Abstract
The sodium-dependent multivitamin transporter (SMVT) is essential for mediating and regulating biotin entry into mammalian cells. In cells, biotin is covalently linked to histones in a reaction catalyzed by holocarboxylase synthetase (HCS); biotinylation of lysine 12-biotinylated histone H4 (K12Bio H4) causes gene silencing. Here, we propose a novel role for HCS in sensing and regulating levels of biotin in eukaryotic cells. We hypothesized that nuclear translocation of HCS increases in response to biotin supplementation; HCS then biotinylates histone H4 at SMVT promoters, silencing biotin transporter genes. Jurkat lymphoma cells were cultured in media containing 0.025, 0.25, or 10 nmol/l biotin. The nuclear translocation of HCS correlated with biotin concentrations in media; the relative enrichment of both HCS and K12Bio H4 at SMVT promoter 1 (but not promoter 2) increased by 91% in cells cultured in medium containing 10 nmol/l biotin compared with 0.25 nmol/l biotin. This increase of K12Bio H4 at the SMVT promoter decreased SMVT expression by up to 86%. Biotin homeostasis by HCS-dependent chromatin remodeling at the SMVT promoter 1 locus was disrupted in HCS knockdown cells, as evidenced by abnormal chromatin structure (K12Bio H4 abundance) and increased SMVT expression. The findings from this study are consistent with the theory that HCS senses biotin, and that biotin regulates its own cellular uptake by participating in HCS-dependent chromatin remodeling events at the SMVT promoter 1 locus in Jurkat cells.
Keywords: Biotin, Chromatin, Holocarboxylase synthetase, Human, Sodium-dependent multivitamin transporter
1. Introduction
Biotin uptake into mammalian cells is a carrier-dependent process [1,2] that is mediated by the sodium-dependent multivitamin transporter (SMVT) [3]. The expression of biotin transporters increases in response to a heightened cellular demand for biotin such as in proliferating cells [4,5] and biotin-deficient cells [6,7]. The activity of biotin transporters is regulated at both the transcriptional and the posttranscriptional level. For example, the transcriptional activity of SMVT reporter genes correlates inversely with biotin supply in human choriocarcinoma cells, and reporter gene activity is a good predictor for abundance of SMVT and transport rates of biotin [7]. The 5′-regulatory region of the human SMVT gene comprises two distinct promoters, each containing regulatory cis elements such as AP1 and Sp1 sites [8]. Some of these elements are known to be biotin-responsive. For example, biotin deficiency is associated with increased DNA-binding activities of transcription factors Fos and Jun, enhancing the activity of reporter gene constructs driven by AP1 [9]. Not withstanding the regulation of biotin transporters at the level of transcription, posttranscriptional events also participate in transporter regulation. For example, SMVT contains two putative protein kinase C phosphorylation sites [10,11]. Activators of protein kinase C inhibit biotin uptake, whereas inhibitors of protein kinase C stimulate biotin uptake in colorectal adenocarcinoma cells [12].
Recently, evidence emerged that covalent modifications of proteins in chromatin are an important mechanism of gene regulation by biotin. Chromatin comprises DNA, histones H1, H2A, H2B, H3, and H4, and non-histone proteins [13]. DNA and histones form repetitive nucleoprotein units, the nucleosomal core particles [13]. The amino terminal tails of histones protrude from the nucleosomal surface; covalent modifications of these tails (e.g., methylation and biotinylation) affect the structure of chromatin and form the basis for gene regulation [14–16]. Site-specific modifications of histones have distinct functions; for example, trimethylation of lysine (K) 4 in histone H3 is associated with transcriptional activation of surrounding DNA [17,18], whereas dimethylation of K9 is associated with transcriptional silencing [19,20].
Biotin is covalently linked to ε-amino groups in distinct lysine residues in histones [21,22]; the binding of biotin to histones is mediated by holocarboxylase synthetase (HCS) and, perhaps, biotinidase [21,23]. The following biotinylation sites have been identified: K9, K13, K125, K127, and K129 in histone H2A [24]; K4, K9, and K18 in histone H3 [25]; and K8 and K12 in histone H4 [26]. Importantly, biotinylation of K12 in histone H4 is associated with heterochromatin structures and gene silencing in human cells [27]. Here, we propose a novel mechanism by which cells maintain biotin homeostasis. The test hypothesis is that HCS senses biotin, and that biotin regulates its own cellular uptake by participating in HCS-dependent chromatin remodeling events at SMVT promoter loci. Specifically, we determined (i) whether nuclear translocation of HCS depends on biotin, (ii) whether the cellular concentration of biotin affects HCS-dependent biotinylation of K12 in histone H4 at SMVT promoter loci, (iii) whether increased biotinylation of histone H4 at SMVT promoter loci is associated with decreased expression of SMVT, and (iv) whether knockdown of HCS disrupts the feedback loop by which cellular biotin regulates uptake of biotin from the extracellular space.
2. Materials and methods
2.1. Cell culture
Jurkat lymphoblastoma cells were obtained from ATCC (Manassas, VA, USA). HCS-deficient Jurkat cells were generated by using the pSilencer 4.1-CMV neo vector according to the manufacturer’s instructions (Ambion, Austin, TX, USA); the following oligonucleotides were used for generating the siRNA-producing HCS knockdown vector (denoted “siRNA-HCS”): sense=5′-GAT CCG GAC TCC ACT CTG AAG GAA TTC AAG AGA TTC CTT CAG AGT GGA GTC CTGA-3′; antisense=5′-AGC TTC AGG ACT CCA CTC TGA AGG AAT CTC TTG AAT TCC TTC AGA GTG GAG TCC G-3′. As controls we used commercial pSilencer 4.1-CMV neo vectors containing glyceraldehyde-3-phosphate dehydrogenase (GAPDH) sequences (denoted “pSilencer 4.1-CMV neo GAPDH”) and a hairpin (denoted “pSilencer 4.1-CMV neo negative control”) with limited homology to sequences in the human genome (Ambion). Jurkat cells were transfected with pSilencer vectors, and stably transformed cells were selected using 0.3 g/l G418 for 10 days. Expression of HCS and GAPDH was quantified by real-time polymerase chain reaction (PCR) and Western blot analysis as described below.
Cells were cultured in the following biotin-defined media for at least 5 weeks before sample collection [6]: 0.025 nmol/l of biotin (denoted “deficient”), 0.25 nmol/l of biotin (“physiological”), and 10 nmol/l of biotin (“pharmacological”). These concentrations represent plasma from biotin-deficient individuals, normal subjects, and users of biotin supplements [6]. Biotin concentrations in media were confirmed by avidin-binding assay [28] with modifications [6].
2.2. Biotin-dependent carboxylases
HCS mediates the covalent attachment of biotin to acetyl-CoA carboxylases α and β, pyruvate carboxylase, propionyl-CoA carboxylase (PCC), and 3-methylcrotonyl-CoA carboxylase in addition to mediating biotinylation of histones [29]. Holocarboxylases are reliable markers for both biotin concentration and HCS activity [29]. Holocarboxylases in cell extracts were probed using gel electrophoresis and streptavidin peroxidase [6]. PCC and 3-methylcrotonyl-CoA carboxylase contain biotinylated and nonbiotinylated subunits; only the biotin-containing α subunits are detectable in streptavidin blots.
2.3. Real-time PCR
The abundance of mRNA coding for HCS and GAPDH in knockdown cells and wild-type controls was quantified by real-time PCR, using primers “HCS Trans” and “GAPDH Trans” (Table 1). The abundance of mRNA coding for SMVT was quantified by using the “SMVT Trans” primers. ABsolute QPCR SYBR Green fluorescein mix (ABgene, Rochester, NY, USA) was used for real-time PCR reactions.
Table 1.
PCR primers used for quantification of DNA from ChIP experiments and transcripts (denoted “Trans”)
| Target | Experiment | Size | Forward primers
|
Reverse primers
|
||
|---|---|---|---|---|---|---|
| Location a | Sequence | Location | Sequence | |||
| Chr1 alpha b | ChIP | 136 bp | N/A | 5′-TCATTCCCACAAACTGCGTTG-3′ | N/A | 5′-TCCAACGAAGGCCACAAGA-3′ |
| GAPDH c | ChIP | 294 bp | +1399 | 5′-CCCAACTTTCCCGCCTCTC-3′ | +1692 | 5′-CAGCCGCCTGGTTCAACTG-3′ |
| GAPDH | Trans | 78 bp | +292 | 5′-TCCACTGGCGTCTTCACC-3′ | +369 | 5′-GGCAGAGATGATGACCCTTT-3′ |
| HCS d | Trans | 102 bp | +1 | 5′-ATGGAAGATAGACTCCACAT-3′ | +102 | 5′-TGAGACCTGATCCTTAACTTCC-3′ |
| SMVT P1 e | ChIP | 226 bp | −5556 | 5′-AATGGCCTGTATCCAGCATC-3′ | −5331 | 5′-TCGCCGTTTGTTCTTTTCTT-3′ |
| SMVT P2 f | ChIP | 129 bp | −4249 | 5′-CATCCGCCCTCAGTAAACAT-3′ | −4121 | 5′-ACACGTGGGTAGCCAGAGC-3′ |
| SMVT g | Trans | 150 bp | +420 | 5′-TGTGCGAGTGTGTGGAAC-3′ | +569 | 5′-TAGACGGTACAGACAATGCC-3′ |
2.4. Western blot analyses
Whole-cell extracts and nuclear extracts were prepared as described [9,30]. Proteins were resolved using 4–12% Bis-tris gels and 18% Tris-glycine gels (Invitrogen, Carlsbad, CA, USA), as described [26,30]. SMVT was probed using polyclonal rabbit antihuman SMVT serum [31]. HCS was probed using antiserum to a peptide spanning amino acids 58–77 in the N-terminus of human HCS (denoted “anti-HCS/N”) [24]. Alternatively, HCS was probed using a novel antiserum to a peptide spanning amino acids 707–726 in the C-terminus of human HCS (denoted “anti-HCS/C”): VHPDGNSFDMLRNLILPKRR. Anti-HCS/C was generated in a commercial facility as described [24]. In control experiments, transblots were probed by using goat antihuman histone H3 (Santa Cruz Biotechnology, Santa Cruz, CA, USA), goat antihuman GAPDH (Santa Cruz Biotechnology), or secondary antibodies in the absence of primary antibodies. The antibody to histone H3 targets an epitope that does not contain any known biotinylation sites; hence, altered biotinylation of histone H3 in HCS- or biotin-deficient cells is an unlikely confounder in control experiments using antihistone H3.
2.5. Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) assays were conducted as described [32], using the following antibodies: (i) rabbit antihuman K12-biotinylated histone H4 (K12Bio H4) serum [26], (ii) rabbit antihuman HCS/C (see above), (iii) affinity-purified rabbit antihuman K9-dimethylated histone H3 (K9Me2 H3) (Abcam; Cambridge, MA, USA), and (iv) affinity-purified rabbit anti-human K4-trimethylated histone H3 (K4Me3 H3) (Abcam). Precipitation of chromatin with bulk rabbit IgG (background control) yielded negligible amounts of DNA and was not considered for further analyses. Total DNA from Jurkat cells served as input control (denoted “input DNA”).
2.6. Reverse-transcriptase PCR
The relative abundance of the following genomic sequences in immunoprecipitated DNA and input DNA was quantified by PCR, using the primers listed in Table 1: SMVT promoters 1 (locus-5705 to locus-5172 relative to the transcriptional start site) and 2 (locus-4276 to locus-4103) [8], alpha satellite repeats in chromosome 1 (heterochromatin), and GAPDH (euchromatin). PCR analyses were conducted as described [33] with the following modifications: genomic DNA was amplified for 10 PCR cycles using the above loci-specific primers; 20 ng of DNA from each of these reactions was used as a template in a second round of PCR amplifications using the same primer pairs. Platinum Taq DNA Polymerase (Invitrogen) was used for all PCR reactions. PCR products from within the exponential phase of amplification were quantified by gel densitometry [33]. The relative enrichment of select genomic sequences (amplicons) is expressed as ratio of antibody-precipitated DNA over an equal amount of input DNA in the starting material.
2.7. Biotin transport
Biotin transport rates were quantified by using a physiological concentration of [3H]biotin (0.475 nmol/l), as described previously [34].
2.8. Statistics
Homogeneous variances were detected by using Bartlett’s test, and data were log-transformed if applicable [35]. Effects of the “HCS expression×biotin concentration” interaction on dependent variables were analyzed by analysis of covariance. Effects of the biotin concentration in culture media on dependent variables were analyzed by one-way analysis of variance; Fisher’s protected least significant difference procedure was used for post hoc testing [35]. Effects of HCS expression on dependent variables were analyzed by paired t test in samples matched for biotin concentration. StatView 5.0.1 (SAS Institute, Cary, NC, USA) was used to perform all calculations. Differences were considered significant at P<.05.
3. Results
Stable transformation of Jurkat cells with the siRNA-HCS vector decreased HCS expression. This observation is important for evaluating the studies of SMVT regulation described below. The abundance of HCS mRNA in knockdown cells decreased to <30% of the abundance in wild-type controls in biotin-matched samples (Fig. 1A). The abundance of HCS mRNA increased significantly in response to supplementation with pharmacological doses of biotin in wild-type cells but not in knockdown cells. Transformation of cells with the control vectors pSilencer 4.1-CMV neo GAPDH and pSilencer 4.1-CMV neo negative control did not affect the abundance of HCS mRNA (data not shown). The abundance of mRNA coding for GAPDH did not decrease in response to transformation with siRNA-HCS (data not shown), suggesting specificity. In contrast, if biotin-normal cells were transformed with the pSilencer 4.1-CMV neo GAPDH vector, the abundance of GAPDH mRNA decreased to 50±2.2% of wild-type controls (P<.05; n=3). The abundance of mRNA coding for biotinidase, a suspected histone biotinyl transferase [21], did not decrease in response to transformation with the siRNA-HCS vector (data not shown).
Fig. 1.
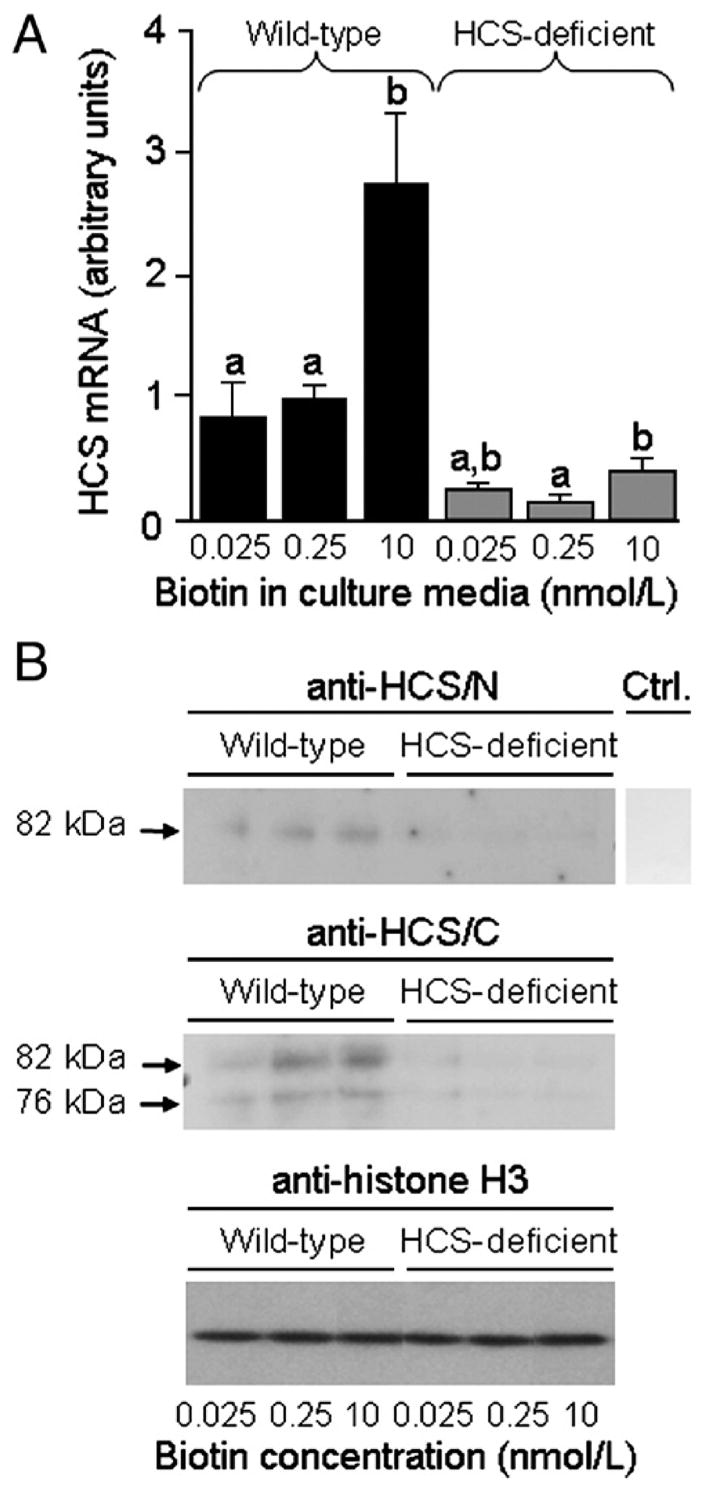
Expression of HCS decreases in response to both biotin deficiency and HCS siRNA in Jurkat cells. Wild-type cells and siRNA-treated cells were cultured in biotin-defined media for 5 weeks. (A) Abundance of HCS mRNA was quantified by real-time PCR. Values are means±S.D. a,bBars not sharing the same superscript are significantly different in cells matched for HCS expression. *P<.05, significantly different from knockdown cells matched for biotin in medium (n=3). (B) HCS protein in whole cell extracts was detected by Western blot analysis: (Top) N-terminus probed with anti-HCS/N. (Middle) C-terminus probed with anti-HCS/C. (Bottom) Equal loading of lanes was confirmed by probing blots with anti-histone H3. Ctrl. indicates control (blot probed with secondary antibody in the absence of anti-HCS/N and anti-HCS/C). Anti-HCS/N and Ctrl. samples were run on the same gel, but the Ctrl lane was processed separately after electroblotting. Samples probed with anti-histone H3 were run on the same gel, but the sequence of lanes was electronically rearranged after autoradiography.
The abundance of HCS protein showed a pattern similar to that observed for HCS mRNA. Full-length HCS (82 kDa) was detectable in bulk extracts from wild-type cells probed with anti-HCS/N, but not in extracts from HCS knockdown cells (Fig. 1B, upper panel). Secondary antibody alone did not produce a detectable signal (negative control). The abundance of 82-kDa HCS in wild-type cells depended on the concentration of biotin in culture media: 0.025<0.25<10 nmol/l biotin. If extracts from wild-type cells were probed with anti-HCS/C, we detected a 76-kDa variant of HCS in addition to 82-kDa, full-length HCS (Fig. 1B, middle panel), consistent with previous observations [36]. Trace amounts of the 76-kDa variant were also detected in HCS knockdown cells. The abundance of both the 76-kDa variant and 82-kDa full-length HCS depended on the concentration of biotin in culture media in wild-type cells: 0.025<0.25<10 nmol/l biotin. The abundance of histone H3 (control) did not depend on HCS or biotin (Fig. 1B, bottom panel).
Previous studies have provided evidence that biotin-dependent carboxylases such as PCC are reliable and sensitive markers for biotin status in human lymphoid cells [37,38]. Here, knockdown of HCS was associated with decreased biotinylation of carboxylases (Fig. 2), consistent with the known role of HCS as a biotin-carboxylase ligase [29]. Likewise, the cellular abundance of PCC correlated with biotin concentrations in culture media, consistent with decreased cellular biotin levels in response to low biotin supply. Note that biotinylated alpha chains of PCC and 3-methylcrotonyl-CoA carboxylase comigrated under the conditions used here and that acetyl-CoA carboxylase was not detectable in Jurkat cells. The abundance of biotinylated carboxylases depended on the concentration of biotin in culture media in both wild-type cells and HCS-deficient cells.
Fig. 2.
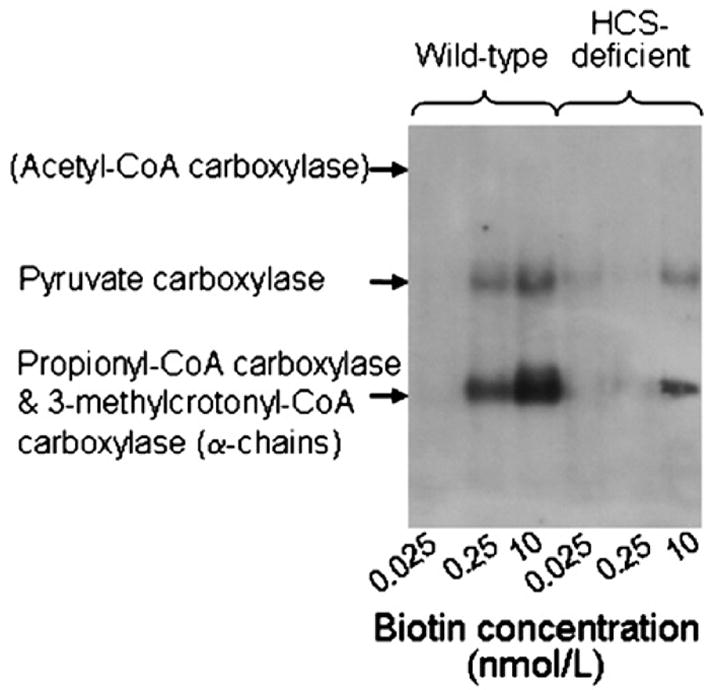
Biotinylation of carboxylases decreases in response to biotin deficiency and HCS deficiency in Jurkat cells. Wild-type cells and HCS-deficient cells were cultured in biotin-defined media for 5 weeks. Cellular proteins were resolved by gel electrophoresis and biotin was probed by using streptavidin peroxidase. Acetyl-CoA carboxylase was not detectable, but its expected location on the gel is indicated.
Supplementation of wild-type cells with pharmacological doses of biotin was associated with decreased expression of SMVT. The abundance of SMVT mRNA in wild-type cells decreased by about 85% in response to biotin supplementation: 1.0±0.05 (0.25 nmol/l biotin) vs. 0.15±0.09 arbitrary units (10 nmol/l biotin) (Fig. 3A). This feedback control mechanism was impaired in HCS knockdown cells, where the abundance of SMVT mRNA decreased by only 56% in response to biotin supplementation: 1.13±0.06 (0.25 nmol/l biotin) vs. 0.49±0.02 arbitrary units (10 nmol/l biotin). Analysis of biotin transport rates corroborated a role for HCS in the regulation of SMVT. Rates of biotin uptake decreased from 2.6±0.4 fmol biotin/(mg protein×min) in wild-type cells in physiological medium to 1.9±0.1 fmol biotin/(mg protein×min) in wild-type cells in pharmacological medium (P<.05; n=5). In contrast, rates of biotin uptake did not decrease significantly in HCS knockdown cells cultured in pharmacological medium [2.7±0.1 fmol biotin/(mg protein×min)] compared with physiological medium [2.9±0.1 fmol biotin/(mg protein×min)] (P=0.52; n=5). Finally, the abundance of SMVT protein was inversely associated with the concentrations of biotin in culture media in wild-type cells but not in HCS knockdown cells (Fig. 3B). Secondary antibody alone did not produce a detectable signal (negative control). Histone H3 was used as a marker to confirm equal loading of lanes (data not shown) (Fig. 1B).
Fig. 3.
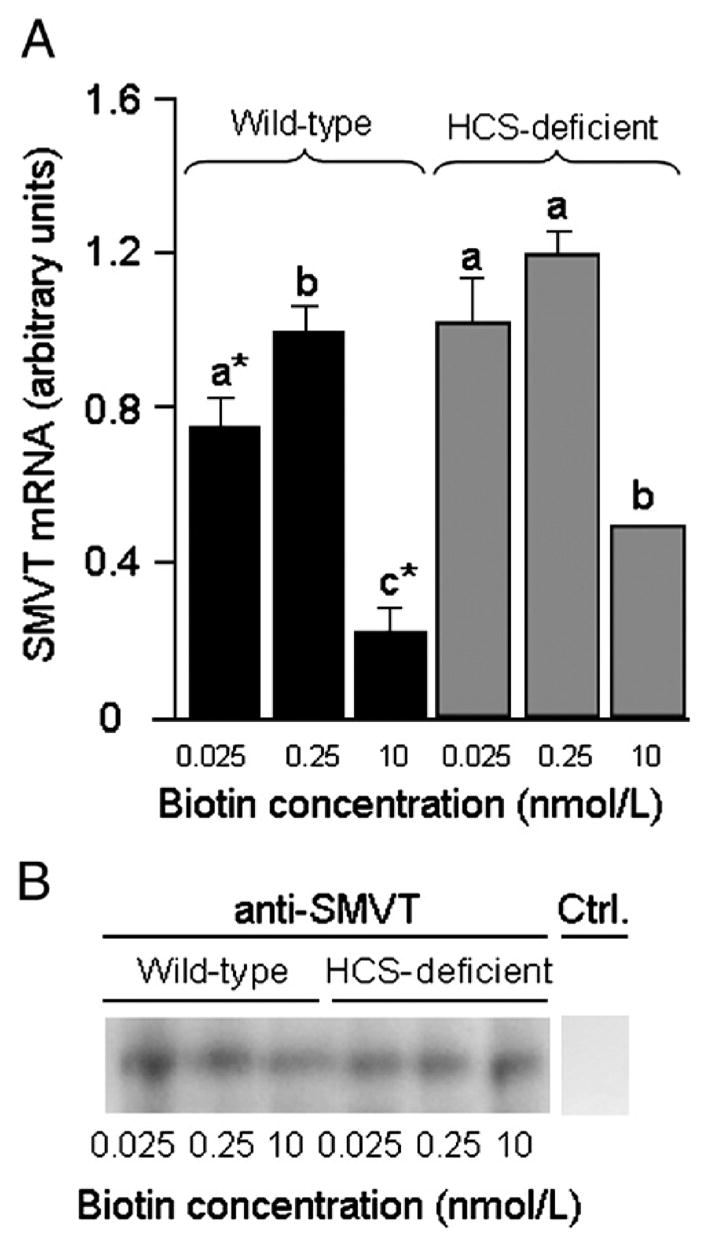
Expression of SMVT decreases in response to biotin supplementation in wild-type cells, but down-regulation is impaired in HCS-deficient cells. (A) Abundance of SMVT mRNA was quantified by real-time PCR. Values are means±S.D. a,b,cBars not sharing the same superscript are significantly different in cells matched for HCS expression. *P<.05, significantly different from knockdown cells matched for biotin (n=3). (B) SMVT protein was probed by Western blot analysis. Ctrl. indicates control (blot probed with secondary antibody in the absence of anti-SMVT). anti-SMVT and Ctrl. samples were run on the same gel, but the Ctrl. lane was processed separately after electroblotting.
Biotin supplementation was associated with increased nuclear translocation of HCS in wild-type cells, consistent with the proposed role of HCS as a biotin sensor (Fig. 4, top panel). Only trace amounts of HCS were detectable in nuclei from knockdown cells. No signal was detectable if cell extracts were probed with secondary antibody alone (compare Fig. 1B). Histone H3 was used as a marker to confirm equal loading of lanes (Fig. 4, bottom panel). Contamination of nuclear extracts with cytoplasmic proteins was quantitatively negligible, as judged by the absence of the cytoplasmic marker GAPDH; whole cell extracts (positive control) produced a strong signal if probed for GAPDH (Fig. 4, bottom panel).
Fig. 4.
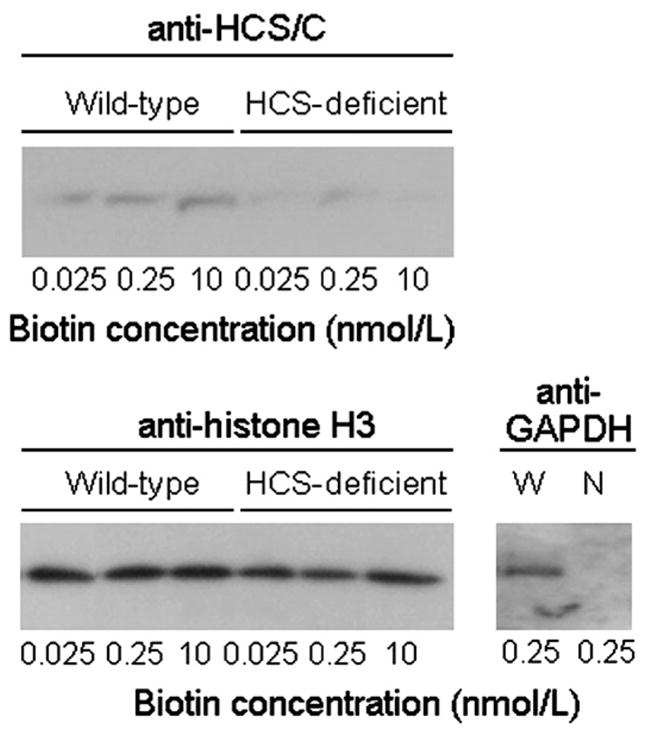
Nuclear translocation of HCS increases in response to biotin supplementation in wild-type cells but not in HCS-deficient cells. (Top) anti-HCS/C was used to probe the C-terminus of HCS in nuclear extracts. (Bottom) Equal loading of lanes was confirmed using histone H3 as a marker. Absence of a meaningful contamination of nuclear extracts (N) with cytoplasmic proteins was confirmed by probing the cytoplasmic marker GAPDH. Whole cell extracts (W) were used as positive control. Samples probed with anti-histone H3 were run on the same gel, but the sequence of lanes was electronically rearranged after autoradiography.
K12Bio H4 increased at SMVT promoter loci in response to biotin supplementation in wild-type cells but not in HCS knockdown cells. For example, if wild-type cells were supplemented with 10 nM biotin, the relative enrichment of SMVT promoter 1 sequences by ChIP with anti-K12Bio H4 increased by 91±6.3% compared with physiological controls (Fig. 5A). In HCS knockdown cells, the abundance of K12Bio H4 at SMVT promoter 1 was less than 41% of that in wild-type cells at 10 nmol/L biotin. The relative magnitudes of effects of HCS knockdown on K12Bio H4 were similar if cells were cultured in media containing 0.025 and 0.25 nmol/L biotin (data not shown). The relative enrichment of SMVT promoter 1 sequences by ChIP with anti-HCS/C was similar to that observed for anti-K12Bio H4 (Fig. 5A). For example, biotin supplementation (10 nmol/L) was associated with a 146±21% increase of HCS at the SMVT promoter 1 locus compared with physiological controls; this increase was abolished in HCS knockdown cells.
Fig. 5.
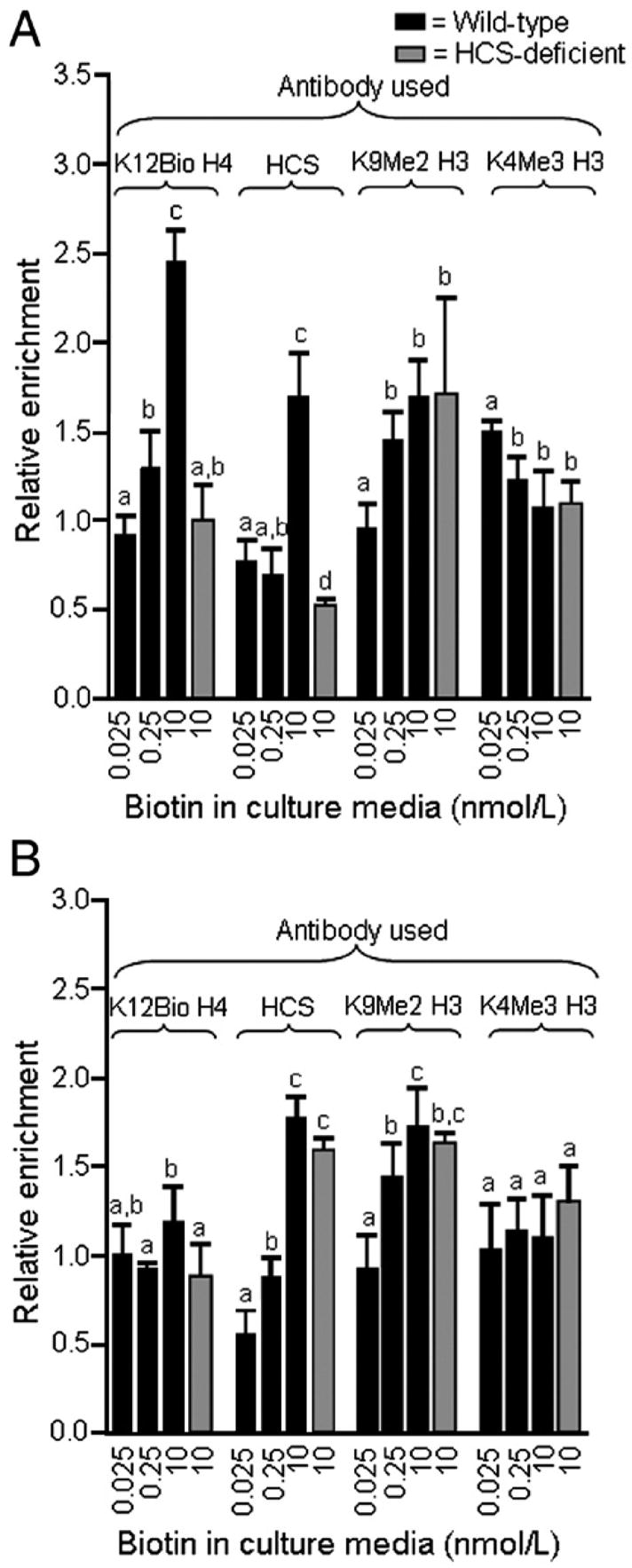
The relative enrichment of K12Bio H4 and HCS at SMVT promoter loci increases in response to biotin supplementation in wild-type cells but not in HCS-deficient cells. Wild-type cells and HCS-deficient cells were cultured in biotin-defined media for 5 weeks. Chromatin was immunoprecipitated using antibodies to K12Bio H4, K9Me2 H3, K4Me3 H3, and HCS (C-terminus), and DNA was quantified by PCR. (A) Relative enrichment of SMVT promoter 1 sequences by immunoprecipitation, using input DNA as denominator. (B) Relative enrichment of SMVT promoter 2 sequences by immunoprecipitation, using input DNA as denominator. Values are means ±S.D. a,b,c,dBars not sharing the same superscript are significantly different for the same antibody (P<.05, n=3–4).
The increase of K12Bio H4 and HCS at the SMVT promoter 1 locus in response to biotin supplementation coincided with an increase of the heterochromatin marker K9Me2 H3 and a decrease of the euchromatin marker K4Me3 H3 (Fig. 5A). HCS knockdown did not affect K9Me2 H3 and K4Me3 H3 compared with biotin-matched wild-type controls, e.g., the relative abundance of K9Me2 H3 and K4Me3 H3 at SMVT promoter 1 in HCS knockdown cells was within 3% of that in wild-type cells at 10 nmol/l biotin (Fig. 5B).
The relative enrichment of K12Bio H4 at SMVT promoter 2 sequences exhibited a relatively weak dependence on biotin supply and HCS expression (Fig. 5B). In contrast, HCS and K9Me2 H3 increased significantly at the SMVT promoter 2 locus in response to supplementation with 10 nM biotin, yielding patterns similar to promoter 1. These findings suggest that histone modifications other than biotinylation of K12 in histone H4 might participate in the regulation of SMVT promoter 2 in Jurkat cells (see Discussion).
In control experiments, we quantified the relative enrichment of histone modifications and HCS in heterochromatin (alpha satellite repeats in chromosome 1) and euchromatin (GAPDH). As expected from our previous studies [27], enrichment of alpha satellite repeat regions was greater if chromatin was immunoprecipitated with anti-K12Bio H4 and anti-K9Me2 H3 compared with anti-K4Bio H3 in wild-type cells (Fig. 6A). K12Bio H4 was 16–31% less abundant in satellite regions from HCS knockdown cells compared with biotin-matched, wild-type controls (Fig. 6A, and data not shown). Immunoprecipitation of chromatin with anti-HCS/C did not result in an enrichment of satellite sequences compared with input DNA (ratio ~1), suggesting a relatively low abundance of HCS in heterochromatin. HCS knockdown did not have a meaningful effect on K9Me2 H3 and K4Me3 H3 in satellite regions, consistent with observations made for SMVT promoters.
Fig. 6.
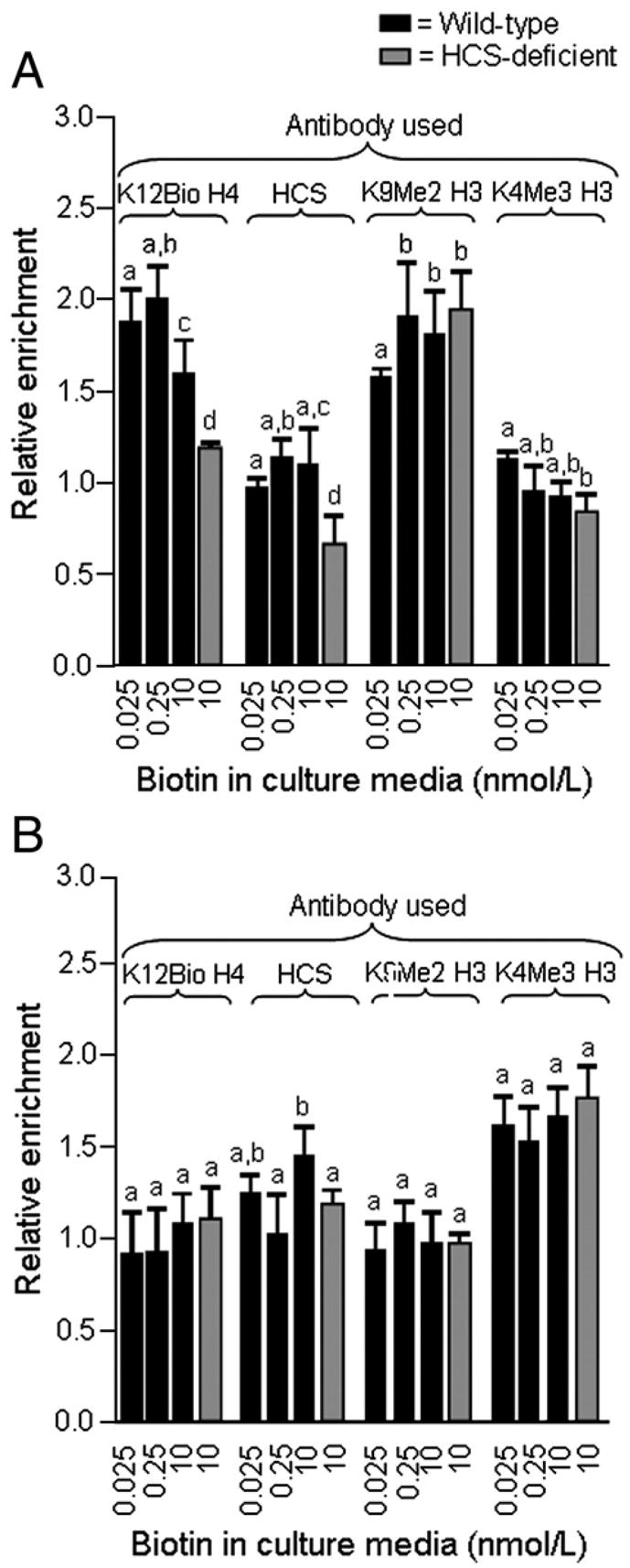
The relative enrichment of K12Bio H4 and HCS at alpha satellite repeats increases in response to biotin supplementation in wild-type cells but not in HCS-deficient cells. Wild-type cells and HCS-deficient cells were cultured in biotin-defined media for 5 weeks. Chromatin was immunoprecipitated using antibodies to K12Bio H4, K9Me2 H3, K4Me3 H3, and HCS (C-terminus), and DNA was quantified by PCR. (A) Relative enrichments of alpha satellite repeats by immunoprecipitation, using input DNA as denominator. (B) Relative enrichment of GAPDH by immunoprecipitation, using input DNA as denominator. Values are means±S.D. a,b,cBars not sharing the same superscript are significantly different in cells matched for HCS expression (P<.05, n=3–4).
Precipitation of chromatin by using anti-K12Bio H4, anti-HCS/C, and anti-K9Me2 H3 was not associated with an enrichment for euchromatin (GAPDH) sequences (Fig. 6B), consistent with previous studies [27]. In contrast, the euchromatin marker K4Me3 H3 was associated with GAPDH sequences. The abundance of K4Me3 H3 at the GAPDH locus did not depend on the expression of HCS or the concentration of biotin in culture media.
4. Discussion
This is the first published report that a nutrient regulates its own cellular uptake by mediating chromatin remodeling at a transporter gene locus. Specifically, the data presented here suggest that nuclear translocation of HCS increases in response to supplementation of cells with high doses of biotin; that nuclear translocation of HCS increases K12Bio H4 at the SMVT promoter 1 locus, and that increased abundance of K12Bio H4 at the SMVT promoter 1 locus decreases the expression of SMVT. Decreased (increased) expression of SMVT in biotin-supplemented (biotin-deficient) cells reduces (increases) the rate of biotin uptake and, hence, the risk for biotin overdose (deficiency) decreases.
HCS plays an essential role as both biotin sensor and chromatin-remodeling enzyme in this regulatory network. Previous studies suggested that biotin deficiency decreases HCS mRNA in human hepatocarcinoma cells [39] and that HCS enters the cell nucleus despite lacking a known nuclear localization sequence [23,24]. Here, we expand these previous findings by demonstrating that the nuclear translocation of HCS depends on biotin. Currently, it is unknown whether the increased nuclear abundance of HCS in biotin-supplemented cells is the result of increased expression of HCS or of increased rates of nuclear import. We hypothesize that the lack of a nuclear localization sequence in HCS is overcome by interaction with other proteins that assist with nuclear import, and we are pursuing the identification of HCS-binding proteins by using yeast two-hybrid assays. It is currently unknown whether HCS is a component of the cell membrane or whether the effects described here are purely intracellular events.
Previous studies have provided evidence that HCS acts as a biotin-histone ligase [23] in addition to its classical role as a biotin-carboxylase ligase [29]. Histones are underbiotinylated in cells from HCS-deficient patients compared with wild-type cells [23,25]. Here, we demonstrate that HCS prefers SMVT promoter loci over other loci (e.g., GAPDH) as targets for biotinylation of histones. It is noteworthy in this context that staining of polytene chromosomes from Drosophila melanogaster with anti-HCS produces a pattern of defined chromosomal bands; this observation is consistent with the theory that HCS is targeted to specific regions in chromatin [40].
The data presented here are less conclusive for SMVT promoter 2 than for promoter 1. Enrichment of K12Bio H4 at promoter 2 depended very weakly on biotin supply, whereas enrichment of HCS and K9M2 H3 at this locus were clearly biotin-dependent. We offer the following not mutually exclusive explanations for this observation. First, SMVT promoter 2 is silenced by K9Me2 H3 rather than K12Bio H4 in response to biotin supplementation. Second, the increased abundance of HCS at the SMVT promoter 2 in response to biotin supplementation is associated with biotinylation of lysine residues other than K12 in histone H4; biotinylation of these residues might mediate silencing of promoter 2. Consistent with this theory, 10 distinct biotinylation sites in human histones have been identified [24–26].
This study offers an exciting model by which HCS acts as a biotin sensor with the ability to mediate chromatin remodeling events at SMVT promoter loci in human cells; SMVT then serves as a gatekeeper to regulate biotin entry into cells. There are striking parallels between this feedback mechanism and the biotin operon in microbes, where biotin regulates its own synthesis. In Escherichia coli and other enteric bacteria, a complex of BirA (the microbial homologue of HCS) and the intermediate biotinyl-AMP binds to promoter regions in the biotin operon, a cluster of genes mediating biotin biosynthesis; binding of biotinyl-AMP/BirA represses the transcription of these genes [41]. This feedback loop prevents excessive biosynthesis of biotin. Human cells cannot synthesize biotin and depend on regulating biotin uptake to maintain homeostasis. Regulation appears to be achieved by biotin-dependent chromatin remodeling events at the gene locus that controls biotin entry into cells.
Abbreviations
- ChIP
chromatin immunoprecipitation
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- HCS
holocarboxylase synthetase
- K
lysine
- K4Me3 H3
K4-trimethylated histone H3
- K9Me2 H3
K9-dimethylated histone H3
- K12Bio H4
K12-biotinylated histone H4
- PCC
propionyl-CoA carboxylase
- SMVT
sodium-dependent multivitamin transporter
Footnotes
A contribution of the University of Nebraska Agricultural Research Division, supported in part by funds provided through the Hatch Act. Additional support was provided by NIH grants DK 063945 and 1R21ES015206, USDA grant 2006-35200-01540, and by NSF EPSCoR grant EPS-0346476.
References
- 1.Bowers-Komro DM, McCormick DB. Biotin uptake by isolated rat liver hepatocytes. In: Dakshinamurti K, Bhagavan HN, editors. Biotin. New York (NY): New York Academy of Sciences; 1985. [DOI] [PubMed] [Google Scholar]
- 2.Said HM, Redha R. A carrier-mediated system for transport of biotin in rat intestine in vitro. Am J Physiol. 1987;252:G52–5. doi: 10.1152/ajpgi.1987.252.1.G52. [DOI] [PubMed] [Google Scholar]
- 3.Prasad PD, Wang H, Kekuda R, Fujita T, Fei Y-J, Devoe LD, et al. Cloning and functional expression of a cDNA encoding a mammalian sodium-dependent vitamin transporter mediating the uptake of pantothenate, biotin, and lipoate. J Biol Chem. 1998;273:7501–6. doi: 10.1074/jbc.273.13.7501. [DOI] [PubMed] [Google Scholar]
- 4.Zempleni J, Mock DM. Mitogen-induced proliferation increases biotin uptake into human peripheral blood mononuclear cells. Am J Physiol Cell Physiol. 1999;276:C1079–84. doi: 10.1152/ajpcell.1999.276.5.C1079. [DOI] [PubMed] [Google Scholar]
- 5.Stanley JS, Griffin JB, Mock DM, Zempleni J. Biotin uptake into human peripheral blood mononuclear cells increases early in the cell cycle, increasing carboxylase activities. J Nutr. 2002;132:1854–9. doi: 10.1093/jn/132.7.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manthey KC, Griffin JB, Zempleni J. Biotin supply affects expression of biotin transporters, biotinylation of carboxylases, and metabolism of interleukin-2 in Jurkat cells. J Nutr. 2002;132:887–92. doi: 10.1093/jn/132.5.887. [DOI] [PubMed] [Google Scholar]
- 7.Crisp SERH, Camporeale G, White BR, Toombs CF, Griffin JB, Said HM, et al. Biotin supply affects rates of cell proliferation, biotinylation of carboxylases and histones, and expression of the gene encoding the sodium-dependent multivitamin transporter in JAr choriocarcinoma cells. Eur J Nutr. 2004;43:23–31. doi: 10.1007/s00394-004-0435-9. [DOI] [PubMed] [Google Scholar]
- 8.Dey S, Subramanian VS, Chatterjee NS, Rubin SA, Said HM. Characterization of the 5′ regulatory region of the human sodium-dependent multivitamin transporter, hSMVT. Biochim Biophys Acta. 2002;1574:187–92. doi: 10.1016/s0167-4781(02)00226-9. [DOI] [PubMed] [Google Scholar]
- 9.Rodriguez-Melendez R, Griffin JB, Sarath G, Zempleni J. High-throughput immunoblotting identifies biotin-dependent signaling proteins in HepG2 hepatocarcinoma cells. J Nutr. 2005;135:1659–66. doi: 10.1093/jn/135.7.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chatterjee NS, Kumar CK, Ortiz A, Rubin SA, Said HM. Molecular mechanism of the intestinal biotin transport process. Am J Physiol. 1999;277:C605–13. doi: 10.1152/ajpcell.1999.277.4.C605. [DOI] [PubMed] [Google Scholar]
- 11.Prasad P, Wang H, Huang W, Fei Y-J, Leibach FH, Devoe LD, et al. Molecular and functional characterization of the intestinal Na+-dependent multivitamin transporter. Arch Biochem Biophys. 1999;366:95–106. doi: 10.1006/abbi.1999.1213. [DOI] [PubMed] [Google Scholar]
- 12.Said HM. Cellular uptake of biotin: mechanisms and regulation. J Nutr. 1999;129:490S–493S. doi: 10.1093/jn/129.2.490S. [DOI] [PubMed] [Google Scholar]
- 13.Wolffe A. San Diego (Calif): Chromatin Academic Press; 1998. [Google Scholar]
- 14.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–80. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 15.Fischle W, Wang Y, Allis CD. Histone and chromatin cross-talk. Curr Opin Cell Biol. 2003;15:172–83. doi: 10.1016/s0955-0674(03)00013-9. [DOI] [PubMed] [Google Scholar]
- 16.Zempleni J. Uptake, localization, and noncarboxylase roles of biotin. Annu Rev Nutr. 2005;25:175–96. doi: 10.1146/annurev.nutr.25.121304.131724. [DOI] [PubMed] [Google Scholar]
- 17.Liang G, Lin JC, Wei V, Yoo C, Cheng JC, Nguyen CT, et al. Distinct localization of histone H3 acetylation and H3-K4 methylation to the transcription start sites in the human genome. Proc Natl Acad Sci U S A. 2004;101:7357–62. doi: 10.1073/pnas.0401866101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–81. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 19.Lehnertz B, Ueda Y, Derijck AA, Braunschweig U, Perez-Burgos L, Kubicek S, et al. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr Biol. 2003;13:1192–200. doi: 10.1016/s0960-9822(03)00432-9. [DOI] [PubMed] [Google Scholar]
- 20.Martens JH, O’Sullivan RJ, Braunschweig U, Opravil S, Radolf M, Steinlein P, et al. The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO J. 2005;24:800–12. doi: 10.1038/sj.emboj.7600545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hymes J, Fleischhauer K, Wolf B. Biotinylation of histones by human serum biotinidase: assessment of biotinyl-transferase activity in sera from normal individuals and children with biotinidase deficiency. Biochem Mol Med. 1995;56:76–83. doi: 10.1006/bmme.1995.1059. [DOI] [PubMed] [Google Scholar]
- 22.Stanley JS, Griffin JB, Zempleni J. Biotinylation of histones in human cells: effects of cell proliferation. Eur J Biochem. 2001;268:5424–9. doi: 10.1046/j.0014-2956.2001.02481.x. [DOI] [PubMed] [Google Scholar]
- 23.Narang MA, Dumas R, Ayer LM, Gravel RA. Reduced histone biotinylation in multiple carboxylase deficiency patients: a nuclear role for holocarboxylase synthetase. Hum Mol Genet. 2004;13:15–23. doi: 10.1093/hmg/ddh006. [DOI] [PubMed] [Google Scholar]
- 24.Chew YC, Camporeale G, Kothapalli N, Sarath G, Zempleni J. Lysine residues in N-and C-terminal regions of human histone H2A are targets for biotinylation by biotinidase. J Nutr Biochem. 2006;17:225–33. doi: 10.1016/j.jnutbio.2005.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kobza K, Camporeale G, Rueckert B, Kueh A, Griffin JB, Sarath G, et al. K4, K9, and K18 in human histone H3 are targets for biotinylation by biotinidase. FEBS J. 2005;272:4249–59. doi: 10.1111/j.1742-4658.2005.04839.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Camporeale G, Shubert EE, Sarath G, Cerny R, Zempleni J. K8 and K12 are biotinylated in human histone H4. Eur J Biochem. 2004;271:2257–63. doi: 10.1111/j.1432-1033.2004.04167.x. [DOI] [PubMed] [Google Scholar]
- 27.Camporeale G, Oommen AM, Griffin JB, Sarath G, Zempleni J. K12-biotinylated histone H4 marks heterochromatin in human lymphoblastoma cells. J Nutr Biochem. 2007 doi: 10.1016/j.jnutbio.2006.12.014. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mock DM. Determinations of biotin in biological fluids. In: McCormick DB, Suttie JW, Wagner C, editors. Methods in enzymology. New York (NY): Academic Press; 1997. [DOI] [PubMed] [Google Scholar]
- 29.Camporeale G, Zempleni J. Biotin. In: Bowman BA, Russell RM, editors. Present knowledge in nutrition. Washington (DC): International Life Sciences Institute; 2006. [Google Scholar]
- 30.Griffin JB, Rodriguez-Melendez R, Zempleni J. The nuclear abundance of transcription factors Sp1 and Sp3 depends on biotin in Jurkat cells. J Nutr. 2003;133:3409–15. doi: 10.1093/jn/133.11.3409. [DOI] [PubMed] [Google Scholar]
- 31.Griffin JB, Stanley JS, Zempleni J. Synthesis of a rabbit polyclonal antibody to the human sodium-dependent multivitamin transporter. Int J Vitam Nutr Res. 2002;72:195–8. doi: 10.1024/0300-9831.72.4.195. [DOI] [PubMed] [Google Scholar]
- 32.Boyd KE, Wells J, Gutman J, Bartley SM, Farnham PJ. c-Myc target gene specificity is determined by a post-DNA binding mechanism. Proc Natl Acad Sci U S A. 1998;95:13887–92. doi: 10.1073/pnas.95.23.13887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zempleni J, Stanley JS, Mock DM. Proliferation of peripheral blood mononuclear cells causes increased expression of the sodium-dependent multivitamin transporter gene and increased uptake of pantothenic acid. J Nutr Biochem. 2001;12:465–73. doi: 10.1016/s0955-2863(01)00162-0. [DOI] [PubMed] [Google Scholar]
- 34.Zempleni J, Mock DM. Uptake and metabolism of biotin by human peripheral blood mononuclear cells. Am J Physiol Cell Physiol. 1998;275:C382–8. doi: 10.1152/ajpcell.1998.275.2.C382. [DOI] [PubMed] [Google Scholar]
- 35.SAS Institute. StatView Reference. Cary (NC): SAS Publishing; 1999. [Google Scholar]
- 36.Hiratsuka M, Sakamoto O, Li X, Suzuki Y, Aoki Y, Narisawa K. Identification of holocarboxylase synthetase (HCS) proteins in human placenta. Biochim Biophys Acta. 1998;1385:165–71. doi: 10.1016/s0167-4838(98)00032-6. [DOI] [PubMed] [Google Scholar]
- 37.Mock DM, Henrich CL, Carnell N, Mock NI, Swift L. Lymphocyte propionyl-CoA carboxylase and accumulation of odd-chain fatty acid in plasma and erythrocytes are useful indicators of marginal biotin deficiency. J Nutr Biochem. 2002;13:462–70. doi: 10.1016/s0955-2863(02)00192-4. [DOI] [PubMed] [Google Scholar]
- 38.Mock DM, Mock NI. Lymphocyte propionyl-CoA carboxylase is an early and sensitive indicator of biotin deficiency in rats, but urinary excretion of 3-hydroxypropionic acid is not. J Nutr. 2002;132:1945–50. doi: 10.1093/jn/132.7.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Solorzano-Vargas RS, Pacheco-Alvarez D, Leon-Del-Rio A. Holocarboxylase synthetase is an obligate participant in biotin-mediated regulation of its own expression and of biotin-dependent carboxylases mRNA levels in human cells. Proc Natl Acad Sci U S A. 2002;99:5325–30. doi: 10.1073/pnas.082097699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Camporeale G, Giordano E, Rendina R, Zempleni J, Eissenberg JC. Drosophila holocarboxylase synthetase is a chromosomal protein required for normal histone biotinylation, gene transcription patterns, lifespan and heat tolerance. J Nutr. 2006;136:2735–42. doi: 10.1093/jn/136.11.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cronan JE., Jr The E coli bio operon: transcriptional repression by an essential protein modification enzyme. Cell. 1989;58:427–9. doi: 10.1016/0092-8674(89)90421-2. [DOI] [PubMed] [Google Scholar]


