumulation or mobility in SDS-PAGE. Analysis of HSV-2 UL13 by mass spectrometry will be needed to definitively demonstrate autophosphorylation at both S118 and S121. Additional minor autophosphorylation sites may exist because autophosphorylation is not completely ablated in the double S118A/S121A mutant. However, because we were not able to employ a purified recombinant enzyme preparation, we cannot completely exclude the possibility that a contaminating cellular kinase phosphorylates HSV-2 UL13 at a low level.
Whereas both the S118A and S121A mutations significantly reduced autophosphorylation, only the S121A mutation significantly reduced the capacity of HSV-2 UL13 to phosphorylate exogenous substrates. This result is consistent with the possibility that autophosphorylation at S121 augments HSV-2 UL13 protein kinase activity, but it is also possible that the S121A mutation altered UL13 conformation and consequently its substrate binding or kinase activity. Nevertheless, phosphorylation of exogenous substrates occurred even with the S121A mutation. Thus, HSV-2 UL13 resembles HCMV UL97 because both autophosphorylate, but their capacities to phosphorylate exogenous substrates are not strictly dependent on autophosphorylation (Baek et al., 2002a).
We originally hypothesized that HSV-2 UL13 recognizes ERK or Cdc2 motifs, or both motifs, based on data showing 1) HSV-2 UL13 mediates phosphorylation of HSV-2 VP22 at serines within overlapping ERK and Cdc2 recognition motifs (Geiss et al., 2004); 2) HSV-2 UL13 autophosphorylates at a serine within overlapping ERK and Cdc2 motifs; and 3) HSV-1 UL13 and the EBV UL13 homologue, BGLF4, recognize a sequence within EF-1δ and CKII-β consistent with the Cdc2 recognition motif (Kawaguchi and Kato, 2003;Kawaguchi et al., 2003). Therefore, we used synthetic peptides to address this hypothesis. Our data showed that HSV-2 UL13 does not mimic ERK or Cdc2 substrate recognition because substitutions within a peptide in the UL13 autophosphorylation sequence at residues critical for recognition by ERK and/or Cdc2 ablated phosphorylation by ERK and Cdc2 but did not markedly affect phosphorylation by HSV-2 UL13. Conversely, HSV-2 UL13 phosphorylation of conventional ERK and Cdc2 peptide substrates was at least 90% less than phosphorylation of the UL13-WT peptide. The Cdc2 peptide substrate CKIIß contains the SPVK sequence recognized by Cdc2, HSV-1 UL13, and EBV BGLF4. However, not all UL13 homologues utilize the Cdc2 motif. The HCMV UL13 homologue UL97, for example, is not proline-dependent (Baek et al., 2002b). Our data demonstrate that HSV-2 UL13 recognizes a novel phosphorylation motif apparently not shared with some other UL13 homologs, although direct comparison of HSV-2 UL13 with UL13 homologs will be needed to explicitly demonstrate that their substrate recognition motifs differ.
The capacity of HSV-2 UL13 to phosphorylate the UL13-WT peptide except when alanine substitutions are made at prolines adjacent to serines suggests that a serine flanked by prolines can serve as an HSV-2 UL13 recognition motif, and the data in Figure 8 are consistent with this prediction. While we cannot discount a potential influence of the glycines and alanines on recognition of the SP or PS peptides, HSV-2 UL13 phosphorylation of the SP peptide was more readily detected than that of the PS peptide despite being surrounded by the same glycine-alanine background. The SP motif appears simplistic based on primary amino acid sequence, but the secondary structure of a protein or peptide can be constrained by this short sequence because a single proline residue is often associated with a beta-turn (Hall et al., 1991;Rose et al., 1985). Sequence alignments of HSV-2 UL13 with other known proline-directed kinases, however, did not show homology at the site thought to form a pocket for a serine phosphoacceptor followed by a proline (not shown).
Although the widely-accepted optimal ERK consensus motif is PXS/TP, it had been suggested that the minimal motif for ERK is S/TP and that the proline at the -2 position in the optimal recognition motif is strongly preferred but not required (Haycock et al., 1992). This hypothesis had been tested using a degenerate peptide library, which demonstrated that ERK could phosphorylate SP-containing peptides with hydrophobic amino acids at the -2 position, although proline was preferred (Songyang et al., 1996). In the present study, we found that ERK also can phosphorylate SP within a uniform, relatively neutral glycine-alanine background. Therefore, although HSV-2 UL13 does not recognize the optimal ERK recognition motif, it does recognize the same minimal SP motif identified for ERK.
SP can be found in a vast number of proteins, but HSV-2 UL13 does not promiscuously phosphorylate proteins in infected cells. Therefore, additional residues or structural features must contribute to HSV-2 UL13 substrate specificity. HSV-2 UL13 substrate recognition may be increased by additional prolines near the phosphoacceptor site. This is suggested because HSV-2 UL13 efficiently phosphorylates UL13 peptides except when substitutions are made at serines or prolines at residues 116, 117, and 120 in the UL13 sequence. Other residues may hinder UL13 recognition, as has been previously shown for some kinases (Pinna et al., 1984). For example, the CKII-β peptide contains a SP, but it is not phosphorylated by HSV-2 UL13. Therefore the amino acid context surrounding the phosphoacceptor site can influence UL13 substrate recognition. In addition, amino acid sequences outside of the immediate recognition motif may convey a large portion of UL13 substrate specificity, presumably sequences which influence binding of the substrate to UL13. Predicting UL13 substrates will therefore require a full understanding of this binding specificity in addition to the recognition motif.
Materials and Methods
Plasmids
UL13 mutant constructs were derived from pSG5-UL13 which contains the UL13 gene of HSV-2 strain 186 downstream of the SV40 early promoter in pSG5 (Stratagene). pSG5-UL13-K176A contains an alanine mutation at lysine 176, resulting in an enzymatically inactive UL13. pSG5-UL13-S118A, pSG5-UL13-S121A, and pSG5-UL13-S118AS121A were constructed in the pSG5-UL13 background. All plasmids contain an influenza hemagglutinin (HA) tag fused to the carboxy terminus of UL13.
Transcription/translation assay
The TNT T7 coupled reticulocyte lysate system (Promega) was used for in vitro transcription/translation according to manufacturer's recommendations. For experiments examining autophosphorylation and protein kinase activity of UL13-mutants, 20 μCi of [35S]methionine (Perkin Elmer; >1,000Ci/mmol) was added during translation.
Peptides and peptide synthesis
UL13, SP, and PS peptides were synthesized by standard F-moc chemistry using a multiple peptide synthesizer (Symphony/Multiplex; Protein Technologies). Following synthesis, peptides were HPLC purified and their purity was assessed by mass spectrometry. The WT-UL13 peptide and UL13 residue substitution peptides contain HSV-2 UL13 wild-type sequence, amino acids 116-124. The alanine upstream of position 116 and the glycines downstream of the WT sequence are arbitrary. Arginines were placed at the end of all peptide sequences to facilitate binding to P81 phosphocellulose.
Immunoprecipitation
Translation products were pre-cleared twice for 30 minutes each with protein A/G agarose beads (Oncogene Research) at 4°C in immunoprecipitation buffer (50 mM Tris HCl [pH 8.0], 150 mM NaCl, 1% NP-40) supplemented with 40 mM sodium fluoride, 1mM sodium orthovanadate, 10mM β-glycerophosphate, 2μg/ml aprotinin, 3μg/ml leupeptin, and 1 mM phenylmethylsulfonyl fluoride, then incubated with protein A/G-agarose beads complexed with anti-HA antibody (3F-10; Roche Diagnostics) for 3 h at 4°C in immunoprecipitation buffer. The immunoprecipitates were washed three times with cold immunoprecipitation buffer, and then used directly in in vitro kinase assays.
Western blot analysis
Proteins were resolved by SDS-PAGE and transferred to polyvinylidene difluoride membranes (Millipore). Target proteins were bound by the 3F-10 antibody and detected with an alkaline phosphatase-conjugated secondary antibody and NBT/BCIP (Promega).
32P-labelling of infected cells and detection of labelled products
Vero cells in 25-cm2 flasks were left infected with 333-UL13-HA at an MOI of 20 or left uninfected. From 3 hr to 4 hr post-infection cells were incubated in phosphate-free medium, then labelled with 200 μCi of 32P-orthophosphate (Perkin Elmer) for 6 hr. Following the labelling period, cells were washed with 1X PBS and lysed in the immunoprecipitation (IP) buffer described above. Radiolabeled lysates were pre-cleared and immunoprecipitated with protein A/G-agarose beads complexed with anti-HA antibody, 3F-10, as described above. Immunoprecipitated proteins were separated by SDS-PAGE and subjected to western blot and phosphorimager analysis.
In vitro kinase assays
Kinase assays were performed with UL13 immobilized on protein A/G beads and peptide substrates added at indicated concentrations in a total volume of 25μl kinase assay buffer (25 mM Tris-HCl [pH7.5], 20μM EGTA) supplemented with Magnesium/ATP cocktail (Upstate) and 10 μCi [γ-32P]ATP (Perkin Elmer; 3000Ci/mmol) at 30°C for 30 min. The reaction was then centrifuged to separate the enzyme bound to beads from substrate in the supernatant. Supernatants were spotted onto P81 phosphocellulose papers (Whatman), and the filters were washed five times for 5 min with 0.75% phosphoric acid then once with acetone. Filters were transferred to vials containing scintillation cocktail and retained radioactivity was determined using a beta counter (Ludlum model 2). All assays were done in triplicate unless otherwise noted. In all experiments (except for Figure 4), values for phosphorylation of the substrates by K176A were designated as background and subtracted from values for UL13 phosphorylation of substrates.
In vitro autophosphorylation assay
35S-labeled UL13 or mutants obtained by in vitro translation were immunoprecipitated and subjected to the in vitro kinase assay in the absence of an exogenous substrate. Products were resolved by SDS-PAGE and phosphorimager analysis was used to quantitate the total amount of enzyme (35S-labeled) (obtained before the addition of 32P), and autophosphorylated enzyme (32P-labeled). The phosphorimager intensity value of 32P-labeled enzyme was divided by the value of 35S-labeled enzyme to normalize for translation efficiency. Values for the UL13 mutants were normalized to the wild-type UL13 value set to 1. In samples containing both 32P and 35S, the 35S signal was shielded by a sheet of exposed X-ray film between the blot and phosphorimager plate (Cao et al., 2005;Tavis et al., 1998). Statistics were obtained by a oneway ANOVA followed by a Bonferroni post hoc and a P-value of 0.05.
MBP in vitro kinase assay
Myelin basic protein (MBP) was used as an exogenous protein substrate in the in vitro kinase assay described above. 17.9 μM MBP (Upstate) was added to the UL13 immunoprecipitates in kinase assay buffer. Recombinant activated ERK 1 (Upstate) was used as a positive control for assays involving MBP. Values for phosphorylation of a substrate by the inactive K176A mutant were designated as background and were subtracted from all other values. To compare phosphorylation of MBP by UL13 and UL13 mutants, the samples were normalized for translation efficiency as described above.
Acknowledgments
We thank Steve Horvath and Paul Allen for peptide synthesis. We thank Joe Baldassare, Peter Zassenhaus, Abdul Waheed, Brian Geiss, Kristine Wylie, and the Leib, Pekosz, Diamond, Wang, Yu, Blithe, and Stuart labs for helpful advice and discussions. This work was supported by AHA 0410096Z and PHS AI061845 to G. C. M. and PHS AI059050 to L.A.M.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alvarez E, Northwood IC, Gonzalez FA, Latour DA, Seth A, Abate C, Curran T, Davis RJ. Pro-Leu-Ser/Thr-Pro Is a consensus prinmary sequence for substrate protein phosphorylation. J Biol Chem. 1991;266:15277–15286. [PubMed] [Google Scholar]
- Baek MC, Krosky PM, Coen DM. Relationship between autophosphorylation and phosphorylation of exogenous substrates by the human cytomegalovirus UL97 protein kinase. J Virol. 2002a;76:11943–11952. doi: 10.1128/JVI.76.23.11943-11952.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek MC, Krosky PM, He Z, Coen DM. Specific phosphorylation of exogenous protein and peptide substrates by the human cytomegalovirus UL97 protein kinase. Importance of the P+5 position. J Biol Chem. 2002b;277:29593–29599. doi: 10.1074/jbc.M202312200. [DOI] [PubMed] [Google Scholar]
- Cao F, Badtke MP, Metzger LM, Yao E, Adeyemo B, Gong Y, Tavis JE. Identification of an essential molecular contact pint on the duck hepatitis B virus reverse transcriptase. J Virol. 2005;79:10164–10170. doi: 10.1128/JVI.79.16.10164-10170.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrera AC, Alexandrov K, Roberts TM. The conserved lysine of the catalytic domain of protein kinases is actively involved in the phosphotransfer reaction and not required for anchoring ATP. Proc Natl Acad Sci. 1993;90:442–446. doi: 10.1073/pnas.90.2.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chee MS, Lawrence GL, Barrell BG. Alpha-, beta- and gammaherpesviruses encode a putative phosphotransferase. J Gen Virol. 1989;70:1151–1160. doi: 10.1099/0022-1317-70-5-1151. [DOI] [PubMed] [Google Scholar]
- Clark-Lewis I, Sanghera JS, Pelech SL. Definition of a consensus sequence for peptide subsrate recoginitionby p44mpk, the Meiosis-activated myelin basic protein kinase. J Biol Chem. 1991;266:15180–15184. [PubMed] [Google Scholar]
- Coulter JL, Moss HWM, Lang J, McGeoch DJ. A mutant of herpes simplex virus type 1 in which the UL13 protein kinase gene is disrupted. J Gen Virol. 1993;74:387–395. doi: 10.1099/0022-1317-74-3-387. [DOI] [PubMed] [Google Scholar]
- Cunningham C, Davison AJ, Dolan A, Frame MC, McGeoch DJ, Meredith DM, Moss HW, Orr AC. The UL13 virion protein of herpes simplex virus type 1 is phosphorylated by a novel virus-induced protein kinase. J Gen Virol. 1992;73:303–311. doi: 10.1099/0022-1317-73-2-303. [DOI] [PubMed] [Google Scholar]
- Daikoku T, Shibata S, Goshima F, Oshima S, Tsurumi T, Yamada H, Yamashita Y, Nishiyama Y. Purification and characterization of the protein kinase encoded by the UL13 gene of herpes simplex virus type 2. Virology. 1997;235:82–93. doi: 10.1006/viro.1997.8653. [DOI] [PubMed] [Google Scholar]
- Dolan A, Jamieson F, Cunningham C, Barnett B, McGeoch DJ. The genome sequence of herpes simplex virus type 2. J Virol. 1998;72:2010–2021. doi: 10.1128/jvi.72.3.2010-2021.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott G, O'Reilly D, O'Hare P. Identification of phosphorylation sites within the herpes simplex virus tegument protein VP22. J Virol. 1999;73:6203–6206. doi: 10.1128/jvi.73.7.6203-6206.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiss BJ, Cano GL, Tavis JE, Morrison LA. Herpes simplex virus 2 VP22 phosphorylation induced by cellular and viral kinases does not influence intracellular localization. Virology. 2004;330:74–81. doi: 10.1016/j.virol.2004.08.034. [DOI] [PubMed] [Google Scholar]
- Geiss BJ, Tavis JE, Metzger LM, Leib DA, Morrison LA. Temporal regulation of herpes simplex virus type 2 VP22 expression and phosphorylation. J Virol. 2001;75:10721–10729. doi: 10.1128/JVI.75.22.10721-10729.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez FA, Raden DL, Davis RJ. Identification of substrate recognition determinants for human ERK1 and ERK2 protein kinases. J Biol Chem. 1991;266:22159–22163. [PubMed] [Google Scholar]
- Hall FL, Braun RK, Mihara K, Fung YK, Berndt N, Carbonaro-Hall DA, Vulliet PR. Characterization of the cytoplasmic proline-directed protein kinase in proliferative cells and tissues as a heterodimer comprised of p34cdc2 and p58cyclin A. J Biol Chem. 1991;266:17430–17440. [PubMed] [Google Scholar]
- Hanks SK, Quinn AM, Hunter T. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science. 1988;241:42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- Haycock JW, Ahn NG, Cobb MH, Krebs EG. ERK1 and ERK2, two microtubule-associated protein 2 kinases mediate the phosphorylation of tyrosine hydroxylase at serine-31 in situ. Proc Natl Acad Sci. 1992;89:2365–2369. doi: 10.1073/pnas.89.6.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z, He YS, Kim Y, Chu L, Ohmstede C, Biron KK, Coen DM. The Human Cytomegalovirus UL97 Protein Is a Protein Kinase That Autopohospohrylates on Serines and Threonines. J Virol. 1997;71:405–411. doi: 10.1128/jvi.71.1.405-411.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer GH, Garrod S, Woods VL, Taylor SS. Catalytic independent functions of a protein kinase revealed by a kinase-dead mutant: study of the Lys72His mutant of CAMP-dependent kinase. J Mol Biol. 2005;351:1110–1122. doi: 10.1016/j.jmb.2005.06.011. [DOI] [PubMed] [Google Scholar]
- Kawaguchi Y, Kato Protein kinases conserved in herpesviruses potentially share a function mimicking the cellular protein kinase cdc2. Rev Med Virol. 2003;13:331–340. doi: 10.1002/rmv.402. [DOI] [PubMed] [Google Scholar]
- Kawaguchi Y, Kato K, Tanaka M, Kanamori M, Nishiyama Y, Yamanashi Conserved protein kinases encoded by herpesviruses and cellular protein kinase cdc2 target the same phosphorylation site in eukaryotic elongation factor 1delta. J Virol. 2003;77:2359–2368. doi: 10.1128/JVI.77.4.2359-2368.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi Y, Matsumoto M, Roizman B, Hirai K. Cellular elongation factor 1delta is modified in cells infected with representative alpha-, beta-, or gammaherpesviruses. J Virol. 1999;77:4456–4460. doi: 10.1128/jvi.73.5.4456-4460.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi Y, Van Sant C, Roizman B. Eukaryotic elongation factor 1delta is hyperphosphorylated by the protein kinase encoded by the U(L)13 gene of herpes simplex virus 1. J Virol. 2001;72:1731–1736. doi: 10.1128/jvi.72.3.1731-1736.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon TK, Lynch J, Hay J, Ruyechan W, Grose C. Varicella-zoster virus ORF47 protein serine kinase: characterization of a cloned, biologically active phosphotransferase and two viral substrates, ORF62 and ORF63. J Virol. 2001;75:8854–8858. doi: 10.1128/JVI.75.18.8854-8858.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litchfield DW, Bosc DG, Slominski E. The protein kinase from mitotic human cells that phosphorylates Ser-209 on the casein kinase II ß-subunit is p34cdc2. Biochim Biophys Acta. 1995;1269:69–78. doi: 10.1016/0167-4889(95)00100-7. [DOI] [PubMed] [Google Scholar]
- Long MC, Leong V, Schaffer PA, Spencer CA, Rice SA. ICP22 and the UL13 protein kinase are both required for herpes simplex virus-induced modification of the large subunit of RNA polymerase II. J Virol. 1999;73:5593–5604. doi: 10.1128/jvi.73.7.5593-5604.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin O, Meggio F, Draetta G, Pinna LA. The consensus sequences for cdc2 kinase and for casein kinase mutually incompatible. A study with peptides derived from the b subunit of casein kinase-2. FEBS Lett. 1992;13:111–114. doi: 10.1016/0014-5793(92)80221-2. [DOI] [PubMed] [Google Scholar]
- McGeoch DJ, Davison AJ. Alphaherpesviruses possess a gene homologous to the protein kinase gene family of eukaryotes and retroviruses. Nucleic Acids Res. 1986;25:1765–1777. doi: 10.1093/nar/14.4.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel D, Kramer S, Hohn S, Schaarschmidt P, Wunderlich K, Mertens T. Amino acids of conserved kinase motifs of cytomegalovirus protein UL97 are essential for autophosphorylation. J Virol. 1999;73:8898–8901. doi: 10.1128/jvi.73.10.8898-8901.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison E, Wang YF, Meredith D. Phosphorylation of structural components promotes dissociation of the herpes simplex virus type 1 tegument. J Virol. 1998;72:7108–7114. doi: 10.1128/jvi.72.9.7108-7114.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng TI, Ogle WO, Roizman B. UL13 protein kinase of herpes simplex virus 1 complexes with glycoprotein E and mediates the phosphorylation of the viral Fc receptor: glycoproteins E and I. Virology. 1998;241:37–48. doi: 10.1006/viro.1997.8963. [DOI] [PubMed] [Google Scholar]
- Nigg EA. The Substrates of the cdc2 kinase. Semin Cell Biol. 1991;4:261–270. [PubMed] [Google Scholar]
- Ogle W, Ng TI, Carter KL, Roizman B. The UL13 protein kinase and the infected cell type are determinants of posttranslational modification of ICP0. Virology. 1997;235:406–413. doi: 10.1006/viro.1997.8710. [DOI] [PubMed] [Google Scholar]
- Overton HA, McMillan DJ, Klavinskis LS, Hope L, Ritchie AJ, Wong-Kai-In P. Herpes simplex virus type 1 gene UL13 encodes a phosphoprotein that is a component of the virion. Virology. 1992;190:184–192. doi: 10.1016/0042-6822(92)91204-8. [DOI] [PubMed] [Google Scholar]
- Pinna LA, Meggio F, Marchiori F, Borin G. Opposite and mutually incompatible structural requirements of type-2 casein kinase and cAMP-dependent protein kinase as visualized with synthetic peptide substrates. FEBS Lett. 1984;171:211–214. doi: 10.1016/0014-5793(84)80490-1. [DOI] [PubMed] [Google Scholar]
- Purves FC, Ogle W, Roizman B. Processing of the herpes simplex virus regulatory protein α22 mediated by the UL13 protein kinase determines the accumulation of a subset of α and γ mRNAs and proteins in infected cells. Proc Natl Acad Sci U S A. 1993;90:6701–6705. doi: 10.1073/pnas.90.14.6701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purves FC, Roizman B. The UL13 gene of herpes simplex virus 1 encodes the functions for posttranslational processing associated with phosphorylation of the regulatory protein alpha-22. Proc Natl Acad Sci. 1992;89:7310–7314. doi: 10.1073/pnas.89.16.7310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roizman B, Sears AE. Herpes simplex viruses and their replication. In: Fields BN, Knipe DM, Howley PM, editors. Fields Virology. Lippencott Raven; Philidelphia, PA: 1996. pp. 2231–2295. [Google Scholar]
- Rose G, Gierasch LM, Smith J. Turns in peptides and proteins. Adv Protein Chem. 1985;37:1–109. doi: 10.1016/s0065-3233(08)60063-7. [DOI] [PubMed] [Google Scholar]
- Shah OJ, Ghosh S, Hunter T. Mitotic regulation of ribosomal S6 kinase 1 involves Ser/Thr, Pro phosphorylation of consensus and non-consensus sites by cdc2. J Biol Chem. 2003;278:16433–16442. doi: 10.1074/jbc.M300435200. [DOI] [PubMed] [Google Scholar]
- Smith RF, Smith TF. Identification of new protein kinase-related genes in three herpes viruses, herpes simplex virus, varicella-zoster virus, and Epstein-Barr virus. J Virol. 1989;63:450–455. doi: 10.1128/jvi.63.1.450-455.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Songyang Z, Lu KP, Kwon YT, Tsai LH, Filhol O, Cochet C, Brickey DS, Soderling TR, Bartleson C, Graves DJ, DeMaggio AJ, Hoekstra MF, Blenis J, Hunter T, Cantley LC. A structural basis for substrate specificities of protein Ser/Thr kinases: primary sequence preference of casein kinases I and II, NIMA, phosphorylase kinase, calmodulin-dependent kinase II, CDK5, and Erk1. Mol Cell Biol. 1996;16:6486–6493. doi: 10.1128/mcb.16.11.6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan J, Koszelak M, Mendelow M, Kwon YG, Lawrence DS. The design of peptide-based substrates for the cdc2 proteins kinase. Biochem J. 1995;309:927–931. doi: 10.1042/bj3090927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Nishiyama Y, Sata T, Kawaguchi Y. The role of protein kinase activity expressed by the UL13 gene of herpes simplex virus 1: the activity is not essential for optimal expression of UL41 and ICP0. Virology. 2005;341:301–312. doi: 10.1016/j.virol.2005.07.010. [DOI] [PubMed] [Google Scholar]
- Tavis JE, Massey B, Gong Y. The duck hepatitis B virus polymerase is activated by its RNA packaging signal, epsilon. J Virol. 1998;72:5789–5796. doi: 10.1128/jvi.72.7.5789-5796.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Wu J. Autophosphorylation kinetics of protein kinases. Biochem J. 2002;368:947–952. doi: 10.1042/BJ20020557. [DOI] [PMC free article] [PubMed] [Google Scholar]