Abstract
Avian H7 influenza viruses from both the Eurasian and North American lineage have caused outbreaks in poultry since 2002, with confirmed human infection occurring during outbreaks in The Netherlands, British Columbia, and the United Kingdom. The majority of H7 infections have resulted in self-limiting conjunctivitis, whereas probable human-to-human transmission has been rare. Here, we used glycan microarray technology to determine the receptor-binding preference of Eurasian and North American lineage H7 influenza viruses and their transmissibility in the ferret model. We found that highly pathogenic H7N7 viruses from The Netherlands in 2003 maintained the classic avian-binding preference for α2–3-linked sialic acids (SA) and are not readily transmissible in ferrets, as observed previously for highly pathogenic H5N1 viruses. However, H7N3 viruses isolated from Canada in 2004 and H7N2 viruses from the northeastern United States isolated in 2002–2003 possessed an HA with increased affinity toward α2–6-linked SA, the linkage type found prominently on human tracheal epithelial cells. We identified a low pathogenic H7N2 virus isolated from a man in New York in 2003, A/NY/107/03, which replicated efficiently in the upper respiratory tract of ferrets and was capable of transmission in this species by direct contact. These results indicate that H7 influenza viruses from the North American lineage have acquired sialic acid-binding properties that more closely resemble those of human influenza viruses and have the potential to spread to naïve animals.
Keywords: hemagglutinin, transmission, receptor binding, animal model
Avian influenza viruses within the H5 and H7 subtype continue to pose a major public health threat. Since 2004, highly pathogenic avian influenza (HPAI) H5N1 viruses have resulted in >380 cases of laboratory-confirmed human infection in 14 countries (1). Despite the high virulence of H5N1 viruses observed in humans and mammalian models (2), human-to-human transmission has been only rarely documented (3–5). Additionally, influenza H7 subtype viruses within both Eurasian and North American lineages have been responsible for multiple outbreaks and human infections since 2002. These include outbreaks of HPAI H7N7 in The Netherlands in 2003 that resulted in >80 cases of human infection and one fatality; HPAI H7N3 in British Columbia, Canada, in 2004 that resulted in two cases of conjunctivitis; a cluster of human infections of low pathogenic avian influenza (LPAI) H7N2 in the United Kingdom in 2007 that resulted in multiple cases of influenza-like illness and conjunctivitis; and a single case of human respiratory infection in New York in 2003 (6–11). The majority of human infections with H7 influenza viruses have resulted in conjunctivitis, but similar to H5N1 viruses, probable human-to-human transmission among family contacts has been rarely documented through molecular diagnosis (7). Representative viruses isolated from these outbreaks were found to replicate efficiently in the mouse and ferret models, and one virus isolated from a fatal respiratory case during the H7N7 Netherlands outbreak, A/NL/219/03, was highly lethal in both mammalian models (12, 13). However, further study is needed to assess the pandemic potential of H7 influenza viruses within this subtype.
Influenza virus attachment to host cells is mediated by the virus HA binding to sialic acid (SA) glycans present on host cell surfaces. Avian influenza viruses predominantly bind α2–3-linked SA, whereas human influenza viruses preferentially bind to α2–6 SA (14). The three influenza pandemic viruses of the last century, causing the pandemics of 1918 (H1N1), 1957 (H2N2), and 1968 (H3N2), each possessed an HA with a human α2–6 SA-binding preference yet are thought to have originated from an avian virus possessing the α2–3 SA-binding preference (15, 16). With few exceptions, avian H5N1 influenza viruses isolated from humans have maintained the classic α2–3 SA binding (17–20). However, the SA-binding preference of recent H7 influenza viruses associated with disease in humans has not been well studied.
The ferret model has been used successfully to study the transmission of human and avian influenza viruses (21–23), because ferrets exhibit a similar distribution of SA as reported in humans with a higher proportion of α2–6 SA glycans on upper respiratory tract epithelial cells and α2–3 SA in the lower respiratory tract (24–27). These studies have shown that avian H5N1 viruses, despite replicating to high titers in the respiratory tract, are not readily transmissible by either respiratory droplet or contact transmission (21). To date, the transmissibility of viruses within the H7 subtype has not been examined experimentally. Here, we use glycan microarray technology to determine the receptor-binding preference of H7 influenza viruses of both Eurasian and North American lineages and assess the transmissibility of selected H7 influenza viruses using the ferret model. Surprisingly, we found that recently isolated H7N2 and H7N3 viruses of the North American lineage possess increased binding to α2–6 SA, with several strains exhibiting preferential binding characteristic of human influenza viruses. One of these was an H7N2 virus, A/NY/107/03, associated with respiratory disease in an adult male, which we found to be capable of efficient direct contact transmission in the ferret model.
Results
Receptor-Binding Preference of Eurasian H7 Influenza Viruses.
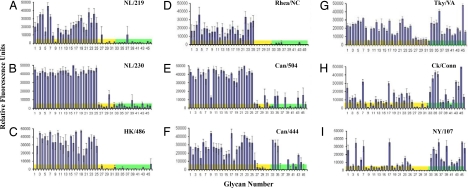
Previous studies have elucidated the molecular basis for the receptor-binding preference of influenza viruses of multiple subtypes, including H1, H2, H3, H5, and H9 viruses (15, 16, 28–30). However, recently isolated H7 influenza viruses have not been comprehensively analyzed for their HA-binding preference. We used a glycan microarray with whole virus to determine the α2–3 and α2–6 SA-binding preference of Eurasian or North American lineage H7 influenza viruses associated with disease in humans or related viruses isolated from birds. Two HPAI H7N7 Eurasian lineage viruses isolated from an outbreak in The Netherlands in 2003 were tested, A/NL/219/03 (NL/219) and A/NL/230/03 (NL/230). NL/219 was isolated from a human with fatal respiratory disease, whereas NL/230 was isolated from an individual with conjunctivitis (6). Both H7N7 viruses exhibited preferential binding specificity toward α2–3 SA (Fig. 1 A and B). This pattern of binding closely resembles the strong α2–3 SA-binding preference observed with HPAI H5N1 viruses isolated from humans, as has been reported and is demonstrated here with A/HK/486/97 (HK/486) virus (Fig. 1C) (29). These results were confirmed by hemagglutination assay, with NL/219, NL/230, and HK/486 viruses binding to turkey red blood cells (RBCs) resialylated with α2–3- but not α2–6-linked sialosides (Table 1). These findings suggest that HPAI H7N7 Eurasian lineage viruses, similar to HPAI H5N1 viruses, have maintained classic avian specificity for α2–3 SA despite causing productive infections in humans.
Fig. 1.
Glycan microarray analysis of Eurasian and North American lineage H7 influenza viruses. Analysis was performed on the following viruses: NL/219 (A), NL/230 (B), HK/486 (H5N1) (C), Rhea/NC (D), Can/504 (E), Can/444 (F), Tky/VA (G), Ck/Conn (H), and NY/107 (I). The glycan microarray was performed by using whole virus with antisera raised against homologous or cross-reactive virus as a primary antibody. Colored bars highlight glycans that contain α2–3 SA (yellow) and α2–6 SA (green). Error bars reflect the standard deviation in the signal for six independent replicates on the array. Structures of each of the numbered glycans are found in Table S1 (SI Text) and for selected glycans in Table 2.
Table 1.
Hemagglutination assay of H7 influenza viruses using differentially sialylated turkey RBCs
| Virus | Presence or absence of hemagglutination |
|||
|---|---|---|---|---|
| TRBC | α2–6 RBC | α2–3 RBC | desial RBC | |
| NL/219 | + | − | + | − |
| NL/230 | + | − | + | − |
| Rhea/NC | + | − | + | − |
| Can/504 | + | + | + | − |
| Can/444 | + | + | + | − |
| NY/107 | + | + | + | − |
| Tky/VA | + | + | + | − |
| Ck/Conn | + | + | + | − |
| HK/486 | + | − | + | − |
| Tx/91 | + | + | − | − |
| PBS | − | − | − | − |
Receptor-Binding Preference of North American H7 Influenza Viruses.
H7N2 subtype viruses have been routinely isolated from the live-bird market system in the northeastern United States since 1994 (31). Glycan-binding analysis of A/Rhea/NC/39482/93 (Rhea/NC), a LPAI H7N1 virus isolated in 1993, exhibited a classic avian α2–3 SA receptor-binding preference (Fig. 1D). However, the more recent H7N2 viruses A/Tky/VA/4529/02 (Tky/VA), which caused a major outbreak among commercial poultry in Virginia and was associated with serologic evidence of human infection (32), and a 2003 H7N2 poultry isolate A/Ck/Conn/260413–2/03 (Ck/Conn), exhibited significantly increased binding to glycans with α2–6 SA (Fig. 1 G and H). A genetically related H7N2 virus isolated from a single case of human respiratory infection in 2003, A/NY/107/03 (NY/107), also exhibited a marked increase in α2–6 SA binding and reduced binding to glycans with α2–3 SA (Fig. 1I). Two H7N3 viruses (A/Canada/504/04 and A/Canada/444/04), associated with human conjunctivitis during an outbreak of HPAI in British Columbia (Fig. 1 E and F), also revealed increased binding to α2–6 SA compared with Eurasian lineage viruses (Fig. 1 A–C). An assay using resialylated erythrocytes independently documented the dual α2–6 and α2–3 SA binding of all H7N3 and H7N2 viruses (Table 1).
More detailed analysis of glycan microarray data revealed that the specificity differences among the H7 viruses was more striking for subclasses of α2–6 and α2–3 glycans as summarized in Table 2. Although neither of the Eurasian viruses nor the Rhea/NC virus bound glycans with α2–6 SA, all of the post-2002 North American viruses exhibited moderate to strong binding to the α2–6 SA of the biantennary N-linked glycans (nos. 34 and 35). Three viruses, Tky/VA, Ck/Conn, and NY/107, exhibited moderate to strong binding to most glycans with α2–6 SA, including a glycan with an internal sialic acid (no. 45) not recognized by the other viruses. Although these three viruses were similar in binding α2–6 SA, they exhibited significant differences in their binding of glycans with α2–3 SA. Tky/VA bound as well to glycans with α2–3 SA as the Eurasian viruses. In contrast, Ck/Conn and NY/107 exhibited strong binding to only 4 of the 32 glycans with α2–3 SA, including two sulfated (nos. 1 and 4), one branched (no. 7), and one linear (no. 17) glycan (Fig. 2 and Table 2). Binding to the remaining glycans with α2–3 SA was significantly reduced, especially for NY/107. The reduced binding to glycans with α2–3 SA is notable, because this was a characteristic of influenza viruses with H1, H2, and H3 HAs when first introduced into the human population (15, 16, 28, 33).
Table 2.
Comparison of the detailed sialoside receptor specificity of H7 influenza viruses
| Virus | α 2–3 sialosides |
α 2–6 sialosides |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Sulfated |
Branched |
Linear |
Fucosylated |
Branched |
Linear |
Internal |
|||
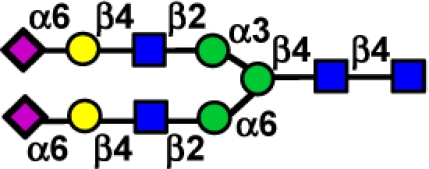 |
|||||||||
| 1,4 | 3,5 | 7 | 17 | 10–12,15,16, 18,19 | 20–25 | 34,35 | 40–44 | 45 | |
| NL/219 | +++ | +++ | +++ | +++ | +++ | +++ | − | − | − |
| NL/230 | +++ | +++ | +++ | +++ | +++ | +++ | +/− | +/− | +/− |
| Rhea/NC | +++ | +++ | +++ | +++ | +++ | +++ | − | − | − |
| Can/504 | +++ | +++ | +++ | +++ | +++ | +++ | ++ | + | − |
| Can/444 | +++ | +++ | +++ | +++ | +++ | +++ | +++ | + | − |
| Tky/VA | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| Ck/Conn | ++ | + | ++ | ++ | + | ++ | +++ | ++ | +++ |
| NY/107 | +++ | + | +++ | +++ | + | + | +++ | ++ | +++ |
Structures shown in symbol form (see key below) are structures of single glycans or composite structures for chemically related glycans represented by the numbers underneath, which correspond to the numbers in the complete structure list (Table S1) and the glycan microarray data in Fig. 2. Error bars reflect the standard deviation in the signal for six independent replicates on the array. The relative binding of the virus to each glycan subclass is qualitatively estimated based on relative strength of the signal for the data shown in Figs. 1 and 2. Strong (+++), moderate (++), weak (+) detectable (+/−), absent (−).  , NeuAc;
, NeuAc;  , Gal;
, Gal;  , GalNAc;
, GalNAc;  , Glc;
, Glc; , GlcNAc;
, GlcNAc; , Man;
, Man; , Fuc
, Fuc
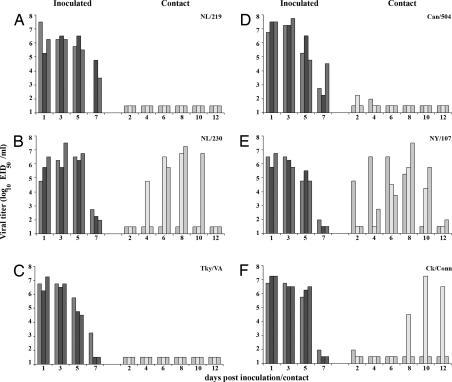
Fig. 2.
Direct contact transmissibility of H7 influenza viruses. Three ferrets were inoculated with 107 EID50 of NL/219 (A), NL/230 (B), Tky/VA (C), Can/504 (D), NY/107 (E), or Ck/Conn (F) virus, and nasal washes were collected from each ferret on the indicated days p.i. (dark bars). A naïve ferret was placed in the same cage as each inoculated ferret 24 h p.i., and nasal washes were collected from each contact ferret on indicated days p.c. (light bars). The limit of virus detection was 101.5 EID50/ml.
Transmissibility of Eurasian H7 Influenza Viruses in Ferrets.
To assess the impact of enhanced α2–6 SA specificity on the transmissibility of the North American H7 viruses, both respiratory droplet and contact transmission experiments were performed as described (21) by using the ferret transmission model. Six ferrets were inoculated intranasally with 107 50% egg infectious doses (EID50), a dose reported to consistently infect ferrets with human or avian influenza viruses (34). Twenty-four hours postinoculation (p.i.), three of the inoculated ferrets were placed in modified cages with a perforated side wall adjacent to a naïve ferret, allowing air exchange between ferrets while preventing direct contact of animals or indirect contact with food or bedding (respiratory droplet transmission). The remaining three inoculated ferrets were each cohoused with a naïve ferret to assess direct contact transmission. Criteria for efficient transmission included detection of virus in nasal washes (NW) of contact ferrets and seroconversion of convalescent sera from contact ferrets, both of which occur during the efficient transmission of human influenza H3N2 viruses as shown in this model system (21).
NL/219 virus has been shown to be highly virulent in the ferret model (13). In the current study, both NL/219 and NL/230 viruses replicated efficiently in the upper respiratory tract of inoculated ferrets, with peak mean nasal wash virus titers of 6.3 ± 0.2 and 6.5 ± 0.9 log10 EID50/ml detected on day 3 p.i., respectively (Fig. 2 A and B). Two of three animals inoculated with NL/219 virus in each experiment were humanely killed 5–7 days p.i. because of severe weight loss or development of hind-limb paralysis. NL/219 virus did not transmit by either direct contact or respiratory droplets, because virus was not isolated from nasal washes of contact ferrets, and seroconversion of contact animals for hemagglutination inhibition (HI) antibody did not occur (Fig. 2A, Table 3, data not shown). Respiratory droplet transmission of NL/230 virus was not observed (data not shown); however, in the direct contact experiment, NL/230 virus was detected in the nasal washes of two of three contact ferrets, with peak NW virus titers >106.5 EID50/ml by day 8 postcontact (p.c.) in these animals (Fig. 2B). Both NL/230 contact ferrets that had virus isolated from NW seroconverted by the end of the experiment (Table 3). The third NL/230 contact ferret did not have detectable virus in NW and did not seroconvert (Fig. 2B, Table 3). This pattern of NL/230 virus transmission by direct contact was confirmed in a duplicate experiment that resulted in seroconversion of only two of three ferrets. Taken together, these results indicate that, despite similar receptor-binding properties as measured by glycan array and resialyation assay, NL/230 virus exhibited an enhanced ability to transmit in the ferret model by direct contact compared with NL/219 virus.
Table 3.
Clinical signs, virus replication, seroconversion, and direct contact transmission in ferrets inoculated with H7 influenza viruses
| Virus | Subtype | No. of inoculated ferrets/total number |
No. of contact ferrets/total number |
||||
|---|---|---|---|---|---|---|---|
| Clinical signs |
Peak mean log10 nasal wash titer, day p.i. | Seroconversion (HI titer range)† | |||||
| Weight loss, %* | Respiratory symptoms, day p.i. | Virus detected in nasal wash | Seroconversion (HI titer range)† | ||||
| NL/219 | H7N7 | 3/3 (18.3) | 3/3 (3) | 6.3 (1,3) | 1/1 (320)‡ | 0/3 | 0/3 |
| NL/230 | H7N7 | 3/3 (6.7) | 1/3 (5) | 6.5 (3,5) | 3/3 (320–640) | 2/3 | 2/3 (320, 640) |
| Can/504 | H7N3 | 3/3 (17.2) | 1/3 (5) | 7.4 (3) | 3/3 (320–640) | 2/3 | 0/3 |
| Tky/VA | H7N2 | 3/3 (6.6) | 2/3 (3) | 6.75 (1) | 3/3 (320–640) | 0/3 | 0/3 |
| NY/107 | H7N2 | 3/3 (6.3) | 1/3 (5) | 6.3 (1) | 3/3 (640–1,280) | 3/3 | 3/3 (1,280–2,560) |
| Ck/Conn | H7N2 | 2/3 (3.8) | 1/3 (7) | 7.1 (1) | 3/3 (320–640) | 1/3 | 1/3 (160) |
*The percentage mean maximum weight loss is shown.
†HI assays were performed with homologous virus and horse RBCs.
‡Only one ferret survived and was tested.
Transmissibility of North American H7 Influenza Viruses in Ferrets.
Next, we assessed the ability of the H7 viruses of the North American lineage to spread to naïve ferrets by either respiratory droplet or contact transmission. Tky/VA virus replicated efficiently in the upper respiratory tract of inoculated ferrets, with peak mean virus titers reaching 6.75 ± 0.5 log10 EID50/ml on day 1 p.i. (Fig. 2C). However, Tky/VA virus did not transmit by direct contact, because virus was not isolated from nasal washes of contact ferrets, and seroconversion of contact ferrets was not detected (Fig. 2C, Table 3). Can/504, a HPAI H7N3 virus, was also found to replicate efficiently in the upper respiratory tract of inoculated ferrets, with peak mean nasal wash virus titers reaching 7.4 ± 0.3 log10 EID50/ml on day 3 p.i. (Fig. 2D). Additionally, substantial weight loss was observed in ferrets inoculated with Can/504 virus (Table 3). Low levels of virus were detected in the nasal washes of two ferrets in direct contact with inoculated animals (1.98–2.25 log10 EID50/ml); however, seroconversion of these contact ferrets did not occur (Table 3). These low virus titers are most likely due to the presence of residual virus on the noses of contact ferrets that was acquired from the environment or from the inoculated ferrets and therefore does not constitute efficient virus transmission, because sustained high titers of virus in the upper respiratory tract were not detected, and seroconversion did not occur. Respiratory droplet transmission of Tky/VA or Can/504 virus was not detected (data not shown).
As discussed above, two H7N2 viruses, NY/107 and Ck/Conn, exhibited enhanced α2–6 SA binding with decreased binding to α2–3 SA in the glycan microarray, with NY/107 showing the most significant decrease. Similar to all other H7 viruses tested, NY/107 and Ck/Conn viruses were detected at high titers in nasal washes of inoculated ferrets, with peak mean virus titers of 6.3 ± 0.5 log10 EID50/ml and 7.1 ± 0.3 log10 EID50/ml, respectively, on day 1 p.i. (Fig. 2 E and F). In contrast with Tky/VA or Can/504 viruses, NY/107 virus transmitted efficiently to three of three ferrets by direct contact, with peak virus titers in nasal washes from each contact ferret reaching ≥105.25 EID50/ml and seroconversion occurring in all contact animals (Fig. 2E, Table 3). Transmission by direct contact occurred by day 2 p.c. in one ferret and by day 6 p.c. in the remaining two contact animals (Fig. 2E). In comparison, Ck/Conn virus transmitted by direct contact in one of three contact ferrets, with peak NW virus titer reaching 107.25 EID50/ml on day 10 p.c. (Fig. 2F). Seroconversion of the remaining two contact ferrets did not occur, indicating that Ck/Conn virus, unlike NY/107 virus, did not transmit efficiently by direct contact (Table 3). Similar to other H7 viruses in this study, respiratory droplet transmission was not observed with either virus (data not shown). These findings demonstrate the ability of an H7 influenza virus isolated from a human, NY/107, to transmit efficiently by direct contact in the ferret model.
Discussion
Like other avian influenza viruses, those within the H7 subtype fall into two geographically distinct lineages, Eurasian and North American (35, 36). H7 viruses within these lineages have caused outbreaks and human infection in recent years and continue to pose a public health threat. To better assess the pandemic potential of H7 influenza viruses, we examined the receptor-binding preference and transmissibility of selected H7 viruses associated with disease in humans. We found that Eurasian lineage HPAI H7 influenza viruses tested in this study closely resemble recent HPAI H5N1 viruses with respect to their binding preference for α2–3 SA receptors. Conversely, we observed an increase in α2–6 binding among North American lineage H7 viruses isolated between 2002 and 2004. Several of these also showed reduced binding of α2–3 SA receptors characteristic of human influenza viruses. The most dramatic shift in receptor specificity was observed for a human H7 influenza virus that was also transmitted efficiently between animals by direct contact.
Previous studies have suggested that Eurasian lineage H7 influenza viruses share receptor-binding properties similar to H5 viruses, because analysis of the HA crystal structure derived from the H7 virus A/Tky/Italy/02 demonstrated specific binding to avian receptor and not human receptor analogues (37). Recent advances in glycan microarray technology allowed us to more closely analyze the fine differences in receptor specificity of viruses between both Eurasian and North American H7 viruses. Here, we used a whole-virus assay that allowed for examination of the binding properties of influenza viruses without the need for generation of recombinant HA. We were particularly interested in the North American H7 viruses, because some of these avian viruses appear to be adapted to the upper respiratory tract of chickens, which have been shown to express more α2–6 SA receptors compared with wild aquatic birds (38); in vivo studies have demonstrated that North American lineage LPAI H7N2 viruses replicate to high titer in the upper respiratory tract of chickens and turkeys compared with the gastrointestional tract (39–41).
All H7 viruses tested replicated to high titer in the upper respiratory tract in inoculated ferrets as shown (13); nevertheless, most isolates tested failed to transmit despite the high titers of virus shed by inoculated animals. Respiratory symptoms such as sneezing and nasal discharge were observed in some ferrets inoculated with each H7 virus (Table 3). However, the frequency and duration of these symptoms in this model more closely resembled those observed in ferrets inoculated with H5N1 viruses, rather than the more pronounced respiratory symptoms observed in ferrets infected with human H3N2 or H1N1 viruses (21, 23). With the exception of NL/219-inoculated ferrets, which exhibit substantial lethargy after infection (13), ferrets inoculated with H7 viruses in this study remained alert and playful for the duration of the experiment, suggesting that frequent interaction between inoculated and contact ferrets is not sufficient for virus transmission to occur. Additionally, the results of this study indicate that increased virus binding to α2–6 SA is not sufficient for transmission of avian influenza viruses to occur, supporting previous studies demonstrating the lack of transmission of an H5N1 virus with an increased α2–6-binding preference (21, 22). Recent studies have highlighted increased complexity of the structural topology among α2–3 and α2–6 SA and suggest that conformational features of the linkage contribute to virus binding and could play a role in virus transmissibility (42). This diversity of SA receptors could in part contribute to the enhanced ability of NL/230 virus to transmit in the ferret model by direct contact compared with NL/219 virus. Although the Eurasian lineage H7N7 viruses analyzed in this study displayed similar receptor-binding properties as measured by glycan array and hemagglutination assay, Munster et al. (43) found differential attachment of NL/219 virus and a virus closely related to NL/230 to tissues in the lower respiratory tract of humans. The enhanced transmissibility observed with NL/230 virus in this study compared with NL/219 virus would additionally suggest subtle differences in the receptor-binding properties between these H7N7 viruses that have yet to be identified.
Unlike most subtypes of influenza, infection with H7 influenza viruses frequently results in conjunctivitis in humans and not respiratory disease (6, 7, 9). Unlike the upper respiratory tract in humans, which contains a high distribution of α2–6 SA, corneal, conjunctivial, and lacrimal duct epithelial cells of the human eye express predominantly α2–3 SA (27, 44, 45). Additionally, the sialylated secretions (mucins) of both surfaces differ in their SA content; mucins in the airway epithelium contain α2–3 SA, whereas ocular secreted mucins contain α2–6 SA (46–48). The high α2–3 SA content of the human ocular surface suggests that avian influenza viruses would be well suited to use this surface as a portal of entry. However, although human infection with H7 influenza viruses frequently results in conjunctivitis, documented cases of ocular disease after H5N1 infection are rare (7, 49, 50). The heterogeneity of SA-binding preference observed between H7 influenza virus lineages suggests that, similar to virus transmissibility, ocular tropism is a complex property that cannot be explained by SA receptor binding alone.
We identified a LPAI H7N2 virus, NY/107, which was associated with human respiratory infection and not ocular disease and was effectively transmitted in the ferret model by direct contact (10). Among all H7 viruses analyzed by glycan microarray, NY/107 displayed the most dramatic increase in α2–6 SA binding along with decreased α2–3 SA binding avidity. Strong α2–6 SA binding appears to be an essential component of conferring transmissibility in human influenza viruses, because H1N1 variant viruses exhibiting the classic avian α2–3-binding preference or dual α2–3- and α2–6-binding preference were unable to transmit efficiently (23). These results suggest that a decrease in α2–3 SA binding may also be needed in addition to α2–6 SA-binding avidity. However, despite similar α2–3 and α2–6 SA binding observed by glycan array with Ck/Conn virus, this virus was transmitted only by direct contact in one of three animals. Efficient contact transmission was also not observed with Tky/VA virus, despite this virus sharing 98.4% HA amino acid identity with NY/107 virus (40). Future studies will allow for a better understanding of the genetic determinants responsible for the heightened transmissibility observed with this virus. NY/107 virus, like all H7 viruses tested in this study, did not transmit by respiratory droplets in the ferret model. However, the efficient NY/107 virus transmission observed by direct contact in ferrets has not been observed with HPAI H5N1 viruses (21, 22) and may indicate the capacity of a NY/107-like virus to acquire properties that would confer efficient transmission by respiratory droplets; this underscores the importance of studying virus transmissibility by both routes.
LPAI H7N2 viruses have been acquiring additional basic amino acids at the HA cleavage site since 1994, resulting in a cleavage site that more closely resembles HPAI viruses (51). These viruses are also characterized by a deletion of 8 aa in the HA1 proximal to the receptor-binding site (31); further study will help elucidate whether this deletion contributes to the enhanced α2–6 SA binding observed among these viruses. The classic avian specificity for α2–3-linked SA observed with Rhea/NC could suggest a possible correlation between the acquisition of α2–6 SA binding and the introduction of LPAI H7N2 viruses into the live bird markets of the northeastern U.S. The finding of enhanced α2–6 SA binding of North American H7 viruses underscores the necessity for continued surveillance and study of these viruses as they continue to resemble viruses with pandemic potential.
Materials and Methods
Viruses.
Virus stocks were grown in the allantoic cavity of 10-day-old embroynated hens' eggs as described (13). The 50% EID50 titer for each virus stock was calculated by the method of Reed and Muench (52), after serial titration in eggs. A/Texas/36/91 (Tx/91) stock was grown on Madin–Darby canine kidney cells containing DMEM, 0.025 M Hepes, 0.3% BSA (Gibco Invitrogen), and N-p-tosyl-l-phenylalanine chloromethyl ketone (TPCK)-treated trypsin (Sigma–Aldrich). All experiments with HPAI viruses were conducted under biosafety level 3 containment, including enhancements required by the U.S. Department of Agriculture and the Select Agent Program (53).
Glycan Microarray Analysis.
Analysis of the receptor specificity of influenza virus using glycan microarrays was done largely as described (33, 54). Custom arrays for influenza research were produced for the Centers for Disease Control and Prevention on National Health Service-activated glass slides (Schott Nexterion) by using a glycan library provided by the Consortium for Functional Glycomics [www.functionalglycomics.org; see supporting information (SI) Table S1 for a list of glycan structures]. Viruses were inactivated by treatment with β-propiolactone (0.001%) overnight at 4°C with virus inactivation confirmed by two rounds of passage in eggs. Virus preparations were diluted to 1 ml into PBS buffer containing 3% (wt/vol) BSA (PBS-BSA) to HA titers of 256–512. Virus suspensions were applied to slides and the slides were incubated in a closed container and subjected to gentle agitation for 1 h. Unbound virus was washed off by dipping slides sequentially in PBS with 0.05% Tween-20 (PBS-T) and PBS. While still wet, slides were overlaid with corresponding primary antibodies diluted in PBS-BSA, either goat antiserum A/FPV/Rostock/34 (H7N1) (1:500) (for NL/219, NL/230, and Ck/Conn viruses), ferret anti-A/Canada/444/04 (H7N3) (1:500) (for Can/504 and Can/444 viruses), ferret anti-A/Turkey/VA/4529/02 (H7N2) (1:500) (for Tky/VA virus), ferret anti-A/NY/107/03 (H7N2) (1:500) (for NY/107 virus), chicken anti-A/Rhea/NC/39482/93 (H7N1) (1:500) (for Rhea/NC virus), or sheep anti-A/Vietnam/1203/04 (H5N1) (1:1,000) (for HK/486 virus) (1 h). Slides were washed briefly with PBS-T/PBS as above followed by application of the appropriate secondary antibody conjugates, either anti-ferret-IgG FITC (1:200), anti-goat-IgG FITC (1:200), goat anti-chicken-IgY-FITC (1:200) (Genway Biotechnology), or anti-sheep-IgG-FITC (1:200) in PBS-BSA were subsequently incubated (1 h) followed by PBS-T/PBS washes and a final wash step in deionized water. After the slides were dried in a steam of nitrogen, they were immediately scanned (ProScanArray HT slide scanner with Autoloader, Perkin–Elmer) followed by image analysis with ImaGene 6.1 software (Biodiscovery).
Ferret Transmission Experiments.
Male Fitch ferrets, 7–10 months of age (Triple F Farms) and serologically negative by HI assay for currently circulating influenza A H1N1, H3N2, and B viruses were used in this study. Ferrets were housed for the duration of each experiment in a Duo-Flo Bioclean mobile clean room (Lab Products). Ferrets were inoculated with 107 EID50 of each virus, and nasal washes were collected on indicated days p.i. as described (2). Respiratory droplet and contact transmission experiments were conducted as described (21), with a total of six ferrets used for each experiment.
Hemagglutination Assays.
Convalescent sera were collected from all ferrets on days 18–21 p.i./p.c. and tested for H7 specific antibodies by HI by using homologous virus and 1% horse RBCs as described (55). Hemagglutination assays using resialyated turkey RBC were performed as described (56, 57) with minor modifications. Turkey RBC were enzymatically desialyated, followed by resialylation using either α2–6-(N)-sialyltransferase (Japan Tobacco) or α2–3-(N)-sialyltransferase (Calbiochem). Assays were performed by using both 4 and 8 hemagglutination units of virus yielding identical results.
Supplementary Material
Acknowledgments.
We thank David Swayne (Southeast Poultry Research Laboratory, U.S. Department of Agriculture, Agricultural Research Service, Athens, GA), Ron Fouchier (Erasmus Medical Center, Rotterdam, The Netherlands), and Yan Li (Canadian Center for Human and Animal Health) for providing some of the viruses used in this study. We also thank Julia Hoffman for early help with the glycan microarray and Anna Crie for assistance with generating the figures. The glycan microarray was produced for the Centers for Disease Control by using a glycan library generously provided by the Consortium for Functional Glycomics funded by National Institute of General Medical Sciences Grant GM62116. J.A.B. received financial support for this work from the Oak Ridge Institute for Science and Education, Oak Ridge, TN. Glycan microarray data presented here will be made available on-line through the Consortium for Functional Glycomics web site upon publication (www.functionalglycomics.org).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0801259105/DCSupplemental.
References
- 1.World Health Organization. Cumulative Number of Confirmed Human Cases of Avian Influenza A/H5N1) Reported to WHO. Geneva: World Health Organization; 2007. [Google Scholar]
- 2.Maines TR, et al. Avian influenza (H5N1) viruses isolated from humans in Asia in 2004 exhibit increased virulence in mammals. J Virol. 2005;79:11788–11800. doi: 10.1128/JVI.79.18.11788-11800.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ungchusak K, et al. Probable person-to-person transmission of avian influenza A (H5N1) N Engl J Med. 2005;352:333–340. doi: 10.1056/NEJMoa044021. [DOI] [PubMed] [Google Scholar]
- 4.Olsen SJ, et al. Family clustering of avian influenza A (H5N1) Emerg Infect Dis. 2005;11:1799–1801. doi: 10.3201/eid1111.050646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kandun IN, et al. Three Indonesian clusters of H5N1 virus infection in 2005. N Engl J Med. 2006;355:2186–2194. doi: 10.1056/NEJMoa060930. [DOI] [PubMed] [Google Scholar]
- 6.Fouchier RA, et al. Avian influenza A virus (H7N7) associated with human conjunctivitis and a fatal case of acute respiratory distress syndrome. Proc Natl Acad Sci USA. 2004;101:1356–1361. doi: 10.1073/pnas.0308352100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koopmans M, et al. Transmission of H7N7 avian influenza A virus to human beings during a large outbreak in commercial poultry farms in the Netherlands. Lancet. 2004;363:587–593. doi: 10.1016/S0140-6736(04)15589-X. [DOI] [PubMed] [Google Scholar]
- 8.Hirst M, et al. Novel avian influenza H7N3 strain outbreak, British Columbia. Emerg Infect Dis. 2004;10:2192–2195. doi: 10.3201/eid1012.040743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tweed SA, et al. Human illness from avian influenza H7N3, British Columbia. Emerg Infect Dis. 2004;10:2196–2199. doi: 10.3201/eid1012.040961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Centers for Disease Control. Update: influenza activity–United States and worldwide, 2003–04 season, and composition of the 2004–05 influenza vaccine. MMWR Morb Mortal Wkly Rep. 2004;53:547–552. [PubMed] [Google Scholar]
- 11.Centers for Disease Control. Avian influenza A/(H7N2) outbreak in the United Kingdom. Euro Surveill. 2007;12:E070531 2. [PubMed] [Google Scholar]
- 12.de Wit E, et al. Protection of mice against lethal infection with highly pathogenic H7N7 influenza A virus by using a recombinant low-pathogenicity vaccine strain. J Virol. 2005;79:12401–12407. doi: 10.1128/JVI.79.19.12401-12407.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Belser JA, et al. Pathogenesis of Avian Influenza (H7) Virus Infection in Mice and Ferrets: Enhanced Virulence of Eurasian H7N7 Viruses Isolated from Humans. J Virol. 2007;81:11139–11147. doi: 10.1128/JVI.01235-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matrosovich MN, et al. Avian influenza A viruses differ from human viruses by recognition of sialyloligosaccharides and gangliosides and by a higher conservation of the HA receptor-binding site. Virology. 1997;233:224–234. doi: 10.1006/viro.1997.8580. [DOI] [PubMed] [Google Scholar]
- 15.Matrosovich M, et al. Early alterations of the receptor-binding properties of H1, H2, and H3 avian influenza virus hemagglutinins after their introduction into mammals. J Virol. 2000;74:8502–8512. doi: 10.1128/jvi.74.18.8502-8512.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Connor RJ, Kawaoka Y, Webster RG, Paulson JC. Receptor specificity in human, avian, and equine H2 and H3 influenza virus isolates. Virology. 1994;205:17–23. doi: 10.1006/viro.1994.1615. [DOI] [PubMed] [Google Scholar]
- 17.Matrosovich M, Zhou N, Kawaoka Y, Webster R. The surface glycoproteins of H5 influenza viruses isolated from humans, chickens, and wild aquatic birds have distinguishable properties. J Virol. 1999;73:1146–1155. doi: 10.1128/jvi.73.2.1146-1155.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shinya K, et al. Characterization of a human H5N1 influenza A virus isolated in 2003. J Virol. 2005;79:9926–9932. doi: 10.1128/JVI.79.15.9926-9932.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gambaryan A, et al. Evolution of the receptor binding phenotype of influenza A (H5) viruses. Virology. 2006;344:432–438. doi: 10.1016/j.virol.2005.08.035. [DOI] [PubMed] [Google Scholar]
- 20.Yamada S, et al. Haemagglutinin mutations responsible for the binding of H5N1 influenza A viruses to human-type receptors. Nature. 2006;444:378–382. doi: 10.1038/nature05264. [DOI] [PubMed] [Google Scholar]
- 21.Maines TR, et al. Lack of transmission of H5N1 avian-human reassortant influenza viruses in a ferret model. Proc Natl Acad Sci USA. 2006;103:12121–12126. doi: 10.1073/pnas.0605134103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yen HL, et al. Inefficient transmission of H5N1 influenza viruses in a ferret contact model. J Virol. 2007;81:6890–6898. doi: 10.1128/JVI.00170-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tumpey TM, et al. A two-amino acid change in the hemagglutinin of the 1918 influenza virus abolishes transmission. Science. 2007;315:655–659. doi: 10.1126/science.1136212. [DOI] [PubMed] [Google Scholar]
- 24.Baum LG, Paulson JC. Sialyloligosaccharides of the respiratory epithelium in the selection of human influenza virus receptor specificity. Acta Histochem Suppl. 1990;40:35–38. [PubMed] [Google Scholar]
- 25.Leigh MW, Connor RJ, Kelm S, Baum LG, Paulson JC. Receptor specificity of influenza virus influences severity of illness in ferrets. Vaccine. 1995;13:1468–1473. doi: 10.1016/0264-410x(95)00004-k. [DOI] [PubMed] [Google Scholar]
- 26.van Riel D, et al. H5N1 Virus Attachment to Lower Respiratory Tract. Science. 2006;312:399. doi: 10.1126/science.1125548. [DOI] [PubMed] [Google Scholar]
- 27.Shinya K, et al. Avian flu: influenza virus receptors in the human airway. Nature. 2006;440:435–436. doi: 10.1038/440435a. [DOI] [PubMed] [Google Scholar]
- 28.Rogers GN, D'Souza BL. Receptor binding properties of human and animal H1 influenza virus isolates. Virology. 1989;173:317–322. doi: 10.1016/0042-6822(89)90249-3. [DOI] [PubMed] [Google Scholar]
- 29.Stevens J, et al. Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus. Science. 2006;312:404–410. doi: 10.1126/science.1124513. [DOI] [PubMed] [Google Scholar]
- 30.Matrosovich MN, Krauss S, Webster RG. H9N2 influenza A viruses from poultry in Asia have human virus-like receptor specificity. Virology. 2001;281:156–162. doi: 10.1006/viro.2000.0799. [DOI] [PubMed] [Google Scholar]
- 31.Suarez DL, Garcia M, Latimer J, Senne D, Perdue M. Phylogenetic analysis of H7 avian influenza viruses isolated from the live bird markets of the Northeast United States. J Virol. 1999;73:3567–3573. doi: 10.1128/jvi.73.5.3567-3573.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Centers for Disease Control. Update: influenza activity - United States, 2003–04 season. MMWR Morb Mortal Wkly Rep. 2004;53:284–287. [PubMed] [Google Scholar]
- 33.Stevens J, et al. Glycan microarray analysis of the hemagglutinins from modern and pandemic influenza viruses reveals different receptor specificities. J Mol Biol. 2006;355:1143–1155. doi: 10.1016/j.jmb.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 34.Hinshaw VS, Webster RG, Easterday BC, Bean WJ., Jr Replication of avian influenza A viruses in mammals. Infect Immun. 1981;34:354–361. doi: 10.1128/iai.34.2.354-361.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rohm C, Horimoto T, Kawaoka Y, Suss J, Webster RG. Do hemagglutinin genes of highly pathogenic avian influenza viruses constitute unique phylogenetic lineages? Virology. 1995;209:664–670. doi: 10.1006/viro.1995.1301. [DOI] [PubMed] [Google Scholar]
- 36.Banks J, Speidel EC, McCauley JW, Alexander DJ. Phylogenetic analysis of H7 haemagglutinin subtype influenza A viruses. Arch Virol. 2000;145:1047–1058. doi: 10.1007/s007050050695. [DOI] [PubMed] [Google Scholar]
- 37.Russell RJ, Stevens DJ, Haire LF, Gamblin SJ, Skehel JJ. Avian and human receptor binding by hemagglutinins of influenza A viruses. Glycoconj J. 2006;23:85–92. doi: 10.1007/s10719-006-5440-1. [DOI] [PubMed] [Google Scholar]
- 38.Gambaryan A, Webster R, Matrosovich M. Differences between influenza virus receptors on target cells of duck and chicken. Arch Virol. 2002;147:1197–1208. doi: 10.1007/s00705-002-0796-4. [DOI] [PubMed] [Google Scholar]
- 39.Tumpey TM, Kapczynski DR, Swayne DE. Comparative susceptibility of chickens and turkeys to avian influenza A H7N2 virus infection and protective efficacy of a commercial avian influenza H7N2 virus vaccine. Avian Dis. 2004;48:167–176. doi: 10.1637/7103. [DOI] [PubMed] [Google Scholar]
- 40.Pappas C, Matsuoka Y, Swayne DE, Donis RO. Development and evaluation of an influenza subtype H7N2 vaccine candidate for pandemic preparedness. Clin Vaccine Immunol. 2007;14:1425–1432. doi: 10.1128/CVI.00174-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Swayne DE, Pantin-Jackwood M. Pathogenicity of avian influenza viruses in poultry. Dev Biol (Basel) 2006;124:61–67. [PubMed] [Google Scholar]
- 42.Chandrasekaran A, et al. Glycan topology determines human adaptation of avian H5N1 virus hemagglutinin. Nat Biotechnol. 2008;26:107–113. doi: 10.1038/nbt1375. [DOI] [PubMed] [Google Scholar]
- 43.Munster VJ, et al. The molecular basis of the pathogenicity of the Dutch highly pathogenic human influenza A H7N7 viruses. J Infect Dis. 2007;196:258–265. doi: 10.1086/518792. [DOI] [PubMed] [Google Scholar]
- 44.Terraciano AJ, et al. Sialyl Lewis X, Lewis X, and N-acetyllactosamine expression on normal and glaucomatous eyes. Curr Eye Res. 1999;18:73–78. doi: 10.1076/ceyr.18.2.73.5377. [DOI] [PubMed] [Google Scholar]
- 45.Paulsen F, et al. Functional anatomy of human lacrimal duct epithelium. Anat Embryol (Berl) 1998;198:1–12. doi: 10.1007/s004290050160. [DOI] [PubMed] [Google Scholar]
- 46.Couceiro JN, Paulson JC, Baum LG. Influenza virus strains selectively recognize sialyloligosaccharides on human respiratory epithelium; the role of the host cell in selection of hemagglutinin receptor specificity. Virus Res. 1993;29:155–165. doi: 10.1016/0168-1702(93)90056-s. [DOI] [PubMed] [Google Scholar]
- 47.Berry M, Ellingham RB, Corfield AP. Polydispersity of normal human conjunctival mucins. Invest Ophthalmol Vis Sci. 1996;37:2559–2571. [PubMed] [Google Scholar]
- 48.Thale A, et al. The efferent lacrimal ducts of the human. Morphological and biochemical studies. Ophthalmologe. 2001;98:67–73. doi: 10.1007/s003470170202. [DOI] [PubMed] [Google Scholar]
- 49.Chan PK. Outbreak of avian influenza A(H5N1) virus infection in Hong Kong in 1997. Clin Infect Dis. 2002;2(34) Suppl:S58–64. doi: 10.1086/338820. [DOI] [PubMed] [Google Scholar]
- 50.Tam JS. Influenza A (H5N1) in Hong Kong: an overview. Vaccine. 2002;2(20) Suppl:S77–81. doi: 10.1016/s0264-410x(02)00137-8. [DOI] [PubMed] [Google Scholar]
- 51.Spackman E, Senne DA, Davison S, Suarez DL. Sequence analysis of recent H7 avian influenza viruses associated with three different outbreaks in commercial poultry in the United States. J Virol. 2003;77:13399–13402. doi: 10.1128/JVI.77.24.13399-13402.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reed LJ, Muench HA. A simple method of estimating fifty per cent endpoints. Am J Hyg. 1938;27:493–497. [Google Scholar]
- 53.Richmond JY, McKinney RW. In: Biosafety in microbiological and biomedical laboratories. 5th ed. Richmond JY, McKinney RW, editors. Atlanta: Centers for Disease Control and Prevention; 2007. [Google Scholar]
- 54.Blixt O, et al. Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proc Natl Acad Sci USA. 2004;101:17033–17038. doi: 10.1073/pnas.0407902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stephenson I, Wood JM, Nicholson KG, Charlett A, Zambon MC. Detection of anti-H5 responses in human sera by HI using horse erythrocytes after MF59-adjuvanted influenza A/Duck/Singapore/97 vaccine. Virus Res. 2004;103:91–95. doi: 10.1016/j.virusres.2004.02.019. [DOI] [PubMed] [Google Scholar]
- 56.Glaser L, et al. A single amino acid substitution in 1918 influenza virus hemagglutinin changes receptor binding specificity. J Virol. 2005;79:11533–11536. doi: 10.1128/JVI.79.17.11533-11536.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Glaser L, et al. Sequence analysis and receptor specificity of the hemagglutinin of a recent influenza H2N2 virus isolated from chicken in North America. Glycoconj J. 2006;23:93–99. doi: 10.1007/s10719-006-5441-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.