Abstract
Background
Kinesin and dynein are the two families of microtubule-based motors that drive much of the intracellular movements in eukaryotic cells. Using a gene knockout strategy, we address here the individual function(s) of four of the 13 kinesin proteins in Dictyostelium. The goal of our ongoing project is to establish a minimal motility proteome for this basal eukaryote, enabling us to contrast motor functions here with the often far more elaborate motor families in the metazoans.
Results
We performed individual disruptions of the kinesin genes, kif4, kif8, kif10, and kif11. None of the motors encoded by these genes are essential for development or viability of Dictyostelium. Removal of Kif4 (kinesin-7; CENP-E family) significantly impairs the rate of cell growth and, when combined with a previously characterized dynein inhibition, results in dramatic defects in mitotic spindle assembly. Kif8 (kinesin-4; chromokinesin family) and Kif10 (kinesin-8; Kip3 family) appear to cooperate with dynein to organize the interphase radial microtubule array.
Conclusion
The results reported here extend the number of kinesin gene disruptions in Dictyostelium, to now total 10, among the 13 isoforms. None of these motors, individually, are required for short-term viability. In contrast, homologs of at least six of the 10 kinesins are considered essential in humans. Our work underscores the functional redundancy of motor isoforms in basal organisms while highlighting motor specificity in more complex metazoans. Since motor disruption in Dictyostelium can readily be combined with other motility insults and stresses, this organism offers an excellent system to investigate functional interactions among the kinesin motor family.
Background
Dictyostelium discoideum is a compact amoeba that spends much of its natural existence crawling through the soil, searching for and ingesting bacteria. When food sources are exhausted, individual amoebae trigger a developmental program that initiates both inter and intracellular signaling, to aggregate ~100,000 amoebae and form a multicellular mass. Each cell within this mass undergoes multiple adhesions and conformational changes, forming a cooperative slug that can migrate to new areas. The slug undergoes further multicellular differentiation to form supportive stalk cells, a rudimentary immuno-like surveillance system, and regenerative spores that resist environmental stresses. This dualistic life cycle and its associated transitions (single cell to metazoan organism) have made Dictyostelium an attractive model in which to study cell motility, signal transduction, and a relatively simple developmental program (reviewed in [1], see also [2]).
Motility-wise, Dictyostelium behaves in a manner similar to that of many vertebrate cells (crawling, sensing, and engulfing targets, robust intracellular movements). Yet, this organism clearly retains a simplicity associated with its relatively small and compact genome, and exhibits features commonly seen in protozoa and fungi (for example, an intranuclear spindle for cell division). Characterization of the actin cytoskeleton in Dictyostelium has led to the identification of actin binding proteins, multiple myosin motors, and signaling cascades whose functions are conserved among eukaryotic cells. Preliminary characterization of the microtubule-associated network has revealed a level of complexity intermediate between some of the simple single-celled eukaryotes and metazoans. For example, the machinery in Dictyostelium that drives movement along microtubules contains 14 motors (13 kinesin ATPases, 1 dynein ATPase, [3,4]); twice the number found in Saccharomyces cerevisiae [5], but less than a quarter of the number encoded in the human genome [6]. Paradoxically, deletions of kinesins whose homologs are essential for vertebrate activities have produced relatively mild phenotypes in Dictyostelium. Are these results reflective of Dictyostelium's unique life cycle? Or do they reveal core functional redundancies and interactions that, like the actin system work, can be utilized to understand microtubule-based motor action in more complex systems?
Because of their homologies to motors with known functions, 12 of the 13 kinesins in Dictyostelium can be placed within existing kinesin families and subdivided into two functional categories (Fig. 1). Four members are closely related to kinesins in metazoans that have organelle transport functions, while eight members are related to motors significant for mitotic events. To date, six individual kinesins have been genetically disrupted by homologous recombination in Dictyostelium (kif's 1, 2, 5, 7, 12, 13), producing varied effects on cell function. Disruption of kif12 (kinesin-6; MKLP family) resulted in significant cytokinetic defects [7,8]. kif12- cells failed to divide in suspension, but were able to undergo non-mitotic cytofission on a surface to enable their propagation. For each of the other five kinesins, single deletions did not produce significant effects on cell development or viability. However, closer examination revealed defects suggestive of redundant or cooperative effects with other motor activities. For example, kif1- cells (kinesin-3, Unc104) showed a 62% decrease in overall organelle movements in vivo [9]. There was a 90% reduction in plus end-directed motility as measured in an in vitro assay, but no demonstrable effects on mitochondrial movement. When examined in detail, kif13- cells (kinesin-5, BimC/Eg5) showed an increased rate and decreased stability of mitotic spindle elongation [10]. When combined with an otherwise viable dynein perturbation (380 K, [11]), kif13-/380 K cells were unable to divide properly. kif2- (kinesin-14, ncd/kar3), kif5- (kinesin-1, KHC), and kif7- (kinesin-1, KHC) cells showed mitotic, actin-filament, and developmental defects respectively, but only when challenged with overexpression or competition assays that further stressed the individual cells [12-14]. In contrast, mammalian homologs of kif12, kif13, kif2 (and kif4, kif8, kif10 in this report) were found to be essential for cell viability [15]. Thus the non-lethal disruption of these genes in Dictyostelium offers us the opportunities to examine basal motor activities and interactions, to further understand the motors functions and regulation. We address here the consequences of individual disruption of four kinesin genes in Dictyostelium, and we contrast the functional redundancies among such motors in single-celled organisms with their functional specificity in metazoan organisms.
Figure 1.
Kinesin Gene Family in Dictyostelium. Schematic representation of the 13 kinesin motors, identified by functional and/or sequence analyses (adapted from ref [3]). The motor domain is indicated in blue, along with the common family name. The remaining neck/linker/tail domains are drawn to scale in green. Both formal and Dictyostelium-specific gene names are listed. Preexisting gene knockouts for six of the kinesins are indicated by the black X's (kif5, kif7, kif1, kif13, kif12, kif2, [7, 10, 12-14, 46]): new gene disruptions reported in this paper are indicated by the red X's.
Results
kif4, kif8, kif10, and kif11 are Not Essential Genes in Dictyostelium
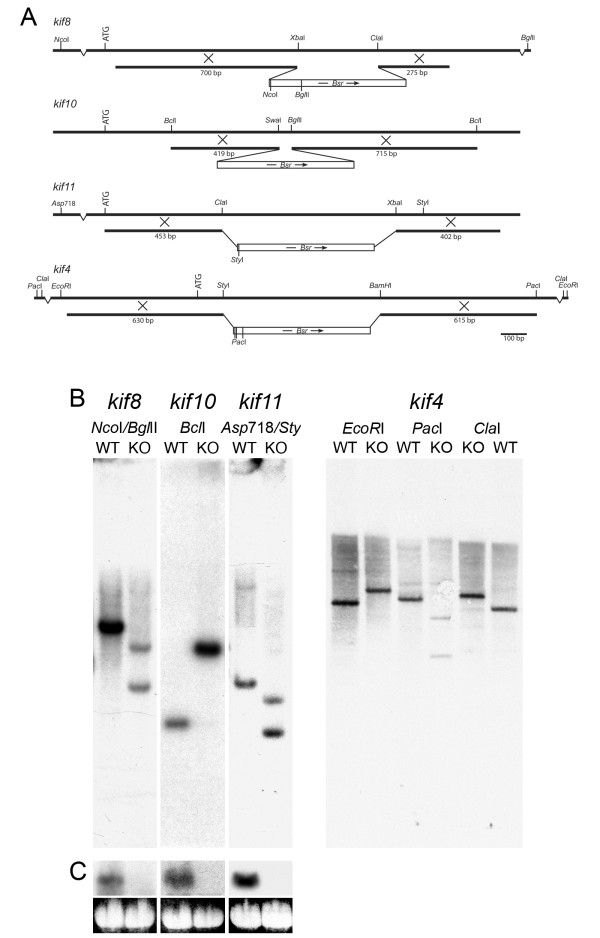
Genomic fragments of kif4, kif8, kif10, and kif11 were isolated by PCR amplification of wild-type AX-2 cell DNA, and were used to generate deletion-mutant alleles for their respective kinesin genes (Fig. 2A, Methods). A similar strategy was followed for each gene. Internal restriction enzyme sites of the amplified fragments were utilized to replace coding sequence with a 1.6-kb blasticidin resistance cassette. Genomic sequences flanking the cassette (275–763 bp) targeted the insertion of these mutant alleles into their wild-type gene by homologous recombination, and thus disrupted transcription of the native gene product. Integration of the mutant alleles at their correct sites was confirmed by PCR and Southern blot analyses (Fig. 2B). Northern blot analysis further confirmed loss of full-length mRNA in kif8-, kif10-, and kif11- transformants (Fig. 2C). There was no evidence for shorter transcripts that would indicate partial expression of the sequence upstream of the integration site for these three clones. Northern blots of kif4- were performed, but we were unable to detect the native message in wild-type cells. Very low message levels of the kif4 gene, undetectable during log phase growth has been previously reported [12], and is consistent with tight cell-cycle regulation of kinesin-7/CENP-E homologs in other organisms [16,17].
Figure 2.
Kinesin Gene Disruptions. (A). Schematics of constructs, showing details of the homologous regions and relevant enzymes used to target recombination and to confirm disruption. The position of the bsrr cassette used for selection is also shown. ATG indicates the start of the protein-coding region. (B). Southern blot comparisons of wild-type AX-2 control (WT), kif8, kif10, kif11, and kif4 knockout (KO) DNAs. DNA was digested with the indicated enzymes and probed with the entire amplified kinesin gene fragment. Since we were unable to detect kif4 mRNA in wild type cells (see text), we include multiple digests in this panel to demonstrate disruption. All resulting DNA fragments are as predicted from the wild-type and recombination sequences. (C). Northern analysis of AX-2, kif8, kif10 and kif11 knockout cells. Top panel shows mRNA hybridization, bottom panel shows a loading control (4.1 kb 26S rRNA). Note the abundant level of kinesin message in wild-type cells, but the complete absence of message in the disrupted clones.
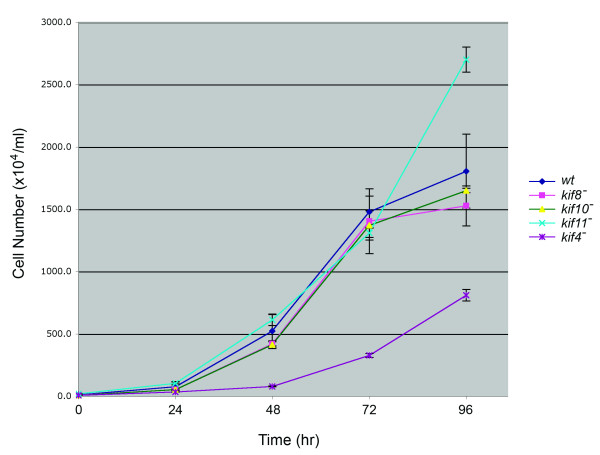
Individual disruptions of all four kinesins resulted in viable cells with no gross morphological defects. Cells were comparable in size with wild-type controls and retained the ability to undergo a complete developmental cycle so as to generate viable spores (data not shown). kif8-, kif10-, and kif11- cells grew at log-phase rates indistinguishable from wild-type AX-2 cells (Fig. 3). Interestingly, kif4- cells grew significantly more slowly than the wild type, or the other three kinesin-null strains. This reduced growth rate was observed both in solution (Fig. 3) and on solid support in Petri dishes. kif11- cells appeared to reach stationary phase at a higher density than the other strains, but otherwise showed no difference in viability or morphology than wild type cells.
Figure 3.
Comparison of Growth Rates. Dictyostelium enters stationary phase at ~1–3 × 107 cells/ml, a point reached here after 4 days of growth. Wild-type, kif8-, kif10-, and kif11- cells exhibit very similar rates of logarithmic phase growth, with a doubling time in axenic medium of 8–9 hrs. Kif4- cells grow significantly more slowly than any of the three kinesin nulls as well as the wild-type control. Kif11- cells appear to delay their entry into stationary phase (96 hrs), but remain within the normal range of maximum cell density reported for Dictyostelium.
Microtubule Distributions Appear Normal in Kinesin Null Cells
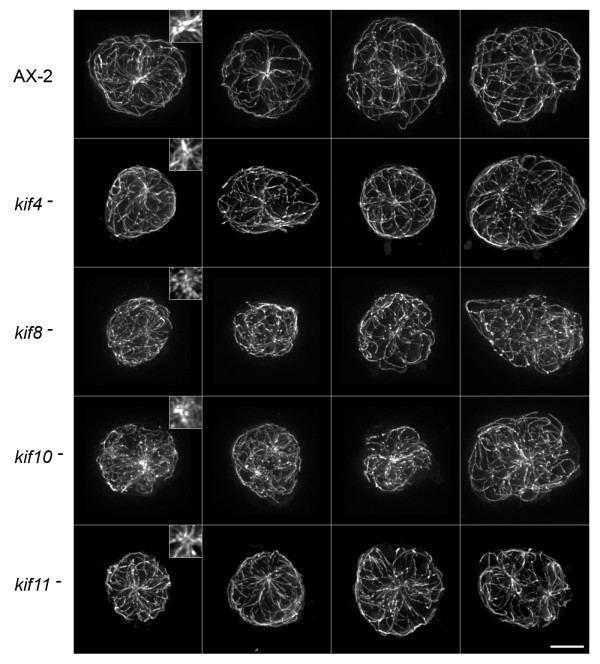
Since these motors interact with and move along microtubules, and since motor activity is responsible for significant microtubule movement in Dictyostelium, we stained fixed cells with tubulin antibodies to determine whether deletions led to any aberrant microtubule distributions. Figure 4 shows a gallery of interphase cells from wild-type AX-2 cells and from the four kinesin mutant lines. All cells display the conventional radial distribution of microtubules that emanate from a centrally located organizing center (MTOC), and, in the few examples shown of binucleate cells, the two centrosomes remain spatially distinct. Interestingly, MTOCs are less distinct in the kif8- and kif10- cell lines than they are in wild-type, kif4-, or kif11- cells. MTOC's are present in the individual image slices of kif8- or kif10- cells, but when summed as projections, the ring like appearance of the Dictyostelium centrosome is either less apparent or the microtubules do not seem as tightly focused into this structure (inserts in Fig. 4). There were no obvious morphological defects in mitotic microtubule arrays in any of the four kinesin null strains (not shown), although this qualitative observation should be examined in greater detail (see below).
Figure 4.
Microtubule Patterns in Interphase Cells. Maximum intensity projections of deconvolved image stacks showing interphase microtubule distributions in wild-type AX-2 and kinesin-null cells (fixed cells imaged by indirect immunofluoresence, using a tubulin antibody). Each row displays four examples of the cell strain indicated on the left. Inserts in the first column show 2X enlargements of the MTOC area. For AX-2, kif4- and kif11- cells, a distinctive ring-like appearance of the centrosome can be seen. This feature is less apparent in the kif8- and kif10- cells, moreover, a convergence of microtubules into the centrosome in the kif10- cell is less obvious. Scale bar = 5 μm.
Kif8, Kif10, and Dynein Cooperate to Organize Interphase Microtubules
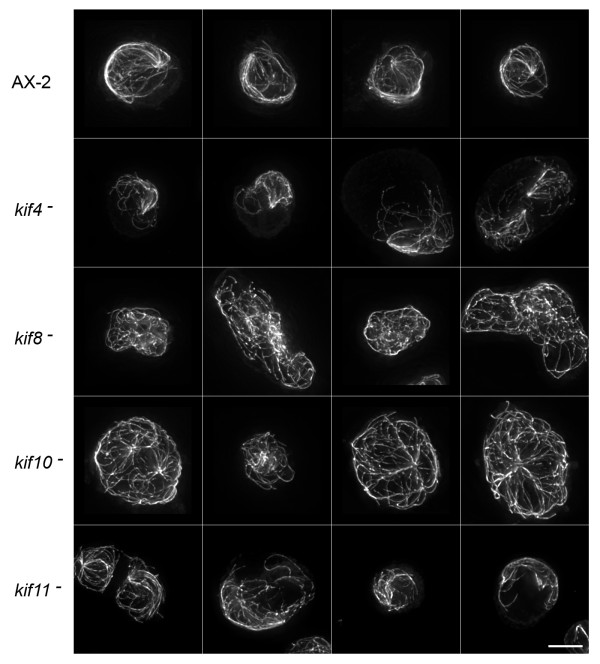
We previously characterized a dominant-negative, dynein-mediated defect whereby the entire microtubule network in interphase cells became motile and circulated throughout the cytoplasm (380 K cells) [11,18,19]. The directionality of such motion suggested a role for a kinesin-like motor that pushes against microtubules. To address whether any of the kinesins examined here could have been responsible for generating that aberrant motility, we expressed the 380-kDa dynein motor fragment in each of the four kinesin null backgrounds. The distinctive comet-like microtubule phenotype was found in 82.3% of the control 380 K cells, and in roughly half of the kif4- and kif11- cell lines (55% and 39.8%, respectively) (Fig. 5, Table 1). However, expression of the 380-kDa polypeptide in either kif8- or kif10- cells, at levels comparable to affect microtubule organization in control cells (Fig. 6, see also [18,20]), failed to create aberrant microtubule arrays (0%, 4.6%, respectively) (Fig. 5, Table 1). These results suggest that dynein, Kif8, and Kif10 cooperate in producing lateral microtubule motions that organize the interphase microtubule distribution.
Figure 5.
Microtubule Patterns in Dynein-Inhibited Interphase Cells. Interphase microtubule distributions, similar to Figure 4 except that each cell line is also transformed with the dynein motor domain expression plasmid (380 K). AX-2, kif4- and kif11-cells display the distinctive 380 K comet-tail phenotype. However, note the relatively normal, radial distribution of microtubules in the kif8- and kif10- backgrounds. Scale bar = 5 μm.
Table 1.
MT Array Morphology in Interphase Cells
| Cell-type | Radial MT | Comet-Tail MT | Comets (%) |
| AX-2 Control | 588 | 19 | 3.1% |
| 380 K | 60 | 279 | 82.3% |
| kif8- | 461 | 1 | 0% |
| kif8-/380 K | 451 | 0 | 0% |
| kif10- | 583 | 12 | 2.0% |
| kif10-/380 K | 475 | 23 | 4.6% |
| kif11- | 212 | 26 | 10.9% |
| kif11-/380 K | 127 | 84 | 39.8% |
| kif4- | 189 | 9 | 4% |
| kif4-/380 K | 68 | 84 | 55% |
Numbers represent individual fixed cells, either containing a normal, radial interphase microtubule array (Radial MT), or a mutant comet-like microtubule array (Comet-Tail MT) resulting from dynein motor overexpression (380 K) in different control or kinesin null cell stains. % Comet refers to the percentage of total cells that showed the distinctive mutant phenotype.
Figure 6.
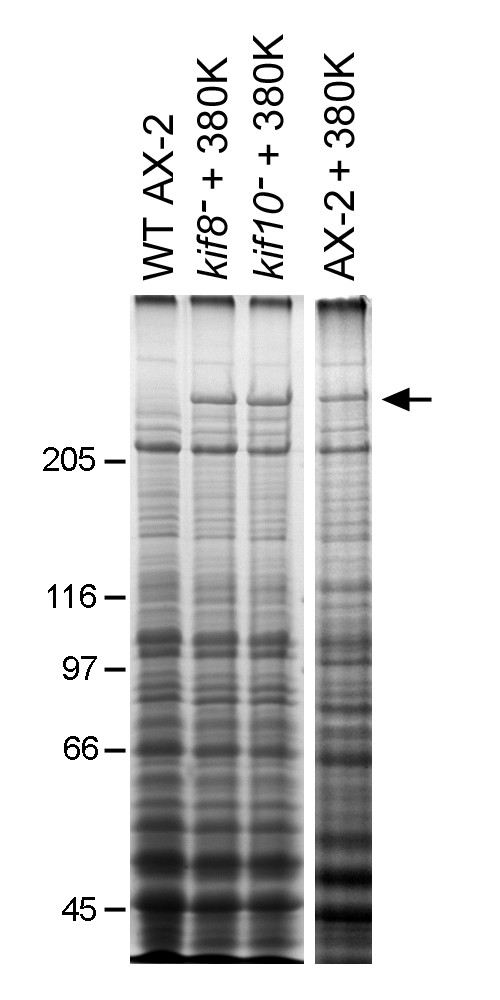
Expression of the Dynein Motor in kif8- and kif10- Cells. Coomassie-stained gel lanes showing high speed supernatants from wild-type control cells, and from kif8-, kif10-, and AX-2 cells that have been transformed with the 380 K expression plasmid. The panel confirms the expression of the 380 kDa dynein motor domain polypeptide in kif8- and kif10- cells (arrow), at a level comparable to the AX-2 cells shown in Figure 5.
Kif4 and Dynein Cooperate in Mitotic Spindle Assembly
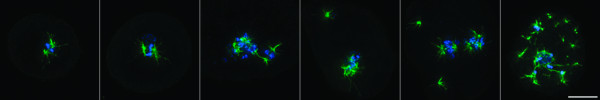
The kif4- deletion is notable, since these cells grow significantly slower than the three other kinesin knockout strains. Interestingly, kif4- cells appear normal in shape and size, indicating that the slow growth is not the result of structurally defective cell division (karyo- or cytokinetic failure). When kif4- cells were transformed with the 380-kDa expression plasmid, the cell growth rate was reduced even further. While cells grown on a solid surface gradually increased in number, suspension cultures of kif4-/380 K cells remained at their initial cell density, even when measured for as long as a week. Although defective mitotic figures were not evident in the kinesin-alone knockouts, there was a significant increase in aberrant spindle formation in kif4-/380 K cells. 66% of the observed mitotic kif4-/380 K cells (10/15) showed division defects of various types, including multi-polar spindles and supernumerary or fragmented centrosomes (Fig. 7). Although normal-appearing spindles were found among the cell population, it is probable that defective spindle assembly plays a major role in the decreased growth rate of these cells. The results described here indicate that Kif4 and dynein cooperate in some aspect of mitotic spindle assembly; closer evaluation of cell division in these cells is in progress.
Figure 7.
Spindle Morphologies in kif4-/380 K cells. Representative examples of normal and aberrant mitotic spindles in kif4-/380 K cultures. The two left-most panels show cells with normal appearing metaphase arrangements, with bipolar spindle MTs (in green) that flank condensed chromosomes (blue). These two figures are indistinguishable from mitotic wild type cells. The remaining four panels show examples of aberrant mitotic figures (multipolar spindles, supernumerary or fragmented spindle poles, asynchronous chromatin condensation) that are not normally found in wild-type, kif4-, or 380 K cells alone. This panel suggests that a combination of dynein and Kif4 (kinesin-7, CENP-E) activity is required for proper spindle assembly. Scale bar = 5 μm.
Discussion
We have presented gene deletions for four of the 13 kinesin family members in Dictyostelium, and have described the effects of these deletions on cell growth and viability. Individually, none of the four gene products is essential for cell viability nor do the proteins play critical roles in this organism's ability to undergo chemotaxis or to develop upon starvation. The knockout strains do, however, show subtle defects suggesting that many of the key forms of intracellular motility essential for Dictyostelium biosynthesis and reproduction are supported by more than one motor protein.
In wild-type Dictyostelium cells, both plus end-directed microtubule pushing, and minus end-directed pulling forces are important for maintenance of centrosome position and the radial distribution of interphase microtubules [18,21]. If minus end-directed dynein motility is impaired, a kinesin-like activity appears to dominate and push both the centrosome and microtubule array throughout the cytoplasm [19]. Here we have identified two kinesins, kif8 (kinesin-4 family) and kif10 (kinesin-8 family), that appear to collaborate with dynein in this organization process. In other eukaryotic cells, kinesin-4 motors participate in a number of diverse activities [22]. One subset of kinesin-4 family members (KIF4) function during mitotic events, with chromatin- and spindle-associated motors that organize bipolar microtubule assemblies and facilitate chromosome alignment [23]. Other subsets of kinesin-4 motors (e.g., KIF21) appear to power interphase organelle transport in cultured cells such as fibroblasts and post-mitotic neurons [24,25]. The single Dictyostelium kinesin-4 (kif8) is a divergent member of this family, the motor domain is most closely homologous with KIF4 subfamily, yet it contains carboxy-terminal WD-40 repeat motifs in the heavy chain tail that are characteristic of the KIF21 subfamily [3,22]. The kinesin-8 family of motors (kif10 in Dictyostelium) is thought to mediate chromosome movements through a combination of translocation and microtubule depolymerization activities (recently reviewed in [26], see also [27,28]. The S. cerevisiae isoform (Kip3) has previously been shown to cooperate with dynein in positioning mitotic spindles through cortically mediated force production and through control of microtubule length [27,29,30]. Deletions of kinesin-8 isoforms in Schizosaccharomyces pombe also suggest a combined force and length control mechanism that positions nuclei and spindles through microtubule-cortex interactions [31,32]. In the absence of either kinesin-4 or kinesin-8 in Dictyostelium, we are unable to induce the distinctive centrosome movements via dynein motor overexpression. It is conceivable that Kif8 and Kif10 counterbalance dynein-mediated forces through force-production or anchoring activities at the cell cortex (e.g. kinesin-8) and via lateral microtubule-microtubule interactions (e.g. kinesin-4) that supply sufficient rigidity to allow plus end-directed motors to effectively push (and not simply bend) microtubules. In wild-type Dictyostelium, the balance between opposing dynein and kinesin motor activities serves to reinforce the centrosome position and help maintain the radial character of the interphase microtubule array as these cells crawl around and change shape.
Disruption of the kinesin-7 motor (CENP-E) in the mouse is embryonic lethal [33]; this motor is thought to be essential for the proper connection between kinetochores of condensed chromosomes and the mitotic spindle [34]. In contrast, neither member of the kinesin-7 family in Dictyostelium (Kif4, Kif11) is essential for mitosis, although removal of Kif4, the isoform that is most homologous to the vertebrate kinetochore CENP-E greatly affects cell growth rate. Preliminary characterization of Kif4 suggests that this motor functions together with dynein in organizing spindle assembly during cell division. While the motor domain of Kif11 is homologous with the kinesin-7 family [3], this polypeptide is significantly shorter and expressed at a much higher level than other CENP-E-like proteins. Outside of a minor enhancement of stationary phase cell density, removal of this motor has no obvious effect on cell viability or function. Closer inspection of each kinesin, and of cells lacking their expression will be required before we can fully understand their individual function(s)
Our study here extends previous work from several laboratories that, taken together, have individually deleted 10 of the total 13 kinesins in Dictyostelium [7,9,10,12-14]. All of these deletions have proven to generate cell lines that can survive over multiple generations of growth, indicating that none of these 10 kinesin motors is immediately required for cell viability. Although the Kif12 disruption (kinesin-6, MKLP) produced significant defects in cytokinesis, mutant cells were still able to undergo some form of division that allows strain propagation [7]. The only, potentially essential, kinesin gene reported so far in Dictyostelium encodes one of the organelle transporter motors, kif3 (kinesin-1 family)[35]. Kif3 can be isolated biochemically and shown capable of powering microtubule gliding, but efforts by Röhlk et al, [35] and in our own lab (Nag, Tikhonenko, and Koonce, unpublished) have not yet yielded viable cells lacking this motor. The resiliency of Dictyostelium to motor disruptions is similar to systematic analyses of kinesin isoforms in S. cerevisiae, where all six kinesin-related motors (and one dynein isoform) can be individually deleted without loss of viability [5]. The yeast work provided a major guiding principle, for it was the first to suggest that high degree of functional redundancy is present among kinesin family members, and that deletion of motor combinations is required to inhibit cell division. Although, to our knowledge, complete survey disruptions have not yet been reported in other simple eukaryotes, there are clear indications of motor redundancy in some cell models such as S. pombe [36], Aspergillus nidulans, [37]and Ustilago maydis [38]. The kinesins in Dictyostelium likewise possess overlapping functions.
The evolutionary transition from very simple single-celled eukaryotes into metazoans generally correlates with an increased number of gene family members. S. cerevisiae contains 6 kinesin genes [5]; S. pombe has 8 [38]; U. maydis has 10 [38]; and A. nidulans has 11 [37]). In contrast, even the primitive metazoan Caenorhabditis elegans has 21 kinesin genes [39]; Drosophila melanogaster has 25 [40], and the human genome codes for 45 kinesins [6] (obviously there are exceptions to this general trend, e.g Giardia lamblia contains as many as 23 kinesin genes [41]). One might expect that larger gene families enhance the opportunity for redundant mechanisms, but at least for kinesins, the opposite seems to hold true (summarized in Table 2). Despite the greater number of motor sequences, human cell lines appear far more sensitive to disruption of individual kinesins than do yeast or Dictyostelium. Cell division and organelle transport is likely more complex in metazoans, entailing open nuclear spindles, strict spindle position and orientation requirements, cell-type specificities, and many transport features that are not required in unicellular organisms. Thus one could argue that in metazoans, each kinesin motor is custom built for one selective function while in simpler eukaryotes, the motors retain a more generic ability to power microtubule-based transport in a variety of contexts. Understanding the functional details of the same kinesin ortholog in different organisms will therefore define sequence motifs that lead to essential and non-essential functions of basically the same engine, and thus will lead to a more complete understanding of how the motor protein operates.
Table 2.
Mitotic kinesin disruptions in simple eukaryotes vs metazoans.
| Kinesin Family | S. cerevisiae | Dictyostelium | C. elegans | Drosophila | Human |
| Mitotic Function | |||||
| Kinesin-4 (Chromokin) | Kif8 | Klp-19 | Klp3A | Kif4A, Kif4B | |
| Kinesin-5 (BimC/Eg5) | Cin8, Kip1 | Kif13 | BMK1 | Klp61F | Eg5 |
| Kinesin-6 (MKLP) | Kif12 | Zen-4 | Pavarotti | MKLP1, MKLP2 | |
| Kinesin-7 (CENP-E) | Kip2 | Kif4, Kif11 | CENP-E Meta | CENP-E | |
| Kinesin- 8 (Kip3) | Kip3 | Kif10 | Klp67A | Kif18 | |
| Kinesin-13 (MCAK) | ? | Klp-7 | KLP10A | Kif2A, 2B, MCAK | |
| Kinesin-14 (NCD/Kar3) | Kar3 | Kif2 | Klp-3,15,16,17 | Ncd | KifC1 |
| Other Function | |||||
| Kinesin-3 (Unc104) | Kif1 | Kif14 | |||
| Kinesin-10 (Nod) | Nod | Kid | |||
| Kinesin12 (Xklp2) | Klp-18 |
Normal text (viable), italicized text (not viable) differentiates the individual kinesin-isoform disruptions (knockout, knockdown, or mutation) and their effects on mitosis. The bold text for the kinesin-6 family members indicates cytokinesis defects. In this case, cells can proceed through one or more divisions, but longer term, the mutated protein is essential for organism viability. References for S.c [5], D.d[7-10, 12-14], C.e [39, 47-51], D.m [40], and H.s [15, 52].
Conclusion
Analysis of the kinesin gene family in Dictyostelium suggests that a significant level of functional redundancy or overlap exists among the organism's motor activities. This result is similar to findings from functional analyses performed in basal organisms such as yeast and fungi, but it contrasts sharply with the roles of individual motors in metazoans. At first glance, most of the kinesins in Dictyostelium can be deleted individually without penalty to growth or viability. Yet, upon closer scrutiny or in cases where we impose under additional stresses, we can discern clear phenotypic changes in the cell that provide insight into motor function that may not be obvious in other organisms. Given its greater complement of motor isoforms, and its greater utility of microtubule function relative to other basal eukaryotes, Dictyostelium offers an interesting model in which to investigate functional interactions and the regulation of multiple motor proteins.
Methods
Molecular Genetics
Kinesin gene sequences were obtained from the dictybase website (see Availability and requirements section). The following primer combinations were used to amplify kinesin gene fragments from AX2 cell genomic DNA; also listed are the downstream kinesin gene-specific primers used for screening recombinants:
kif4 (DDB0191404)
Forward: 5'CGCAAGCTTAGCCACCAAGACCATTACTTGGACCA 3' (-501 to -476)
Reverse: 5'CGCGAGCTCTTAAACTACCACCAATTATTGCGTCATT 3' (+1318 to +1345)
Screen: 5'CATCATCATCCTCTTCACCACTACTATT 3' (+1501 to +1528)
kif8 (DDB0191403)
Forward: 5'CGCGGATCCGGGTTGCATTAAGAGTTAGACCC 3' (+44 to +66)
Reverse: 5'CCCAAGCTTGAATCGGCAGGACTAACACATGC 3' (+ 1302 to +1324)
Screen: 5'GATTGGTTAATACACACCTAATTG 3' (+1381 to +1404)
kif10 (DDB0215386)
Forward 5'CGCGGATCCTGATCAATATGCAACTCAAGAAGAAG 3' (+249 to +274)
Reverse 5'CCCAAGCTTGATCATTGTCATCATCATCATC 3' (+1408 to +1429)
Screen: 5'GTATCATTGATTCATCATTATCCCT 3' (+1501 to +1525)
kif11 (DDB0201556)
Forward: 5'CGCGGATCCGAATGAACGAGAATATATCGGTTAGC 3' (-2 to +24)
Reverse: 5'CCCAAGCTTCCATTACCACTACCACTACCACCT 3' (+1497 to +1520)
Screen: 5'TGACTTGGTGAAACAAATGTTGATC 3' (+1532 to +1556)
+1 of the numbering scheme refers to the position A of the ATG start codon. Restriction enzyme sites were engineered into the ends of each primer (BamH1, HindIII or Sac1, shown in bold type) to facilitate cloning of the amplified DNA into a pUC19 host plasmid, and (in most cases) to excise the DNA construct for transformation. Each construct was sequenced to confirm the identity of the kinesin fragment. Native restrictions sites (Fig. 2) were used to excise and replace an internal fragment of the kinesin sequences (47–669 bp) with a 1.6-kb blasticidin resistance cassette (Bsrr) (SmaI digest) from pLRBLP [42], obtained from the Dictyostelium Stock Center (see Availability and requirements section for URL). Final constructs were again sequenced to determine the orientation of the Bsrr cassette (diagramed in Fig. 2). The kif8 construct was designed to terminate message coding at S202; kif10 at N223; kif11 at S151; and kif4 at W45. In all cases, these disruptions occur upstream of the microtubule-binding domain of the motor.
Standard molecular biology procedures were followed for DNA isolation, manipulation, and blotting. RNA was isolated using the RNeasy kit from Qiagen, following the manufacturer's instructions. kif8, kif10, and kif11 blots were probed with 32P-labeled DNA. the kif4 Southern blot was performed using chemiluminescence procedures (ECL, Amersham Biosciences). All blots (Southern and Northern) were probed with the initial amplified genomic target corresponding to the relevant kinesin clone, as indicated above and in Figure 2A.
Cell Transformation
A calcium phosphate procedure was used to transform Dictyostelium AX-2 cells, with 15 μg of linearized DNA per near confluent 10-cm dish (107 cells) [43]. Transformants were selected with 5 μg/ml blasticidin. Individual colonies were picked with a pipette into 24 well plates, and were screened by PCR for homologous recombination. Amplification of a 1.6-kb target with a primer internal to the Bsrr marker (5' GAATGGCAAGTTAGTCAAAACTACG 3') and a primer downstream of the recombination site (indicated above for each kinesin sequence) was used to initially identify positive recombinants. Cells from positive colonies were further purified by serial dilution, and were again confirmed by PCR with downstream and upstream primer combinations. For dynein disruptions, we introduced a motor domain expression plasmid (aa 1384–4725), into kinesin null cells by either a CaPO4 or an electroporation method [44]. kif-/380 K expressing cells were selected with 10 μg/ml G-418 (geneticin, Sigma Chemical Co).
Light Microscopy
Cells were flattened on glass coverslips using an agarose sheet, fixed with formaldehyde, labeled with a tubulin antibody [45], and in some cases Hoechst 33342, as described in [11]. Z-series of images were obtained on a DeltaVision light microscopy workstation and were deconvolved using softWoRx 2.5 (Applied Precision, Issaquah, WA). Maximum intensity projections were compiled using ImageJ (NIH); figures were assembled in Adobe Photoshop. For cell growth measurements, triplicate 100-ml cultures were seeded with 9 × 104 cells/ml, shaken at 200 rpm at RT, and counted with a hemocytometer every 24 hr. Growth curves were calculated and displayed with Microsoft Excel; error bars indicate standard deviation.
Availability and requirements
The Dictybase website: http://dictybase.org/
Dictyostelium Stock Center: http://dictybase.org/StockCenter/StockCenter.html
Authors' contributions
DKN designed the knockout constructs and participated in the molecular genetic studies. IT participated in the gene knockout and screening work, performed cell culture analyses and 380 K dynein expression analyses. IS participated in the molecular genetic analysis. MPK performed the light microscopy, participated in the molecular genetic studies, assembled the figures, and wrote the manuscript. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
We are grateful to the efforts at http://dictybase.org/ to archive and annotate Dictyostelium sequence information, and to the Dictyostelium Stock Center Resource for plasmids. Drs. Alexey Khodjakov and Conly Rieder provided valuable discussion and assistance with the light microscopy. We appreciate the use of Wadsworth Center's Molecular Genetics Core for DNA sequencing. This work was supported in part by the NSF (MCB-0542731 to MPK).
Contributor Information
Dilip K Nag, Email: nagd@wadsworth.org.
Irina Tikhonenko, Email: tikhonen@wadsworth.org.
Ikko Soga, Email: ikkosoga@gmail.com.
Michael P Koonce, Email: koonce@wadsworth.org.
References
- Kessin RH. Dictyostelium. Evolution, Cell Biology, and the Development of Multicellularity. Cambridge University Press; 2001. [Google Scholar]
- Chen G, Zhuchenko O, Kuspa A. Immune-like Phagocyte Activity in the Social Amoeba. Science. 2007;317:678–681. doi: 10.1126/science.1143991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollmar M, Glockner G. Identification and phylogenetic analysis of Dictyostelium discoideum kinesin proteins. BMC Genomics. 2003;4:47. doi: 10.1186/1471-2164-4-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonce MP, Grissom PM, McIntosh JR. Dynein from Dictyostelium: primary structure comparisons between a cytoplasmic motor enzyme and flagellar dynein. J Cell Biol. 1992;119:1597–1604. doi: 10.1083/jcb.119.6.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt ER, Hoyt MA. Mitotic motors in Saccharomyces cerevisiae. Biochim Biophys Acta (BBA) – Mol Cell Res. 2000;1496:99–116. doi: 10.1016/S0167-4889(00)00012-4. [DOI] [PubMed] [Google Scholar]
- Miki H, Setou M, Kaneshiro K, Hirokawa N. All kinesin superfamily protein, KIF, genes in mouse and human. Proc Nat Acad Sci (USA) 2001;98:7004–7011. doi: 10.1073/pnas.111145398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshmikanth GS, Warrick HM, Spudich JA. A mitotic kinesin-like protein required for normal karyokinesis, myosin localization to the furrow, and cytokinesis in Dictyostelium. Proc Natl Acad Sci USA. 2004;101:16519–16524. doi: 10.1073/pnas.0407304101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Lakshmikanth GS, Spudich JA, De Lozanne A. The Localization of Inner Centromeric Protein (INCENP) at the Cleavage Furrow Is Dependent on Kif12 and Involves Interactions of the N Terminus of INCENP with the Actin Cytoskeleton. Mol Biol Cell. 2007;18:3366–3374. doi: 10.1091/mbc.E06-10-0895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock N, de Hostos EL, Turck CW, Vale RD. Reconstitution of membrane transport powered by a novel dimeric kinesin motor of the Unc104/KIF1A family purified from Dictyostelium. J Cell Biol. 1999;147:493–506. doi: 10.1083/jcb.147.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikhonenko I, Nag DK, Martin N, Koonce MP. Kinesin-5 contributes to mitotic spindle stability in Dictyostelium. 2008. under review.
- Koonce MP, Samsó M. Overexpression of cytoplasmic dynein's globular head causes a collapse of the interphase microtubule network in Dictyostelium. Mol Biol Cell. 1996;7:935–948. doi: 10.1091/mbc.7.6.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hostos EL, McCaffrey G, Sucgang R, Pierce DW, Vale RD. A Developmentally Regulated Kinesin-related Motor Protein from Dictyostelium discoideum. Mol Biol Cell. 1998;9:2093–2106. doi: 10.1091/mbc.9.8.2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwai S, Ishiji A, Mabuchi I, Sutoh K. A Novel Actin-bundling Kinesin-related Protein from Dictyostelium discoideum. J Biol Chem. 2004;279:4696–4704. doi: 10.1074/jbc.M308022200. [DOI] [PubMed] [Google Scholar]
- Iwai S, Suyama E, Adachi H, Sutoh K. Characterization of a C-terminal-type kinesin-related protein from Dictyostelium discoideum. FEBS Letters. 2000;475:47–51. doi: 10.1016/S0014-5793(00)01619-7. [DOI] [PubMed] [Google Scholar]
- Zhu C, Zhao J, Bibikova M, Leverson JD, Bossy-Wetzel E, Fan JB, Abraham RT, Jiang W. Functional analysis of human microtubule-based motor proteins, the kinesins and dyneins, in mitosis/cytokinesis using RNA interference. Mol Biol Cell. 2005;16:3187–3199. doi: 10.1091/mbc.E05-02-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KD, Coulson RM, Yen TJ, Cleveland DW. Cyclin-like accumulation and loss of the putative kinetochore motor CENP-E results from coupling continuous synthesis with specific degradation at the end of mitosis. J Cell Biol. 1994;125:1303–1312. doi: 10.1083/jcb.125.6.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen TJ, Li G, Schaar BT, Szilak I, Cleveland DW. CENP-E is a putative kinetochore motor that accumulates just before mitosis. Nature. 1992;359:536–539. doi: 10.1038/359536a0. [DOI] [PubMed] [Google Scholar]
- Koonce MP, Kohler J, Neujahr R, Schwartz JM, Tikhonenko I, Gerisch G. Dynein motor regulation stabilizes interphase microtubule arrays and determines centrosome position. EMBO J. 1999;18:6786–6792. doi: 10.1093/emboj/18.23.6786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito DA, Strauss J, Magidson V, Tikhonenko I, Khodjakov A, Koonce MP. Pushing forces drive the comet-like motility of microtubule arrays in Dictyostelium. Mol Biol Cell. 2005;16:3334–3340. doi: 10.1091/mbc.E05-01-0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonce MP. Dictyostelium, a model organism for microtubule-based transport. Protist. 2000;151:17–25. doi: 10.1078/1434-4610-00004. [DOI] [PubMed] [Google Scholar]
- Koonce MP, Khodjakov A. Dynamic microtubules in Dictyostelium. J Muscle Res Cell Motil. 2002;23:613–619. doi: 10.1023/A:1024446821701. [DOI] [PubMed] [Google Scholar]
- Miki H, Okada Y, Hirokawa N. Analysis of the kinesin superfamily: insights into structure and function. Trends Cell Biol. 2005;15:467–476. doi: 10.1016/j.tcb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Kurasawa Y, Earnshaw WC, Mochizuki Y, Dohmae N, Todokoro K. Essential roles of KIF4 and its binding partner PRC1 in organized central spindle midzone formation. EMBO J. 2004;23:3237–3248. doi: 10.1038/sj.emboj.7600347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Midorikawa R, Takei Y, Hirokawa N. KIF4 Motor Regulates Activity-Dependent Neuronal Survival by Suppressing PARP-1 Enzymatic Activity. Cell. 2006;125:371–383. doi: 10.1016/j.cell.2006.02.039. [DOI] [PubMed] [Google Scholar]
- Sekine Y, Okada Y, Noda Y, Kondo S, Aizawa H, Takemura R, Hirokawa N. A novel microtubule-based motor protein (KIF4) for organelle transports, whose expression is regulated developmentally. J Cell Biol. 1994;127:187–201. doi: 10.1083/jcb.127.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpff J, Wordeman L. Chromosome Congression: The Kinesin-8-Step Path to Alignment. Current Biology. 2007;17:R326–R328. doi: 10.1016/j.cub.2007.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta ML, Carvalho P, Roof DM, Pellman D. Plus end-specific depolymerase activity of Kip3, a kinesin-8 protein, explains its role in positioning the yeast mitotic spindle. Nat Cell Biol. 2006;8:913–923. doi: 10.1038/ncb1457. [DOI] [PubMed] [Google Scholar]
- Varga V, Helenius J, Tanaka K, Hyman AA, Tanaka TU, Howard J. Yeast kinesin-8 depolymerizes microtubules in a length-dependent manner. Nat Cell Biol. 2006;8:957–962. doi: 10.1038/ncb1462. [DOI] [PubMed] [Google Scholar]
- Cottingham FR, Hoyt MA. Mitotic Spindle Positioning in Saccharomyces cerevisiae Is Accomplished by Antagonistically Acting Microtubule Motor Proteins. J Cell Biol. 1997;138:1041–1053. doi: 10.1083/jcb.138.5.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeZwaan TM, Ellingson E, Pellman D, Roof DM. Kinesin-related KIP3 of Saccharomyces cerevisiae Is Required for a Distinct Step in Nuclear Migration. J Cell Biol. 1997;138:1023–1040. doi: 10.1083/jcb.138.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran PT, Marsh L, Doye V, Inoue S, Chang F. A Mechanism for Nuclear Positioning in Fission Yeast Based on Microtubule Pushing. J Cell Biol. 2001;153:397–412. doi: 10.1083/jcb.153.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West RR, Malmstrom T, Troxell CL, McIntosh JR. Two Related Kinesins, klp5+ and klp6+, Foster Microtubule Disassembly and Are Required for Meiosis in Fission Yeast. Mol Biol Cell. 2001;12:3919–3932. doi: 10.1091/mbc.12.12.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putkey FR, Cramer T, Morphew MK, Silk AD, Johnson RS, McIntosh JR, Cleveland DW. Unstable Kinetochore-Microtubule Capture and Chromosomal Instability Following Deletion of CENP-E. Developmental Cell. 2002;3:351–365. doi: 10.1016/S1534-5807(02)00255-1. [DOI] [PubMed] [Google Scholar]
- McEwen BF, Chan GKT, Zubrowski B, Savoian MS, Sauer MT, Yen TJ. CENP-E Is Essential for Reliable Bioriented Spindle Attachment, but Chromosome Alignment Can Be Achieved via Redundant Mechanisms in Mammalian Cells. Mol Biol Cell. 2001;12:2776–2789. doi: 10.1091/mbc.12.9.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohlk C, Rohlfs M, Leier S, Schliwa M, Liu X, Parsch J, Woehlke G. Properties of the Kinesin-1 motor DdKif3 from Dictyostelium discoideum. European Journal of Cell Biology. doi: 10.1016/j.ejcb.2007.11.001. Corrected Proof. [DOI] [PubMed] [Google Scholar]
- Grishchuk EL, McIntosh JR. Microtubule depolymerization can drive poleward chromosome motion in fission yeast. EMBO J. 2006;25:4888–4896. doi: 10.1038/sj.emboj.7601353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rischitor PE, Konzack S, Fischer R. The Kip3-Like Kinesin KipB Moves along Microtubules and Determines Spindle Position during Synchronized Mitoses in Aspergillus nidulans Hyphae. Eukaryotic Cell. 2004;3:632–645. doi: 10.1128/EC.3.3.632-645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuchardt I, Assmann D, Thines E, Schuberth C, Steinberg G. Myosin-V, Kinesin-1, and Kinesin-3 Cooperate in Hyphal Growth of the Fungus Ustilago maydis. Mol Biol Cell. 2005;16:5191–5201. doi: 10.1091/mbc.E05-04-0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnichsen B, Koski LB, Walsh A, Marschall P, Neumann B, Brehm M, Alleaume AM, Artelt J, Bettencourt P, Cassin E, et al. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature. 2005;434:462–469. doi: 10.1038/nature03353. [DOI] [PubMed] [Google Scholar]
- Goshima G, Vale RD. The roles of microtubule-based motor proteins in mitosis: comprehensive RNAi analysis in the Drosophila S2 cell line. J Cell Biol. 2003;162:1003–1016. doi: 10.1083/jcb.200303022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison HG, McArthur AG, Gillin FD, Aley SB, Adam RD, Olsen GJ, Best AA, Cande WZ, Chen F, Cipriano MJ, et al. Genomic Minimalism in the Early Diverging Intestinal Parasite Giardia lamblia. Science. 2007;317:1921–1926. doi: 10.1126/science.1143837. [DOI] [PubMed] [Google Scholar]
- Faix J, Kreppel L, Shaulsky G, Schleicher M, Kimmel AR. A rapid and efficient method to generate multiple gene disruptions in Dictyostelium discoideum using a single selectable marker and the Cre-loxP system. Nucl Acids Res. 2004;32:e143. doi: 10.1093/nar/gnh136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelhoff TT, Titus MA, Manstein DJ, Ruppel KM, Spudich JA. Molecular genetic tools for study of the cytoskeleton in Dictyostelium. In: Vallee RB, editor. Methods in Enzymology. Vol. 196. Academic Press; 1991. pp. 319–334. [DOI] [PubMed] [Google Scholar]
- Knecht DA, Jung J, Matthews L. Quantification of transformation efficiency using a new method for clonal growth and selection of axenic Dictyostelium cells. Developmental Genetics. 1990;11:403–409. doi: 10.1002/dvg.1020110513. [DOI] [PubMed] [Google Scholar]
- Piperno G, Fuller MT. Monoclonal antibodies specific for an acetylated form of alpha-tubulin recognize the antigen in cilia and flagella from a variety of organisms. J Cell Biol. 1985;101:2085–2094. doi: 10.1083/jcb.101.6.2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock N, Koonce MP, de Hostos EL, Vale RD. In vitro microtubule-based organelle transport in wild-type Dictyostelium and cells overexpressing a truncated dynein heavy chain. Cell Motil Cytoskeleton. 1998;40:304–314. doi: 10.1002/(SICI)1097-0169(1998)40:3<304::AID-CM8>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Bishop JD, Han Z, Schumacher JM. The Caenorhabditis elegans Aurora B Kinase AIR-2 Phosphorylates and Is Required for the Localization of a BimC Kinesin to Meiotic and Mitotic Spindles. Mol Biol Cell. 2005;16:742–756. doi: 10.1091/mbc.E04-08-0682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers J, Bossinger O, Rose D, Strome S, Saxton W. A nematode kinesin required for cleavage furrow advancement. Current Biology. 1998;8:1133–1136. doi: 10.1016/S0960-9822(98)70470-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers J, Rose DJ, Saunders A, Dunkelbarger S, Strome S, Saxton WM. Loss of KLP-19 polar ejection force causes misorientation and missegregation of holocentric chromosomes. J Cell Biol. 2004;166:991–1001. doi: 10.1083/jcb.200403036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders AM, Powers J, Strome S, Saxton WM. Kinesin-5 acts as a brake in anaphase spindle elongation. Current Biology. 2007;17:R453–R454. doi: 10.1016/j.cub.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segbert C, Barkus R, Powers J, Strome S, Saxton WM, Bossinger O. KLP-18, a Klp2 Kinesin, Is Required for Assembly of Acentrosomal Meiotic Spindles in Caenorhabditis elegans. Mol Biol Cell. 2003;14:4458–4469. doi: 10.1091/mbc.E03-05-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning AL, Ganem NJ, Bakhoum SF, Wagenbach M, Wordeman L, Compton DA. The Kinesin-13 Proteins Kif2a, Kif2b, and Kif2c/MCAK Have Distinct Roles during Mitosis in Human Cells. Mol Biol Cell. 2007;18:2970–2979. doi: 10.1091/mbc.E07-02-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]