Abstract
Enteroinvasive Escherichia coli (EIEC) causes dysentery; however, it is less widely reported than other etiological agents in studies of diarrhea worldwide. Between August 2003 and July 2005, stool samples were collected in case-control studies in 22 rural communities in northwestern Ecuador. Infection was assessed by PCR specific for LT and STa genes of enterotoxigenic E. coli (ETEC), the bfp gene of enteropathogenic E. coli (EPEC), and the ipaH gene of both enteroinvasive E. coli and Shigellae. The pathogenic E. coli most frequently identified were EIEC (3.2 cases/100 persons) and Shigellae (1.5 cases/100 persons), followed by ETEC (1.3 cases/100 persons), and EPEC (0.9 case/100 persons). EIEC exhibited similar risk-factor relationships with other pathotypes analyzed but different age-specific infection rates. EIEC was the predominant diarrheagenic bacteria isolated in our community-based study, a unique observation compared with other regions of the world.
INTRODUCTION
Enteroinvasive Escherichia coli (EIEC) was first shown in 1971 to cause diarrheal disease in otherwise healthy volunteers.1 It is known to cause shigellosis-like symptoms in both adults and children. Despite its acknowledged status as a human pathogen, very little research has been conducted to identify individual risk factors for infection, possible reservoirs, or even infection rates. Other pathotypes, such as enteropathogenic (EPEC), enterohemorrhagic (EHEC), and enterotoxic (ETEC) E. coli, as well as Shigellae, have received much more scientific attention.
A potential contributor to the lack of attention to the epidemiology of EIEC is that it is often observed to be an infrequent cause of diarrhea relative to other diarrhea-causing E. coli. In a Medline search of studies testing for the presence of EIEC, we identified 42 articles. Of these studies, 35% (15) found no EIEC (contact authors for specific citations) and 40% (17) found EIEC to be a minor strain,2-18 i.e., representing less than 4% and fewer than 10 isolated cases and of the collected stool samples. There were, however, notable exceptions. In 1989, 15 EIEC samples were identified in 221 cases of childhood diarrhea in a Beijing hospital19; in 1985, 17 cases of EIEC re observed in 410 children with diarrhea in a Bangkok hospital20; and in 1982-1986, 17 cases of EIEC were observed in 912 infants with diarrhea in Chile.21 More recently, in the mid-1990s, EIEC was identified in 87 of 1579 stool samples from patients with travel-associated diarrhea.22 In the late 1990s, 16 EIEC-positive isolates were identified from 279 Senegalese individuals,23 and EIEC was the predominant enteropathogen during a 2-month period of increased diarrhea episodes in the Jordan Valley.24 These and four additional studies25-28 represented all studies identified that reported 10 or more diarrheal cases positive for EIEC (7 of 42, or 22% of studies reviewed). These studies are widely distributed geographically, including Europe, Central and South America, the Middle East, western Africa, and southeastern Asia. In over half of the studies that isolated EIEC, EIEC was identified as a possible etiologic agent of diarrhea.
Thus, EIEC is seldom identified; when it is found, it tends to be in small numbers. EIEC infection rates have never been reported for Ecuador. We report here EIEC as the predominant E. coli pathotype identified from both cases and controls in a community-based case-control study in northern Ecuador. Patterns of EIEC infection are compared with infections with Shigellae as well as enterotoxigenic (ETEC) and enteropathogenic (EPEC) E. coli.
PARTICIPANTS, MATERIALS, AND METHODS
Study population
The study area is located in the northern Ecuadorian province of Esmeraldas in the canton Eloy Alfaro, which comprises approximately 150 villages. The study reported here was carried out in 22 communities, all located within the drainage system of three rivers: the Cayapas, the Santiago, and the Onzole. Borbón is situated at the confluence of the rivers, is the largest of the study communities, and is the main population center of the region (pop. ∼ 5000). A random sample of 200 households in Borbón was selected and enrolled into the study. In the 21 smaller villages, all households were eligible to be enrolled into the study, and > 98% consented to participate. Four of these villages are located along a road. The remaining 17 villages are primarily accessed by river: two are downstream from Borbón, and 15 are upstream from Borbón. Oral consent for participation was obtained at both the village and household levels. IRB committees at the Universidad San Francisco de Quito and University of California, Berkeley, approved all protocols. Details on the region can be accessed elsewhere.29-31
Study design
In Borbón, one 15-day case-control study was conducted in July 2005. Each of the 21 smaller study villages was visited three times between August 2003 and June 2005. During each visit, a 15-day case-control study was conducted in which fecal specimens were collected for every case of diarrhea in the community. For each case, three asymptomatic control specimens were also collected: one from a member of the case’s household and two randomly selected from the community. A case was defined as an individual that had three or more loose stools in a 24-hour period. A control was defined as someone with no symptoms of diarrhea.
Pathogenic E. coli identification
For each stool sample, five lactose-fermenting colonies were isolated on a MacConkey agar plate. The five colonies were pooled, resuspended in 300 μL of sterile distilled water, and boiled for 10 min to release the DNA. The resulting supernatant was used for PCR testing. Identification of E. coli pathovars was performed by PCR, with primers designed to amplify the bfp gene of EPEC, the LT and Sta genes of ETEC, and the ipaH gene of EIEC.15 Non-lactose-fermenting colonies that were identified by API 20E (BioMèrieux, Marcy l’Etoile, France) as Shigellae or E. coli were subsequently analyzed by PCR with primers designed to amplify the ipaH gene. The primer sequences and the amplification protocols were published previously.15 Briefly, a 2.5 μL aliquot of DNA suspension was amplified with PuRe Taq Ready-To-Go PCR beads (Amersham Biosciences, Piscataway, NJ). The 2.5 μL solution added to the beads contained 0.08 μM of each appropriate oligonucleotide primer. The cycling parameters were as follows: 30 cycles of denaturation at 94°C for 1 min, annealing at 56°C for 2 min, and extension at 72°C for 1 min. The PCR products were resolved by 1.6% agarose gel electrophoresis and visualized by UV transillumination after ethidium bromide staining. Positive and negative control strains for PCR tests were kindly provided by Lee W. Riley, University of California, Berkeley.
PFGE typing
All of the E. coli isolates identified by PCR as EIEC or Shigellae were subjected to pulsed field gel electrophoresis typing (PFGE). Briefly, an overnight cell culture was resuspended in SE buffer (75 mM NaCl, 25 mM EDTA, pH 8.0) to an OD610 of ∼ 0.7. An aliquot of 200 μL of this suspension was mixed with an equal volume of a solution containing 10 μL of proteinase K (20 mg/mL), 1% SDS, and 1% agarose (pulsed field certified agarose, Bio-Rad Laboratories, Hercules, CA). This mixture was dispensed into disposable plug molds. After solidification, the agarose plugs were transferred to tubes containing 1.5 mL of lysis buffer (50 mM Tris-HCl, pH 8.0; 50 mM EDTA, pH 8.0; 1% N-laurylsarcosine; and 0.1 mg/mL of proteinase K), and lysis was carried out overnight at 54°C with constant shaking. After lysis, the agarose plugs were washed five times with 10 mL of warm (50°C) TE buffer (10 mM Tris; 1 mM EDTA, pH 8.0) and stored in TE at 4°C. Slices of the agarose plugs were digested with 60 U of XbaI (New England Biolabs, Boston, MA) overnight at 37°C, in accordance with the manufacturer’s instructions.
Restriction fragments were separated in a 1% pulsed field certified agarose (Bio-Rad Laboratories) gel by PFGE using a CHEF electrophoresis cell (Bio-Rad Laboratories). The gels were run in 0.5× TBE buffer with 100 μM thiourea.32 Running conditions were 14°C at 6 V/cm, with an initial pulse time of 2.2 s that was increased to 54.2 s over the course of 23 h. Gels were stained with ethidium bromide and visualized with UV light.
Images of PFGE electrophoretic patterns were imported and analyzed with GelCompar II, version 2.0 (Applied Maths, Kortrijk, Belgium). From the electrophoretic curves, a distance matrix was calculated using the Pearson correlation algorithm implemented by the GelCompar program. A dendrogram was generated from the distance matrix by the neighbor-joining method. The PFGE fingerprint patterns of isolates that appeared to cluster together on the dendrograms were then visually examined to confirm their identity.
Invasion cell assay
The invasive phenotype of bacterial isolates was confirmed by inoculation of a confluent monolayer of HeLa cells. Isolates were grown in LB media to an OD600 of 0.4-0.6, washed twice with sterile PBS, and then resuspended to an OD600 of 0.5; 25 μL of this suspension was added to PBS-washed HeLa cells and 1 mL of DMEM. This mixture was centrifuged and then incubated in 5% CO2 at 37°C for 3 h. Cells were washed three times with PBS and then incubated at 37°C for 1 h with 1.5 mL of DMEM containing 100 μL/mL gentamicin. Cells were lysed by pipetting after the addition of 1 mL of 0.1% Triton X-100 in PBS and shaking for 5 min. Serial dilutions were plated to LB media and grown overnight at 37°C. Colonies were counted, and bacteria were visualized microscopically with Giemsa stain.
Statistical analysis
All data were analyzed with Stata 8.0 (StataCorp. LP, College Station, TX). Prevalence of infection was estimated as a weighted sum of cases and controls assuming that all cases were identified during each 15-day visit to a community and that the controls were randomly sampled. Because wage income is relatively uncommon in the study area, socioeconomic status was assessed through ownership of material goods. Surveys were conducted in each case and control household to determine the number and type of consumer goods each household possessed, and a standard of living index (SLI) was calculated by weighting and summing these results. Sanitation was defined as either improved (pit latrine or septic tank) or unimproved (river or open ground); water source was defined as improved (well or piped) or unimproved (surface). Food consumption habits were reported for the week prior to stool collection.
RESULTS
Detection and prevalence of pathogenic E. coli
A total of 4220 individuals from 21 villages and 877 from Borbón were enrolled in this study. Between August 2003 and July 2005, 342 cases of diarrhea were identified, and 970 asymptomatic controls were selected (three for each diarrhea sample). From these cases and controls, 915 stool samples (236 cases, 679 controls) were subjected to further analysis.
Lactose-fermenting enterobacterial colonies were evaluated by PCR from all 915 stool samples. Non-lactose fermenting colonies were also isolated from 355 fecal samples and evaluated by PCR. Forty-three of these isolates were identified as EIEC (21 cases, 21 controls, 1 unknown). Seven isolates were lactose fermenters (lac+) and further identified by PCR to contain the ipaH gene. Thirty-six isolates were lactose non-fermenters (lac-) that were further identified by biochemical tests as E. coli and by PCR to contain the ipaH gene. A random sample of 10 of these isolates (5 cases, 4 controls, 1 unknown) was further analyzed by a tissue culture invasion assay, and 80% (5 cases, 2 controls, 1 unknown) were confirmed to be invasive.
Ninety-one pathogenic E. coli strains or Shigellae were identified in 88 samples. Of the three co-infections, one individual was co-infected with EIEC and Shigellae, one with ETEC and EIEC, and the other with EPEC and Shigellae. The prevalence of each pathotype stratified by location and case versus control status is shown in Table 1. The pathogenic bacteria most frequently identified were EIEC (3.2 cases/100 persons) and Shigellae (1.5 cases/100 persons), followed by ETEC (1.3 cases/100 persons), and EPEC (0.9 case/100 persons).
TABLE 1.
Isolation of E. coli pathotypes and Shigellae by geographic region and case (D+) versus control (D-) status
| Region | Tested (N) | EIEC+ (D+, D-) | Prevalence (D+, D-) | OR [95% CI] | Shigellae+ (D+, D-) | Prevalence (D+, D-) | OR [95% CI] |
|---|---|---|---|---|---|---|---|
| Borbón | 169 | 25 (9, 16) | 13.2 (20.5, 12.8) | 1.8 [0.6, 4.6] | 7 (0, 7) | 5.3 (0, 5.6) | NA |
| Communities | 746 | 17 (12, 5) | 1.0 (6.3, 0.9) | 7.3 [2.4, 26.8] | 5 (2, 3) | 0.6 (1.0, 0.5) | 1.9 [0.2, 17.0] |
| Total | 915 | 42 (21, 21) | 3.2 (8.9, 3.1) | 3.1 [1.6, 6.0] | 12 (2, 10) | 1.5 (0.9, 1.5) | 0.6 [0.06, 2.7] |
| Region | Tested (N) | ETEC+ (D+, D-) | Prevalence (D+, D-) | OR [95% CI] | EPEC+ (D+, D-) | Prevalence (D+, D-) | OR [95% CI] |
|---|---|---|---|---|---|---|---|
| Borbón | 169 | 7 (4, 3) | 2.7 (9.1, 2.4) | 4.1 [0.7, 28.7] | 2 (1, 1) | 0.9 (2.3, 0.8) | 2.9 [0.04, 228.1] |
| Communities | 746 | 19 (14, 5) | 1.0 (7.3, 0.9) | 8.6 [2.9, 31.0] | 8 (3, 5) | 0.9 (1.6, 0.9) | 1.7 [0.3, 9.0] |
| Total | 915 | 26 (18, 8) | 1.3 (7.6, 1.2) | 6.9 [2.8, 18.6] | 10 (4, 6) | 0.9 (1.7, 0.9) | 1.9 [0.4, 8.2] |
For communities, estimates are based on the average of three 15-day case control studies across all 21-study villages. Borbón estimates are based on one 15-day case control study. Prevalence is based on a weighted average of infection in cases and controls.
Geographic distribution of pathotypes
All pathotypes had a higher prevalence in Borbón than in the smaller communities. In Borbón, EIEC was the dominant pathotype for both cases and controls (21 and 13 cases/100 persons, respectively). In the communities, EIEC and ETEC were the dominant pathotypes in the diarrhea cases (6.3 and 7.3 cases/100 persons, respectively). In the community controls, however, the prevalence of all pathotypes was ∼ 1 case/100 persons.
Only EIEC and ETEC infections were significantly associated with diarrheal disease in the communities. Although prevalence of infection was higher in Borbón than in the communities, infection was not significantly associated with disease in Borbón for any of the pathotypes.
Age distribution of pathotypes
In general, prevalence dropped off in the > 20 year age group (Table 2). This was less evident in EIEC (RR = 1.5 comparing 0-5 year olds with those > 20 years old; 95% CI: 1.1 to 2.0) than in Shigellae (RR = 3.1; 95% CI: 2.1 to 4.5), EPEC (RR = 2.9; 95% CI: 2.1 to 4.1), and ETEC (RR = 3.7; 95% CI: 2.4 to 5.6). Specifically, eight EIEC infections (2.5 cases/100 persons) were identified in the > 20 year old age group, 7 of which were asymptomatic individuals. In contrast, the prevalence of ETEC and Shigellae in the > 20 year age group was 0.4 and 0.7 case/100 persons, respectively.
TABLE 2.
Isolation of E. coli pathotypes and Shigellae by age; prevalence is based on a weighted average of infection in cases (D+) and controls (D-)
| Age | Tested (N) | EIEC+ (D+, D-) | Prevalence (D+, D-) | OR [95% CI] | Shigellae+ (D+, D-) | Prevalence (D+, D-) | OR [95% CI] |
|---|---|---|---|---|---|---|---|
| < 5 years | 228 | 14 (11, 3) | 3.7 (8.0, 3.3) | 2.5 [0.6, 14.4] | 4 (2, 2) | 2.2 (1.5, 2.2) | 0.6 [0.05, 9.1] |
| 5-20 years | 312 | 17 (6, 11) | 4.0 (21.4, 3.9) | 6.8 [1.9, 22.1] | 6 (0, 6) | 2.1 (0, 2.1) | 0 [0, 6.6] |
| > 20 years | 318 | 8 (1, 7) | 2.5 (2.8, 2.5) | 1.1 [0.02, 9.2] | 2 (0, 2) | 0.7 (0, 0.7) | 0 [0, 15.3] |
| Unknown | 57 | 3 (3, 0) | 0.7 (8.8, 0) | NA [0.5, NA] | 0 | 0 | NA |
| Total | 915 | 42 (21, 21) | 3.2 (8.9, 3.1) | 3.1 [1.6, 6.0] | 12 (2, 10) | 1.5 (0.9, 1.5) | 0.6 [0.06, 2.7] |
| Age | Tested (N) | ETEC+ (D+, D-) | Prevalence (D+, D-) | OR [95% CI] | EPEC+ (D+, D-) | Prevalence (D+, D-) | OR [95% CI] |
|---|---|---|---|---|---|---|---|
| < 5 years | 228 | 11 (10, 1) | 1.6 (7.3, 1.1) | 7.0 [0.95, 305.0] | 4 (1, 3) | 3.1 (0.7, 3.3) | 0.2 [0.004, 2.7] |
| 5-20 years | 312 | 8 (2, 6) | 2.1 (7.1, 2.1) | 3.6 [0.3, 21.1] | 0 | 0 | NA |
| > 20 years | 318 | 5 (4, 1) | 0.4 (11.1, 0.4) | 35.1 [3.3, 1735.2] | 3 (0.3) | 1.1 (0, 1.1) | 0 [0, 10.2] |
| Unknown | 57 | 2 (2, 0) | 0.4 (5.9, 0) | NA [0.4, NA] | 3 (3, 0) | 0.7 (8.8, 0) | NA [0.5, NA] |
| Total | 915 | 26 (18, 8) | 1.3 (7.6, 1.2) | 6.9 [2.8, 18.6] | 10 (4, 6) | 0.9 (1.7, 0.9) | 1.9 [0.4, 8.2] |
Risk-factor analysis
Aggregating all E. coli and Shigellae isolates showed no association between infection and water source, sanitation, or food consumption (Table 3). The SLI was protective for infection (RR = 0.91; 95% CI: 0.86 to 0.97). Living with an infected case did not pose a significant risk of asymptomatic infection (OR = 2.2; 95% CI: 0.5 to 8.1), although it did have the highest point estimate. These results were generally consistent across all E. coli pathotypes and Shigellae though not always statistically significant, possibly due to small sample size.
TABLE 3.
Bivariate risk factor analysis aggregating infections of E. coli pathotypes and Shigellae, where (+) is positive for at least one of the following pathogens: EIEC, Shigellae, ETEC, or EPEC
| (+) | (-) | RR [95% CI] | |
|---|---|---|---|
| Socioeconomic status (Standard of Living index) | 7.4 | 8.8 | 0.91 [0.86, 0.97] |
| No. in household | 6.2 | 6.8 | 0.94 [0.86, 1.02] |
| Age (continuous) | 14.5 | 20.8 | 0.98 [0.97, 0.995] |
| (+) | (-) | OR [95% CI] | ||
|---|---|---|---|---|
| Living with a (+) case (these observations are pairs) | Yes | 5 | 34 | 2.19 [0.52, 8.13] |
| No | 8 | 119 | ||
| Unimproved sanitation | 17 | 149 | 0.98 [0.51, 1.81] | |
| Improved | 47 | 404 | ||
| Unimproved water source | 47 | 506 | 0.81 [0.45, 1.51] | |
| Improved | 19 | 166 | ||
| Chicken consumption | Yes | 55 | 527 | 1.12 [0.64, 2.05] |
| No | 19 | 204 | ||
| Beef/pork consumption | Yes | 43 | 416 | 1.06 [0.63, 1.79] |
| No | 30 | 307 | ||
| Fish consumption | Yes | 59 | 630 | 0.64 [0.34, 1.26] |
| No | 15 | 102 | ||
| Wild animal consumption | Yes | 17 | 236 | 0.63 [0.33, 1.12] |
| No | 57 | 495 | ||
Pulsed field gel electrophoresis (PFGE) typing
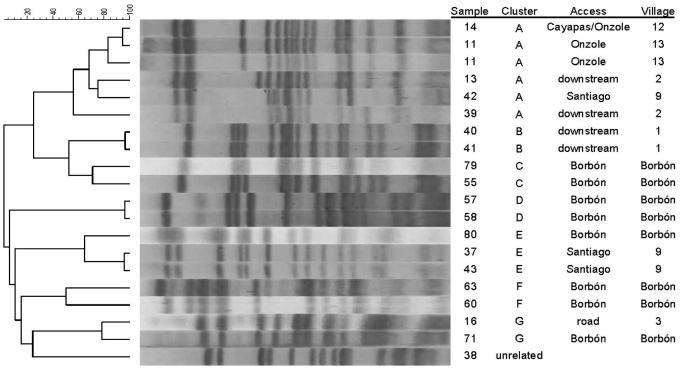
PFGE analysis of the 43 EIEC isolates identified one five-member cluster, one three-member cluster, and five two-member clusters (Figure 1). Four of the five two-member clusters were within-village clusters, three of them within Borbón. All other clusters linked individuals in different villages: one connected a road community with Borbón; one connected a Santiago river community with Borbón; and one connected communities along two river basins (the Santiago and the Onzole), as well as a community downstream from Borbón. These clusters represent 18 of 43 isolates. The remaining 25 isolates were not related to each other or to any of the seven clusters, suggesting that the presence of EIEC is likely due to the appearance of multiple clones in the communities rather than to a single source outbreak.
Figure 1.
XbaI PFGE patterns for selected clonal groups (A-G). Area refers to whether the isolate comes from (1) a village that is on an upstream river basin (Cayapas, Onzole, Santiago); (2) a village that is downstream from Borbón; (3) a village along a road; or (4) within Borbón. Dendrogram was inferred based on a cosine coefficient similarity matrix generated by GelCompar II using the unweighted pair group method with arithmetic averages (UPGMA).
DISCUSSION
The high prevalence levels of EIEC infection in diarrheal cases observed in this study (20.5 cases/100 persons in Borbón and 6.3 cases/100 persons in the communities) are unprecedented in both hospital- and community-based studies of E. coli. EIEC was isolated two to three times more often than ETEC, EPEC, and Shigellae. This result contradicts the opposite finding more commonly seen in the literature on pathogenic E. coli. For example, estimates of an ETEC incidence of 2.45 cases/child-year versus an EIEC incidence of 0.29 case/child-year in Ghana have been made.17 Of the studies reviewed in the introduction, 75% found few or no EIEC infections. The two most notable exceptions were a 1997 travelers’ diarrhea study in which 6% of the stool samples analyzed detected EIEC22 and a 1989 study in Beijing in which 7% of the stool samples from children with diarrhea were positive for EIEC.19
There is no clear explanation for the elevated prevalence of EIEC relative to other pathotypes in this region of Ecuador. One possibility would be that the EIEC isolates were from a single point-source outbreak. A number of EIEC outbreaks have been reported,33-37 many of which were food-borne.38-41 Previous studies have shown the potential for using molecular tools to identify EIEC outbreak clusters.42,43 Our PFGE results, however, indicate that these EIEC isolates were not from any single source; i.e., there were clearly multiple sources of EIEC within our study region. Interestingly, the larger cluster (A) shown in Figure 1 suggests that transmission occurred between two river basins (the Santiago and the Onzole) as well as downstream from Borbón. The most likely explanation is that the source was Borbón. The lack of evidence for a single source of infection suggests that there may be specific risk factors that promote the transmission of EIEC. Unfortunately, very little is known about the epidemiology of EIEC. Our small sample size precluded us from making any firm conclusions about the risks associated with the 43 identified infections. After aggregating all E. coli infections (Table 3), higher socioeconomic status (SES) was found to be protective (OR = 0.91 [95% CI = 0.86, 0.97]). No other significant associations with regard to water, sanitation, or food risks were found. When disaggregated, the protective relationship with SES was maintained with EIEC and Shigellae but was no longer significant with ETEC and EPEC (Table 4). Additional samples are needed to improve our understanding of EIEC transmission patterns within the region.
A number of methodological issues exist that may partially explain why low levels of EIEC are found in other studies. Specifically, microbiological analyses used in other studies often do not distinguish between EIEC and Shigellae (see, for example, refs. 9, 17, 21, and 44). Differentiating between EIEC and Shigellae is difficult because of their genetic similarities. The four species of Shigellae are often considered to be types of E. coli and are most similarly related to EIEC45; these bacteria are characterized by a large virulence plasmid (220 kb) and by their ability to invade epithelial cells and disseminate from cell to cell.45 Depending on the design of the particular study, this may result in underestimates of EIEC or of Shigellae.46
Another methodological issue is that EIEC can be either lactose-positive or lactose-negative,46 an unusual trait among E. coli. In this study, 36 EIEC isolates were lactose-negative and only seven were lactose-positive. Many other studies only screen for lactose-positive E. coli strains (see, for example, refs. 4, 6, 12, 16, 20, and 47) and thus may underestimate the prevalence of EIEC infection.
From a clinical perspective, distinguishing between these two bacteria is unnecessary because treatment of the two infections is the same. From a public health intervention perspective, however, the distinction may be more important. Although these organisms are closely related, EIEC and Shigellae have important differences relating to transmission. The minimum infectious inoculum of EIEC is higher than at least two of the four Shigellae species,48,49 and some studies suggest that the mode of transmission may differ. Shigellae has primarily been associated with transmission via personal contact,50 whereas EIEC has principally been associated with contaminated food and water33,38 although cases of person-to-person transmission of EIEC have been noted.34 In our study, there was some evidence of the similarity and divergence between the epidemiology of EIEC and Shigellae. For example, both exhibited a similar relationship to SES levels and to crowding, and both exhibited insignificant relationships with water source, sanitation level, and food consumption. In contrast, with respect to age-stratified infection rates, EIEC was found more often in older age groups than was Shigellae, suggesting that adults were exposed to EIEC more than to Shigellae. The EIEC isolated from adults was more likely to be from controls, suggesting that EIEC is highly endemic, i.e., that exposure occurs in younger age groups, resulting in immune adults. These analyses, however, are limited because of the small sample size. Given the potential importance of this invasive pathogen, more work should be focused on why EIEC is highly prevalent in this study region. In addition, studies that properly distinguish EIEC from Shigellae would help determine if this high prevalence is also common in other areas of the world. Additional studies that provide samples representative of communities would provide valuable information on the EIEC epidemiology patterns. Understanding these patterns of EIEC infection and transmission would provide important information on how best to design intervention and control strategies targeted at both EIEC and Shigellae.
Acknowledgments
The authors thank the EcoDESS field team for their invaluable contribution in collecting the data.
Financial support: This study was supported by a grant from the National Institute of Allergy and Infectious Disease (NIAID; grant RO1-AI050038).
REFERENCES
- 1.DuPont HL, Formal SB, Hornick RB, Snyder MJ, Libonati JP, Sheahan DG, LaBrec EH, Kolas JP. Pathogenesis of Escherichia coli diarrhea. N Engl J Med. 1971;285:1–9. doi: 10.1056/NEJM197107012850101. [DOI] [PubMed] [Google Scholar]
- 2.Akinyemi KO, Oyefolu AO, Opere B, Otunba-Payne VA, Oworu AO. Escherichia coli in patients with acute gastroenteritis in Lagos, Nigeria. East Afr Med J. 1998;75:512–515. [PubMed] [Google Scholar]
- 3.Katouli M, Jaafari A, Farhoudi-Moghaddam AA, Ketabi GR. Aetiological studies of diarrhoeal diseases in infants and young children in Iran. J Trop Med Hyg. 1990;93:22–27. [PubMed] [Google Scholar]
- 4.Kim KH, Suh IS, Kim JM, Kim CW, Cho YJ. Etiology of childhood diarrhea in Korea. J Clin Microbiol. 1989;27:1192–1196. doi: 10.1128/jcm.27.6.1192-1196.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee CL, Chiou SI, Liu TP, Pan TM. Pathogenic strains of Escherichia coli in Taiwan. Zhonghua Min Guo Wei Sheng Wu Ji Mian Yi Xue Za Zhi. 1997;30:55–59. [PubMed] [Google Scholar]
- 6.Okeke IN, Lamikanra A, Steinruck H, Kaper JB. Characterization of Escherichia coli strains from cases of childhood diarrhea in provincial southwestern Nigeria. J Clin Microbiol. 2000;38:7–12. doi: 10.1128/jcm.38.1.7-12.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Orlandi PP, Silva T, Magalhaes GF, Alves F, de Almeida Cunha RP, Durlacher R, da Silva LH. Enteropathogens associated with diarrheal disease in infants of poor urban areas of Porto Velho, Rondonia: a preliminary study. Mem Inst Oswaldo Cruz. 2001;96:621–625. doi: 10.1590/s0074-02762001000500005. [DOI] [PubMed] [Google Scholar]
- 8.Pabst WL, Altwegg M, Kind C, Mirjanic S, Hardegger D, Nadal D. Prevalence of enteroaggregative Escherichia coli among children with and without diarrhea in Switzerland. J Clin Microbiol. 2003;41:2289–2293. doi: 10.1128/JCM.41.6.2289-2293.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rappelli P, Folgosa E, Solinas ML, Dacosta JL, Pisanu C, Sidat M, Melo J, Cappuccinelli P, Colombo MM. Pathogenic enteric Escherichia coli in children with and without diarrhea in Maputo, Mozambique. FEMS Immunol Med Microbiol. 2005;43:67–72. doi: 10.1016/j.femsim.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 10.Regua-Mangia AH, Gomes TA, Vieira MA, Andrade JR, Irino K, Teixeira LM. Frequency and characteristics of diarrhoeagenic Escherichia coli strains isolated from children with and without diarrhoea in Rio de Janeiro, Brazil. J Infect. 2004;48:161–167. doi: 10.1016/s0163-4453(03)00138-5. [DOI] [PubMed] [Google Scholar]
- 11.Shehabi AA, Bulos NK, Hajjaj KG. Characterization of diarrhoeagenic Escherichia coli isolates in Jordanian children. Scand J Infect Dis. 2003;35:368–371. doi: 10.1080/003655403100009086. [DOI] [PubMed] [Google Scholar]
- 12.Sunthadvanich R, Chiewsilp D, Seriwatana J, Sakazaki R, Echeverria P. Nationwide surveillance program to identify diarrhea-causing Escherichia coli in children in Thailand. J Clin Microbiol. 1990;28:469–472. doi: 10.1128/jcm.28.3.469-472.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taylor DN, Houston R, Shlim DR, Bhaibulaya M, Ungar BL, Echeverria P. Etiology of diarrhea among travelers and foreign residents in Nepal. JAMA. 1988;260:1245–1248. [PubMed] [Google Scholar]
- 14.Teng LJ, Hsueh PR, Liaw SJ, Ho SW, Tsai JC. Genetic detection of diarrheagenic Escherichia coli isolated from children with sporadic diarrhea. J Microbiol Immunol Infect. 2004;37:327–334. [PubMed] [Google Scholar]
- 15.Tornieporth NG, John J, Salgado K, de Jesus P, Latham E, Melo MC, Gunzburg ST, Riley LW. Differentiation of pathogenic Escherichia coli strains in Brazilian children by PCR. J Clin Microbiol. 1995;33:1371–1374. doi: 10.1128/jcm.33.5.1371-1374.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Torres ME, Pirez MC, Schelotto F, Varlea G, Parodi V, Allende F, Falconi E, Dell’Acqua L, Gaione P, Mendez MV, Ferrari AM, Montano A, Zanetta E, Acuna AM, Chiparelli H, Ingold E. Etiology of children’s diarrhea in Montevideo, Uruguay: associated pathogens and unusual isolates. J Clin Microbiol. 2001;39:2134–2139. doi: 10.1128/JCM.39.6.2134-2139.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Valentiner-Branth P, Steinsland H, Fischer TK, Perch M, Scheutz F, Aaby P, Molbak K, Sommerfelt H. Cohort study of Guinean children: incidence, pathogenicity, conferred protection, and attributable risk for enteropathogens during the first 2 years of life. J Clin Microbiol. 2003;41:4238–4245. doi: 10.1128/JCM.41.9.4238-4245.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Youssef M, Shurman A, Bougnoux M, Rawashdeh M, Bretagne S, Strockbine N. Bacterial, viral and parasitic enteric pathogens associated with acute diarrhea in hospitalized children from northern Jordan. FEMS Immunol Med Microbiol. 2000;28:257–263. doi: 10.1111/j.1574-695X.2000.tb01485.x. [DOI] [PubMed] [Google Scholar]
- 19.Kain KC, Barteluk RL, Kelly MT, He X, de Hua G, Ge YA, Proctor EM, Byrne S, Stiver HG. Etiology of childhood diarrhea in Beijing, China. J Clin Microbiol. 1991;29:90–95. doi: 10.1128/jcm.29.1.90-95.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taylor DN, Echeverria P, Sethabutr O, Pitarangsi C, Leksomboon U, Blacklow NR, Rowe B, Gross R, Cross J. Clinical and microbiologic features of Shigella and enteroinvasive Escherichia coli infections detected by DNA hybridization. J Clin Microbiol. 1988;26:1362–1366. doi: 10.1128/jcm.26.7.1362-1366.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Faundez G, Figueroa G, Troncoso M, Cabello FC. Characterization of enteroinvasive Escherichia coli strains isolated from children with diarrhea in Chile. J Clin Microbiol. 1988;26:928–932. doi: 10.1128/jcm.26.5.928-932.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Altwegg M, Perschil I, Gruner E. Molecular biology detection and antibiotic sensitivities of Shigella spp. and enteroinvasive Escherichia coli (EIEC) in patients returning from the tropics. Schweiz Rundsch Med Prax. 1997;86:348–351. [PubMed] [Google Scholar]
- 23.Gassama-Sow A, Sow PS, Gueye M, Gueye-N’diaye A, Perret JL, M’boup S, Aidara-Kane A. Characterization of pathogenic Escherichia coli in human immunodeficiency virus-related diarrhea in Senegal. J Infect Dis. 2004;189:75–78. doi: 10.1086/380489. [DOI] [PubMed] [Google Scholar]
- 24.Meqdam MM, Youssef MT, Rawashdeh MO, Al-khdour MS. Non-seasonal viral and bacterial episode of diarrhoea in the Jordan Valley, West of Jordan. FEMS Immunol Med Microbiol. 1997;18:133–138. doi: 10.1111/j.1574-695X.1997.tb01038.x. [DOI] [PubMed] [Google Scholar]
- 25.Cortes-Ortiz IA, Rodriguez-Angeles G, Moreno-Escobar EA, Tenorio-Lara JM, Torres-Mazadiego BP, Montiel-Vazquez E. Outbreak caused by Escherichia coli in Chalco, Mexico. Salud Publica Mex. 2002;44:297–302. [PubMed] [Google Scholar]
- 26.Nguyen TV, Le Van P, Le Huy C, Gia KN, Weintraub A. Detection and characterization of diarrheagenic Escherichia coli from young children in Hanoi, Vietnam. J Clin Microbiol. 2005;43:755–760. doi: 10.1128/JCM.43.2.755-760.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ratchtrachenchai OA, Subpasu S, Hayashi H, Ba-Thein W. Prevalence of childhood diarrhoea-associated Escherichia coli in Thailand. J Med Microbiol. 2004;53:237–243. doi: 10.1099/jmm.0.05413-0. [DOI] [PubMed] [Google Scholar]
- 28.Svenungsson B, Lagergren A, Ekwall E, Evengard B, Hedlund KO, Karnell A, Lofdahl S, Svensson L, Weiantraub A. Enteropathogens in adult patients with diarrhea and healthy control subjects: a 1-year prospective study in a Swedish clinic for infectious diseases. Clin Infect Dis. 2000;30:770–778. doi: 10.1086/313770. [DOI] [PubMed] [Google Scholar]
- 29.Rival L. The meanings of forest governance in Esmeraldas, Ecuador. Oxf Dev Stud. 2003;31:479–501. [Google Scholar]
- 30.Sierra R. Traditional resource-use systems and tropical deforestation in a multi-ethnic region in North-west Ecuador. Environ Conserv. 1999;26:136–145. [Google Scholar]
- 31.Whitten NE. Black Frontiersmen: A South American Case. Schenkman Pub. Co.; Cambridge, MA: 1974. [Google Scholar]
- 32.Zhang Y, Yakrus MA, Graviss EA, Williams-Bouyer N, Turenne C, Kabani A, Wallace RJ., Jr Pulsed-field gel electrophoresis study of Mycobacterium abscessus isolates previously affected by DNA degradation. J Clin Microbiol. 2004;42:5582–5587. doi: 10.1128/JCM.42.12.5582-5587.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gordillo ME, Reeve GR, Pappas J, Mathewson JJ, DuPont HL, Murray BE. Molecular characterization of strains of enteroinvasive Escherichia coli O143, including isolates from a large outbreak in Houston, Texas. J Clin Microbiol. 1992;30:889–893. doi: 10.1128/jcm.30.4.889-893.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harris JR, Mariano J, Wells JG, Payne BJ, Donnell HD, Cohen ML. Person-to-person transmission in an outbreak of enteroinvasive Escherichia coli. Am J Epidemiol. 1985;122:245–252. doi: 10.1093/oxfordjournals.aje.a114095. [DOI] [PubMed] [Google Scholar]
- 35.Ozbonfil D, Cohen D, Ohad E, Sechter I. An outbreak of enteritis associated with enteroinvasive E. coli in an Israeli military base. Public Health Rev. 1990;18:171–177. [PubMed] [Google Scholar]
- 36.She SL. An outbreak of diarrhoea in newborns caused by EIEC. Zhonghua Liu Xing Bing Xue Za Zhi. 1989;10:333–336. [PubMed] [Google Scholar]
- 37.Sobotkova J, Hausner O, Aldova E, Jiskrova R, Maskova L. An epidemic of diarrhea caused by enteroinvasive serotypes of Escherichia coli O28 ac (Ca 792) Cesk Epidemiol Mikrobiol Imunol. 1984;33:315. [PubMed] [Google Scholar]
- 38.Marier R, Wells JG, Swanson RC, Dalthan W, Mehlman IJ. An outbreak of enteropathogenic Escherichia coli foodborne disease traced to imported French cheese. Lancet. 1973;ii:1376–1378. [Google Scholar]
- 39.Snyder JD, Wells JG, Yashuk J, Puhr N, Blake PA. Outbreak of invasive Escherichia coli gastroenteritis on a cruise ship. Am J Trop Med Hyg. 1984;33:281–284. doi: 10.4269/ajtmh.1984.33.281. [DOI] [PubMed] [Google Scholar]
- 40.Yamamura K, Sumi N, Egashira Y, Fukuoka I, Motomura S, Tsuchida R. Food poisoning caused by enteroinvasive Escherichia coli (O164:H-)—a case in which the causative agent was identified. Kansenshogaku Zasshi. 1992;66:761–768. doi: 10.11150/kansenshogakuzasshi1970.66.761. [DOI] [PubMed] [Google Scholar]
- 41.Yang SZ. An outbreak of food poisoning due to enteroinvasive E. coli. Zhonghua Liu Xing Bing Xue Za Zhi. 1986;7:129–131. [PubMed] [Google Scholar]
- 42.Beutin L, Gleier K, Kontny I, Echeverria P, Scheutz F. Origin and characteristics of enteroinvasive strains of Escherichia coli (EIEC) isolated in Germany. Epidemiol Infect. 1997;118:199–205. doi: 10.1017/s0950268897007413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Llovet T, Coll P, March F, Montserrat I, Atela I, Mirelis B, Prats G. Comparison of macrorestriction analysis of genomic DNA by pulsed-field gel electrophoresis and ribotyping with conventional methods for differentiation of Escherichia coli O124 isolates. Clin Microbiol Infect. 1995;1:127–133. doi: 10.1111/j.1469-0691.1995.tb00457.x. [DOI] [PubMed] [Google Scholar]
- 44.Jallat C, Livrelli V, Darfeuille-Michaud A, Rich C, Joly B. Escherichia coli strains involved in diarrhea in France: high prevalence and heterogeneity of diffusely adhering strains. J Clin Microbiol. 1993;31:2031–2037. doi: 10.1128/jcm.31.8.2031-2037.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pupo GM, Lan R, Reeves PR. Multiple independent origins of Shigella clones of Escherichia coli and convergent evolution of many of their characteristics. Proc Natl Acad Sci USA. 2000;97:10567–10572. doi: 10.1073/pnas.180094797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nataro JP, Kaper JB. Diarrheagenic Escherichia coli. Clin Microbiol Rev. 1998;11:142–201. doi: 10.1128/cmr.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sarantuya J, Nishi J, Wakimoto N, Erdene S, Nataro JP, Sheikh J, Iwashita M, Mango K, Tokuda K, Yoshinaga M, Miyata K, Kawano Y. Typical enteroaggregative Escherichia coli is the most prevalent pathotype among E. coli strains causing diarrhea in Mongolian children. J Clin Microbiol. 2004;42:133–139. doi: 10.1128/JCM.42.1.133-139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.DuPont HL, Levine MM, Hornick RB, Formal SB. Inoculum size in shigellosis and implications for expected mode of transmission. J Infect Dis. 1989;159:1126–1128. doi: 10.1093/infdis/159.6.1126. [DOI] [PubMed] [Google Scholar]
- 49.Hsia RC, Small PL, Bavoil PM. Characterization of virulence genes of enteroinvasive Escherichia coli by TnphoA mutagenesis: identification of invX, a gene required for entry into HEp-2 cells. J Bacteriol. 1993;175:4817–4823. doi: 10.1128/jb.175.15.4817-4823.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Niyogi SK. Shigellosis. J Microbiol. 2005;43:133–143. [PubMed] [Google Scholar]