Abstract
Conventional cytogenetic analysis of an aggressive angiomyxoma of the rectal wall of a 72-year-old female revealed a translocation between the long arms of chromosomes 12 and 21 [46,XX,t(12;21)(q15;q21.1)]. Involvement of the HMGA2 gene locus (12q15) was confirmed by fluorescence in situ hybridization (FISH) using an HMGA2 breakpoint flanking probe set performed on metaphase and interphase cells from an in situ culture of fresh lesional tissue. Karyotypic rearrangements of 12q13-15 are considered recurrent in aggressive angiomyxoma, although reported in only five previous cases. Translocation partner chromosome 21 is novel to the current case.
Introduction
Aggressive angiomyxoma, first described by Steeper and Rosai in 1983 [1] and now also referred to as “deep angiomyxoma” by the WHO [2], is a rare benign slow growing hypocellular soft tissue neoplasm composed of cytologically bland stellate and spindle shaped cells in a myxoid background, with a conspicuous vascular component. It is most commonly found in the vulvovaginal region of white females of reproductive age [3-5], but has also been reported in the scrotal and perineal/perianal region of males.
Clinical misdiagnosis, most commonly as a Bartholin’s cyst, is a common problem, reported in as many as 82% of cases [6]. Though aggressive angiomyxoma is benign, it is locally infiltrative with a high rate of recurrence (71% at three years in one study) [7], with intervals from 9 to 180 months [1,8]. The high rate of recurrence is thought to be secondary to inadequate primary excision [8]. Two cases of metastatic aggressive angiomyxoma have been reported [9,10].
Abnormal cytogenetic findings of only seven cases of aggressive angiomyxoma have been reported to date [11-19]. Here, we report the conventional cytogenetic, molecular cytogenetic, and immunohistochemical findings of an aggressive angiomyxoma arising in the rectal wall of a 72 year-old female.
Materials and Methods
Clinical History
A 72 year-old woman with a history of hypothyroidism, hypertension, coronary artery disease, and diverticulosis presented to her local physician with complaints of abdominal pain. A perirectal mass (3.1 × 1.5 cm) was detected radiographically and the patient was subsequently referred to the University of Nebraska Medical Center (UNMC). At presentation to UNMC, the patient reported a 6 month history of dull intermittent abdominal pain accompanied by nausea, but without vomiting, localized to the left lower quadrant with occasional pain radiating down the left leg. The pain reportedly lasted for hours at a time, was worse at night, and was not improved or aggravated by any specific activity or position. Review of systems included weight loss (20lb over the prior 6 months), constipation, and urinary frequency, urgency, and incontinence. Her past surgical history included TAH/BSO (details unknown), appendectomy, and cholecystectomy. The patient’s family history included a sister who died of lymphoma and grandmother who died of breast cancer.
Physical examination was remarkable for a palpable soft mobile mass on rectal exam. CT and MRI of the abdomen and pelvis showed diverticulosis and a 3-cm mass in the left ischiorectal fossa, without involvement of the rectum. Colonoscopy demonstrated multiple diverticula but no evidence of the mass. A fine-needle biopsy of the mass was obtained. Histopathologic examination of the biopsy specimen demonstrated a hypocellular lesion consisting of cytologically bland spindle-shaped cells in a myxoid background (Figure1A). No significant nuclear pleomorphism or mitotic activity was present. The neoplastic cells were immunoreactive for desmin and negative for S-100 protein and estrogen receptor. The differential diagnosis included myxoma and aggressive angiomyxoma. The latter diagnosis was favored because of the desmin immunoreactivity.
Figure 1.
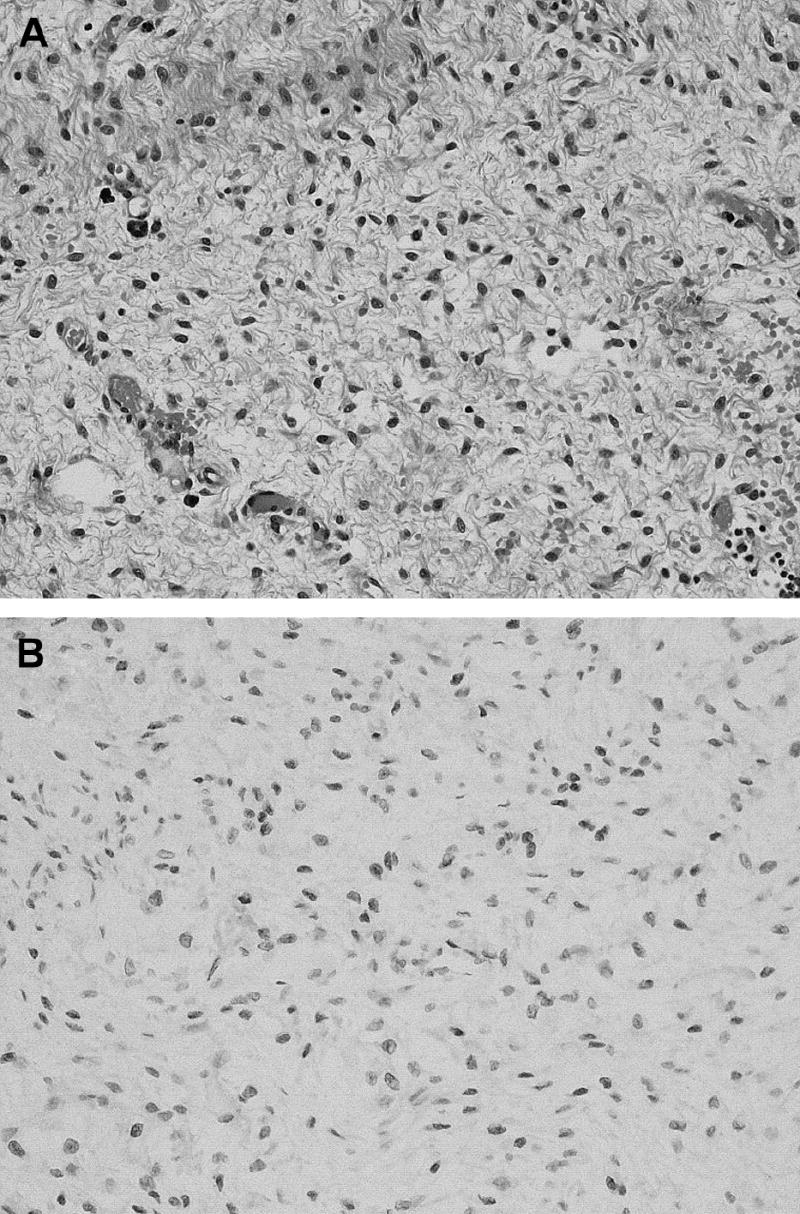
A, The hypocellular perirectal mass is composed of cytologically bland spindle-shaped cells in a myxoid background (H & E 200x). B, Tumor cells demonstrating HMGA2 immunoreactivity (200x).
Subsequently, the patient underwent a surgical excision of the mass followed by radiation therapy. The surgically excised specimen measuring 7.0 × 3.0 × 1.0 cm was tan-pink and rubbery with an attached hemorrhagic cyst (1.6 cm). Serial sectioning revealed tan-white gelatinous cut surfaces. A portion of the specimen was sterilely submitted for cytogenetic analysis. The frozen section diagnosis was consistent with aggressive angiomyxoma with extension to the margin. Additional tissue was consequently removed by the surgeon.
The permanent histologic sections of the surgically excised specimen demonstrated a low grade myxoid neoplasm irregularly infiltrating adjacent fibroadipose tissue. This myxoid neoplasm included a relatively uniform population of spindled to stellate cells embedded in a myxoid background with scattered thin and thick walled vessels. The bland spindled to stellate cells stained positively for desmin, estrogen receptor (the initial biopsy was negative for ER) and HMGA2 (Figure 1B), but were negative for AE1/AE3 and S-100 protein.
Cytogenetic Analysis
Standard culture and harvesting procedures were performed, as described previously [20]. Metaphase cells were banded with Wright trypsin, and the karyotypes were expressed according to the International System for Human Cytogenetic Nomenclature 2005 [21].
Molecular Cytogenetic Analysis
Bicolor fluorescence in situ hybridization (FISH) analysis with an HMGA2 breakpoint flanking probe set composed of CTD-2276H2 and CTD-2328M22 (bacterial artificial chromosome (BAC) clones proximal to HMGA2) and RP11-1050J21 and RP11-118B13 (BAC clones distal to HMGA2) was performed on metaphase and interphase cells from an in situ culture. Probes were directly labeled with either fluorescein-12-dUTP or rhodamine-5-dUTP (Enzo Life Sciences, Farmingdale, NY) by nick translation according to the manufacturer’s protocol (Vysis, Abbott Laboratories, Des Plaines, IL, USA). Prior to hybridization, the GTW stained in situ slide was de-stained with consecutive 5 minute incubations in 70% ethanol, 90% ethanol and methanol, followed by a brief rinse in 3:1 methaonl:acetic acid. The slide was then incubated in 2x saline sodium citrate (SSC) at 37°C for 15 min, dehydrated in an ethanol series (70%, 85%, and 100%) at room temperature for 1 min each and air-dried. The cells and probes were codenatured at 73°C for 2 min and incubated at 37°C overnight using the HYBrite™ denaturation/hybridization system (Vysis).
Posthybridization washing was performed in 0.4x SSC/0.3% NP-40 at 72°C for 2 min, followed by 2x SSC/0.1% NP-40 at room temperature for 2 min. The slide was air-dried in the dark and counterstained with 4’,6’-diamino-2-phenylindole (DAPI).
FISH analysis was also performed on destained metaphase and interphase cells with the LSI TEL/AML1 ES Dual Color Translocation Probe (Vysis, Abbott Laboratories, Des Plaines, IL) according to the manufacturer’s instructions. Hybridization signals were assessed in 3 karyotypically abnormal metaphase cells and 10 interphase cells for each probe set.
Results
Standard cytogenetic analysis revealed the following abnormal chromosomal complement: 46,XX,t(12;21)(q15;q21.1) in all twenty metaphase cells examined (Figure 2A). Fluorescence in-situ hybridization with an HMGA2 breakpoint flanking probe set revealed loss of the probe signal just distal to the HMGA2 gene locus consistent with a rearrangement of HMGA2, (Figure 2B). FISH analysis with the LSI TEL/AML1 ES Dual Color Translocation Probe confirmed translocation of the AML1 gene locus (21q22) from the derivative chromosome 21 to the derivative chromosome 12. The following nomenclature encompasses these FISH findings: 46,XX,t(12;21)(q15;q21.1)(HMGA2r+, HMGA2g-,AML1+; AML1-).
Figure 2.
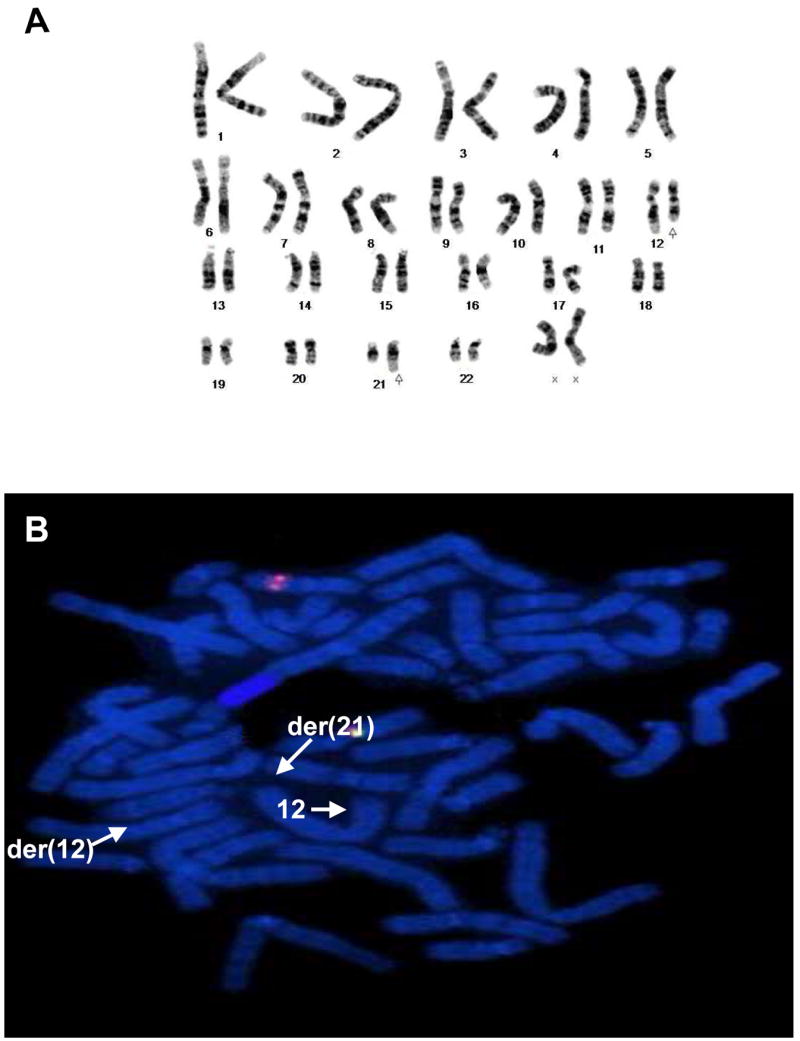
A, Representative karyotype illustrating the 12;21 translocation (arrows). B, Loss of the probe signal distal to the HMGA2 locus can be seen consistent with an HMGA2 rearrangement.
Discussion
Aggressive angiomyxoma is a rare benign, but locally aggressive, hypocellular soft tissue neoplasm found in the vulvovaginal region of females of reproductive age and less commonly in the perineal/perianal region of males. Cytogenetic abnormalities have been identified in seven cases of aggressive angiomyxoma to date, Table 1 [11-19]. Abnormalities of chromosome 12 were detected in six of these cases with specific involvement of the 12q13-15 region in five.
Table 1.
Karyotypic Findings in Aggressive Angiomyxoma
| Reference: | Age/Sex | Karyotype | HMGA2 Rearrangement by FISH | HMGA2 Immunoperoxidase | |
|---|---|---|---|---|---|
| 1 | Horsman 1991 [11] | 42/F | 45-50,XX,-12,-5,+der(5)t(5q15;?),-16,+der(16)t(5q;16q),-15,+der(15)t(15ql3;?)(l-4),+/-frag | NP | NP |
| 2 | Betz 1995 [14] | 32/F | 46,XX,t(7;12)(q22-31;q13-14)[14] | NP | NP |
| 3 | Kazmierczak 1995 [12] & 1998 [13] | 44/F | 46,XX/46,XX,t(5;12)(q31;p11.2),der(12)inv(12)(p11.2;q14)t(5;12)(q31;p11.2)[50] | Positive | NP |
| 4 | Kenny-Moynihan 1996 [15] | 16/F | 45,X,-X[8] | NP | NP |
| 5 | Nucci 1999 &Nucci 2001 [16, 17] | 18*/F | 46,XX,t(8;12)(p12;q15) | Negative | Positive |
| 6 | Micci 2006 [18] | 44/F | 46,XX,t(11;12)(q23;q15)[16] | Negative | Positive |
| 7 | Rabban 2006 [38] | 34/F | NP | Positive | Negative |
| 8 | Medeira 2007 [19] | 53/F | 46, XX,t(1;12)(p32;q15)[20] | Positive | NP |
| 9 | 49/F | 46, XX | Negative | NP | |
| 10 | Current Case | 72/F | 46,XX,t(12;21)(q15;q21.1)[20] | Positive | Positive |
NP = not performed;
Patient initially presented at age 18, but study material for cytogenetics was obtained on resection of a second recurrence at age 24
Notably, the HMGA2 (high mobility group AT-Hook2) gene, previously called HMGIC, a non-histone architectural transcription factor normally expressed in fetal tissues, and in adult lung and kidney [22] is localized to 12q15. The HMGA2 gene contains three DNA-binding domains (exons 1-3) and an acidic domain (exons 4 and 5) separated by a 140kb untranslated region. The acidic domain has an inhibitory effect on the DNA-binding domains.
The 140kb untranslated region between exons 3 and 4 is the site of the breakpoint observed in lipomas which separates the negatively acting acidic domains from the DNA-binding domains thus resulting in increased HMGA2 activity. [23, 24] Fusion of the HMGA2 gene with other genes resulting in increased HMGA2 activity has been noted in lipoma [25, 26], pulmonary chondroid hamartoma [23], and uterine leiomyoma [23, 24] among other neoplasms. Aberrant HMGA2 expression has been identified in benign mesenchymal tumors, including lipoma [27-29], uterine leiomyoma [30], inflammatory myofibroblastic tumor [31], and chondroma [32], and in malignant tumors, including well differentiated liposarcoma [33], uterine leiomyosarcoma [34], osteosarcoma [35] and acute lymphoblastic leukemia [36].
The chromosomal translocation partner has varied among the five previous cases of aggressive angiomyxoma exhibiting 12q13-15 rearrangements. Betz et al. reported a translocation between chromosomes 7 and 12 involving the 12q13-14 region [t(7;12)(q22-31;q13-14)] in an aggressive angiomyxoma of the vaginal wall in a 32 year old woman [14]. In contrast, Kazmierczak et al. detected a chromosome 12 inversion in an aggressive angiomyxoma in the vulvovaginal region of a 44 year old woman resulting in an aberrant transcript containing exons 1-3 of HMGA2 with a fused ectopic sequence [12]. The protein product was effectively truncated, leaving the DNA-binding domains to function unencumbered by the inhibitory 3’ acidic domain, thus resulting in a hyperfunctional HMGA2 product, analogous to other mesenchymal tumors.
Interestingly, two cases of aggressive angiomyxoma have been described that cytogenetically involve 12q15, but the breakpoint at the molecular level is lying outside of the HMGA2 gene locus. However, there still appears to be increased expression of the protein product. Nucci et al. described an aggressive angiomyxoma of the vulva in a 18 year old woman exhibiting a translocation 3’ to HMGA2, t(8;12)(p12;q15), resulting in an intact transcript (confirmed by RT-PCR), but aberrant HMGA2 protein expression (confirmed by immunohistochemistry) [17]. Similarly, Micci et al. reported an aggressive angiomyxoma of the right labium majus in a 44 year old woman characterized by a t(11;12)(q23;q15) in which the HMGA2 gene locus was negative for a rearrangement by FISH (using a probe set consisting of cosmid clones specific for exons 1-2 and 4-5), but showed aberrant production of intact HMGA2 product (demonstrable by RT-PCR using primers specific for exons 1-3 and 1-5) [18].
The findings in these two cases of aggressive angiomyxoma demonstrated aberrant activity of the HMGA2 gene with a seemingly different molecular mechanism than that described by Kazmierczak [13]: deregulated expression resulting from extragenic rearrangement in the former versus fusion transcript resulting from intragenic rearrangement in the latter.
Conversely, Rabban et al. utilizing FISH analysis with similar cosmid clones, identified an HMGA2 rearrangement in an aggressive angiomyxoma, but HMGA2 expression was absent as assessed by immunohistochemistry [37]. The authors postulated that the rearranged protein product was present, but was either too low in quantity to be detected, or too altered in conformation to be recognized by the immunohistochemical antibodies used. These findings suggest that anti-HMGA2 antibody may not be useful in all cases of aggressive angiomyxoma [37].
Most recently, Madeiras et al. reported the immunohistochemical and FISH findings of 42 cases of aggressive angiomyxoma [19]. Two cases were cytogenetically evaluated and showed a normal karyotype in one and a t(1;12)(p32;q15) in another. The latter case also showed an HMGA2 rearrangement by FISH, while the former did not. Somewhat surprisingly, only 33% of the aggressive angiomyxoma cases (14/42) demonstrated HMGA2 rearrangements by FISH, even when using larger flanking probes in an effort to detect translocations adjacent to the HMGA2 gene. These findings suggest a limited utility of FISH in the diagnosis of aggressive angiomyxoma. HMGA2 immunohistochemical analysis was not performed in this study.
In summary, the current case of aggressive angiomyxoma demonstrated a 12;21 translocation with FISH confirmation of an HMGA2 rearrangement and immunohistochemical confirmation of HMGA2 overexpression. These findings lend further support to the identification 12q15 rearrangements to involve the HMGA2 locus as characteristic of a subset of aggressive angiomyxomas, and suggest that cytogenetic, FISH and/or immunohistochemical studies may be considered adjunctive in its identification.
Acknowledgments
The authors would like to thank Ms. Patti Cattano and Ms. Chestnut Livermore for their technical assistance and Dr. Christopher Fletcher MD FRCPath (Brigham & Women’s Hospital) for performing the HMGA2 immunohistochemical studies. This work was supported in part by UNMC College of Medicine Educational Support Grant, State of Nebraska LB595, and NIH/NCI P30 CA 36727.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Steeper TA, Rosai J. Aggressive angiomyxoma of the female pelvis and perineum. Report of nine cases of a distinctive type of gynecologic soft-tissue neoplasm. Am J Surg Pathol. 1983;7:463–75. doi: 10.1097/00000478-198307000-00009. [DOI] [PubMed] [Google Scholar]
- 2.Tavassoli FA, Devilee P, editors. World Health Organization Classification of Tumours: Pathology and Genetics of Tumours of the Breast and Female Genital Organs. IARC Press; Lyon: 2003. pp. 305–329. [Google Scholar]
- 3.Simo M, Zapata C, Esquius J, Domingo J. Aggressive angiomyxoma of the female pelvis and perinenm. Report of two cases and review of the literature. Br J Obstet Gynaecol. 1992;99:925–27. doi: 10.1111/j.1471-0528.1992.tb14446.x. [DOI] [PubMed] [Google Scholar]
- 4.Boudhraa K, Oueslati H, Chtourou M, Neji K, Barouni M, Horchani A, Ben Romdhane K, Reziga H. “Aggressive” angiomyxoma of the vagina. A case report and review of the literature. Tunis Med. 2003;81:140–4. Article in French. [PubMed] [Google Scholar]
- 5.Papachristou DJ, Batistatou A, Paraskevaidis E, Agnantis NJ. Aggressive angiomyxoma of the vagina: a case report and review of the literature. Eur J Gynaecol Oncol. 2004;25:519–21. [PubMed] [Google Scholar]
- 6.Smith OH, Worrel RV, Smith AY, Docin MH, Rosenberg RD, Bartow SA. Aggressive angiomyxoma of the female pelvis and perineum: review of the literature. Gynecol Oncol. 1991;42:79–85. doi: 10.1016/0090-8258(91)90235-w. [DOI] [PubMed] [Google Scholar]
- 7.Chan YM, Hon E, Ngai SW, Ng TY, Wong LC. Aggressive angiomyxoma in females: is radical resection the only option? Acta Obstet Gynecol Scand. 2000 Mar;79(3):216–20. Erratum in: Acta Obstet Gynecol Scand 2000;79:432. Chan IM [corrected to Chan YM] [PubMed] [Google Scholar]
- 8.Begin LR, Clement PB, Kirk ME, Jothy S, McCaughey WT, Ferenczy A. Aggressive angiomyxoma of pelvic soft parts: A clinicopathologic study of nine cases. Hum Pathol. 1985;16:621–628. doi: 10.1016/s0046-8177(85)80112-x. [DOI] [PubMed] [Google Scholar]
- 9.Siassi RM, Papadopoulos T, Matzel K. Metastasizing aggressive angiomyxoma. Engl J Med. 1999;2:1772. doi: 10.1056/nejm199912023412315. [DOI] [PubMed] [Google Scholar]
- 10.Blandamura S, Cruz J, Faure Vergara L, Machado Puerto I, Ninfo V. Aggressive angiomyxoma: a second case of metastasis with patient’s death. Hum Pathol. 2003;34:1072–4. doi: 10.1053/s0046-8177(03)00419-2. [DOI] [PubMed] [Google Scholar]
- 11.Horsman DE, Berean KW, Salski CB, Clement PB. Aggressive angiomyxoma of the pelvis: Cytogenetic findings in a single case. Cancer Genet and Cytogenet. 1991;56:130. ABSTRACT. [Google Scholar]
- 12.Kazmierczak B, Rohen C, Wanschura S, Schoenmakers E, Van de Ven W, Bullerdick J. Involvement of the MAR region in two rare benign mesenchymal tumors: A hamartoma of the breast and an aggressive angiomyxoma. Cancer Genet Cytogenet. 1995;84:148. doi: 10.1016/0165-4608(95)00060-7. ABSTRACT. [DOI] [PubMed] [Google Scholar]
- 13.Kazmierczak B, Dal Cin P, Wanschura S, Bartnitzke S, Van den Berghe H, Bullerdiek J. Cloning and molecular characterization of part of a new gene fused to HMGIC in mesenchymal tumors. Am J Pathol. 1998;152:431–5. [PMC free article] [PubMed] [Google Scholar]
- 14.Betz JL, Meloni AM, U’ren LA, Moore GE, Sandberg AA. Cytogenetic findings in a case of angiomyxoma of the vaginal wall. Cancer Genet Cytogenet. 1995;84:157. ABSTRACT. [Google Scholar]
- 15.Kenny-Moynihan MB, Hagen J, Richman B, McIntosh DG, Bridge JA. Loss of an X chromosome in aggressive angiomyxoma of female soft parts: a case report. Cancer Genet Cytogenet. 1996;89:61–4. doi: 10.1016/0165-4608(95)00350-9. [DOI] [PubMed] [Google Scholar]
- 16.Nucci MR, Weremowicz S, Sornberger K, Tallini G, Morton CG, Quade BJ. Chromosomal translocation t(8;12) induces aberrant HMGIC expression in aggressive angiomyxoma of the vulva. Mod Path. 1999;12:700. doi: 10.1002/gcc.1179. ABSTRACT. [DOI] [PubMed] [Google Scholar]
- 17.Nucci MR, Weremowicz S, Neskey DM, Sornberger K, Tallini G, Morton CC, Quade BJ. Chromosomal translocation t(8;12) induces aberrant HMGIC expression in aggressive angiomyxoma of the vulva. Genes Chromosomes Cancer. 2001;32:172–6. doi: 10.1002/gcc.1179. [DOI] [PubMed] [Google Scholar]
- 18.Micci F, Panagopoulos I, Bjerkehagen B, Heim S. Deregulation of HMGA2 in an aggressive angiomyxoma with t(11;12)(q23;q15) Virchows Arch. 2006;448:838–42. doi: 10.1007/s00428-006-0186-5. Epub 2006 Mar 28. [DOI] [PubMed] [Google Scholar]
- 19.Medeiros F, Erickson-Johnson MR, Keeney GL, Clayton AC, Nascimento AG, Wang X, Oliveira AM. Frequency and characterization of HMGA2 and HMGA1 rearrangements in mesenchymal tumors of the lower genital tract. Genes Chromosomes Cancer. 2007;46:981–90. doi: 10.1002/gcc.20483. Epub 2007 Jul 25. [DOI] [PubMed] [Google Scholar]
- 20.Bridge JA, Liu J, Qualman SJ, et al. Genomic gains and losses are similar in genetic and histologic subtypes of rhabdomyosarcoma, whereas amplification predominates in embryonal with anaplasia and alveolar subtypes. Genes Chromosomes Cancer. 2002;33:310–321. doi: 10.1002/gcc.10026. [DOI] [PubMed] [Google Scholar]
- 21.Shaffer LG, Tommerup N, editors. ISCN 2005. An International System for Human Cytogenetics Nomenclature. Basel: Karger; 2005. [Google Scholar]
- 22.Gattas GJ, Quade BJ, Nowak RA, Morton CC. HMGIC expression in human adult and fetal tissues and in uterine leiomyomata. Genes Chromosomes Cancer. 1999;25:316–22. [PubMed] [Google Scholar]
- 23.Kazmierczak B, Pohnke Y, Bullerdiek J. Fusion transcripts between the HMGIC gene and RTVL-H-related sequences in mesenchymal tumors without cytogenetic aberrations. Genomics. 1996;38:223–226. doi: 10.1006/geno.1996.0619. [DOI] [PubMed] [Google Scholar]
- 24.Mine N, Kurose K, Nagai H, Doi D, Ota Y, Yoneyama K, Konishi H, Araki T, Emi M. Gene fusion involving HMGIC is a frequent aberration in uterine leiomyomas. J Hum Genet. 2001;46:408–412. doi: 10.1007/s100380170059. [DOI] [PubMed] [Google Scholar]
- 25.Petit MM, Schoenmakers EF, Huysmans C, Geurts JM, Mandahl N, Van de Ven WJ. LHFP, a novel translocation partner gene of HMGIC in a lipoma, is a member of a new family of LHFP-like genes. Genomics. 1999;57:438–41. doi: 10.1006/geno.1999.5778. [DOI] [PubMed] [Google Scholar]
- 26.Schoenmakers EF, Geurts JM, Kools PF, Mols R, Huysmans C, Bullerdiek J, Van den Berghe H, Van de Ven WJ. A 6-Mb yeast artificial chromosome contig and long-range physical map encompassing the region on chromosome 12q15 frequently rearranged in a variety of benign solid tumors. Genomics. 1995;29:665–78. doi: 10.1006/geno.1995.9952. [DOI] [PubMed] [Google Scholar]
- 27.Fedele M, Battista S, Manfioletti G, Croce CM, Giancotti V, Fusco A. Role of the high mobility group A proteins in human lipomas. Carcinogenesis. 2001;22:1583–91. doi: 10.1093/carcin/22.10.1583. [DOI] [PubMed] [Google Scholar]
- 28.Matsui Y, Hasegawa T, Kubo T, Goto T, Yukata K, Endo K, Bando Y, Yasui N. Intrapatellar tendon lipoma with chondro-osseous differentiation: detection of HMGA2-LPP fusion gene transcript. J Clin Pathol. 2006;59:434–6. doi: 10.1136/jcp.2005.026393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nilsson M, Mertens F, Hoglund M, Mandahl N, Panagopoulos I. Truncation and fusion of HMGA2 in lipomas with rearrangements of 5q32-->q33 and 12q14-->q15. Cytogenet Genome Res. 2006;112:60–6. doi: 10.1159/000087514. [DOI] [PubMed] [Google Scholar]
- 30.Ingraham SE, Lynch RA, Surti U, Rutter JL, Buckler AJ, Khan SA, Menon AG, Lepont P. Identification and characterization of novel human transcripts embedded within HMGA2 in t(12;14)(q15;q24.1) uterine leiomyoma. Mutat Res. 2006;602:43–53. doi: 10.1016/j.mrfmmm.2006.07.007. Epub 2006 Oct 12. [DOI] [PubMed] [Google Scholar]
- 31.Kazmierczak B, Dal Cin P, Sciot R, Van den Berghe H, Bullerdiek J. Inflammatory myofibroblastic tumor with HMGIC rearrangement. Cancer Genet Cytogenet. 1999;112:156–60. doi: 10.1016/s0165-4608(98)00268-4. [DOI] [PubMed] [Google Scholar]
- 32.Dahlen A, Mertens F, Rydholm A, Brosjo O, Wejde J, Mandahl N, Panagopoulos I. Fusion, disruption, and expression of HMGA2 in bone and soft tissue chondromas. Mod Pathol. 2003;16:1132–40. doi: 10.1097/01.MP.0000092954.42656.94. [DOI] [PubMed] [Google Scholar]
- 33.Trahan S, Erickson-Johnson MR, Rodriguez F, Aubry MC, Cheville JC, Myers JL, Mederios AM. Formation of the 12q14-q15 amplicon precedes the development of a well-differentiated liposarcoma arising from a nonchondroid pulmonary hamartoma. Am J Surg Pathol. 2006;30:1326–9. doi: 10.1097/01.pas.0000213257.69478.2f. [DOI] [PubMed] [Google Scholar]
- 34.Cho YL, Bae S, Koo MS, Kim KM, Chun HJ, Kim CK, Ro DY, Kim JH, Lee CH, Kim YW, Ahn WS. Array comparative genomic hybridization analysis of uterine leiomyosarcoma. Gynecol Oncol. 2005;99:545–51. doi: 10.1016/j.ygyno.2005.07.017. Epub 2005 Aug 24. [DOI] [PubMed] [Google Scholar]
- 35.Kools PF, Van de Ven WJ. Amplification of a rearranged form of the high-mobility group protein gene HMGIC in OsA-CI osteosarcoma cells. Cancer Genet Cytogenet. 1996;91:1–7. doi: 10.1016/s0165-4608(96)00109-4. [DOI] [PubMed] [Google Scholar]
- 36.Pierantoni GM, Santulli B, Caliendo I, Pentimalli F, Chiappetta G, Zanesi N, Santoro M, Bulrich F, Fusco A. HMGA2 locus rearrangement in a case of acute lymphoblastic leukemia. Int J Oncol. 2003;23:363–7. [PubMed] [Google Scholar]
- 37.Rabban JT, Dal Cin P, Oliva E. HMGA2 rearrangement in a case of vulvar aggressive angiomyxoma. Int J Gynecol Pathol. 2006;25:403–7. doi: 10.1097/01.pgp.0000209572.54457.7b. [DOI] [PubMed] [Google Scholar]


