Abstract
Quantitative MRI assessment of leukoencephalopathy is difficult because the MRI properties of leukoencephalopathy significantly overlap those of normal tissue. This report describes the use of an automated procedure for longitudinal measurement of tissue volume and relaxation times to quantify leukoencephalopathy. Images derived by using this procedure in patients undergoing therapy for acute lymphoblastic leukemia (ALL) are presented. Five examinations from each of five volunteers (25 examinations) were used to test the reproducibility of quantitated baseline and subsequent, normal-appearing images; the coefficients of variation were less than 2% for gray and white matter. Regions of leukoencephalopathy in patients were assessed by comparison with manual segmentation. Two radiologists manually segmented images from 15 randomly chosen MRI examinations that exhibited leukoencephalopathy. Kappa analyses showed that the two radiologists’ interpretations were concordant (κ = 0.70) and that each radiologist’s interpretations agreed with the results of the automated procedure (κ = 0.57 and 0.55).The clinical application of this method was illustrated by analysis of images from sequential MR examinations of two patients who developed leukoencephalopathy during treatment for ALL. The ultimate goal is to use these quantitative MR imaging measures to better understand therapy-induced neurotoxicity, which can be limited or even reversed with some combination of therapy adjustments and pharmacological and neurobehavioral interventions.
Keywords: quantitative MR, leukoencephalopathy, neurotoxicity, high-dose methotrexate
When cancer treatment specifically targets the immature brain, treatment efficacy must be balanced against potential chronic neurotoxicity and its associated impact on survivors’ quality of life. Acute lymphoblastic leukemia (ALL) is the most common childhood cancer: approximately 3500 new cases are diagnosed each year in the United States. The 3-year event-free survival estimate for pediatric patients with ALL is approximately 90%. Treatment of pediatric ALL must include central nervous system (CNS)-directed therapy to control occult CNS leukemia. Historically, CNS prophylaxis most commonly involved the administration of intermediate-dose or high-dose methotrexate (HDMTX), with or without cranial radiation therapy (CRT). More recently, efforts to avoid the adverse neurological and neuropsychological effects of CRT have prompted the elimination of CRT and the intensification of CNS chemotherapy.
Some researchers have argued that methotrexate may in some circumstances exert a protective effect on the CNS (1). However, several other reports have shown a significant association between treatment with HDMTX and development of leukoencephalopathy (2-5). The term leukoencephalopathy is used to designate white-matter damage in the CNS that is identified by hyperintense signal in T2-weighted MR imaging after therapy. In addition to the volumetric changes associated with leukoencephalopathy, T1 and T2 relaxation rates are also changed in response to treatment-induced damage of white matter.
Historically, most studies of leukoencephalopathy have investigated its occurrence in patients with ALL who were treated with HDMTX and CRT. Ochs et al. (2) used quantitative CT measures within regions of interest to detect areas of decreased density of white matter in patients with ALL. Although quantitative, their study did not report the size or magnitude of density change with respect to normal-appearing white matter. Subsequent studies using MRI to examine patients with ALL have thus far reported only qualitative assessments of leukoencephalopathy (3-5). In those studies, the intensity of the leukoencephalopathy was rated on a subjective grading scale as mild, moderate, or severe; no clear guidelines were presented for discrimination between the intensity grades and intra- and interobserver variance was not evaluated. This subjective scale precludes comparison of therapy-induced leukoencephalopathy between institutions or trials.
The qualitative studies demonstrated that quantitative MRI-based measures are needed to establish the extent and degree of leukoencephalopathy that develops during treatment for ALL. The imaging characteristics of leukoencephalopathy are similar to those of nonenhancing multiple sclerosis lesions. Vinitski et al. (6,7) used T1, T2, proton density (PD), and either magnetization transfer or postcontrast T1 images to create a 3D (6) and later a 4D (7) feature map segmentation of the CNS to detect multiple sclerosis lesions. Their studies showed significant, progressive improvement in segmentation quality across 2D, 3D, and 4D feature maps. Traditional or modified k-nearest neighbor algorithms were used as a basis for segmentation; these algorithms required the manual selection of 20-50 seed points per tissue class and resulted in 8% intraobserver variability. The features of the selected pixels (samples) served as the sole basis of all segmentation.
More recently, Kikinis and colleagues (8,9) reported a procedure for automated segmentation of 2D feature maps (T2 and PD); the procedure used a multichannel, multi-class, nonparametric algorithm based on a self-adaptive expectation-maximization algorithm. All tissues were segmented into four classes: white matter, gray matter, cerebrospinal fluid (CSF), and white matter lesions. Inaccuracy in the segmentation of partial volumes was resolved by using morphologic operators and connectivity principles. The authors reported a 1.4% intrarater and 1.1% interrater coefficient of variation for the detection of segmented white matter lesions. Partial volume correction using a priori anatomical knowledge reduced the estimate of total brain lesions by over 80%. This type of false-positive lesion is caused primarily by partial voluming of CSF and brain parenchyma. Errors in lesion detection can be attributed to a combination of systematic errors (i.e., white matter lesions classified as gray matter) and systematic random errors (i.e., gray matter classified as white matter lesions). The authors suggested that both errors could be resolved by using a white matter mask.
Udupa et al. (10) proposed another approach to automated segmentation that was based on fuzzy-connectedness principles for segmentation in a 2D feature space (T2 and PD). Like the method used by Kikinis and colleagues, the algorithm used here relied on operator-selected samples of white matter, gray matter, and CSF. However, the segmentation procedure developed by Udupa et al. identified pixels outside their tissue classes as potential lesions and the user had to manually edit every section.
The present study examined the validity of using a white matter mask combined with an automated, hybrid neural network to segment regions of leukoencephalopathy and normal-appearing brain tissue. Quantitative measures of T1 relaxation rates were combined with the segmentation to objectively measure both the volume and intensity of leukoencephalopathy regions. The purpose of this research was to develop quantitative MR imaging measures that can be used to better understand therapy-induced neurotoxicity, which can be limited or even reversed.
MATERIALS AND METHODS
This method of quantitative assessment of leukoencephalopathy was specifically designed for patients treated for ALL with regimens containing HDMTX. All patients included in this study were less than 18 years of age and were receiving treatment for ALL on an institutional protocol that included seven courses of HDMTX with targeted systemic exposure to eliminate individual differences caused by variability in clearance (11). MRI examinations were performed after the first, fourth, and seventh course of HDMTX.
MRI
Asato et al. (4) have shown that regions of HDMTX-induced leukoencephalopathy in patients with ALL are not associated with abnormal contrast-enhancement effects in any part of the brain. Leukoencephalopathy is best visualized with a T2-weighted sequence, preferably with CSF attenuated. Patients underwent a clinical imaging protocol after informed consent had been obtained from the patient, parent, or guardian, as appropriate. The imaging protocol was designed to simultaneously yield raw images for the segmentation procedures and images necessary for the clinical evaluation of the patients. All MRI examinations were performed without contrast agent on a 1.5T Vision (Siemens Medical Systems, Iselin, NJ) whole-body imager with a standard circular polarized volume head coil.
A set of T1-weighted sagittal images was collected as part of the diagnostic examination. The midline sagittal image was used to define an oblique transverse-imaging plane, defined by the most inferior extent of the genu and the splenium of the corpus callosum on the midline sagittal image, used on all subsequent sequences. All images were 3-mm thick contiguous slices acquired as two interleaved sets of 19 and 18 slices with a 3-mm gap to avoid cross-talk between slice excitations. T1-weighted images were acquired with a multiecho inversion recovery imaging sequence (repetition time between spin excitations [TR], 8000 ms; echo time [TE], 20 ms; time between inversion and the excitation pulse [TI], 300 ms; one acquisition; seven echoes). PD- and T2-weighted images were acquired simultaneously with a dual spin-echo sequence (TR/TE1/TE2 = 3500/17/102 ms; one acquisition). Fluid-attenuated inversion recovery (FLAIR) images were acquired with a multiecho inversion recovery sequence (TR/TE/TI = 9000/119/2470 ms; one acquisition; seven echoes).
Quantitative T1 imaging was performed by using an adapted precise and accurate inversion-recovery (PAIR) sequence (12-16). Although the complete imaging set was used in the segmented volumetric studies, only two sections were used to determine quantitative relaxation measures. This restriction in coverage was used to accurately assess relaxation times because it balanced the benefit of additional quantitative measures with the disadvantage of long acquisition times. The first section, which was at the level of the basal ganglia, included the genu and splenium of the corpus callosum and generally showed the putamen and the lateral ventricle. The second section was selected to sample the centrum semiovale. Each of these sections was sampled with a single 3-mm thick slice acquired with a phase-sensitive, multiecho inversion-recovery sequence (TR, 2500 ms; TE, 29 ms; TI, 100, 500, 900, and 2389 ms; one acquisition; seven echoes).
Quantitative T2 imaging was performed with a series of spin-echo images. Two sections were sampled to match the sections selected for the quantitative T1 imaging. Each of these sections was sampled with 3-mm thick slices acquired with a multiple spin-echo imaging sequence (TR = 2000 ms; 16 echoes evenly spaced from 22.5 to 360 ms; one acquisition).
Positioning and Registration
Manufacturer-installed standard positioning beams on the magnet and head immobilization devices built into the head coil were sufficient to ensure adequate positioning and immobilization of the head in these studies. Registration methods developed by Ostuni et al. (17) were used. In brief, intensity gradients served as the basis for a robust, automatic volume registration of the three imaging sequences used to acquire the segmentation data. This method consists of a matching algorithm that uses iterative techniques to find the correspondence of the closest voxels containing a high 3D intensity gradient magnitude. The registration software aligns stacks of 2D original MR images covering the entire head without requiring removal of regions of extrameningeal tissues. An adjustable threshold (gradient multiplier) allows the user to focus the registration on voxels containing the largest 3D gradients, which would include any edge in the images in which the signal intensity changes abruptly. Because the algorithm looks for the closest correspondence of these gradient voxels, if the size of the ventricles changes then the level of correspondence decreases; however, the maximum correspondence still registers the images because of the much larger proportion of gradient voxels at the bone interfaces.
In addition to registering different imaging sets obtained during the same examination, images obtained during subsequent examinations of each patient were also registered to the baseline images; thus, this registration facilitated the longitudinal assessment of tissue volume changes in brain parenchyma regardless of slight variations in head tilt between examinations.
Processing of Quantitative T1 Maps
After acquisition, the four PAIR images corresponding to each section were transferred to an offline workstation and T1 relaxation times were calculated by using a Levenberg-Marquardt algorithm to fit signal intensity (SI) values (13). A parametric T1 map was produced on which pixel grayscale values were equivalent to the T1 relaxation time in msec. Error of the fit was determined on a pixel-by-pixel basis and further analysis excluded those pixels identified as having error above a statistical threshold (14).
Processing of Quantitative T2 Maps
As in the quantitative T1 analysis, after acquisition the 16 spin-echo images corresponding to each section were transferred to an offline workstation and T2 relaxation times were calculated using a Levenberg-Marquardt algorithm to fit signal intensity (SI) values to a monoexponential model. The curve-fitting procedure returned a T2 value for each pixel; this value was used to produce a parametric T2 map wherein pixel grayscale values were equivalent to the T2 relaxation time in msec.
Segmentation Design
The image set submitted to the segmentation procedure consisted of the index slice (Fig. 1) and the adjacent superior and inferior sections. Because the images were acquired with the head centered in the transverse field-of-view, a recursive seed-growing algorithm was used to select all pixels above a given threshold and located around the center of the head on the PD image. The region of interest, which was determined by analysis of the PD image, was also applied to the T1, T2, and FLAIR images at the same slice position. This technique effectively eliminated excessive background signals; therefore, it reduced the execution time of the overall segmentation and classification process and eliminated the need for manual removal of regions representing extrameningeal tissues.
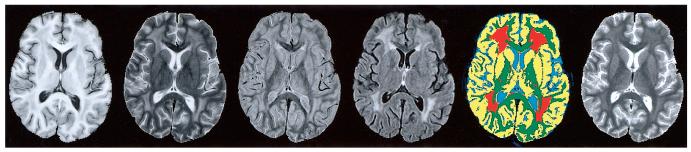
FIG. 1.
Illustration of the four conventional MR images from a single section in one patient, the resulting segmentation map, and the quantitative T1 map. The four MR images, from left to right, are T1-, T2-, PD-, and FLAIR-weighted. The sensitivity of the FLAIR imaging sequence is demonstrated. The resulting pseudocolor segmentation map is used for volumetric analyses and to mask tissues for subsequent analysis of relaxation time. Masks of orange identify regions of leukoencephalopathy and masks of green identify regions of normal-appearing white matter. Yellow areas indicate gray matter and blue areas indicate cerebrospinal fluid. In the corresponding quantitative T1 relaxation map (far right), the signal intensity of each pixel represents the relaxation rate calculated at that position.
Next, volumes of brain parenchyma on MR images were quantitatively assessed by using an adaptation of a previously published automated hybrid neural network segmentation and classification method (18,19). The segmentation procedure uses a Kohonen self-organizing map (SOM) (20,21). For this application, a four-unit input vector was composed of the nonnormalized T1, T2, FLAIR, and PD SIs for a single pixel in the MR image. The SOM configuration was a single layer of 16 neurons arranged in a four-by-four topology.
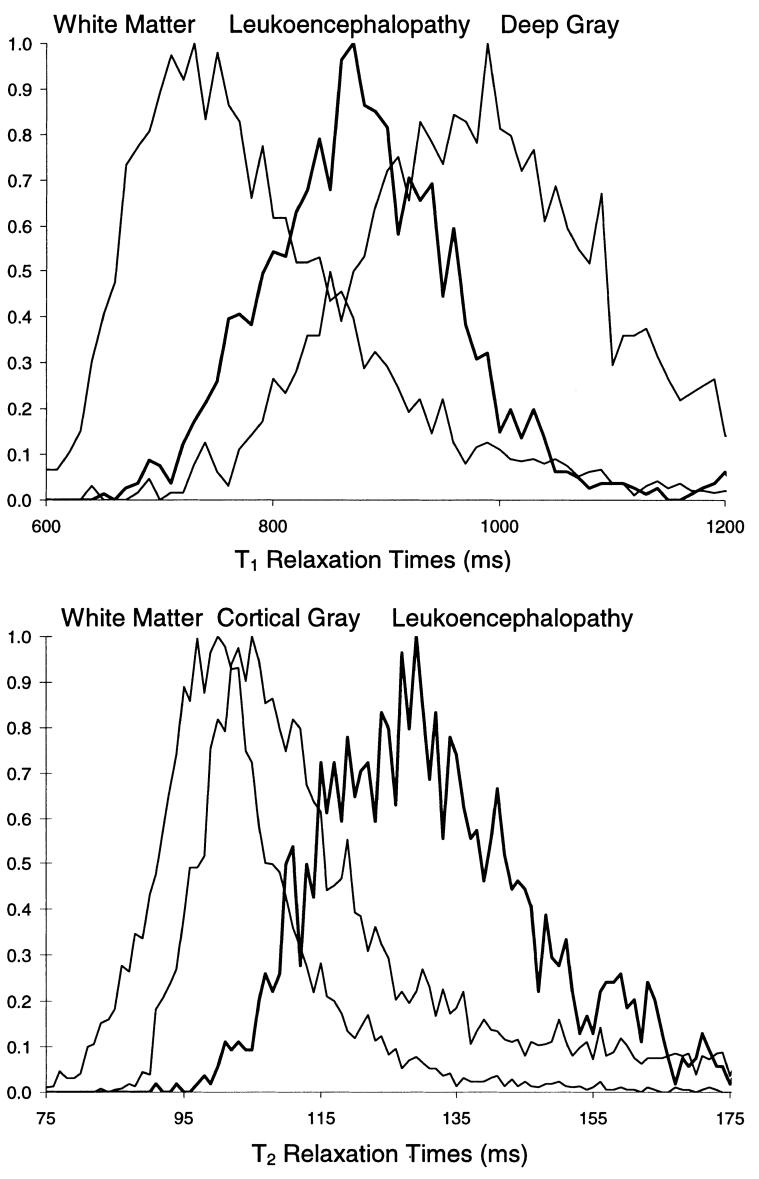
MRI properties of regions of leukoencephalopathy significantly overlap those of regions of normal tissue, as illustrated in Fig. 2. The quantitative T1 and T2 relaxation rates are shown for regions of white matter, gray matter, and leukoencephalopathy. The segmentation design uses four different types of images with different tissue contrast (T1-, T2-, PD-weighted, and FLAIR) to help differentiate leukoencephalopathy. To further address the problem of discerning leukoencephalopathic regions from normal regions, a mask was generated from the segmented and classified white matter of the baseline images. Baseline images were obtained during the first MRI examination, which was performed after only one course of HDMTX; at that time, images were usually normal in appearance. After images from the subsequent examinations were registered to those from the baseline examination, the white matter mask was used to identify regions for segmentation (i.e., normal-appearing white matter or leukoencephalopathy) and the segmented regions of leukoencephalopathy were then excluded from segmentation of the normal-appearing brain tissues.
FIG. 2.
Normalized histograms of T1 (top) and T2 (bottom) relaxation rates demonstrate the overlap of the MR characteristics of leukoencephalopathy and those of normal tissues in an image from a single patient. In general, both relaxation rates are lower for white matter than for gray matter; the relaxation rate of deep gray structures (i.e., putamen, caudate, and thalamus) is lower than that of cortical gray matter but higher than those of white matter. The T1 relaxation rates of leukoencephalopathic tissue (bold line) lies directly between and completely overlaps those of white matter and deep gray matter. As would be expected, the T2 relaxation rate of leukoencephalopathic tissue is, on average, higher than those of cortical gray matter and white matter.
Once the SOM process had been trained on the input vectors within the region of interest, all input vectors from slices covering the entire brain were presented to the trained SOM for a final analysis. In this last step of the segmentation process, all input vectors were assigned to one of the 16 SOM neurons.
Classification Design
After segmentation was completed, each of the 16 levels in the segmented images was classified according to tissue type. Initially, the preliminary data were manually classified referring to the original images (primarily T2-weighted and FLAIR) in consultation with the radiologists to create a training set. The classification neural network was a four-layer feed-forward network trained with error back-propagation (18,19). The input layer contained five neurons corresponding to the patient’s age at the time of the examination and the four components of the prototypical weight vector associated with each neuron of the SOM network.
Two different neural networks were trained. The first network discriminated leukoencephalopathy from normal-appearing white matter and was used after the initial segmentation of only those pixels inside the white matter masks. Therefore, this network had only three possible outputs: normal-appearing white matter, leukoencephalopathy, and partial volumes between the two tissue classes. The second network classified the segmented normal-appearing tissues from the entire imaging set, excluding those pixels previously classified as leukoencephalopathy. This network had seven possible outputs: white matter, partial gray and white matter, gray matter, partial gray matter and CSF, CSF, blood vessels, and background. For both networks the regions that contained partial volumes of different tissues were redistributed by setting to zero each prototypical vector associated with those regions; this procedure produced an equivalent null vector. The final analysis of segmentation was repeated with the resulting set of prototypical vectors and each pixel was assigned to the nearest output neuron classified as one of the pure tissue types. All tissues were then color-coded and displayed as shown in Fig. 1.
Evaluating Tissue Volumetry
Volumetric analysis of the pseudocolor maps from each transverse section was performed in the automated region of interest, which contained only brain parenchyma. However, this automated process becomes less reliable as the separation between brain parenchyma and other tissues becomes more difficult to discern, as is usually the case when the images begin to include eyes, brain stem, and cerebellum. In addition, the most superior images near the vertex of the brain begin to exhibit increased partial volume effects and decreased white matter volumes; these images are susceptible to B1 field inhomogeneity. For these reasons, a set of images extending from 6 mm superior to the ventricles to 6 mm inferior to the corpus callosum on the midsagittal section was analyzed. A study of images from five patients (ages 2, 5, 7, 12, and 18 years) revealed that the mean automated volume of interest sampled was 66% (range, 61%-72%) of the white matter in the full cerebrum, as determined by manual selection. Volumes of classified tissue in the automated volume of interest were determined for each imaging slice by using the histogram to measure the number of pixels in each color and multiplying this number by the pixel volume.
Masking for Quantitative Relaxation Measures
As described previously, the quantitative T1 maps were calculated for oblique sections at the level of the basal ganglia and the level of the centrum semiovale. Using the segmented pseudocolor maps at the same positions, regions were identified as leukoencephalopathy, normal-appearing white matter, cortical gray matter, and deep gray structures in these images. The averaged T1 relaxation times and their standard deviations were determined for each tissue type by using the color masks generated from the segmented maps to identify the regions of interest in each slice (Fig. 1).
Quantifying Images From the Baseline Examination
Processing and analysis of the baseline images was relatively straightforward. The process began with the collection of the MRI images, which are usually normal in appearance and form the base imaging set. These images were registered, segmented, and classified for normal-appearing brain. Tissue volumes were measured and these measurements were used to generate masks for identifying regions of interest in the relaxation maps. The quantitative T1 maps were calculated from the raw MR images as described above and regional values were determined on the basis of the pseudocolor maps. In addition, white matter masks were generated for every section for use in the processing of images from subsequent examinations.
Quantifying Images From Subsequent Examinations
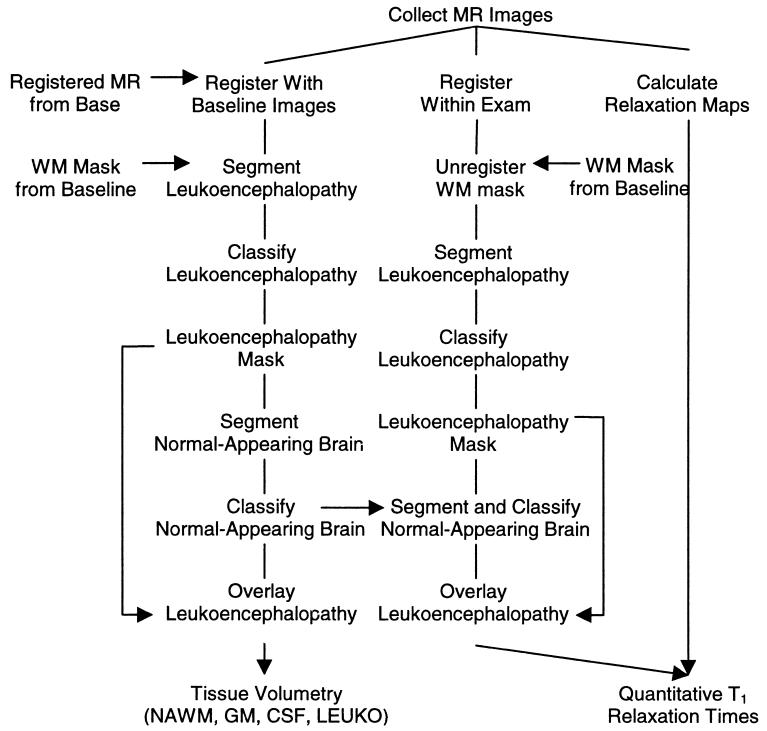
The process for quantifying images from subsequent examinations began with the collection of the MR images. If the images were normal in appearance, the processing and analysis were similar to those used for the baseline images. Processing and analysis of images demonstrating leukoencephalopathy were more complex and a dual processing procedure was used (Fig. 3). Processing of tissue volumes was done according to the procedure described in the left branch of the flow chart, whereas processing of quantitative relaxation measures was done as described in the procedures in the middle and right branches. Tissue volumes were determined by registering the MR imaging sequences to the baseline images. The white matter masks from the baseline images were used to identify regions for segmenting and classifying leukoencephalopathy in the entire volume of interest. These leukoencephalopathy pixels were then excluded from the segmentation and classification of all normal-appearing tissues. After a pseudocolor map of the normal-appearing tissues was created, the leukoencephalopathy pixels were overlaid as regions of orange. All volumetric measures were then performed for the volume of interest.
FIG. 3.
Flow chart of procedures used in longitudinal analysis of MR images to quantify the development of leukoencephalopathy. The left branch represents the segmentation and classification procedures for tissue volumetry. The center branch represents the procedure for segmenting and classifying tissues as a mask for determining the quantitative relaxation measures. The right branch represents the procedure for assessing relaxation rates.
The quantitative T1 maps were calculated from the raw MR images as described above. The MR imaging sequences within the examination were then registered and the white matter mask from the baseline examination was unregistered by using the inverse of the transformation matrix. As in the volumetry procedure, the white matter mask was used to identify regions for segmenting and classifying normal-appearing white matter and leukoencephalopathy. After the regions of leukoencephalopathy had been segmented and classified for the whole volume of interest, the leukoencephalopathy pixels were excluded from the segmentation and classification of the normal-appearing tissues. The prototypical vectors from the volumetry procedure were used in the relaxation procedure to reduce the complexity of the processing. After the pseudocolor map of the normal-appearing tissues had been created, the leukoencephalopathy pixels were overlaid as regions of orange. The complete pseudocolor map with leukoencephalopathy was then used to mask tissues on the T1 relaxation maps.
Test of Method on Normal Volunteers
As a test of the reproducibility of the proposed method for quantitative volumetry of normal brain tissues, five healthy volunteers, ages 23-33 years, were examined. Each volunteer underwent imaging on five separate occasions. The first examination was processed as a baseline image set and the other four were processed as subsequent normal-appearing images. Because the quantitative relaxation methods have been previously validated, images were acquired only for tissue volumetry. Quantitative volumes of white matter, gray matter, and CSF for the full volume of interest were analyzed and a one-way repeated measures ANOVA was performed.
Test of Method for Differentiating Leukoencephalopathy
Although the processing procedure for normal-appearing baseline and subsequent images could be evaluated with volunteers, the performance of the procedure for processing images that contained regions of leukoencephalopathy could be assessed only by using images from patients. MR images exhibiting leukoencephalopathy were collected from 15 patients with ALL after they had completed seven courses of HDMTX treatment. Two single sections, one from the T2 and one from the FLAIR imaging set from each patient, were presented twice in a random order to two radiologists. Blinded to patient identity, the radiologists manually selected regions of leukoencephalopathy on each image set. Kappa analyses were used to assess agreement between the findings of the automated technique and those of each radiologist and between the findings of the two radiologists.
Longitudinal Studies of Therapy-Induced Leukoencephalopathy
Images from a 5-year-old girl (Subject 1) and a 10-year-old boy (Subject 2) treated for ALL were collected after the first, fourth, and seventh courses of HDMTX (5.0 g/m2) and near the end of the therapy regimen. The second examination was performed approximately 3 months after the baseline examination; the third, approximately 8 months after the baseline examination; and the final examination, approximately 2.5 years after the baseline examination. The first examination was processed as baseline images and the remaining three were processed as subsequent images with leukoencephalopathy. The presence of leukoencephalopathy was determined by the radiologist at the time of the examinations.
RESULTS
Test of Method on Normal Volunteers
Images from each examination were registered, segmented, and classified for all subjects and volumetric measures were assessed. A one-way repeated-measures ANOVA was performed for volumes of white matter, gray matter, and CSF from the volume of interest (Table 1). The P values in Table 1 indicate that there were no significant within subject variances for either of the two tissue types or for CSF. The average coefficient of variance across the five subjects was less than 2% for gray and white matter.
Table 1.
Reproducibilitya of Quantitative MR Assessment in Normal Volunteers
| Tissue | Mean CV (%)b | F (4,16)c | P |
|---|---|---|---|
| White matter | 2 | 0.170 | 0.951 |
| Gray matter | 2 | 0.158 | 0.957 |
| Cerebrospinal fluid | 10 | 2.378 | 0.095 |
One-way, repeated-measures ANOVA of tissues in the volumes of interest from 25 examinations of five volunteers (five examinations each).
CV, coefficient of variation; the mean CV of each tissue type across the five subjects was calculated by dividing the SD of the measured tissue volumes by the mean for the five examinations of each volunteer.
Degrees of freedom are indicated in parenthesis.
Test of Method for Differentiating Leukoencephalopathy
After reviewing age-appropriate control images and discussing the precise imaging criteria for defining abnormality, the two radiologists reached a consensus about the regions to be considered abnormal. However, although the radiologists had paired images (T2-weighted and FLAIR images) to examine, both expressed concern about their ability to accurately delineate the boundaries of abnormal regions. Kappa analyses of the radiologists’ manual assessments revealed that the agreement within observers was κ = 0.73 and κ = 0.76, and the agreement between observers was κ = 0.70 (0.13 standard deviation). The kappa scores indicating agreement between the findings of the automated procedure and those of the individual radiologists (κ = 0.57 and κ = 0.55; 0.24 and 0.19 SD, respectively) were similar to the score indicating the agreement between radiologists.
Longitudinal Studies of Therapy-Induced Leukoencephalopathy
The white matter volume, defined as regions of normal appearing white matter and leukoencephalopathy, remained constant in the full volume of interest throughout the longitudinal studies of both patients (Fig. 4 and Table 2); the white matter volume demonstrated with a 9% coefficient of variation. However, the proportion of white matter classified as leukoencephalopathic increased from 0% on both subjects’ baseline image sets to 17% and 18%, respectively, on the second image sets and 35% and 38% on the third. The regions of leukoencephalopathy resolved to only 15% and 12% on the image sets from the last examination at the end of therapy (Table 2).
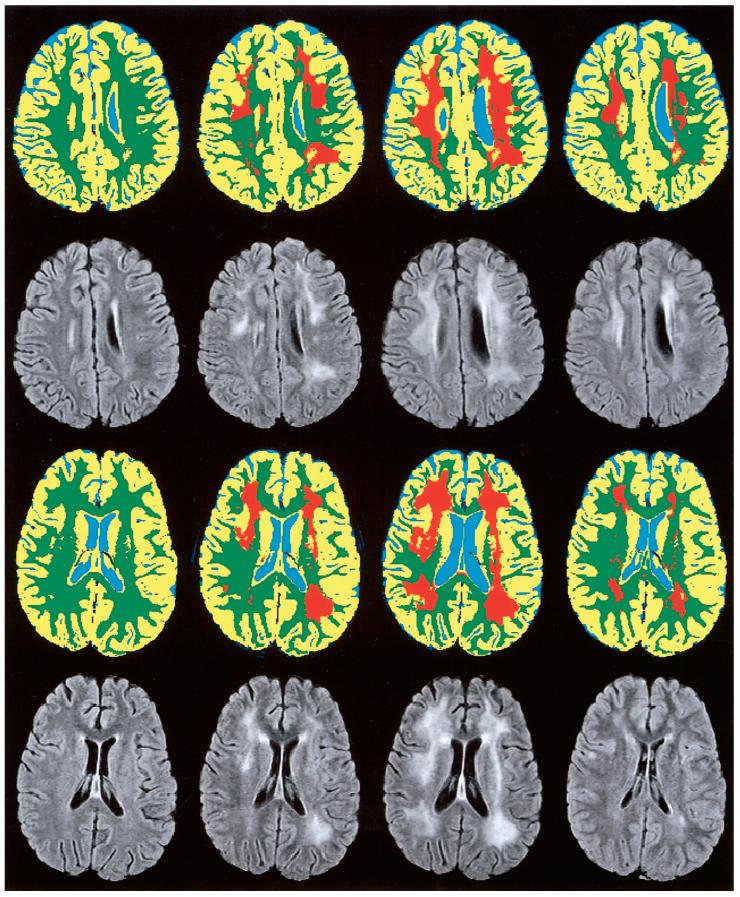
FIG. 4.
MR assessment of leukoencephalopathy in a 5-year-old girl (Subject 1, top rows) and a 10-year-old boy (Subject 2, bottom rows) treated for ALL. MR examinations (from left to right) were performed after the first, fourth, and seventh courses of high-dose methotrexate, which was administered as part of the chemotherapy regimen, and again near the end of therapy (far right). Temporal changes in leukoencephalopathy (orange) are shown on segmented and classified images of a single section. Corresponding FLAIR images are shown for reference.
Table 2.
Longitudinal MR Assessment of Leukoencephalopathy in Two Patients Treated for ALL
| Age (years) | Protocol week | Tissue volumea |
Relaxation time |
|||||
|---|---|---|---|---|---|---|---|---|
| NAWM (cc) | LEUKO (cc) | Affected white matter (%)b | NAWM (ms) | LEUKO (ms) | T1 increase (%)c | |||
| Exam 1 | ||||||||
| Subject 1 | 5.5 | 7 | 214 | — | 0 | 736 | — | 0 |
| Subject 2 | 10.4 | 5 | 298 | — | 0 | 764 | — | 0 |
| Exam 2 | ||||||||
| Subject 1 | 5.7 | 18 | 215 | 42 | 17 | 767 | 842 | 10 |
| Subject 2 | 10.6 | 19 | 247 | 54 | 18 | 773 | 838 | 8 |
| Exam 3 | ||||||||
| Subject 1 | 6.2 | 44 | 145 | 77 | 35 | 751 | 904 | 20 |
| Subject 2 | 11.1 | 43 | 192 | 117 | 38 | 789 | 885 | 12 |
| Exam 4 | ||||||||
| Subject 1 | 7.9 | 132 | 187 | 32 | 15 | 758 | 935 | 23 |
| Subject 2 | 12.8 | 132 | 220 | 31 | 12 | 789 | 810 | 3 |
Abbreviations: NAWM, normal-appearing white matter; LEUKO, leukoencephalopathy.
The full volume of interest encompassed two-thirds of the cerebrum.
The proportion of affected white matter was calculated as LEUKO/(LEUKO+NAWM).
The proportion of increase in T1 relaxation time was calculated as (LEUKO-NAWM)/NAWM.
CSF volumes on the second image sets were decreased in Subject 1 and stable in Subject 2. However, on the third image sets both subjects exhibited increased CSF volumes over baseline. By the fourth imaging sets, CSF volumes in Subject 2 had returned to below baseline but Subject 1 remained elevated. As would be expected with approximately constant intracranial volumes, changes in CSF had corresponding changes in brain parenchyma (R > 0.99; both subjects). These changes in CSF volumes were primarily compensated by changes in gray matter volumes (R = 0.91; both subjects).
The T1 relaxation rates of leukoencephalopathic regions were higher than that of normal-appearing white matter on both subjects’ image sets from their second examinations (10% and 8% higher) and third examinations (20% and 12% higher). However, on the fourth examination the images from the two patients showed dissimilar T1 relaxation rates. The T1 relaxation rate of Subject 1 remained 23% higher than that of normal-appearing white matter, but that of Subject 2 resolved to only 3% higher than normal-appearing white matter.
DISCUSSION AND CONCLUSION
The present study combined the use of a white matter mask with an automated hybrid neural network method for segmentation to distinguish regions of leukoencephalopathy from those of normal-appearing brain tissue. Quantitative measures of T1 relaxation rates were combined with the segmentation procedure to provide objective measures of both the volume and intensity of leukoencephalopathy. The intensity of leukoencephalopathy was determined by the difference between the elevated T1 relaxation times of regions of leukoencephalopathy and the T1 relaxation times of regions of normal-appearing white matter. Although quantitative assessment of leukoencephalopathy is difficult because the MRI properties of normal and leukoencephalopathic tissues significantly overlap, the longitudinal nature of this processing procedure and the inclusion of the white matter masks minimized the problem.
The automated procedure exhibited excellent reproducibility for quantifying normal-appearing images; repeated assessments of the gray and white matter of control subjects resulted in coefficients of variation less than 2%. Furthermore, the automated procedure was able to discern regions of leukoencephalopathy from regions of normal-appearing white matter, as demonstrated by kappa analyses of agreement between the results of the automated procedure and those of manual assessments by two radiologists. Unlike manual selection, the automated analyses are deterministic; thus, no intra- or interobserver variance occurs. The only variation in the analysis of repeated measures results from patient positioning and registration of the examinations, which has been shown to induce less than 2% variance in normal-appearing images. The average age of ALL diagnosis is 4.5 years, requiring the majority of patients to be sedated for the MR examinations. Therefore, imaging of patients with leukoencephalopathy on consecutive days to assess reproducibility of leukoencephalopathy measures is not feasible, because the benefit does not justify the risk of the procedure to the patient.
To demonstrate the clinical performance of the method, images from four sequential MR examinations of two patients who developed leukoencephalopathy during treatment for ALL were assessed. In both cases the intensity of leukoencephalopathy and the proportion of affected white matter increased in proportion to additional courses of HDMTX. Between the last course of HDMTX and near the end of therapy, the proportion of affected white matter decreased but individual differences in the intensity of remaining leukoencephalopathy were seen between patients.
One limitation of the quantitative procedure is the possibility that increased ventricular volume may distort the brain beyond the white matter mask. Because the white matter mask is derived from images obtained during the first examination, any large changes in ventricular size could force ventricular CSF into the area of the white matter mask. In studies thus far, this problem has not arisen. Because the CSF in the ventricles is attenuated on the FLAIR images, even if the ventricles extend into the white matter mask, they are not segmented as leukoencephalopathy.
Another limitation of this procedure is that many patients are very young at the time of their first MR examination. The MR properties of unmyelinated white matter are similar to those of gray matter; thus, on the baseline images of very young patients, unmyelinated white matter is excluded from the white matter mask. If leukoencephalopathy were to develop in these regions, this procedure would not be able to quantify the change. However, it is believed that some therapy, such as HDMTX, may not harm unmyelinated white matter in these terminal zones.
A variety of methods can be used for quantitative T1 measures. The most recent developments have focused on fast multislice measurements based on echoplanar techniques (22). However, these techniques are not feasible in this application because susceptibility artifacts in the bone-air interfaces of the sinuses cause nonlinear warping of the frontal and prefrontal regions. The Fast-PACE (phase acquisition of composite echoes) technique (23) for fast multislice T1 measurements (23) may be a feasible alternative for whole-head measures. This technique is not as rapid as the echoplanar methods, but it provides improved coverage and can be volume-registered with the segmented tissue maps. We are currently investigating the accuracy and precision of this technique for our application.
In conclusion, this study has described a novel approach for analyzing MR images to measure therapy-induced leukoencephalopathy. This automated assessment of MR images of the brain yields objective, quantitative measures of the extent (volume) and degree (increase in quantitative T1 relaxation time) of leukoencephalopathy. Because of its sensitivity to abnormal white matter hyperintensities, quantitative T2 measurements are also being investigated as a possible additional measure of degree of leukoencephalopathy. The clinical significance of therapy-induced leukoencephalopathy for long-term survivors is unknown. However, when combined with individualized pharmacokinetic studies and longitudinal neurocognitive testing, this MR method could substantially advance the understanding of leukoencephalopathy and its impact on the quality of life of survivors of ALL. Ongoing clinical investigations using the developed methodologies should be completed soon for a group of more than 40 patients treated for ALL. Our ultimate goal is to use this method to quantify early therapy-induced neurotoxicity, which is potentially reversible through adjustment of therapy or through individually tailored neurobehavioral and pharmacological interventions.
ACKNOWLEDGMENTS
The authors thank Rhonda Simmons for data analysis and Angela McArthur, Ph.D., for editorial consultation. We also thank Ching-Hon Pui, M.D., for advice and encouragement.
Grant sponsor: Cancer Center Support (CORE); Grant number: P30CA21765; Grant sponsor: National Cancer Institute; Grant number: R01CA90246; Grant sponsor: American Lebanese Syrian Associated Charities (ALSAC).
REFERENCES
- 1.Balsom WR, Bleyer WA, Robison LL, Heyn RM, Meadows AT, A Children’s Cancer Study Group Study Intellectual function in long-term survivors of childhood acute lymphoblastic leukemia: protective effects of pre-irradiation methotrexate? Med Pediatr Oncol. 1991;19:486–492. doi: 10.1002/mpo.2950190607. [DOI] [PubMed] [Google Scholar]
- 2.Ochs JJ, Parvey LS, Whitaker JN, Bowman WP, Ch’ien L, Campbell M, Coburn T. Serial cranial computed-tomography scans in children with leukemia given two different forms of central nervous system therapy. J Clin Oncol. 1983;1:793–798. doi: 10.1200/JCO.1983.1.12.793. [DOI] [PubMed] [Google Scholar]
- 3.Wilson DA, Nitschke R, Bowman ME, Chaffin MJ, Sexauer CL, Prince JR. Transient white matter changes on MR images in children undergoing chemotherapy for acute lymphocytic leukemia: correlation with neuropsychologic deficiencies. Radiology. 1991;180:205–209. doi: 10.1148/radiology.180.1.2052695. [DOI] [PubMed] [Google Scholar]
- 4.Asato R, Akiyama Y, Ito M, Kubota M, Okumura R, Miki Y, Konishi J, Mikawa H. Nuclear magnetic resonance abnormalities of the cerebral white matter in children with acute lymphoblastic leukemia and malignant lymphoma during and after central nervous system prophylactic treatment with intrathecal methotrexate. Cancer. 1992;70:1997–2004. doi: 10.1002/1097-0142(19921001)70:7<1997::aid-cncr2820700732>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 5.Mahoney DH, Jr, Shuster JJ, Nitschke R, Lauer SJ, Steuber CP, Winick NJ, Camitta B. Acute neurotoxicity in children with B-precursor acute lymphoid leukemia: an association with intermediate-dose intravenous methotrexate and intrathecal triple therapy — a pediatric oncology group study. J Clin Oncol. 1998;16:1712–1722. doi: 10.1200/JCO.1998.16.5.1712. [DOI] [PubMed] [Google Scholar]
- 6.Vinitski S, Gonzalez C, Mohamed F, Iwanaga T, Knobler RL, Khalili K, Mack J. Improved intracranial lesion characterization by tissue segmentation based on a 3D feature map. J Magn Reson Imag. 1997;37:457–469. doi: 10.1002/mrm.1910370325. [DOI] [PubMed] [Google Scholar]
- 7.Vinitski S, Gonzalez C, Knobler RL, Andrews D, Iwanaga T, Curtis M. Fast tissue segmentation based on a 4D feature map in characterization of intracranial lesions. J Magn Reson Imag. 1999;9:768–776. doi: 10.1002/(sici)1522-2586(199906)9:6<768::aid-jmri3>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 8.Kikinis R, Guttman CRG, Metcalf D, Wells WM, III, Ettinger MS, Weiner HL, Jolesz FA. Quantitative follow-up of patients with multiple sclerosis using MRI: technical aspects. J Magn Reson Imag. 1999;9:519. doi: 10.1002/(sici)1522-2586(199904)9:4<519::aid-jmri3>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 9.Guttman CRG, Kikinis R, Anderson MC, Jakab M, Warfield S, Killiany RJ, Weiner HL, Jolesz FA. Quantitative follow-up of patients with multiple sclerosis using MRI: reproducibility. J Magn Reson Imag. 1999;9:509. doi: 10.1002/(sici)1522-2586(199904)9:4<509::aid-jmri2>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 10.Udupa JK, Samarasekera S, Miki Y, van Buchem MA, Grossman RI. Multiple sclerosis lesion quantification using fuzzy-connectedness principles. IEEE Trans Med Imag. 1997;16:598–609. doi: 10.1109/42.640750. [DOI] [PubMed] [Google Scholar]
- 11.Evans WE, Relling MV, Rodman JH, Crom WR, Boyett JM, Pui C-H. Conventional compared with individualized chemotherapy for childhood acute lymphoblastic leukemia. N Engl J Med. 1998;338:499–505. doi: 10.1056/NEJM199802193380803. [DOI] [PubMed] [Google Scholar]
- 12.Steen RG, Gronemeyer SA, Kingsley PB, Reddick WE, Langston JW, Taylor JS. Precise and accurate measurement of proton T1 in human brain in vivo: validation and preliminary clinical application. J Magn Reson Imag. 1994;4:681–691. doi: 10.1002/jmri.1880040511. [DOI] [PubMed] [Google Scholar]
- 13.Kingsley PB, Ogg RJ, Reddick WE, Steen RG. Correction of errors caused by imperfect inversion pulses in MR imaging measurement of T1 relaxation times. Magn Reson Imag. 1998;16:1049–1055. doi: 10.1016/s0730-725x(98)00112-x. [DOI] [PubMed] [Google Scholar]
- 14.Reddick WE, Ogg RJ, Steen RG, Taylor JS. Statistical error mapping for reliable quantitative T1 imaging. J Magn Reson Imag. 1996;6:244–249. doi: 10.1002/jmri.1880060143. [DOI] [PubMed] [Google Scholar]
- 15.Steen RG, Gronemeyer SA, Taylor JS. Age-related changes in proton T1 values of normal brain. J Magn Reson Imag. 1995;5:43–48. doi: 10.1002/jmri.1880050111. [DOI] [PubMed] [Google Scholar]
- 16.Cho S, Jones D, Reddick WE, Ogg RJ, Steen RG. Establishing norms for age-related changes in proton T1 of human brain tissue in vivo. Magn Reson Imag. 1997;15:1133–1143. doi: 10.1016/s0730-725x(97)00202-6. [DOI] [PubMed] [Google Scholar]
- 17.Ostuni JL, Levin RL, Frank JA, DeCarli C. Correspondence of closest gradient voxels — a robust registration algorithm. J Magn Reson Imag. 1997;7:410–415. doi: 10.1002/jmri.1880070227. [DOI] [PubMed] [Google Scholar]
- 18.Reddick WE, Mulhern RK, Elkin TD, Glass JO, Merchant TE, Langston JW. A hybrid neural network analysis of subtle brain volume differences in children surviving brain tumors. Magn Reson Imag. 1998;16:413–421. doi: 10.1016/s0730-725x(98)00014-9. [DOI] [PubMed] [Google Scholar]
- 19.Reddick WE, Glass JO, Cook EN, Elkin TD, Deaton R. Automated segmentation and classification of multispectral magnetic resonance images of brain using artificial neural networks. IEEE Trans Med Imaging. 1997;16:911–918. doi: 10.1109/42.650887. [DOI] [PubMed] [Google Scholar]
- 20.Liang Z, MacFall JR, Harrington DP. Parameter estimation and tissue segmentation from multispectral MR images. IEEE Trans Med Imag. 1994;13:441–449. doi: 10.1109/42.310875. [DOI] [PubMed] [Google Scholar]
- 21.Zijdenbos AP, Dawant BM, Margolin RA, Palmer AC. Morphometric analysis of white matter lesions in MR images: method and validation. IEEE Trans Med Imag. 1994;13:716–724. doi: 10.1109/42.363096. [DOI] [PubMed] [Google Scholar]
- 22.Haselgrove J, Moore J, Wang Z, Traipe E, Bilaniuk L. A method for fast multislice T1 measurement: feasibility studies on phantoms, young children, and children with Canavan’s disease. J Magn Reson Imag. 2000;11:360–367. doi: 10.1002/(sici)1522-2586(200004)11:4<360::aid-jmri3>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 23.Ropele S, Stollberger R, Kapeller P, Hartung H-P, Fazekas F. Fast multislice T1 and T1sat imaging using a phase acquisition of composite echoes (PACE) technique. Magn Reson Med. 1999;42:1089–1097. doi: 10.1002/(sici)1522-2594(199912)42:6<1089::aid-mrm13>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]