Abstract
Studies have shown that modification of critical cysteine residues in proteins leads to the regulation of protein function. These modifications include disulfide bond formation, glutathionylation, sulfenic and sulfinic acid formation, and S-nitrosation. The biotin switch assay was developed to specifically detect protein S-nitrosation. In this assay, proteins are denatured with SDS in the presence of methyl methanethiosulfonate (MMTS) to block free thiols. After acetone precipitation or Sephadex G25 separation to remove excess MMTS, HPDP-biotin and 1 mM ascorbate are added to reduce the S-nitrosothiol bonds and label the reduced thiols with biotin. The proteins are then separated on a non-reducing SDS PAGE and detected using either streptavidin-HRP or anti-biotin HRP conjugate. Our examination of this labeling scheme has revealed that the extent of labeling depends on the buffer composition and importantly, on the choice of metal ion chelator (DTPA vs. EDTA). Unexpectedly, using purified S-nitrosated albumin, we have found that “contaminating” copper is required for the ascorbate-dependent degradation of S-nitrosothiol; this is consistent with the fact that ascorbate itself does not rapidly reduce S-nitrosothiols. Removal of copper from buffers by DTPA and other copper chelators preserves approximately 90% of the S-nitrosothiol, while the inclusion of copper and ascorbate completely eliminates the S-nitrosothiol in the preparation and increases the specific biotin labeling. These biotin switch experiments were confirmed using triiodide-based and copper-based reductive chemiluminescence. Additional modifications of the assay using NEM (N-ethylmaleimide) for thiol blockade, ferricyanide pretreatment to stabilize S-nitrosated hemoglobin, and cyanine dye labeling instead of biotin, are presented for the measurement of cellular and blood S-nitrosothiols. These results indicate that degradation of S-nitrosothiol in the standard biotin switch assay is metal ion-dependent and that experimental variability in S-nitrosothiol yields using this assay occurs secondary to the inclusion of metal ion chelators in reagents and variable metal ion contamination of buffers and labware. The addition of copper to ascorbate allows for a simple assay modification that dramatically increases sensitivity while maintaining specificity.
Introduction
Nitric oxide (NO) is a diatomic molecule which plays a critical role in the maintenance of basal vascular tone and in regulating blood vessel homeostasis, neuronal signaling, and immune-function. Nitric oxide and oxidized derivatives of NO such as nitrite are capable of post-translational modification of protein via formation of iron-nitrosyls, nitrated lipids, N-nitrosamines, oxidized thiols and S-nitrosothiols (RSNO) [1-11] Studies have shown that modification of critical cysteine residues in proteins leads to the regulation of protein function. These modifications include disulfide bonds (e.g. glutathionylation), sulfenic and sulfinic acids, and S-nitrosothiols [5, 12-17]. Jaffrey et al. have developed a method, the biotin switch assay, to specifically detect protein S-nitrosation [18]. In this assay, proteins are denatured with sodium dodecyl sulfate (SDS) in the presence of methyl methanethiosulfonate (MMTS) to block free thiols. The SDS, combined with an incubation temperature of 50°C, is used to expose and facilitate modification of thiols buried in the interior of proteins. After acetone precipitation, or Sephadex G25 separation, to remove excess MMTS and other small molecules, 1 mM ascorbate and biotin-HPDP (N-[6-(biotinamido)hexyl]-3′-(2′-pyridyldithio)propionamide) are added to reduce the S-N bond and label the reduced thiol with biotin respectively. The proteins are then separated by SDS PAGE and detected using either streptavidin-HRP or anti-biotin HRP conjugate. This assay is the first to allow sensitive and specific identification of S-nitrosated proteins using gel-based techniques and further allows isolation of modified protein for identification by mass spectrometry.
While the biotin switch assay is now widely used among different laboratories, many groups have had to modify the method by either increasing the ascorbate concentration or incubation time [19-22]. The reason for this is that ascorbate alone is a very poor reducer of RSNO [23-25] and 50 mM ascorbate will minimally reduce S-nitrosated bovine serum albumin (BSA-SNO) during 3 hours incubation at 37 °C [20]. However, there is concern that increasing the ascorbate concentration will diminish the selectivity of this assay. Higher concentrations of ascorbate can reduce not only RSNO, but may also reduce some disulfides, as shown for tubulin, tau and microtubule associated protein-2 [21]. The fact that ascorbate does not have the reductive capacity to rapidly reduce RSNO raises the provocative question of how the biotin switch assay works.
This study was conducted in order to rationalize our anecdotal observation that the “clean experiment” with carefully prepared buffers, Sephadex G25 columns washed with buffer containing diethylenetriaminepentaacetic acid (DTPA), and inclusion of DTPA in buffers (all typically used to stabilize RSNO and minimize other oxidative metal-ion dependent processes) always resulted in diminished or zero biotin labeling, while the “dirty experiment”, in which these precautions were not taken, increased the yield. We have found that different levels of biotin labeling are obtained for the same S-nitrosated protein using different buffers and that the labeling is decreased significantly with the inclusion of different chelators (DTPA, EDTA or Chelex 100). These observations are consistent with many anecdotal (though rarely published) variable results obtained using the assay [26]. We now report that ascorbate/HPDP-biotin, at the levels used in the biotin switch assay protocol [18] does not reduce and label S-nitrosothiol in the absence of contaminating metal ions, thus explaining the aforementioned paradoxical observations. Addition of copper ions to the labeling cocktail results in reproducible and consistent labeling of S-nitrosothiols with biotin. Reduced copper (Cu+) is required for the degradation of RSNO to free thiol for subsequent biotinylation. Similarly, the inclusion of copper chelators in buffer significantly decreases SNO decomposition and biotin labeling which can be reversed by addition of copper to the ascorbate/HPDP-biotin step. This finding explains the variable results observed with this method in different laboratories using varied conditions, chelators and buffers. We also present additional modifications that allow for the extension of this assay to the measurement of S-nitrosated heme proteins, such as hemoglobin, and more sophisticated cyanine dye labeling techniques.
Materials and Methods
Chemicals
All the chemicals and reagents were obtained from Sigma-Aldrich, unless otherwise indicated. Anti-biotin antibodies were obtained from Jackson ImmunoResearch (West Grove, PA). Anti-nitrotyrosine antibodies were obtained from Upstate (Lake Park, NY). Biotin-HPDP was obtained from Pierce Biotechnology (Rockford, IL.). Cy5 maleimide dye was obtained from GE Healthcare.
Protocol
The study protocol for blood acquisition was approved by the Institutional Review Board of the National Heart, Lung and Blood Institute, and all normal volunteers provided written, informed consent. Blood was processed for these studies using methodologies to limit artifactual ex-vivo hemolysis and levels of plasma hemoglobin were consistent with previous studies [27, 28]. Briefly, blood was collected from artery or vein using large bore catheters (18 gauge). The first 3 mL of blood was discarded and the blood then slowly drawn into heparinized syringes. Blood was then spun at 750 X g for 5 minutes without braking and plasma removed. Plasma was then spun at 14,000 X g for 10 minutes to eliminate residual erythrocytes and platelets.
Synthesis of S-nitrosated proteins
Human and bovine serum albumin (HSA and BSA) were purchased from Sigma and dissolved in PBS buffer. Albumin was treated with dithiothreitol (DTT) at a molar ratio of 3:1 DTT:albumin for 30 min. The albumin was dialyzed at 4°C against PBS containing 0.1 mM DTPA with 3 changes of buffer. The dialyzed albumin was mixed with S-nitroso-glutathione (GSNO, Cayman Chemical, Ann Arbor, MI) at molar ratio of 10:1 GSNO:albumin. The reaction was carried out in the dark at room temperature for 45 min. At the end of the reaction, 10 mM N-ethylmaleimide (NEM) was included to stop the reaction and block all possible free thiols. The albumin-SNO solution was dialyzed against PBS/DTPA at 4°C with 3 changes of buffer. The concentration of albumin-SNO was determined using triiodide-based reductive chemiluminescence (radical purger and NO Analyzer; Sievers, CO).
Hemoglobin used in this study was obtained from Sigma or prepared fresh from human red blood cells. S-nitrosocysteine (CSNO) was prepared as previously described using equimolar additions of acidified L-cysteine and nitrite in EDTA buffer [27]. S-nitroso-hemoglobin (Hb-SNO) was prepared by incubation of hemoglobin and CSNO at a molar ratio of 1:3 heme:CSNO at 4°C for 60 min in the dark. At the end of the reaction, NEM was added to block unreacted cysteine residues. The Hb-SNO was either passed through a G25 column or dialyzed against PBS with 0.1 mM DTPA. The concentration of S-nitrosothiol (RSNO) was determined by using triiodide-based reductive chemiluminescence and hemoglobin concentration was measured by Drabkin’s method (all concentrations reported in terms of heme).
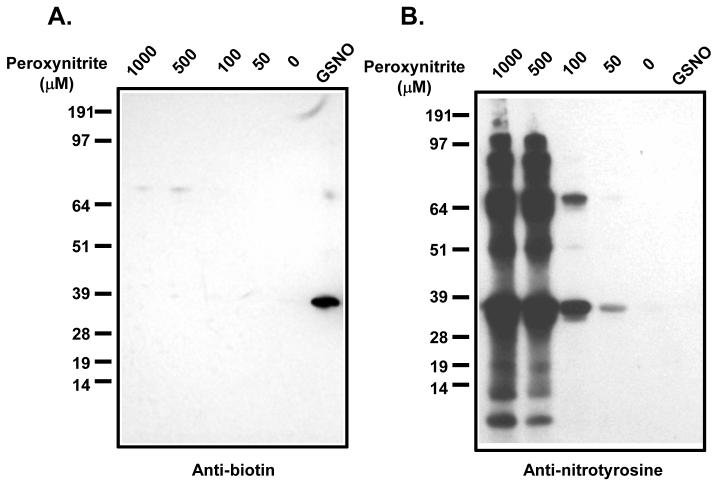
Purified erythrocyte GAPDH was obtained from Sigma. The purified GAPDH was treated with DTT (5 mM), and then passed through a Sephadex G25 column (Amersham). The reduced GAPDH was treated with peroxynitrite or GSNO at the concentrations indicated at room temperature for 30 min in dark. At the end of the reactions, NEM was included and incubation continued for 10 min. The biotin switch assay was performed and proteins were separated by SDS PAGE and transferred to a nitrocellulose membrane. Immunoblots were performed with anti-biotin and anti-nitrotyrosine antibodies.
Detection of S-nitrosated proteins by biotinylation (biotin switch assay)
We used the method of Jaffrey et al. [18] with some modifications. To 20 μl of protein sample or nitrosated protein, 80 μl of blocking buffer was added and mixed. The blocking solution contained 250 mM HEPES, pH 7.7, 1 mM EDTA, 0.1 mM neocuproine (HEN buffer), 1% SDS and either 10 mM MMTS or 10 mM NEM. Amber tubes were used in all these procedures and the reactions were carried out in dark to protect RSNO from light. When whole blood, red cells or hemoglobin were used, 4 mM ferricyanide was included to stabilize Hb-SNO during the SDS blocking step (see results). The reaction was carried out at either 50°C, or at 40°C when NEM was used for blocking step, for 30 min with occasional vortex mixing. Then the reaction mixture was passed 3 times through microspin G25 columns (Amersham Biosciences, Piscataway, NJ 08855-1327). In some situations, acetone was used to precipitate proteins, and the precipitant was washed with 70% acetone and resuspended in 100 μl of PBS. Biotin-HPDP (10 μl of 4 mM) was added to the reaction and 1 mM ascorbate (final concentration) was included, and mixed by gently vortexing. The biotinylation reaction was carried out at room temperature for 60 min. The fresh reactions were then run on non-reducing SDS PAGE or stored at -20°C for later use.
When copper dependence was tested in the biotin switch assay, a saturated CuCl solution was prepared in HPLC grade water by vortexing for 1 min. This saturated CuCl solution was assumed to be 100 μM [29, 30]. The CuCl solution was centrifuged for 1 min at 13000 rpm and was then CuCl was diluted and added to the biotinylation reaction.
To detect S-nitrosated proteins now labeled with HPDP-biotin, western blot was carried out using peroxidase conjugated anti-biotin antibodies (Jackson ImmunoResearch, West Grove, PA).
Nitric oxide detection by triiodide-based reductive gas phase chemiluminescence assay
We followed the standard procedure in our lab to measure S-nitrosothiol by triiodide-based reductive chemiluminescence assay [31-33]. Samples were treated with one tenth volume of 5% acidified sulfanilamide (5% sulfanilamide in 1 N HCl) for 3 min and injected into a triiodide containing vessel actively purged with a helium stream in line with an NO chemiluminescence analyzer (Sievers; Boulder, CO). To discriminate S-nitrosothiol (mercury labile) and iron-nitrosyl (mercury stable) complexes, samples were treated with and without HgCl2 (5 mM) for 2 min, followed by a 3 min treatment with acidified sulfanilamide prior to injection of the sample into the tri-iodide solution.
When RSNO was measured after the biotinylation reaction, samples were prepared as described in the biotin switch assay. The reaction mixture from the biotin switch assay was treated with acidified sulfanilamide and injected into the triiodide containing vessel in a parallel experiment to confirm successful switch of S-nitrosothiol (which gives a signal in tri-idodide reductive chemiluminescence) to a biotin conjugate (which produces no signal in tri-iodide reductive chemiluminescence). The same samples were simultaneously run on non-reducing SDS PAGE, and immunoblots were performed. This experiment allowed us to test the ability of the biotin switch assay to reduce S-nitrosothiol and label with biotin using two complimentary methodologies (one that detects RSNO and the second that detects replacement of RSNO with biotin label).
Copper/cysteine assay
The copper/cysteine assay (2C assay) was performed as reported [30]. The assay was carried out in a purge vessel using cysteine (1 mM) and CuCl (100 μM, pH adjusted to 6.5) at 50°C. To compare signals, as measured by the area under the curve (AUC) of the chemiluminescence trace, obtained from the triiodide and copper/cysteine assays, we used HSA-SNO and Hb-SNO. Hb-SNO (60, 120, 240, 480 pmoles RSNO) was treated with 10 mM K3Fe(CN)6 and KCN in the dark for 10 min prior to injection (this blocks heme autocapture of NO released in the purge vessel from SNO). HSA-SNO (55, 110, 220 pmoles RSNO) was injected without any pre-treatment.
Peroxynitrite treatment of GAPDH
GAPDH (Sigma) was dissolved in PBS and treated with 5 mM DTT. After passing through a Sephadex G25 column to remove excess DTT, GAPDH was treated with the indicated concentrations of peroxynitrite. Peroxynitrite was obtained from Cayman Chemical (Ann Arbor, MI). The concentration of peroxynitrite was determined by spectrophotometry (λmax = 302 nm, ε:1670).
Synthesis of S-Thiolated HSA
HSA was treated with low molecular weight disulfides in order to generate protein mixed disulfides. A 10 mg/ml solution of HSA (in 50 mM phosphate buffer with 1 mM DTPA pH 7.4) was incubated overnight at 4° C either alone (control) or with 2 mM cystine (CSSC) or 2 mM glutathione disulfide (GSSG). NEM (5 mM) was added in order to stop the reaction and block any remaining free thiols. HSA was then separated from excess low molecular weight disulfides and NEM by G25 sephadex column chromatography.
HPLC Detection of Low Molecular Weight Thiols
Protein S-thiolation was determined using the 7-Fluorobenzofurazan-4-sulfonic acid (SBDF) HPLC method. This method employs tributylphosphine (TBP) to reduce protein mixed disulfides. The nascent low molecular weight thiols are then fluorescently labeled with SBDF and separated by HPLC[34]. In detail, S-thiolated HSA (in 40 μl volume) was reduced by the addition of 10 μl of 15% (vol:vol) TBP in N,N-dimethylformamide (DMF) and incubated for 30 minutes at 4° C. After reduction, HSA was precipitated by the addition of 50 μl of 10% (vol:vol) trichloroacetic acid (TCA) and spun down by centrifugation at 2000 x g for 5 minutes. Low molecular weight thiols in 25 μl of supernatant were then derivatized with 75 μl of SBDF (0.67 mg/ml 2.5 M borate buffer pH 9.5) at 60° C for 60 minutes. Samples were analyzed by HPLC with fluorescence detection[34]. Cysteine and GSH that were bound to HSA were quantified based on standard curves generated for each of the compounds. Values were normalized to report low molecular weight thiol quantities in moles per mole of HSA.
Culture of Normal Human Bronchial Epithelial Cells
Primary normal human bronchial epithelial (NHBE) cells were purchased from Cambrex. Cells were maintained in Bronchial Epithelial Basal Medium (BEBM) containing growth supplements (bovine pituitary extract, hydrocortisone, hEGF, epinephrine, insulin, triiodothyronine, transferrin, and retinoic acid) from Cambrex. Cells were incubated at 37° C and 5% CO2 in a humidified incubator and subcultured or treated at 80% confluence. CSNO treatments were added in Hank’s balanced salt solution (HBSS) containing 10 mM HEPES. Following treatments, cells were lysed by sonication in HEN buffer (250 mM HEPES, 1 mM DTPA, 0.1 mM neocuproine) with 50 mM NEM and protease inhibitor cocktail (1:100).
CyDye labeling of modified protein thiols
An adaptation of the biotin switch assay was used to label reversible thiol modifications on cytosolic proteins from control and CSNO-treated HNBE cells [7]. In this method, we replaced the biotin tag with a thiol-reactive fluorescent dye (Cy5 maleimide).
Briefly, free thiols were blocked in lysis buffer containing 250 mM HEPES, 1 mM DTPA, 0.1 mM neocuproine, 50 mM NEM and 2.5% SDS for 1 hour at 50°C with occasional vortexing. Excess NEM was removed by ethanol precipitation of the proteins. Protein pellets were resuspended in a HEPES buffer containing 1% SDS either with or without metal chelators as appropriate for the reduction condition. Reduction using 1mM ascorbate +/- 10 μM copper sulfate was allowed to proceed in the presence of 10 pmoles of maleimide CyDye per μg protein for 1 hour at room temperature. Reduction using 1 mM DTT, however, was performed in the absence of the CyDye tag, for 30 minutes at room temperature, and was followed by a second ethanol precipitation step to remove excess DTT prior to the labeling step. Samples were diluted in Laemmli sample buffer (BioRad) contaiing 5% β-medcaptoethanol and proteins were separated by SDS-PAGE under reducing conditions in a gel cast in low fluorescence glass. Following electrophoresis, the gel image was acquired using a Typhoon Trio scanner.
Results
Detection of S-nitrosated proteins by the classic biotin switch assay results in different results using different laboratory buffers
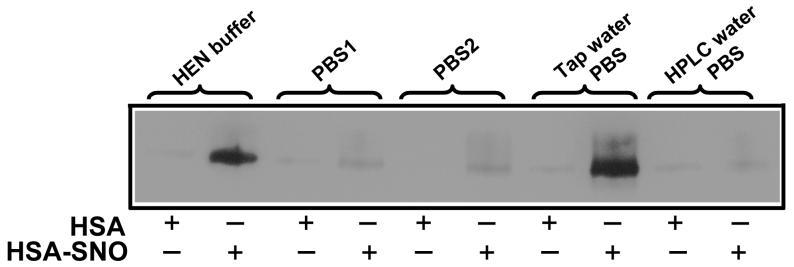
While the biotin switch developed by Jaffrey et al. [18] can specifically and sensitively detect S-nitrosation of proteins, the method has been modified in different laboratories, suggesting the detection of S-nitrosated proteins using this method exhibits unpredictable variability (based as we will now show on unpredictable variability in copper contamination) [19-22]. Indeed, in our own laboratory we found that the assay produced variable results for the same protein standards depending on the use of different labware and standard buffers. For example, in Figure 1 we examine the use of different buffers in the biotin switch assay (using 1 mM ascorbate as the reducing agent) to biotin-label a purified single S-nitrosated protein, S-nitrosated human serum albumin (HSA-SNO). As shown in Figure 1, the yields were quite variable, and surprisingly, the best biotin labeling of S-nitrosated thiol occurred with the use of PBS prepared with unfiltered and unchelated tap-water. PBS1 (BioWhittaker, catolog No.: 17-516F, lot No.:01109706) and PBS2 (Gibco, catalog No.:10010-023, lot No.:1295337) showed lower sinal. An old (8-month shelf life) HEN buffer produced an intermediate signal and freshly prepared PBS using HPLC-grade water produced minimal biotin labeling. This experiment highlights the variability we have encountered in two independent laboratories using this assay under assorted conditions.
Figure 1. Paradoxical different results were obtained with various buffers used in the biotin switch assay.
Equal amounts of HSA-SNO and HSA were subjected to the biotin switch assay. All the biotin switch assays were performed the same way except for the choice of buffers, however the SNO detection was highly variable. PBS1: PBS from BioWhittaker, catolog No.: 17-516F, lot No.:01109706; PBS2: PBS from Gibco, catalog No.:10010-023, lot No.:1295337; Tap water PBS: PBS made with regular tap water in the lab; HPLC water PBS: PBS made with HPLC grade water.
Metal-ion chelation diminished, and copper(I) enhanced the detection of RSNO in the biotin switch assay
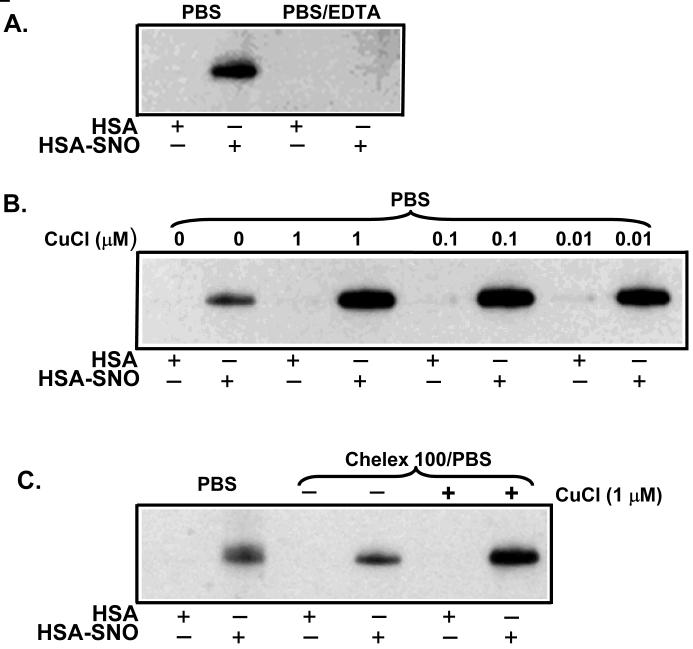
The diminished labeling in the biotin switch assay observed with “cleaner” buffers and the known fact that ascorbate-dependent RSNO reduction is not kinetically facile [23-25] suggested that metal contamination of the biotinylation buffer might represent an important, unidentified, and uncontrolled factor necessary for the biotin switch experiment. According to this hypothesis, the ascorbate may reduce an intermediate metal ion, such as copper(II), to form copper(I) which then reduces the S-nitrosothiol to form thiol and nitric oxide. The reduction of S-nitrosothiols by copper(I) is a well known reaction [35]. Copper and iron are ubiquitous contaminants of labware and buffers; for this reason experiments where the redox chemistry of such metal ions could compromise the validity of results are usually performed with the most effective transition-metal ion chelators (namely DTPA) in the reagents. In regular tap water, copper is often present in the micromolar range[36-39]. Therefore, the variation of biotin switch results between laboratories and even within laboratories may relate to the variable contamination of copper or other transition metal ions, and the variable incorporation of metal-ion chelators. To test this hypothesis, we examined the effectiveness of biotin labeling in the presence of chelating agents and added copper ions. As shown in Figure 2A, EDTA significantly inhibited the biotin switch labeling of HSA-SNO. Furthermore, the addition of different concentrations of CuCl greatly enhanced the biotin labeling (Figure 2B). Treating PBS with Chelex 100 decreased the biotin labeling while addition of CuCl to Chelex 100 treated PBS enhanced the biotin signal (Figure 2C). These data reveal the crucial importance of redox-cycling transition metal ions in for successful biotin labeling.
Figure 2. Copper chelation diminished the detection of RSNO and copper addition enhanced the signal.
Equal amounts of HSA-SNO and HSA were subjected to the biotin switch assay. All the biotin switch assays were performed the same way except for the addition of metal chelators (EDTA or chelex) or copper(CuCl) to the buffers, however the SNO detection was highly variable. (A) Inclusion of 1 mM EDTA in PBS buffer eliminated the biotin switch signal. (B) In the biotinylation reaction, HSA or HSA-SNO (as indicated) were treated with either PBS or PBS with indicated concentrations of CuCl. The addition of CuCl increased biotin labeling (i.e. SNO detection) (C) The biotinylation reaction was performed in either PBS or Chelex 100 (BioRad) treated PBS or Chelex 100 treated PBS with indicated CuCl added.
Combination of ascorbate with CuCl degraded RSNO
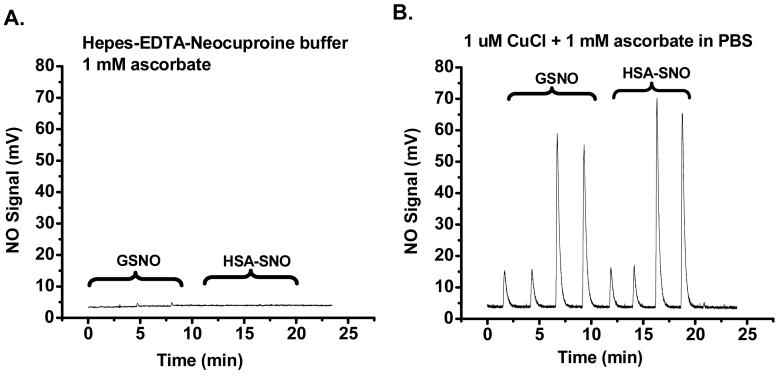
To examine under what conditions ascorbate can reduce S-nitrosothiols to liberate NO, we examined NO formation using an ozone-based chemiluminescence assay. In Figure 3A, both GSNO (20 and 100 pmol, duplicate injections) and HSA-SNO (22 and 110 pmol, duplicate injections) were injected into a purge vessel containing HEN buffer and ascorbate (1 mM). These conditions approximate the reduction conditions employed in the biotin switch assay. Any generated NO is carried to the reaction chamber of an NO analyzer by helium gas. As can be seen (Figure 3A) there was no NO released from the S-nitrosothiols in the ascorbate/HEN buffer. In contrast (Figure 3B), if the purge vessel contained PBS, ascorabate (1 mM) and CuCl (1 μM), robust NO generation was detected. These results were consistent with the greater biotin labeling of the ascorbate-copper buffers shown in Figure 2.
Figure 3. Assay by ozone-based chemiluminescence nitric oxide analyzer revealed that CuCl is required for RSNO decomposition.
Twenty and 100 pmol of S-nitroso-glutathione (GSNO) and 22 and 110 pmol of HSA-SNO were injected into a glass vessel containing the indicated solutions with 1 mM ascorbate (A) and 1 mM ascorbate plus 1 μM CuCl (B). As shown in panel (A), 1 mM ascorbate in HEN buffer does not release NO from either GSNO or HSA-SNO. However, the combination of ascorbate and CuCl facilitates the decomposition of RSNO, releasing NO (B).
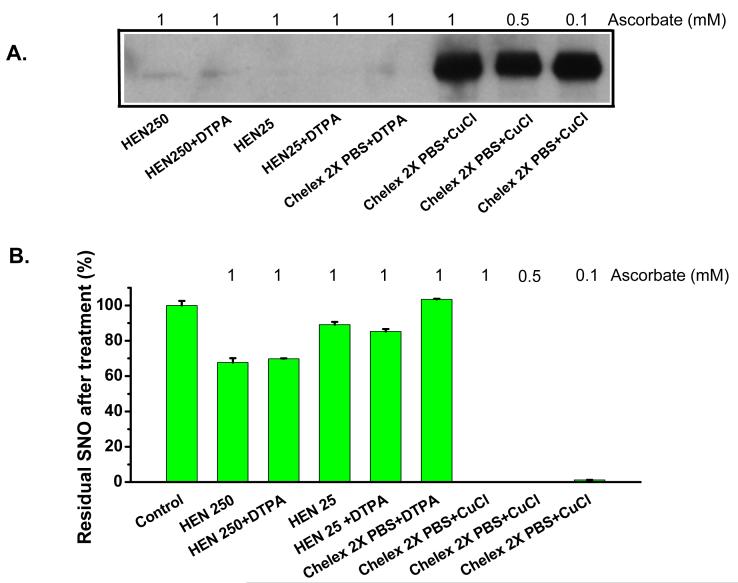
We next measured the RSNO content in the reaction mixtures from the biotin switch assay by triiodide chemiluminescence to confirm successful reduction of S-nitrosothiol. The same samples were run on non-reducing SDS PAGE, and immunoblots were performed. This experiment allows us to test the ability of the biotin switch assay conditions to decompose S-nitrosothiols and label with biotin using complementary methodologies. Figure 4A and B, show the biotinylation intensity and the residual RSNO detected by triiodide-based chemiluminescence after incubation with 1 mM ascorbate in the indicated buffers. Using 250 mM Hepes-containing HEN buffer (HEN250 on Figure 4A) about 70% RSNO remains, while if the HEN buffer was formulated with 25 mM Hepes (HEN25), the residual RSNO was 85% to 89%. Twice Chelex 100-treated PBS totally preserved RSNO did not support ascorbate-mediated RSNO decay. In all of these situations, no protein biotinylation was observed. In contrast, when CuCl was added to Chelex 100-treated PBS, RSNO decomposition was complete and strong biotinylation was observed. In fact, both loss of RSNO and efficient biotinylation were accomplished at even lower ascorbate concentrations (0.5 and 0.1 mM) than recommended for the biotin switch assay (Figure 4). We also performed an alternative assay, the 2C assay, in order to validate the triiodide-based chemiluminescence assay. Comparison of the AUCs obtained from the S-nitrosated proteins using the tri-iodide-based chemiluminescence assay and the 2C assay [29, 30] showed almost identical results, with an R2 value greater than 0.99 (figure 1 supplementary online material).
Figure 4. Copper and ascorbate decompose RSNO in biotinylation buffer.
The Biotin switch assay was carried out with HSA-SNO. HSA-SNO was resuspended in the buffers indicated in biotinylation step. Panel (A) is the immunoblot with anti-biotin peroxidase conjugate. Panel (B) is the residual RSNO measured at the end of the biotinylation reaction by injection of sample into triiodide-based ozone chemiluminescence. Only in the conditions with added copper was the HSA-SNO labeled with biotin (A) and were the NO signals not detected (indicating a replacement of a SNO group with biotin) (B).
Specificity of biotin switch
To verify that this assay is specific for S-nitrosation we examined peroxynitrite-treated and GSNO-treated GAPDH using the copper-modified biotin switch assay. Previous elegant studies have suggested that S-nitrosation of GAPDH transduces apoptotic cell death by nuclear translocation following Siah1 binding[40]. The rationale for this experiment is that peroxynitrite should oxidise protein thiol groups to disulfides and higher oxides whereas GSNO should generate S-nitroso-GAPDH. As shown in Figure 5A, the modified biotin switch assay produced a biotinylated protein band with GSNO treated GAPDH. Whereas peroxynitrite treated GAPDH resulted in minimal protein biotinylation, with only a very weak band observed at higher molecular weight. In order to show that peroxynitrtite was indeed modifying the protein, we examined protein tyrosine nitration by Western analysis (Figure 5B). Nitrotyrosine formation was observed at all peroxynitrite concentrations, and at higher concentrations, significant fragmentation and aggregation of the nitrated proteins was observed. These results suggest that while peroxynitrite can nitrate, fragment ang aggregate GAPDH, it is unable to generate thiol modifications that are detectable in the biotin switch method. This suggests that, at least, for GAPDH, the assay has specificity for S-nitrosation of other thiol oxidative modifications.
Figure 5. Specificity of the biotin switch for S-nitrosated GAPDH.
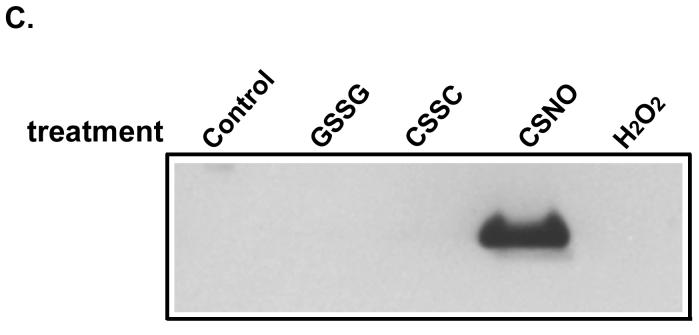
GAPDH (Sigma) was treated with 5 mM DTT and purified by passing through Sephadex G25. This reduced GAPDH was then treated with the indicated concentrations of peroxynitrite or GSNO (50 μM). At end of the reaction, 5 mM NEM was added and the reactions were used for the biotin switch assay. Samples were probed with anti-biotin (A) and anti-nitrotyrosine (B) antibodies. Panel (C) showed Results from biotin switch assay with HSA pre-treated with different chemicals. Cystine and GSSG treated HSA consisted of the same preparations as shown in supplementary figure 2. HSA was also treated with 1 mM H2O2 and 1 mM CSNO respectively at room temperature in dark for 30 min. Biotin switch assay was carried out with CuCl (0.1 μM) included in the biotinylation step.
To further explore whether the combination of ascorbate and CuCl can break disulfide bonds in S-thiolated proteins, HSA was treated with cystine and GSSG to increase the level of protein S-thiolation. It should be noted that HSA preparations are extensively (up to 50%) S-thiolated. Using fluorescence HPLC, it was determined that cystine-treatment increased cysteinylation from 0.34 to 0.5 cystine/HSA and glutathionylation from 0.02 to 0.05 GSH/HSA (supplementary figure 2). These S-thiolated HSA preparations were subjected to the biotin switch assay together with hydrogen peroxide-treated HSA (to form sulfenic acid) and CSNO-treated HSA (to form S-nitroso HSA). As shown in figure 5C, only the CSNO-treated protein gave a positive signal in the biotin switch assay. This data shows that the Cu/ascorbate treatment could selectively detect S-nitroso HSA over S-thiolated HSA and HSA-sulfenic acid. While we have shown evidence of selectivity for S-nitrosation in the case of both GAPDH and HSA, it should be cautioned that protein disulfides and sulfenic acids exist in a variety of protein environments, and that the possibility of false-positives need to be investigated on a protein by protein basis.
Modification of assay to detect Hb-SNO
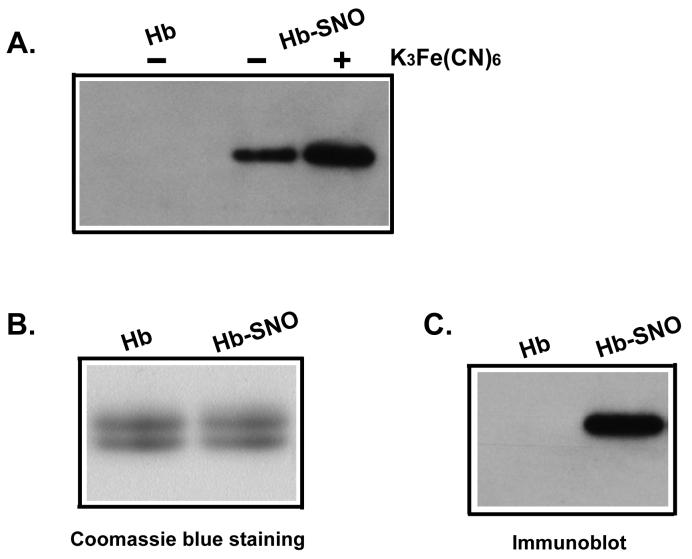
Based on our previous finding that ferricyanide treatment stabilizes Hb-SNO in a reductive milieu and in SDS, we examined if such treatment wound enhance the ability of the biotin switch assay to detect this modification. Pre-treatment with ferricyanide (4 mM concentration was used for red cells, 1 mM for Hb-SNO preparations) enhanced the Hb-SNO signal in the biotin switch assay (Figure 6A). We did not make artifactual RSNO using this ferricyanide/NEM procedure as previously reported [27, 33]. We also confirmed that that the β-chain of Hb is preferentially S-nitrosated using SDS-PAGE (Figure 6B and 6C). These results are consistent with previous findings that cysβ93 is the only solvent exposed cysteine on hemoglobin (there is one buried cysteine in both the alpha and the beta chains of human hemoglobin A). This result from S-nitrosated β chain also reinforces the observation that S-nitrosated cysβ93 in oxygenated R-state hemoglobin is not buried and is certainly accessible to solvent and reduction [27], an observation that refutes arguments based on X-ray crystallographic studies suggesting that only the T-state (deoxy conformation) is accessible to solvent [41].
Figure 6. Detection of Hb-SNO by the biotin switch assay.
(A) Ferricyanide pre-treatment stabilized S-nitrosated hemoglobin in the biotin switch assay. Biotin switch was performed with Hb and Hb-SNO in the presence or absence of ferricyanide (1 mM) as indicated. Panel (B) and (C) showed hemoglobin β chain was S-nitrosated. After performing the biotin switch assay and running the samples on 2 separate non-reducing SDS PAGE gels, the gels were either stained with Coomassie brilliant blue (B) or subjected to immunoblot to probe biotin (C). The comparison of (B) and (C) indicates specific S-nitrosation of the β chains of hemoglobin.
Sensitivity of the biotin switch assay with purified S-nitrosated albumin and hemoglobin using modified biotin switch assay (Copper/Ferricyanide/NEM biotin switch)
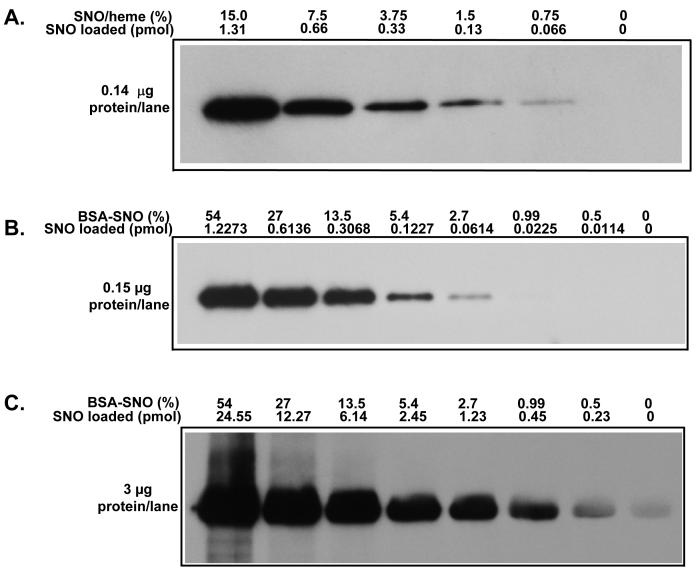
We next examined the sensitivity of the modified biotin switch using purified S-nitrosated albumin and hemoglobin. For these experiments, S-nitrosated protein was mixed with protein that was not nitrosated so that the total protein concentration was the same for each standard dilution. As shown in Figure 7A, by loading 0.14 μg hemoglobin protein in each lane, 0.066 pmol Hb-SNO could be detected (this amount corresponds to 1.29 μM SNO in the sample). When 0.15 μg albumin protein was loaded 0.061 pmol RSNO was detected (Figure 7B; this amount corresponds to 1.64 μM SNO in the sample); if 3 μg albumin protein was loaded then 0.23 pmol was detected (Figure 7C; this amount corresponds to 0.3 μM SNO in the sample). Since the biotin switch is a semiquantitative assay and the sensitivity is related to how much protein is loaded on the gel, our results suggest that 0.23 pmol RSNO might be a limiting detection amount in this system.
Figure 7. Sensitivity of the biotin switch with purified single proteins.
Hemoglobin (Hb) and S-nitroso-hemoglobin (Hb-SNO) (A) or Albumin and S-nitroso-albumin (BSA-SNO) (B, and C) were mixed to maintain equal total protein concentrations in all reactions, and then subjected to the biotin switch assay. RSNO concentrations and protein loading are indicated.
Detection of RSNO in plasma and red cells
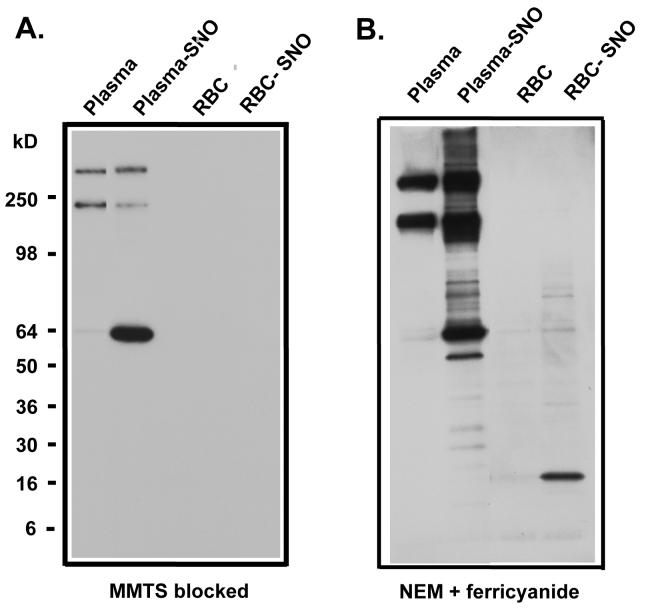
We next evaluated the biotin switch assay for detection of S-nitrosated plasma protein and hemoglobin in biological samples [27]. First, we treated human whole blood with CSNO, separated the plasma from red cells by centrifugation and measured the RSNO in these samples. With CSNO exposure, albumin is S-nitrosated and easily detectable. Interestingly, we were unable to detect Hb-SNO after CSNO treatment if we used MMTS in the blocking step. However, if we incubated samples in the first blocking step with NEM and ferricyanide we were able to detect Hb-SNO (Figure 8A and B). It should be noted that there is a strong false positive signal for immunoglobin in plasma. Furthermore, under basal conditions without CSNO treatment the labeling of albumin and hemoglobin is no greater than other proteins in plasma (immunoglobulin) and the eythrocyte (other high molecular weight proteins) suggesting that these background signals may derive from a failure to completely block all thiols with NEM or MMTS. Note that the band intensities for both albumin and hemoglobin were at the limits of detection in baseline blood. In blood samples analyzed from these volunteers we were unable to measure differences in the levels of biotin-labeled proteins between artery and venous plasma or red cells (data not shown).
Figure 8. Detection of RSNO in plasma and red cells.
Blood was treated as detailed in Materials and Methods. Plasma and red cells were separated. The biotin switch was carried out with plasma, CSNO-treated plasma (plasma-SNO), red cells and CSNO-treated red cells (RBC-SNO) using MMTS blocking (A) and NEM/ferricyanide blocking treatment (B).
CyDye labeling of reversible thiol modifications
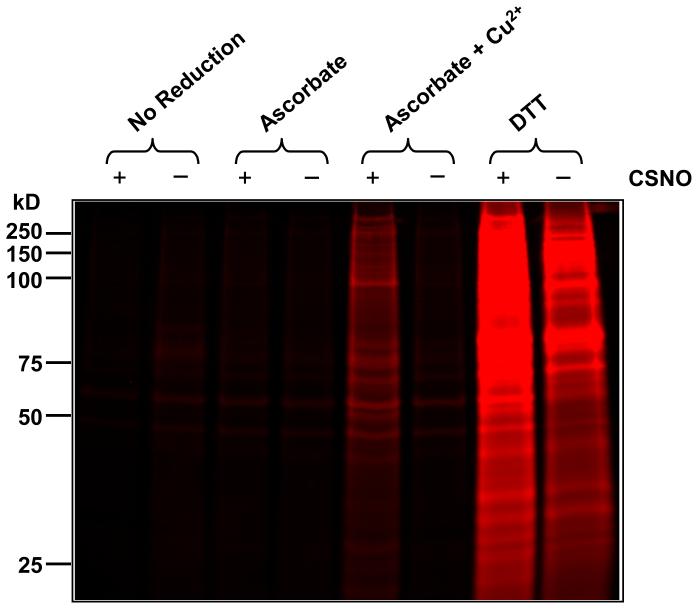
Normal human bronchial epithelial (NHBE) cells were treated with 10 μM CSNO in Hank’s buffered salt solution (HBSS), or HBSS alone, for 30 minutes at 37°C. After lysis, cellular proteins were fluorescently tagged using the CyDye switch protocol using various reduction conditions (indicated above each lane in Figure 9). Control and CSNO-treated samples (1.5 μg total protein per lane) were separated by SDS PAGE. Following electrophoresis, the gel was scanned for Cy5 fluorescence. Negative controls were included in the first two lanes, where no reducing agent was used and very little labeling was observed indicating that thiol blockade with NEM was efficient. As shown in figure 9, ascorbate alone was used as the reducing agent in the presence of the metal ion chelator DTPA. This also resulted in very little labeling and no observable difference between samples from CSNO-treated and control cells. This suggests that ascorbate alone is not sufficient to reduce CSNO-mediated protein thiol modifications. Strikingly, the presence of 10 μM copper (II) sulfate along with ascorbate in the absence of metal ion chelators significantly increased the number of bands observed in the sample from CSNO-treated cells. However, this increase was not detected in the sample from control cells subjected to identical reduction conditions. These results indicate that CSNO treatment induced the formation of protein thiol modifications that are reducible by a mixture of ascorbate and copper (II). This also indicates that the ascorbate/copper (II) mixture does not reduce endogenous disulfides and can also rule out the possibility of interference from mixed disulfides formed during CSNO treatment (see Figure 5C). As shown in figure 9, reducing agent, DTT, was used as positive controls. In this case, as expected, both the control and treated samples are labeled, however the fluorescence intensity was greater in CSNO treated sample, indicating that CSNO treatment increased the number of tagged thiols.
Figure 9. CyDye labeling of reversible thiol modifications.
Normal human bronchial epithelial (NHBE) cells were treated with 10 μM CSNO or buffer alone for 30 minutes. Lysate proteins with reversible thiol modifications were fluorescently tagged via the CyDye switch protocol using multiple reduction conditions (indicated above each lane) and separated by SDS-PAGE. Each well contained 1.5 μg of total protein. Following electrophoresis, the gel was scanned for Cy5 fluorescence.
This experiment demonstrates that in the complex milieu of cellular proteins the ascorbate/copper reduction mixture only reduces thiols that are modified as a result of CSNO exposure and does not reduce endogenous disulfides. Although we have shown S-thiolated HSA is not reduced under these conditions (Figure 5), the reduction of all cellular S-thiolated proteins or other thiol oxidations such as sulfenic and sulfinic acids cannot be completely ruled out, and would need to be assessed on a protein-by-protein basis. Note for example the case of immunoglobulin in plasma described earlier.
Discussion
The biotin switch assay has been widely used to detect S-nitrosated proteins in a number of experimental systems [7]. The strength of this assay is that it can determine which specific protein(s) have been S-nitrosated. This represents an advantage over other assays, such as ozone-based chemiluminescence after photolytic or chemical displacement, which can only report total S-nitrosation [31, 32]. We have previously demonstrated that ascorbate alone at the concentrations used in the original protocol (1 mM) is insufficient to reduce RSNO [20] and we show here that metal ions, including copper are necessary “contaminants” required for RSNO reduction and biotin labeling. This finding helps explain the variability of this assay in different laboratories using copper chelators and allows for a simple assay modification that dramatically increases sensitivity while maintaining specificity.
Copper is very common in the environment. The U.S. Environmental Protection Agency (U.S. EPA) has determined that copper levels in drinking water should not exceed 1300 μg/L. It has been shown that copper is one of the most common metal contaminants in regular laboratories. It appears that the success of the biotin switch assay is quite dependent on the contamination of this and other metals in the solutions. It is known that reduced copper can reduce RSNO [25, 29, 30, 42, 43]. The mechanism for the decomposition of S-nitrosothiols by Cu(I) ions likely involves the one-electron reduction of the RSNO group to give the free thiol, NO and Cu(II). Ascorbate can then re-reduce the Cu(II) ion to Cu(I), completing the catalytic cycle. A combination of copper with reducing reagents, such as cysteine or ascorbate, has been used for detection of RSNO [29, 31, 44]. Our results show that ascorbate alone at 1 mM concentration, without contaminating metal ions or added copper, will not sufficiently and quickly reduce RSNO, a fact that is predictable from published rate constants for this reaction [23-25]. When a chelator is present, ascorbate is unable to reduce the SNO to allow biotin labeling, while addition of copper greatly enhanced the RSNO reduction and biotin labeling. As shown in Figure 2B, as little as 10 nM copper is sufficient to reduce the RSNO in the presence of ascorbate, allowing the biotinylation of the reduced thiol to take place. This indicates that copper is acting catalytically. RSNO breakdown in our system by this trace amount of copper suggests that different amounts of copper and other metal ion contamination in different solutions is the reason for different detection efficiencies.
As illustrated by the intense immunoglobin bands in plasma without S-nitrosothiol exposure, some proteins are strongly biotin labeled in this procedure, independent of S-NO content. Thus care must be taken to ensure that a measured band actually represents an S-nitrosothiol and not an artifact of non-blocked thiol [26, 45]. The thiol blocking step defines the signal:noise ratio of this assay. From a simple theoretical basis, if thiol blocking is 99% efficient, then the assay can not detect less that 1% of thiol modification with a signal to noise ratio of 1:1. Endogenous levels of RSNO are much lower than 1%, and so blocking efficiency needs to be much higher. Blocking efficiency can be easily examined in control experiments to reveal thiol groups that are resistant to blockade by the thiol-blocking agent used.
Using direct fluorescent labeling rather than biotinylation it is possible to examine thiol modifications directly on-gel without the need for Western blotting [7]. Using maleimido Cy-5, we show that ascorbate alone, at a concentration of 1 mM is only able to reduce S-nitrosothiols in the presence of copper ions (Figure 9). This indicates that the role of copper in enhancing S-nitrosothiol detection is independent of the labeling modality used.
It has been shown that high concentrations of ascorbate reduce not only S-nitrothiols [26], but also some disulfides [21]. Therefore, including copper to limit ascorbate concentration and designing controls for the biotin switch assay is essential for identifying S-nitrosated proteins in biological samples with this assay. It is important to ensure that there is enough S-nitrosothiol in the sample under study to make the biotinylation results meaningful. It is very straightforward to measure the total RSNO content of a protein sample by displacement-chemiluminescence techniques using triiodide or another reductant [29, 31, 46]. One can then assess the intensity of the bands observed on gel to see if these results quantitatively agree with the total RSNO level measured in the same original volume of sample by chemiluminescence. An excellent recent example of this approach has been published by Burwell and colleagues who measured mitochondrial protein fractions, separated by blue native gel electrophoresis or Superose 6 column chromatography, using both the biotin switch assay and triiodide-based reductive chemiluminescence [47]. Another important control is to reduce the RSNO-containing sample with dithiothreitol (5 mM dithiothreitol will destroy high levels of RSNO within 30 min) and then subject this sample to the biotin-switch assay. A negative result for this control would prove that thiol blocking was sufficient. Another potential problem is that the thiol has been oxidized to a sulfenic or sulfinic acid or has formed a mixed disulfide, and is not in fact an S-nitrosothiol. Attempts can be made to control for these by exposing cells or protein to oxidants like hydrogen peroxide, or to S-nitrosothiols like S-nitrosoglutathione, and determine which exposure produces the observed biotinylation. An excellent example of how the assay can be used in such a fashion is the recent study published by Hara and colleagues [40]. In using this assay, these investigators employ a number of positive and negative controls, including different NO donors, oxidants, and iNOS knockouts.
The data shown in Figure 9 indicate that the ascorbate/copper system is unable to reduce endogenous disulfides as no increase in labeling was observed between the ascorbate/copper reduced sample and the unreduced control in the absence of CSNO. It is not clear though if mixed disulfides formed from CSNO treatment are being detected. We show that our test protein, HSA does not give positive signals when S-cysteinylated, S-glutathionylated or when treated with hydrogen peroxide (figure 5C). While this significantly rules out false positives for this particular protein, the ease of reduction of disulfides is not uniform, and such controls need to be performed with each identified protein before positive assignment of S-nitrosation can be made used reduction techniques.
In conclusion, our previous report [20] that ascorbate, under the conditions used in the original biotin switch assay (Jaffrey et al) was unable to reduce protein S-nitrosothiols has been confirmed. In addition, we have shown that the removal of metal ion chelating agents from the buffers and the addition of catalytic levels of copper ions (in either the +1 or the +2 oxdiation state) allows low levels of ascorbate to facilitate S-nitrosothiol reduction. This likely explains the variability and uncertainty of this assay and the small modification of adding copper with ascorbate should allow much more consistent results to be obtained from laboratory to laboratory.
Supplementary Material
Acknowledgment
This study was partially supported by National Institute of General Medicine grant GM55792 (NH) and intramural support from National Heart, Lung and Blood Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Bryan NS, Fernandez BO, Bauer SM, Garcia-Saura MF, Milsom AB, Rassaf T, Maloney RE, Bharti A, Rodriguez J, Feelisch M. Nitrite is a signaling molecule and regulator of gene expression in mammalian tissues. Nat Chem Biol. 2005;1:290–297. doi: 10.1038/nchembio734. [DOI] [PubMed] [Google Scholar]
- [2].Luchsinger BP, Rich EN, Gow AJ, Williams EM, Stamler JS, Singel DJ. Routes to S-nitroso-hemoglobin formation with heme redox and preferential reactivity in the beta subunits. Proc Natl Acad Sci U S A. 2003;100:461–466. doi: 10.1073/pnas.0233287100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Marletta MA. Mammalian synthesis of nitrite, nitrate, nitric oxide, and N-nitrosating agents. Chem Res Toxicol. 1988;1:249–257. doi: 10.1021/tx00005a001. [DOI] [PubMed] [Google Scholar]
- [4].Stamler JS, Simon DI, Osborne JA, Mullins ME, Jaraki O, Michel T, Singel DJ, Loscalzo J. S-nitrosylation of proteins with nitric oxide: synthesis and characterization of biologically active compounds. Proc Natl Acad Sci U S A. 1992;89:444–448. doi: 10.1073/pnas.89.1.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Biswas S, Chida AS, Rahman I. Redox modifications of protein-thiols: emerging roles in cell signaling. Biochem Pharmacol. 2006;71:551–564. doi: 10.1016/j.bcp.2005.10.044. [DOI] [PubMed] [Google Scholar]
- [6].Hogg N. The biochemistry and physiology of S-nitrosothiols. Annu Rev Pharmacol Toxicol. 2002;42:585–600. doi: 10.1146/annurev.pharmtox.42.092501.104328. [DOI] [PubMed] [Google Scholar]
- [7].Kettenhofen NJ, Broniowska KA, Keszler A, Zhang Y, Hogg N. Proteomic methods for analysis of S-nitrosation. J Chromatogr B Analyt Technol Biomed Life Sci. 2007 doi: 10.1016/j.jchromb.2007.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Levonen AL, Landar A, Ramachandran A, Ceaser EK, Dickinson DA, Zanoni G, Morrow JD, Darley-Usmar VM. Cellular mechanisms of redox cell signalling: role of cysteine modification in controlling antioxidant defences in response to electrophilic lipid oxidation products. Biochem J. 2004;378:373–382. doi: 10.1042/BJ20031049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Foster MW, McMahon TJ, Stamler JS. S-nitrosylation in health and disease. Trends Mol Med. 2003;9:160–168. doi: 10.1016/s1471-4914(03)00028-5. [DOI] [PubMed] [Google Scholar]
- [10].Huang Z, Shiva S, Kim-Shapiro DB, Patel RP, Ringwood LA, Irby CE, Huang KT, Ho C, Hogg N, Schechter AN, Gladwin MT. Enzymatic function of hemoglobin as a nitrite reductase that produces NO under allosteric control. J Clin Invest. 2005;115:2099–2107. doi: 10.1172/JCI24650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Wright MM, Schopfer FJ, Baker PR, Vidyasagar V, Powell P, Chumley P, Iles KE, Freeman BA, Agarwal A. Fatty acid transduction of nitric oxide signaling: nitrolinoleic acid potently activates endothelial heme oxygenase 1 expression. Proc Natl Acad Sci U S A. 2006;103:4299–4304. doi: 10.1073/pnas.0506541103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Gow AJ, Farkouh CR, Munson DA, Posencheg MA, Ischiropoulos H. Biological significance of nitric oxide-mediated protein modifications. Am J Physiol Lung Cell Mol Physiol. 2004;287:L262–268. doi: 10.1152/ajplung.00295.2003. [DOI] [PubMed] [Google Scholar]
- [13].Georgiou G, Masip L. Biochemistry. An overoxidation journey with a return ticket. Science. 2003;300:592–594. doi: 10.1126/science.1084976. [DOI] [PubMed] [Google Scholar]
- [14].Forman HJ, Fukuto JM, Torres M. Redox signaling: thiol chemistry defines which reactive oxygen and nitrogen species can act as second messengers. Am J Physiol Cell Physiol. 2004;287:C246–256. doi: 10.1152/ajpcell.00516.2003. [DOI] [PubMed] [Google Scholar]
- [15].Biteau B, Labarre J, Toledano MB. ATP-dependent reduction of cysteine-sulphinic acid by S. cerevisiae sulphiredoxin. Nature. 2003;425:980–984. doi: 10.1038/nature02075. [DOI] [PubMed] [Google Scholar]
- [16].Woo HA, Chae HZ, Hwang SC, Yang KS, Kang SW, Kim K, Rhee SG. Reversing the inactivation of peroxiredoxins caused by cysteine sulfinic acid formation. Science. 2003;300:653–656. doi: 10.1126/science.1080273. [DOI] [PubMed] [Google Scholar]
- [17].Rhee SG, Chae HZ, Kim K. Peroxiredoxins: a historical overview and speculative preview of novel mechanisms and emerging concepts in cell signaling. Free Radic Biol Med. 2005;38:1543–1552. doi: 10.1016/j.freeradbiomed.2005.02.026. [DOI] [PubMed] [Google Scholar]
- [18].Jaffrey SR, Erdjument-Bromage H, Ferris CD, Tempst P, Snyder SH. Protein S-nitrosylation: a physiological signal for neuronal nitric oxide. Nat Cell Biol. 2001;3:193–197. doi: 10.1038/35055104. [DOI] [PubMed] [Google Scholar]
- [19].Forrester MT, Foster MW, Stamler JS. Assessment and application of the biotin switch technique for examining protein S-nitrosylation under conditions of pharmacologically induced oxidative stress. J Biol Chem. 2007 doi: 10.1074/jbc.M609684200. [DOI] [PubMed] [Google Scholar]
- [20].Zhang Y, Keszler A, Broniowska KA, Hogg N. Characterization and application of the biotin-switch assay for the identification of S-nitrosated proteins. Free Radic Biol Med. 2005;38:874–881. doi: 10.1016/j.freeradbiomed.2004.12.012. [DOI] [PubMed] [Google Scholar]
- [21].Landino LM, Koumas MT, Mason CE, Alston JA. Ascorbic acid reduction of microtubule protein disulfides and its relevance to protein S-nitrosylation assays. Biochem Biophys Res Commun. 2006;340:347–352. doi: 10.1016/j.bbrc.2005.12.013. [DOI] [PubMed] [Google Scholar]
- [22].Foster MW, Stamler JS. New insights into protein S-nitrosylation. Mitochondria as a model system. J Biol Chem. 2004;279:25891–25897. doi: 10.1074/jbc.M313853200. [DOI] [PubMed] [Google Scholar]
- [23].Smith JN, Dasgupta TP. Kinetics and mechanism of the decomposition of S-nitrosoglutathione by l-ascorbic acid and copper ions in aqueous solution to produce nitric oxide. Nitric Oxide. 2000;4:57–66. doi: 10.1006/niox.2000.0272. [DOI] [PubMed] [Google Scholar]
- [24].Kashiba-Iwatsuki M, Yamaguchi M, Inoue M. Role of ascorbic acid in the metabolism of S-nitroso-glutathione. FEBS Lett. 1996;389:149–152. doi: 10.1016/0014-5793(96)00560-1. [DOI] [PubMed] [Google Scholar]
- [25].Holmes AJ, Williams DLH. Reaction of ascorbic acid with S-nitrosothiols: clear evidence for two distinct reaction pathways. J. Chem. Soc., Perkin Trans. 2000;2:1639–1644. [Google Scholar]
- [26].Huang B, Chen C. An ascorbate-dependent artifact that interferes with the interpretation of the biotin switch assay. Free Radic Biol Med. 2006;41:562–567. doi: 10.1016/j.freeradbiomed.2006.03.006. [DOI] [PubMed] [Google Scholar]
- [27].Gladwin MT, Wang X, Reiter CD, Yang BK, Vivas EX, Bonaventura C, Schechter AN. S-Nitrosohemoglobin is unstable in the reductive erythrocyte environment and lacks O2/NO-linked allosteric function. J Biol Chem. 2002;277:27818–27828. doi: 10.1074/jbc.M203236200. [DOI] [PubMed] [Google Scholar]
- [28].Wang X, Tanus-Santos JE, Reiter CD, Dejam A, Shiva S, Smith RD, Hogg N, Gladwin MT. Biological activity of nitric oxide in the plasmatic compartment. Proc Natl Acad Sci U S A. 2004;101:11477–11482. doi: 10.1073/pnas.0402201101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Doctor A, Platt R, Sheram ML, Eischeid A, McMahon T, Maxey T, Doherty J, Axelrod M, Kline J, Gurka M, Gow A, Gaston B. Hemoglobin conformation couples erythrocyte S-nitrosothiol content to O2 gradients. Proc Natl Acad Sci U S A. 2005;102:5709–5714. doi: 10.1073/pnas.0407490102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Fang K, Ragsdale NV, Carey RM, MacDonald T, Gaston B. Reductive assays for S-nitrosothiols: implications for measurements in biological systems. Biochem Biophys Res Commun. 1998;252:535–540. doi: 10.1006/bbrc.1998.9688. [DOI] [PubMed] [Google Scholar]
- [31].Macarthur PH, Shiva S, Gladwin MT. Measurement of circulating nitrite and S-nitrosothiols by reductive chemiluminescence. J Chromatogr B Analyt Technol Biomed Life Sci. 2007 doi: 10.1016/j.jchromb.2006.12.012. [DOI] [PubMed] [Google Scholar]
- [32].Yang BK, Vivas EX, Reiter CD, Gladwin MT. Methodologies for the sensitive and specific measurement of S-nitrosothiols, iron-nitrosyls, and nitrite in biological samples. Free Radic Res. 2003;37:1–10. doi: 10.1080/1071576021000033112. [DOI] [PubMed] [Google Scholar]
- [33].Wang X, Bryan NS, MacArthur PH, Rodriguez J, Gladwin MT, Feelisch M. Measurement of nitric oxide levels in the red cell: validation of tri-iodide-based chemiluminescence with acid-sulfanilamide pretreatment. J Biol Chem. 2006;281:26994–27002. doi: 10.1074/jbc.M603953200. [DOI] [PubMed] [Google Scholar]
- [34].Toyo’oka T, Imai K. High-performance liquid chromatography and fluorometric detection of biologically important thiols, derivatized with ammonium 7-fluorobenzo-2-oxa-1,3-diazole-4-sulphonate (SBD-F) J Chromatogr. 1983;282:495–500. doi: 10.1016/s0021-9673(00)91626-1. [DOI] [PubMed] [Google Scholar]
- [35].Dicks PA, Swift HR, Williams DLH, Butler AR, Al-Sa’doni HH, Cox BG. Identification of Cu+ as the effective reagent in nitric oxide formation from S-nitrosothiols (RSNO) J. Chem. Soc., Perkin Trans. 1996;2:481–487. [Google Scholar]
- [36].Buettner GR. Use of ascorbate as test for catalytic metals in simple buffers. Methods Enzymol. 1990;186:125–127. doi: 10.1016/0076-6879(90)86100-a. [DOI] [PubMed] [Google Scholar]
- [37].Asplund KU, Jansson PJ, Lindqvist C, Nordstrom T. Measurement of ascorbic acid (vitamin C) induced hydroxyl radical generation in household drinking water. Free Radic Res. 2002;36:1271–1276. doi: 10.1080/1071576021000036425. [DOI] [PubMed] [Google Scholar]
- [38].Pinto JJ, Moreno C, Garcia-Vargas M. A simple and very sensitive spectrophotometric method for the direct determination of copper ions. Anal Bioanal Chem. 2002;373:844–848. doi: 10.1007/s00216-002-1403-y. [DOI] [PubMed] [Google Scholar]
- [39].Lunvongsa S, Tsuboi T, Motomizu S. Sequential determination of trace amounts of iron and copper in water samples by flow injection analysis with catalytic spectrophotometric detection. Anal Sci. 2006;22:169–172. doi: 10.2116/analsci.22.169. [DOI] [PubMed] [Google Scholar]
- [40].Hara MR, Agrawal N, Kim SF, Cascio MB, Fujimuro M, Ozeki Y, Takahashi M, Cheah JH, Tankou SK, Hester LD, Ferris CD, Hayward SD, Snyder SH, Sawa A. S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat Cell Biol. 2005;7:665–674. doi: 10.1038/ncb1268. [DOI] [PubMed] [Google Scholar]
- [41].Angelo M, Singel DJ, Stamler JS. An S-nitrosothiol (SNO) synthase function of hemoglobin that utilizes nitrite as a substrate. Proc Natl Acad Sci U S A. 2006;103:8366–8371. doi: 10.1073/pnas.0600942103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Stubauer G, Giuffre A, Sarti P. Mechanism of S-nitrosothiol formation and degradation mediated by copper ions. J Biol Chem. 1999;274:28128–28133. doi: 10.1074/jbc.274.40.28128. [DOI] [PubMed] [Google Scholar]
- [43].Singh RJ, Hogg N, Joseph J, Kalyanaraman B. Mechanism of nitric oxide release from S-nitrosothiols. J Biol Chem. 1996;271:18596–18603. doi: 10.1074/jbc.271.31.18596. [DOI] [PubMed] [Google Scholar]
- [44].Cha W, Lee Y, Oh BK, Meyerhoff ME. Direct detection of S-nitrosothiols using planar amperometric nitric oxide sensor modified with polymeric films containing catalytic copper species. Anal Chem. 2005;77:3516–3524. doi: 10.1021/ac048192u. [DOI] [PubMed] [Google Scholar]
- [45].Gladwin MT, Wang X, Hogg N. Methodological vexation about thiol oxidation versus S-nitrosation -- a commentary on “An ascorbate-dependent artifact that interferes with the interpretation of the biotin-switch assay”. Free Radic Biol Med. 2006;41:557–561. doi: 10.1016/j.freeradbiomed.2006.05.025. [DOI] [PubMed] [Google Scholar]
- [46].Feelisch M, Rassaf T, Mnaimneh S, Singh N, Bryan NS, Jourd’Heuil D, Kelm M. Concomitant S-, N-, and heme-nitros(yl)ation in biological tissues and fluids: implications for the fate of NO in vivo. Faseb J. 2002;16:1775–1785. doi: 10.1096/fj.02-0363com. [DOI] [PubMed] [Google Scholar]
- [47].Burwell LS, Nadtochiy SM, Tompkins AJ, Young S, Brookes PS. Direct evidence for S-nitrosation of mitochondrial complex I. Biochem J. 2006;394:627–634. doi: 10.1042/BJ20051435. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.