Abstract
Two minichromosomes (α and δ) in addition to two other aberrant chromosomes (β and γ) were found in a transgenic Arabidopsis plant produced by an in planta vacuum infiltration technique. The minichromosomes were successfully separated by successive crossing and selfing and added to wild-type Columbia (Col-0) as a supernumerary chromosome. FISH indicated that both of the two minichromosomes originated from the short arm of chromosome 2. The mini α chromosome contained the whole short-arm 2S and a truncated centromere (180-bp repeat cluster), whereas mini δ lacked the terminal region including telomere repeats. Pachytene FISH clearly revealed that mini δ comprised a ring chromosome carrying two copies of the region from the 180-bp repeat cluster to BAC-F3C11. Both of the 180-bp clusters (each ≈500 kb in length) were thought to possess normal centromere functions because the centromere-specific histone H3 variant (HTR12) was detected on both clusters. Notwithstanding this dicentric and ring form, mini δ was stably transmitted to the next generations, perhaps because of its compact size (<4 Mb). Chromosome β also comprised a dicentric-like structure, with one of the two 180-bp repeat sites derived from chromosome 1 and the other from chromosome 2. However, the latter was quite small and failed to bind HTR12. The data obtained in this study indicated that 500 kb of the 180-bp array of the chromosome 2 centromere, from the edge of the 180-bp array on the short-arm side, is sufficient to form a functional domain.
Keywords: minichromosomes, ring chromosome
The centromere is a multifunctional complex in eukaryotic chromosomes that is involved with the accurate segregation of chromatids to opposite poles during mitosis and with the segregation of half-bivalents and chromatids at the first and second divisions of meiosis, respectively. This function also involves kinetochore formation, microtubule attachment, chromosome movement, heterochromatin establishment, and mitotic checkpoint control. In many higher eukaryotes, tandemly repetitive DNA sequences have been found as a main component of the centromere (1, 2). In Arabidopsis thaliana, these repeats are comprised of a 180-bp repeat family, with units mainly of 178 bp in length (3–5). The cluster size of the 180-bp cores has been estimated to be 2.7–3 Mb in all five chromosomes (6). This size of the repeat was once thought to be critical in maintaining the centromeres, but proteins such as HTR12 (Arabidopsis CENP-A homologue), AtCENP-C, and AtMIS12 have been shown to assemble in only limited parts of the 180-bp repeat clusters (7–10). This indicated that short arrays of centromeric DNA repeats are sufficient to bind kinetochore proteins.
To identify the minimum functional unit that confers centromere function, minichromosomes carrying truncated centromeres have been investigated in Drosophila (11), humans (12), and maize (13). Recently, we found a minichromosome in A. thaliana derived from the short arm of chromosome 4 (14). The size of this “mini 4S” chromosome and the amount of centromeric major satellite (180-bp family) were determined to be ≈7.5 and 1 Mb, respectively. The initial aim of the present study was to introduce marker genes into this mini 4S for subsequent manipulation. Rather than producing T-DNA-tagged mini 4S chromosome, other minichromosomes were produced after T-DNA insertion in the centromeric region of chromosome 2. Here, we describe two types of minichromosomes (α and δ) as well as other aberrant ones (β and γ). Chromosomes δ and β display special features in that the former is a stable dicentric ring minichromosome, whereas the latter is a pseudodicentric. This phenomenon of T-DNA insertion-induced centromere breakage may provide a powerful tool for manipulating plant chromosomes and facilitating the analysis of the functional structures of centromeres.
Results
Chromosomal Aberrations Induced by T-DNA Insertions.
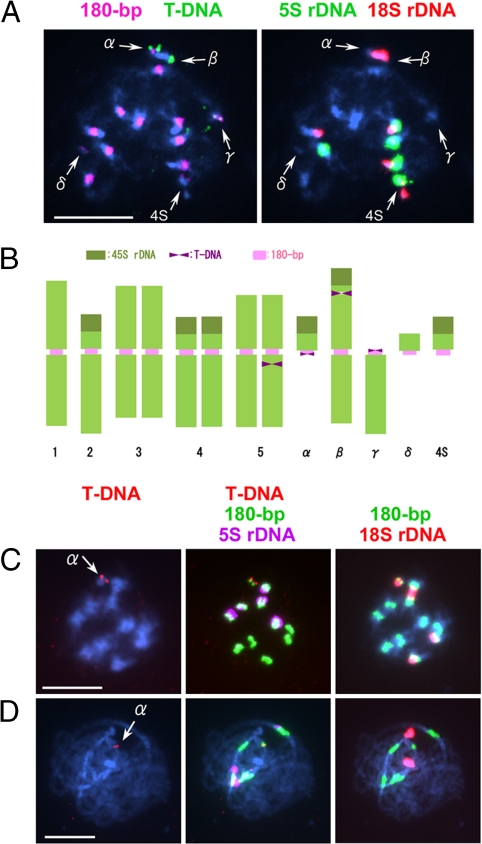
Approximately 50 transformants (T1) were produced in planta and then subjected to FISH analysis by using pBGF101 [supporting information (SI) Fig. S1] and a 180-bp repeat DNA as probes. Among these, one transformant, referred to as G40, was found to contain a T-DNA-tagged minichromosome (Fig. 1A). Subsequent FISH using 18S and 5S rDNA probes revealed that four different types of aberrant chromosomes were involved in this plant, which possessed a total of 13 chromosomes. These aberrant chromosomes were named α, β, γ, and δ (α, minichromosome with T-DNA insertion; β, large chromosome with T-DNA; γ, a small chromosome with T-DNA; δ, the smallest minichromosome without T-DNA but with short 180-bp clusters) (Fig. 1B). Because the plants carrying a pair of mini 4S (14) were used for transformation, a mini 4S chromosome also appeared in the transformant. An additional T-DNA insertion was found in chromosome 5 (Fig. 1 A and B).
Fig. 1.
Cytological analysis of a transgenic plant, G40 (T1 generation) and its offspring carrying minichromosome α. (A) A metaphase cell of G40 with four different aberrant chromosomes (α, β, γ, and δ) in addition to mini 4S. (B) Schematic representation of chromosome constitution of G40 estimated from A and other FISH images. (C) Mitotic metaphase. (D) Pachytene nucleus of a Tr α plant with a mini α (arrow) as a supernumerary chromosome. The centromere-specific 180-bp repeat, 5S and 18S rDNA, and pBGF101 DNA to detect T-DNA insertion sites were used as probes. (Scale bars: 5 μm.)
The transformant plant G40 was fertile, and after selfing, T2 seeds were collected and germinated for further analyses. The plants obtained (G40-1, -2 etc.) were crossed with ecotype Columbia (Col-0) to investigate the T-DNA insertion and chromosome conformation.
Structure and Origin of Two Minichromosomes.
At the back-cross generation, B1 = Col-0(♀)xT2(♂), two types of minichromosomes, with and without T-DNA insertion, referred to as “α” and “δ,” respectively, were separated from the other aberrant chromosomes, and each was added to wild-type Col-0 as a supernumerary in the subsequent generations. The minichromosomes were considerably smaller than the normal chromosomes and hence easily distinguishable. FISH confirmed that α carried 18S but no 5S rDNA (Fig. 1 C and D), suggesting that it had originated from the short arm of chromosome 2 (2S) because the other 18S rDNA cluster on the short arms of chromosome 4 is associated with 5S rDNA (5, 15). FISH using all BAC clones from the short arm of chromosome 2 yielded signals on chromosome α (data not shown), thus confirming the proposed origin. The minichromosome was designated as “mini α,” and the line carrying it was designated as “Tr α.” However, it was later found that the terminal tip from the top arm of chromosome 1 is attached to the centromeric region of mini α. This indicates that the rearrangement involves 2S and the top arm of chromosome 1. Compared with the wild-type (Col-0), no distinct phenotypic changes appeared in these plants except the presence of kanamycin-resistance and green fluorescence in the root tips. These two features are due to expression of T-DNA.
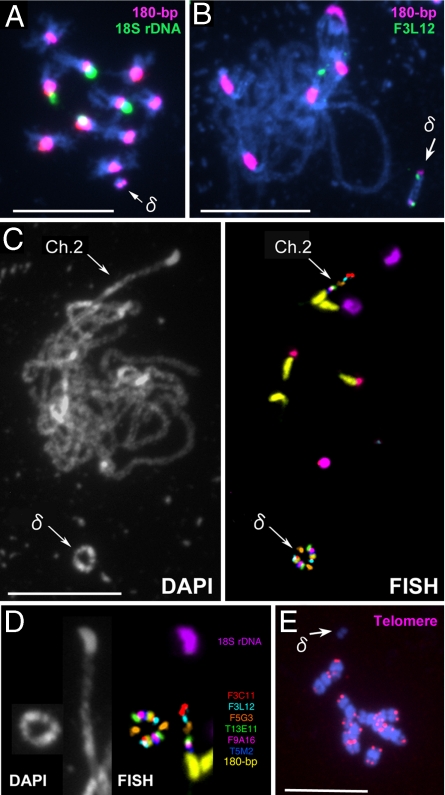
Mini δ also segregated as an additional chromosome to the normal set of 10 chromosomes (2n = 10 + δ) (Fig. 2A). It was smaller than mini α, and no T-DNA insertion was detected in it. BAC clones derived from the pericentric region of chromosome 2S, but not the terminal BAC clones nor 18S rDNA, yielded FISH signals on mini δ. Thus, mini δ also originated from the short arm of chromosome 2 but lacks the terminal region. Pachytene chromosomes probed with the centromere-specific 180-bp repeats and with pericentric BAC clones revealed 180-bp repeats at both ends of mini δ, each associated with a FISH signal of the BAC clone F3L12 (Fig. 2B), suggesting a circular structure for mini δ. Indeed, some pachytene cells revealed a distinct ring shape for this chromosome (Fig. 2 C and D). FISH using the 180-bp repeats and six BAC clones clearly demonstrated that this ring chromosome is dicentric and comprises a head-to-tail duplication, which encloses a region from the 180-bp repeats of CEN2 up to the F3C11 sequence. The weaker signal for F3C11 on mini δ compared with that of wild-type chromosome 2 suggests a truncation of the F3C11 sequence that became fused to the 180-bp repeats, forming a ring that eventually doubled in size after a cross-over event. As expected, no telomere sequences were detected on mini δ (Fig. 2E).
Fig. 2.
FISH images of Tr δ nuclei with mini δ (arrow) as a supernumerary chromosome. (A) A mitotic metaphase cell probed with 180-bp repeats (pink) and 18S rDNA (green). (B) Pachytene chromosomes probed with 180-bp repeats (pink) and F3L12-BAC (green). (C) DAPI and FISH images of pachytene chromosomes probed with 180-bp repeats (yellow), 18S rDNA (purple), 5S rDNA (red), and six different BAC clones; F3C11 (light red, AGI-map position on chromosome 2:1190978–1294851), F3L12 (light blue, 1288088–1404236), F5G3 (orange, 1829395–1953720), T13E11 (green, 3006966–3102924), F9A16 (deep red, 3112063–3214311), and T5M2 (deep blue, 3214009–3308310). (D) Side-by-side comparison of chromosomes 2 and mini δ; the images were derived from C. (E) A mitotic metaphase cell probed with PCR-amplified telomere DNA (5′-TTTAGGG-3′) (pink). (Scale bars: 5 μm.)
Pseudodicentric and Telocentric Chromosomes.
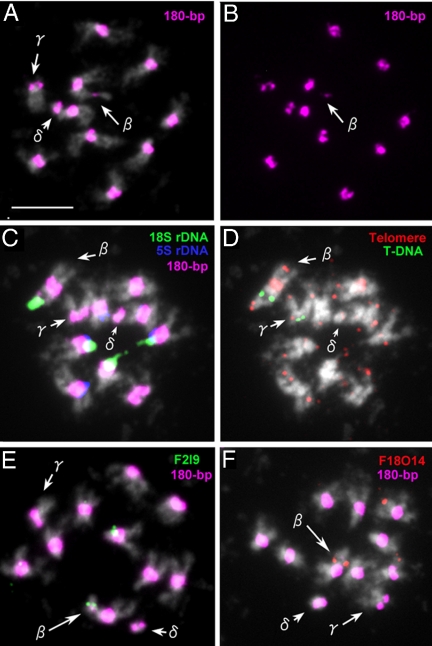
The two aberrant chromosomes β and γ could not be separated from each other, possibly because they contain large portions of chromosomes that compensate each other. Metaphase cells of plant (G40-18-6.7.3) carrying aberrant chromosomes β, γ, and mini δ are shown in Fig. 3. Chromosome β has two 180-bp repeat sites, one of which is much shorter than the other (Fig. 3 A and B). As in the original G-40 plant, T-DNA insertions were found in chromosomes β and γ (Fig. 3D). Chromosome β is longer than γ and carries 18S rDNA at one terminus (Fig. 3C). Interestingly, a gap appeared between the rDNA and the T-DNA insertion, and a 180-bp repeat FISH signal, although very weak, appeared to correspond to this gap (Fig. 3 C and D). This indicated that the gap region is part of the primary constriction of chromosome 2. The region distal to the gap could represent a remnant of 2S after excision of that part that became mini δ. In the region of the large 180-bp repeat cluster, a long stretch of telomeric DNA, typical for the centromeric region of chromosome 1 (16, 17), was detected (Fig. 3D). Chromosome β is most likely a pseudodicentric resulting from a translocation between chromosomes 1 and 2S, probably at their T-DNA insertion sites. This idea was confirmed by FISH with BAC clones F2I9 from chromosome 2S and F18O14 from the top arm of chromosome 1 (Fig. 3 E and F).
Fig. 3.
Cytological analysis of a G40 offspring containing chromosomes β, γ, and mini δ. (A) A mitotic metaphase cell probed with 180-bp repeats (pink). (B) The same cell showing only 180-bp repeat signals (pink) to clearly visualize the weak signal on chromosome β. (C) A mitotic metaphase cell probed with 180-bp repeats (pink), 18S rDNA (green), and 5S rDNA (blue). (D) The same cell probed with telomeric DNA (red) and T-DNA (green). (Note: a weak telomere signal on mini δ is from an adjoining chromosome.) (E) A mitotic metaphase cell probed with 180-bp repeats (pink) and F2I9-BAC (green, AGI-map position of chromosome 2:173440–276564). (F) A mitotic metaphase cell probed with 180-bp repeats (pink) and F18O14-BAC (red, Altman-map of chromosome 1: 29205). (Scale bar: 2 μm.)
Chromosome γ, which is smaller than chromosome β, had a size that was close to the long arm of chromosome 2 (2L) (Fig. 3A). Because the other arm (2S) of chromosome 2 had been identified as mini α, it was highly probable that chromosome γ originated from the centromere breakage of chromosome 2. This assumption was confirmed by FISH with BAC clones from 2L (data not shown).
T-DNA Insertion in the 180-bp Array Coincides with the Translocation Break Point.
In the original transgenic plant G40, four different T-DNA insertion sites were detected. Among these, only the insertion in chromosome 5 was not associated with chromosomal aberrations. The other three insertion sites were either within the centromeric 180-bp repeats of mini α (2S) and γ (2L) or within β between the rDNA of chromosome 2S and the remaining top arm of chromosome 1. The insertions found on mini α and γ apparently result from a single insertion event into the centromeric region of chromosome 2.
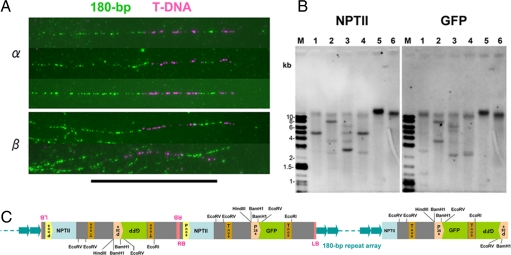
The insertion on mini α was investigated by DNA-fiber FISH and Southern blot analyses (Fig. 4 A and B). DNA-fiber FISH clearly revealed bipartite T-DNA inserts within the 180-bp cluster, much longer than expected for a single T-DNA sequence (≈8 kb) (Fig. 4A). Southern blot analysis showed that three copies of T-DNA (two complete and one truncated) had probably been inserted in the centromeric satellite 180-bp repeat cluster (Fig. 4 B and C). Expression of both GFP and NPTII genes of the T-DNA was suppressed in some of the progeny, probably due to RNA interference, because two complete copies of T-DNA were inserted in a head-to-head fashion in the centromeric region.
Fig. 4.
DNA-fiber FISH and genomic Southern blot analyses. (A) DNA-fiber FISH with chromatin from a Tr α plant and a G40-derived plant carrying β and mini δ chromosomes, probed with T-DNA of pBGF101 (pink) and 180-bp repeats (green). (B) Southern blot analysis of genomic DNA isolated from a Tr α plant, probed with NPTII (neomycin phosphotransferase gene) and GFP (green fluorescence protein gene), M: DNA marker, lanes 1–6: BamHI, EcoRI, EcoRV, HindIII, SmaI, and XbaI digests. (C) A schematic pattern of T-DNA insertions estimated from A and B. (Scale bar: 10 μm.)
A T-DNA insertion site on chromosome β was also detected by DNA-fiber FISH (Fig. 4A), and the insertion pattern was found to be identical with that of mini α. This suggested that chromosomes mini α and β represent derivatives of one and the same chromosome harboring an insertion.
Centromere Size and Activity in Aberrant Chromosomes.
The centromeric regions of mini α, γ, and mini δ and the distal region of β were found to be truncated. Chromosomes β and mini δ possessed two 180-bp repeat clusters, and therefore, appeared as dicentric. An investigation of the repeat cluster size and centromere activity was then undertaken.
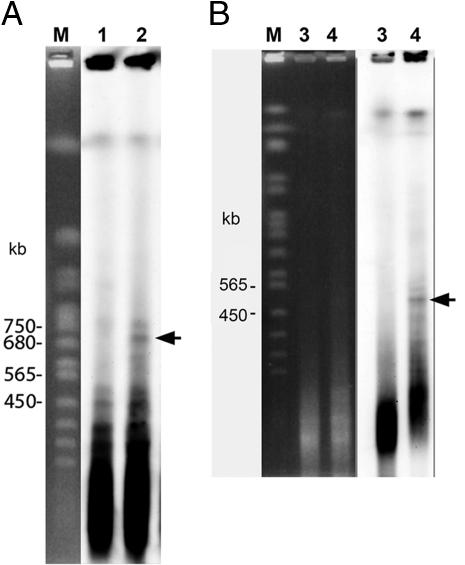
The sizes of the 180-bp repeat clusters were estimated by pulsed-field gel electrophoresis (PFGE). Because mini α and mini δ were added to the Col ecotype as supernumerary chromosomes, agarose plugs were made from lines Tr α and Tr δ. Of the 10 different restriction enzymes used, only HpaII could distinguish plants with and without minichromosomes (α or δ) (Fig. 5 A and B). HpaII cleaves 5′-CCGG-3′ but not 5′-CmCGG (CpG-methylation), and therefore, has been used to examine cytosine methylation in the centromeres of Arabidopsis (18, 19). Specific bands of 700 and 500 kb were detected in plants carrying mini α and mini δ, respectively. These are markedly shorter than the size reported for chromosome 2 (≈3 Mb) (6). The size of two 180-bp clusters on mini δ was similar because only one 500-kb band was detected.
Fig. 5.
PFGE/Southern blot analysis for plants carrying mini α and mini δ for size determination of the 180-bp arrays. (A) PFGE pattern of plants without (lane 1) and with mini α (lane 2). (B) PFGE pattern of plants without (lane 3) and with mini δ (lane 4); M: DNA marker from Saccharomyces cerevisiae. Genomic DNA was digested with HpaII. The 180-bp repeat was used as a probe. Specific bands corresponding to ≈700 and 500 kb from plants containing mini α and mini δ, respectively, are marked with arrows.
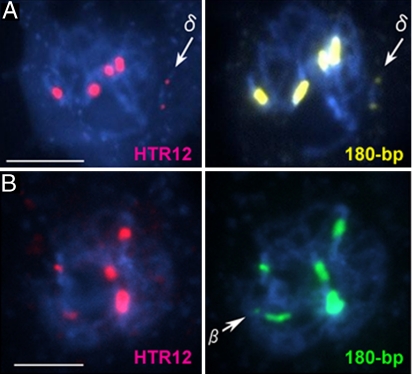
To examine centromere activity of the 180-bp repeat clusters detected by FISH, we used an antibody against HTR12, the centromere-specific histone H3 (CENH3) of Arabidopsis (20), because CENH3 is a basic kinetochore protein that is bound only to functional centromeres (21). CENH3-specific immunosignals appeared on the centromeric regions of mini α and γ, as well as on all normal chromosomes (data not shown). On the ring-shaped mini δ, both 180-bp arrays revealed signals, despite their small size (≈500 kb) (Fig. 6A and Fig. S2A), indicating that both 180-bp sites can function as a centromere. This notion was also supported by the observation of lagging of mini δ, on the equatorial plates, when pulled from the two opposite poles (Fig. S3).
Fig. 6.
HTR12-immunostaining and FISH analysis. (A) A pachytene cell of a Tr δ plant carrying mini δ after fluorescent immunolabeling by using anti-HTR12 antibodies (pink) and FISH using 180-bp repeats (yellow). The arrows indicate mini δ. (B) A pachytene cell of a plant carrying chromosome β after fluorescent immunolabeling by using anti-HTR12 antibodies (pink) and FISH using 180-bp repeats (green) to the pachytene chromosomes. The arrow indicates the small 180-bp sites on chromosome β. (Scale bars: 5 μm.)
Chromosome β also revealed two 180-bp repeat sites, one of which was much smaller than the other (Fig. 3). As expected, the large 180-bp site from chromosome 1 showed a distinct HTR12 immunosignal, whereas no signals were detected on the small 180-bp repeat site (Fig. 6B and Fig. S2B). A constriction was retained at the small 180-bp-repeat site (Fig. 3A). Chromosome β was quite stable, and no anaphase bridges were observed in cells containing this chromosome. This observation indicates that chromosome β is a pseudodicentric and that only the large 180-bp site acts as a centromere.
Discussion
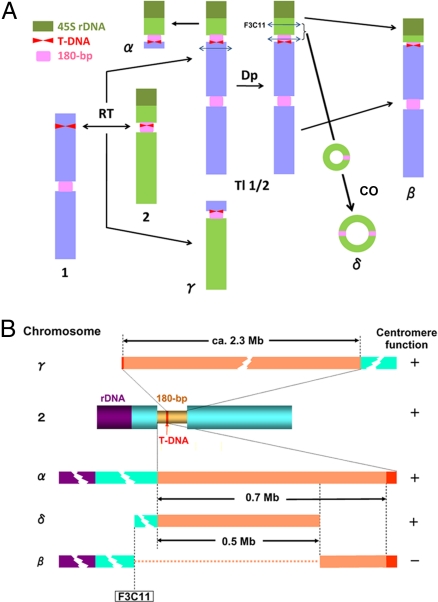
Based on the FISH data, we interpreted the origin of aberrant chromosomes as shown in Fig. 7A. The processes involved include the following.
A translocation event between the two T-DNA insertion sites in the top arm of chromosome 1 and in the centromeric 180-bp array of chromosome 2.
This reciprocal translocation resulted in chromosomes tl1/2 and γ. The former chromosome was not maintained in the original transgenic plant G40 because it was dicentric and thus unstable. In the subsequent cell division, chromosome tl1/2 was probably duplicated through nondisjunction.
Mini α originated from a break in the top arm region of chromosome 1 as one of the duplicated copies of tl1/2 formed an anaphase bridge. Consequently, mini α contains the short arm of chromosome 2 and a small region of chromosome 1 at the opposite side of the centromeric region from chromosome 2. The larger part of tl1/2, representing most of chromosome 1, was apparently lost.
From the other copy of tl1/2, mini δ originated because of two breakages, one within F3C11 and the other within the 180-bp sequences, and subsequent joining of the internal break ends generating a ring chromosome that comprised part of the 180-bp array derived from chromosome 2. This structure was confirmed by PCR using primers from the 180-bp repeat and F3C11 sequences. The remaining two parts of tl1/2, the distal portion of chromosome 2S containing 45S rDNA and the remnant of the 180-bp array from chromosome 2 at the top of chromosome 1, were also joined to generate chromosome β. Because the short cluster of the 180-bp repeats on chromosome β possessed no centromere activity and the other large cluster behaved as a centromere, chromosome β was stably transmitted.
As indicated by McClintock (22), a ring chromosome can double in size and possess two centromeres by sister chromatid exchange after replication. Mini δ must have originated in this manner. Mini δ was rather stable despite its dicentric form (≈33% transmission rate through male but 0% from female side).
Fig. 7.
Interpretation of the origin of aberrant chromosomes generated by T-DNA insertions in the centromere of chromosome 2 and subsequent chromosome rearrangements. (A) Presumed mechanism for generating minichromosomes and other aberrant chromosomes involving T-DNA insertions. RT, reciprocal translocation; Dp, duplication; CO, crossing-over; 1, 2, α, β, γ, δ, and tl1/2 are chromosome designations. (B) Centromere structure and size of chromosomes 2, mini α, β, γ, and mini δ.
The in planta technique for plant transformation is thought to be superior to other techniques, such as those involving root culture, because somaclonal and chromosomal variations, which are common in plant cultured cells, seem not to occur with the former technique. When in planta transformation is applied to A. thaliana, Agrobacterium-mediated transformation occurs preferentially in female gametes 5–10 days before anthesis (23). In this study, a reciprocal translocation involved two T-DNA-insertion loci on chromosomes 1 and 2. This translocation apparently occurred in a somatic cell during early flower development (after female and male differentiation) and premeiotic DNA synthesis because it is otherwise difficult to account for all four chromosomal aberrations (mini α, β, γ, and mini δ) being transmitted through meiosis in a single individual. If the translocation occurred immediately before meiosis, a quadrivalent (ring-of-four) structure between chromosomes 1, 2, tl1/2, and γ (Fig. S4) could have formed at metaphase I. Despite the presence of five active centromeres, a daughter nucleus comprising chromosomes tl1/2 and γ could have formed. During the second meiotic division, nondisjunction of tl1/2 yielded two nuclei containing either two tl1/2 and one γ or no tl1/2 and one γ, respectively. During subsequent cell divisions, chromosome rearrangements involving the two tl1/2 chromosomes yielded mini α, β, and mini δ (Fig. 7A).
To date, only one line carrying a minichromosome had been established in A. thaliana (14). This mini 4S originated from the short arm of chromosome 4. The total size was estimated to be ≈7.5 Mb and included 1 Mb of centromeric 180-bp repeats. In this study, two minichromosomes (α and δ) were identified. Mini α contained the entire short arm of chromosome 2 of ≈7 Mb (5, 24), in addition to ≈700 kb of 180-bp repeats and ≈300 kb from chromosome 1 (unpublished data). Hence, the total size of mini α is ≈8 Mb, which is similar to that of mini 4S and is consistent with the hypothesis proposed by Schubert (25) that stable minichromosome transmission through meiosis requires ≈5% of the genome size.
Mini δ comprises part of the short arm of chromosome 2 (T5E7 to F3C11 and corresponds to ≈1.35 Mb) and 500 kb of a 180-bp repeat cluster, both in duplicate. It is known that a mitochondrial (mt) DNA sequence of ≈270 kb is inserted into the pericentric region on the short arm of chromosome 2 of ecotype Columbia (5, 24). However, Stupar et al. (26) claimed that the size of the insertion was 2.3 times greater (618 ± 42 kb) because of sequence duplications. Therefore, the total size of mini δ would be 3.7 Mb, or 4.5 Mb if the duplication of mtDNA had occurred. Because no duplication of the inserted mtDNA was detectable during pachytene on chromosomes 2 or mini δ (Fig. 2 C and D), 3.7 Mb seems to be a more accurate estimate. Although this size is inconsistent with Schubert's hypothesis (25), it may be generated through its ring structure without telomeres.
A T-DNA insertion within the centromere of chromosome 2 eventually led to four aberrant chromosomes (mini α, β, γ, and mini δ) with truncated centromeres. These centromeres allow for an estimate of the minimal region, which encompasses the functional domain of the centromere of chromosome 2 (Fig. 7B). DNA insertion and subsequent or simultaneous translocation with another T-DNA inserted on chromosome 1 split the 3-Mb centromere (180-bp repeat cluster) into two fragments comprising 0.7 and 2.3 Mb. The former was retained in mini α, whereas the latter was retained in chromosome γ, and both were functional. Although the dicentric mini δ had smaller centromere repeat arrays (500 kb each) than mini α, each of the two centromeres was functional, indicating that the 500-kb region comprising 180-bp repeats encompassed the functional domain(s) of the centromere.
Chromosome β had two 180-bp repeat clusters, the original derived from chromosome 1 and another derived from chromosome 2. The latter might contain only 0.2 Mb because ≈500 kb of the 0.7-Mb fragment was excised during formation of mini δ (Fig. 7 A and B) if no amplification had occurred. This short array was not longer in terms of centromere function, although it originated from the central part of the original chromosome-2 centromere and retained a constriction (Fig. 3A). From these results, we conclude that a 180-bp repeat array >500 kb is required for centromere function. However, we cannot disregard possible differences between the 500- and 200-kb sequences on the centromere of chromosome 2. Epigenetic modifications might differentiate the 180-bp repeat clusters. Inactivation of one centromere on dicentric chromosomes has been reported for humans (27, 28), maize (29), and for barley centromeres in wheat chromosomes (30). Although the mechanism of centromere inactivation remains unclear, these phenomena imply that the DNA sequences alone are insufficient to trigger inactivation and that epigenetic marks are necessary for maintaining centromere function.
Although ring chromosomes have been found in both animals and plants, in general, they are not stably transmitted to the subsequent generations because of mitotic and meiotic recombination processes. The rare inheritance of human ring chromosomes has been reported (31). Mini δ is transmissible to about one-third of the selfed progeny, notwithstanding its ring and dicentric structure, and is now being maintained as a partial trisomic (2n = 10 + δ). The small size (<4 Mb) could reduce recombination frequency and subsequent anaphase bridge formation. During nuclear divisions, the two 180-bp arrays behaved like a single centromere, although interlocked large-sized ring chromosomes lagging on equatorial plates appeared occasionally (≈5%). Similarly, a ring minichromosome containing human alphoid sequences was shown to be transmissible to the offspring through mouse spermatogenesis (32). Recently, meiotic transmission of circular centromeric DNA, which was introduced into embryonic tissues, was reported for maize, although neither circularity nor kinetochore formation has been proved (33). Nevertheless, our studies indicate that small ring-shaped chromosomes might be useful as chromosomal vectors.
Methods
Plant Material and Transformation.
Three- to 4-week-old plants of the Tr4SCo5 line of A. thaliana (Col-0) carrying a pair of mini4S chromosomes (14) were transformed by using the binary vector pBGF101 (Fig. S1) according to the in planta vacuum infiltration method (34).
Cytological and Molecular Analyses.
FISH was essentially the same as described previously (8, 14, 15) but with minor modifications. Briefly, after rinsing with milli-Q water, fixed flower buds were digested with enzyme solution [2% (wt/vol) Cellulase Onozuka RS (Yakult) and 0.5% (wt/vol) Pectolyase Y23 (Kikkoman) in citric buffer (pH 4.5)] at 37°C for 1 h. The digested tissues were fixed and dropped on cold slides. Air-dried slides were denatured in 70% formamide in 2× SSC at 76°C for 1 min and then placed in 70% ethanol and stored at −20°C. For the detection of centromeres, the 180-bp repeats amplified by PCR were labeled by using the FITC High Prime kit (Roche) (8). Other Arabidopsis probes including BAC clones from chromosomes 1 and 2, the ≈500-bp 5S rDNA repeat unit (15), and the 18S rDNA (8) were labeled either with biotin-7-dATP (GIBCO–BRL) or with digoxigenin-11-dUTP (Roche) by nick translation and detected with 5 μg/ml (1:100 dilution) streptavidin-Cy5 conjugate (GE Healthcare) and 4 μg/ml anti-digoxigenin Rhodamine Fab fragments (Roche), respectively. Slides were stained with 0.1 μg/ml DAPI and examined by using a fluorescent microscope (Axioskop; Carl Zeiss). Images were captured by using a digital CCD camera (AxioCam HRm; Carl Zeiss).
For the multiprobed FISH, slides were reprobed one to three times. After detection by using the first probe, slides were placed in PBS solution to remove the coverslips, fixed again for 10 min, and then air-dried. Denaturation and hybridization were as above. The BAC clones containing unique DNA sequences were used first, followed by repetitive DNA sequences. Captured images were overlaid by using Axio Vision 3.1 (Carl Zeiss). For the indirect immunolabeling of the centromere-specific histone H3 variant (CENH3), anti-HTR12 antibodies were raised against a synthetic peptide designed from the first 20 N-terminal amino acids as reported previously (20) and applied to pachytene chromosomes prepared from pollen mother cells fixed in 4% (vol/vol) paraformaldehyde for 30 min at room temperature. Immunofluorescence signals were detected as described previously (8).
DNA fiber FISH was performed according to Fransz et al. (35). PFGE and genome Southern blotting were performed by using the CHEF-DRII system (Bio-Rad) as described previously (3).
Supplementary Material
Acknowledgments.
We thank K. Nagaki for useful discussions and the Arabidopsis Biological Resource Center at Ohio State University for providing the Arabidopsis BAC clones used in this study. This work was supported by Core Research for Evolutional Science and Technology/Japan Science and Technology Agency, Japan, and a special educational study, Ministry of Education, Culture, Sports, Science and Technology, Japan.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0802828105/DCSupplemental.
References
- 1.Choo KHA. The Centromere. Oxford: Oxford Univ Press; 1997. pp. 97–142. [Google Scholar]
- 2.Murata M. Telomeres and centromeres in plants. Curr Genomics. 2002;3:527–538. [Google Scholar]
- 3.Murata M, Ogura Y, Motoyoshi F. Centromeric repetitive sequences in Arabidopsis thaliana. Jpn J Genet. 1994;69:361–370. doi: 10.1266/jjg.69.361. [DOI] [PubMed] [Google Scholar]
- 4.Copenhaver GP, et al. Genetic definition and sequence analysis of Arabidopsis centromeres. Science. 1999;286:2468–2474. doi: 10.1126/science.286.5449.2468. [DOI] [PubMed] [Google Scholar]
- 5.The Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 6.Hosouchi T, Kumekawa N, Tsuruoka H, Kotani H. Physical map-based sizes of the centromeric regions of Arabidopsis thaliana chromosomes 1, 2, and 3. DNA Res. 2002;9:117–121. doi: 10.1093/dnares/9.4.117. [DOI] [PubMed] [Google Scholar]
- 7.Nagaki K, et al. Chromatin immunoprecipitation reveals that the 180-bp satellite repeat is the key functional DNA element of Arabidopsis thaliana centromeres. Genetics. 2003;163:1221–1225. doi: 10.1093/genetics/163.3.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shibata F, Murata M. Differential localization of the centromere-specific proteins in the major centromeric satellite of Arabidopsis thaliana. J Cell Sci. 2004;117:2963–2970. doi: 10.1242/jcs.01144. [DOI] [PubMed] [Google Scholar]
- 9.Ogura Y, Shibata F, Sato H, Murata M. Characterization of a CENP-C homolog in Arabidopsis thaliana. Genes Genet Syst. 2004;79:139–144. doi: 10.1266/ggs.79.139. [DOI] [PubMed] [Google Scholar]
- 10.Sato H, Shibata F, Murata M. Characterization of a Mis12 homologue in Arabidopsis thaliana. Chromosome Res. 2005;13:827–834. doi: 10.1007/s10577-005-1016-3. [DOI] [PubMed] [Google Scholar]
- 11.Sun X, Le HD, Wahlstrom JM, Karpen GH. Sequence analysis of a functional Drosophila centromere. Genome Res. 2003;13:182–194. doi: 10.1101/gr.681703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Auriche C, Donini P, Ascenzioni F. Molecular and cytological analysis of a 5.5 Mb minichromosome. EMBO Rep. 2001;2:102–107. doi: 10.1093/embo-reports/kve018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kato A, et al. Minichromosomes derived from the B chromosome of maize. Cytogenet Genome Res. 2005;109:156–165. doi: 10.1159/000082395. [DOI] [PubMed] [Google Scholar]
- 14.Murata M, Shibata F, Yokota E. The origin, meiotic behavior, and transmission of a novel minichromosome in Arabidopsis thaliana. Chromosoma. 2006;115:311–319. doi: 10.1007/s00412-005-0045-1. [DOI] [PubMed] [Google Scholar]
- 15.Murata M, Heslop-Harrison JS, Motoyoshi F. Physical mapping of the 5S ribosomal RNA genes in Arabidopsis thaliana by multi-color fluorescence in situ hybridization with cosmid clones. Plant J. 1997;12:31–37. doi: 10.1046/j.1365-313x.1997.12010031.x. [DOI] [PubMed] [Google Scholar]
- 16.Richards EJ, Goodman HM, Ausubel FM. The centromere region of Arabidopsis thaliana chromosome 1 contains telomere-similar sequences. Nucleic Acids Res. 1991;19:3351–3357. doi: 10.1093/nar/19.12.3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Armstrong SJ, Jones GH. Meiotic cytology and chromosome behaviour in wild-type Arabidopsis thaliana. J Exp Bot. 2003;54:1–10. doi: 10.1093/jxb/erg034. [DOI] [PubMed] [Google Scholar]
- 18.Vongs A, Kakutani T, Martienssen RA, Richards EJ. Arabidopsis thaliana DNA methylation mutants. Science. 1993;260:1926–1928. doi: 10.1126/science.8316832. [DOI] [PubMed] [Google Scholar]
- 19.Richards EJ, Dawe RK. Plant centromeres: structure and control. Curr Opin Plant Biol. 1998;1:130–135. doi: 10.1016/s1369-5266(98)80014-9. [DOI] [PubMed] [Google Scholar]
- 20.Talbert PB, Masuelli R, Tyagi AP, Comai L, Henikoff S. Centromeric localization and adaptive evolution of an Arabidopsis histone H3 variant. Plant Cell. 2002;14:1053–1066. doi: 10.1105/tpc.010425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Warburton PE. Chromosomal dynamics of human neocentromere formation. Chromosome Res. 2004;12:617–626. doi: 10.1023/B:CHRO.0000036585.44138.4b. [DOI] [PubMed] [Google Scholar]
- 22.McClintock B. The production of homozygous deficient tissues with mutant characteristics by means of the aberrant mitotic behavior of ring-shaped chromosomes. Genetics. 1938;23:315–376. doi: 10.1093/genetics/23.4.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Desfeux C, Clough SJ, Bent AF. Female reproductive tissues are the primary target of Agrobacterium-mediated transformation by the Arabidopsis floral-dip method. Plant Physiol. 2000;123:895–904. doi: 10.1104/pp.123.3.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin X, et al. Sequence and analysis of chromosome 2 of the plant Arabidopsis thaliana. Nature. 1999;402:761–768. doi: 10.1038/45471. [DOI] [PubMed] [Google Scholar]
- 25.Schubert I. Alteration of chromosome numbers by generation of minichromosomes—Is there a lower limit of chromosome size for stable segregation? Cytogenet Cell Genet. 2001;83:175–181. doi: 10.1159/000056981. [DOI] [PubMed] [Google Scholar]
- 26.Stupar RM, et al. Complex mtDNA constitutes an approximate 620-kb insertion on Arabidopsis thaliana chromosome 2: Implication of potential sequencing errors caused by large-unit repeats. Proc Natl Acad Sci USA. 2001;98:5099–5103. doi: 10.1073/pnas.091110398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sullivan BA, Schwartz S. Identification of centromeric antigens in dicentric Robertsonian translocations: CENP-C and CENP-E are necessary components of functional centromeres. Hum Mol Genet. 1995;4:2189–2197. doi: 10.1093/hmg/4.12.2189. [DOI] [PubMed] [Google Scholar]
- 28.Fisher AM, et al. Centromeric inactivation in a dicentric human Y;21 translocation chromosome. Chromosoma. 1997;106:199–206. doi: 10.1007/s004120050240. [DOI] [PubMed] [Google Scholar]
- 29.Han F, Lamb JC, Birchler JA. High frequency of centromere inactivation resulting in stable dicentric chromosomes of maize. Proc Natl Acad Sci USA. 2006;103:3238–3243. doi: 10.1073/pnas.0509650103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nasuda S, Hudakova S, Schubert I, Houben A, Endo TR. Stable barley chromosomes without centromeric repeats. Proc Natl Acad Sci USA. 2005;102:9842–9847. doi: 10.1073/pnas.0504235102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Le Caignec C, et al. Inherited ring chromosome 8 without loss of subtelomeric sequences. Ann Genet. 2004;47:289–296. doi: 10.1016/j.anngen.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 32.Voet T, Liebe B, Labaere C, Marynen P, Scherthan H. Telomere-independent homologue pairing and checkpoint escape of accessory ring chromosomes in male mouse meiosis. J Cell Biol. 2003;162:795–807. doi: 10.1083/jcb.200305065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carlson SR, et al. Meiotic transmission of an in-vitro assembled autonomous maize minichromosome. PLoS Genet. 2007;3:1965–1974. doi: 10.1371/journal.pgen.0030179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bechtold N, Ellis J, Pelletier G. In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci Paris, Life Sci. 1993;316:1194–1199. [Google Scholar]
- 35.Fransz PF, et al. High-resolution physical mapping in Arabidopsis thaliana and tomato by fluorescence in situ hybridization to extended DNA fibres. Plant J. 1996;9:421–430. doi: 10.1046/j.1365-313x.1996.09030421.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.