Abstract
Site-directed spin labeling has qualitatively shown that a key event during activation of rhodopsin is a rigid-body movement of transmembrane helix 6 (TM6) at the cytoplasmic surface of the molecule. To place this result on a quantitative footing, and to identify movements of other helices upon photoactivation, double electron–electron resonance (DEER) spectroscopy was used to determine distances and distance changes between pairs of nitroxide side chains introduced in helices at the cytoplasmic surface of rhodopsin. Sixteen pairs were selected from a set of nine individual sites, each located on the solvent exposed surface of the protein where structural perturbation due to the presence of the label is minimized. Importantly, the EPR spectra of the labeled proteins change little or not at all upon photoactivation, suggesting that rigid-body motions of helices rather than rearrangement of the nitroxide side chains are responsible for observed distance changes. For inactive rhodopsin, it was possible to find a globally minimized arrangement of nitroxide locations that simultaneously satisfied the crystal structure of rhodopsin (Protein Data Bank entry 1GZM), the experimentally measured distance data, and the known rotamers of the nitroxide side chain. A similar analysis of the data for activated rhodopsin yielded a new geometry consistent with a 5-Å outward movement of TM6 and smaller movements involving TM1, TM7, and the C-terminal sequence following helix H8. The positions of nitroxides in other helices at the cytoplasmic surface remained largely unchanged.
Keywords: DEER, G protein-coupled receptor, photoreceptor, spin labeling
G protein-coupled receptors (GPCRs) constitute a large family of membrane proteins that mediate cellular responses to a variety of both physical and chemical signals. Interaction of a GPCR with a cognate signal produces a conformational change leading to an activated receptor. The activated state subsequently binds to and activates a corresponding heterotrimeric G protein, which in turn triggers events that ultimately modulate an amplified cellular response (for recent reviews, see refs. 1 and 2). The nature of the conformational switch or switches that underlie receptor activation is a subject of much interest (3, 4). Crystal structures of an inactive state of the GPCRs rhodopsin (5–9) and β2-adrenergic receptor (10, 11) have been reported, and they provide a crucial foundation for designing and interpreting the results of spectroscopic experiments aimed at revealing conformational changes associated with activation.
Site-directed spin-labeling (SDSL) studies of rhodopsin provided the first direct structural evidence for an outward tilt of the cytoplasmic end of transmembrane helix 6 (TM6) as a major event in the activation of rhodopsin, both in solutions of dodecyl maltoside (DM) and in phospholipid bilayers (12–15). Data from site-directed fluorescent labeling and chemical reactivity (16), UV spectroscopy (17), zinc cross-linking of histidines (18), and disulfide cross-linking (13) offered support for this mechanism. Recent solid-state NMR studies of Smith and coworkers (19) revealed a separation of opposing residues in TM6 and TM3 at the level of the chromophore upon activation. Taken together, the NMR and SDSL data suggest that a significant length of TM6 of rhodopsin is involved in the conformational change. There is considerable evidence that the activating conformational change in other GPCRs, at least at the cytoplasmic surface, has features common with that of rhodopsin (3, 4).
The highly conserved proline in TM6 of the rhodopsin class of GPCRs (Pro-267 in rhodopsin) may serve as a hinge for motion of TM6 (20, 21) triggered by a “rotamer toggle switch” (22). Schwartz (3) has elaborated a “global toggle switch” model for receptor activation that incorporates motion of TM6 at the cytoplasmic surface together with proposed concerted motions of other helices at the extracellular surface.
In contrast to the evidence outlined above, a recent crystal structure at 4.1-Å resolution of a putative photoactivated rhodopsin showed little change in position of any of the TM helices relative to the inactive state (23).
In previous SDSL studies of rhodopsin directed at detecting conformational changes via distance measurements (13, 24, 25), the short range (<20 Å) of the continuous-wave (CW) dipolar interaction method used (26, 27) imposed constraints on the experimental design that could limit the effective resolution. In particular, it was necessary to place spin labels at buried sites in the molecule in order for them to be sufficiently close for interaction, raising the possibility that spin-label perturbation and/or rearrangement of the spin-label side chain itself contributed to the distance changes observed upon activation. This problem can now be overcome by using the time domain method of double electron–electron resonance (DEER) spectroscopy (28) that enables long-range (20–60 Å) distance measurements between pairs of spin labels with a resolution better than 1 Å. This extended range allows the spin labels to be placed at sites on the solvent-exposed surface of the protein, where perturbations to the fold are absent, and where the local environment remains unaltered during a protein conformational change, ensuring that changes in spin-label conformation that could contribute to distance changes do not occur. Under these conditions, changes in interspin distance reflect corresponding motion of the backbone to which R1 is attached.
In the present study, 16 pairs of the nitroxide side chain R1 (Fig. 1Inset) were introduced into rhodopsin, one pair at a time. Sites on the outer surfaces of the TM helices at the cytoplasmic surface were chosen (Fig. 1), and interspin distances were measured in the inactive (R) and light-activated state (R*), using DEER spectroscopy. A global analysis of the data localized individual spins in the structure, and the results for R are in excellent agreement with the rhodopsin crystal structure from Li et al. [Protein Data Bank (PDB) entry 1GZM] (8) and models of R1 based on the known structure of the side chain (29–32). For R*, the data reveal an outward motion of R1 in TM6 by 5 Å and smaller movements of R1 in TM1, TM7, and in the C-terminal domain following helix H8. In addition, there are increases in the widths of the spatial distributions for the nitroxide at these same sites, suggesting increased local disorder. R1 residues in TM2–TM4 remain fixed upon R* formation, consistent with these helices forming part of a stable core (33).
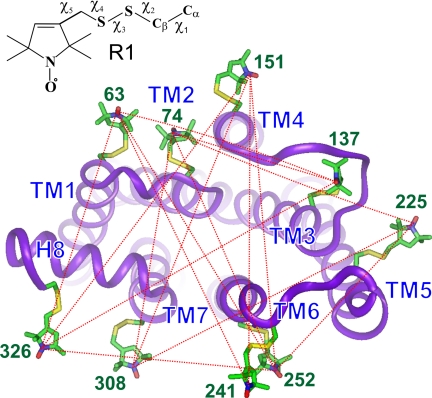
Fig. 1.
View of the cytoplasmic face of inactive rhodopsin (PDB entry 1GZM) showing modeled R1 side chains for all sites studied here. The dotted lines connect pairs where the interspin distance distribution was determined by using DEER. Each distribution was measured before and after light-activation. The R1 side chains are modeled from known conformations (see Methods). (Inset) Structure of R1 with the dihedral angles defined.
The results presented here are at odds with the crystallographic studies of Salom et al. (23) that found little structural change relative to the inactive state for a putative photoactivated species.
Experimental Design and Results
As in most previous SDSL studies, results presented here were obtained for rhodopsin in solutions of DM, a detergent that apparently retains the structure and functionality of R and R* in the native membrane (14, 15, 35, 36).
Fig. 1 shows a view of R looking down at the cytoplasmic surface that recognizes the G protein transducin. Models of the R1 nitroxide side chain (see Methods) are shown at each of the sites selected on the outer surfaces of the TM helices and site 326 in the C terminus. The EPR spectra for the selected sites show little or no change upon photoactivation, as previously reported (12, 37–40). Because the spectra are sensitive to changes in backbone dynamics (41, 42) and local interactions within the protein (29–31, 43), the lack of spectral change upon photoactivation strongly suggests that the conformation of R1, the immediate local environment, and the secondary structure to which it is attached remain the same under rhodopsin activation. Thus, changes in interspin distances between pairs of these nitroxides report relative motion of the structures to which they are attached. Pairs of R1 residues for interspin distance measurements were selected from the set shown in Fig. 1 such that each R1 residue was involved in multiple distance determinations from several directions; this is critical to identifying the relative position of each R1 in the structure and to determining the direction as well as the magnitude of its motion upon photoactivation. The dotted traces in Fig. 1 show the 16 distances measured by using DEER spectroscopy.
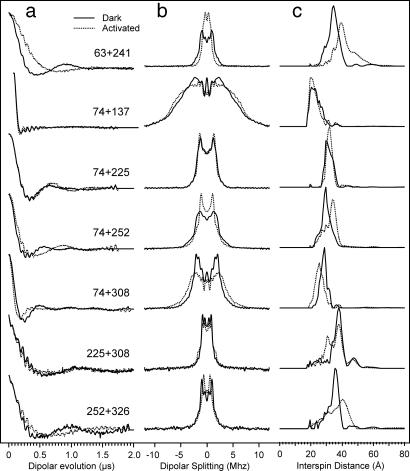
The primary datum obtained in DEER spectroscopy is the amplitude of an electron spin echo as a function of time corresponding to the application of a pulse that inverts the magnetization of an interacting spin partner (28). Subtraction of an exponential background due to random intermolecular magnetic dipolar interactions gives the dipolar evolution function, which is a sum of oscillatory functions with frequencies corresponding to the dipolar interaction energies for the distances represented. The distribution of distances between the spins is obtained by fitting the dipolar evolution function or its Fourier transform (the dipolar interaction spectrum) to a model of interacting spin pairs that have a random orientation with respect to each other (28). Fig. 2 shows representative data for the dipolar evolution functions, the dipolar interaction spectra and distance distributions obtained for spin pairs in R (solid traces) and R* (dotted traces). The examples were selected to illustrate the cases observed; namely, decreases, no change, or increases in distance upon photoactivation.
Fig. 2.
Representative selection of DEER results in the R (solid line) and R* states (dotted line). (a) Normalized dipolar evolution after removal of the exponential background. The high-frequency signal is due to proton modulations (28). (b) DC centered Fourier transform of the data in a. (c) Distance distribution calculated from the data in b by using Tikhonov regularization.
Geometry Optimization and Probable Localization of Individual Nitroxides in the Dark State (R).
To find the relative positions of the nitroxides consistent with the complete set of interspin distances determined experimentally, a global geometry optimization was carried out. The global probability Ω of a given configuration of spins is taken as Ω = ΠPj(r), where the product is over all measured spin pairs j, and Pj(r) is the probability of finding the spin pair j at a distance r. If P(r) is taken as the normalized experimental distance distribution, Ω approaches 1 if all distances correspond to the most probable value for all measured pairs. The optimized geometry is one that maximizes Ω with respect to variation in the coordinates of each spin. The initial locations of the spins were taken as those determined from models of the R1 side chain in rhodopsin as described above. Optimization using a modified Levenberg–Marquardt algorithm resulted in a slight repositioning of the spins in the neighborhood of the starting position. Except for position 326, the optimized positions of the spins are compatible with preferred rotamers of the R1 side chain and give essentially exact agreement with experimental values of the interspin distances. The models of Figs. 1 and 3 represent the optimized geometry for the inactive state.
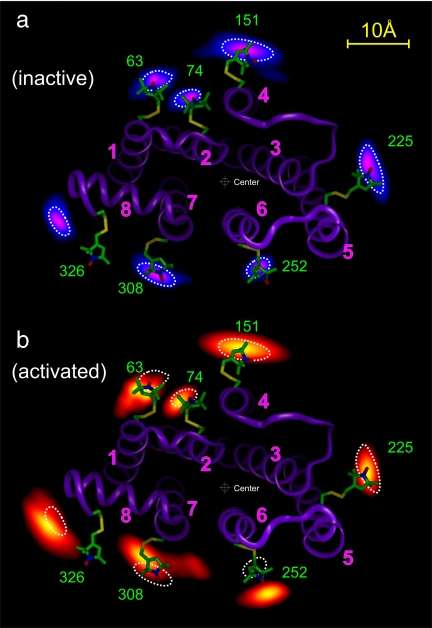
Fig. 3.
Projection contours of the spin locations calculated from the measured distance distributions for representative sites (see text for details). (a) Probable locations of the nitroxide spins in R shown in a gradient from blue to purple and overlaid on a ribbon model of R from the crystal structure (PDB entry 1GZM). (b) Corresponding locations in R* shown in a gradient from red to yellow, and, for reference, the ribbon model and typical contours (dotted traces) for R overlaid. The center used to obtain the data in Fig. 4 is shown.
Starting with the optimized geometry, the probability distribution of individual spins can be estimated in a first approximation by calculating Ω for a particular spin i at a position displaced from the optimized geometry, with all other spins fixed at their optimized positions. For each site, Ω is calculated for points in an xy plane parallel to the membrane surface and passing through nitroxide i with z fixed at the equilibrium position. This procedure for determining relative probability distributions for the spins is aided by the fact that all pairwise distances between 74, 225, and 252 have a narrow distribution (Fig. 2) and form a sharply defined basis triangle. Because each pairwise distance distribution is the convolution of the positional distribution of each partner along the interspin vector, a wider distribution involving only one of these sites must be caused by a wider positional spread of the other site.
Fig. 3a presents the probability distribution for the individual spins in R determined by this procedure. The probability for any given nitroxide is coded with a color gradient from blue (low) to purple (high). The distributions are anisotropic and generally elongated along the protein circumference. This tangential elongation reflects the limited angular spread of the distances determined, and the tangential width of the distribution is likely overestimated. Residue 241R1 is not included in Fig. 3a because of overlap with the data of nearby 252R1. Residue 137 has a relatively poorly defined location and is also not shown. The distributions along the radial direction are more accurate and closely represent the true spin distributions in that dimension. The radial distances and distributions for all residues except 137R1 are shown in Fig. 4(solid traces).
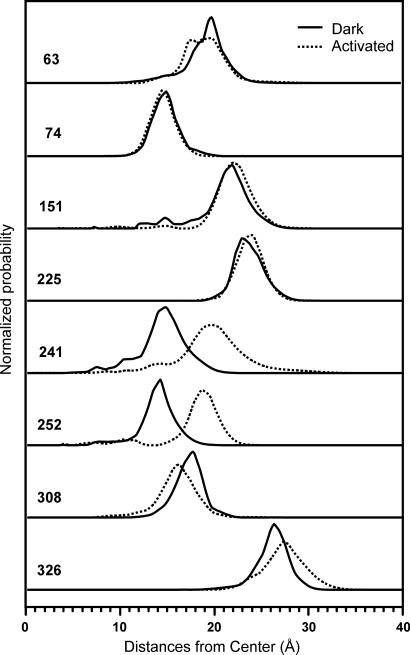
Fig. 4.
After defining a central reference point halfway between the dark location of 74 and 252 (see Fig. 3b), the radial distribution profiles along a line connecting the center and the most probable location is calculated for each site. Each trace incorporates data from two to four measured interspin distance distributions.
A key point to be gleaned from Fig. 3a is that the location of each nitroxide in R determined by DEER agrees remarkably well with that based on the 1GZM structure and models of R1. The only exception is position 326, and the reason for the difference will be discussed below.
Changes Due to Photoactivation.
Upon light-activation, a pattern of R1 movement is reflected in interspin distance changes. Representative examples of the changes as revealed in the raw data are shown in Fig. 2. The new positional distribution of the spins was determined by using the same global analysis described above for the inactive state, and the results are summarized in Fig. 3b. The data for R* are coded with a color gradient from red (low) to yellow (high). For reference, the data are superposed on the R structure, and dotted traces representing the nitroxide positions in R are provided. The distance changes along the radial direction are presented graphically in Fig. 4.
As is evident in Figs. 3b and 4, the activation of rhodopsin is accompanied by a radial outward movement of both 252R1 and 241R1 (Fig. 4 only) in TM6 by 5 Å, a small outward motion of 326R1 by ≈1 Å, and a complex motion of 308R1 in TM7 that has an inward radial component of ≈2 Å. The movement of 63R1 is primarily in the tangential direction with little change in radial position. There is a significant increase in the breadth of the distributions for R1 at 63, 241, 252, 308, and 326 relative to the inactive state. An interpretation of these nitroxide movements in terms of protein structural changes is discussed below.
Discussion
The long-range capability of DEER makes it possible to select solvent-exposed surface sites for placement of R1 residues used in distance determination. The use of surface sites is important for two reasons. First, crystallographic studies show that R1 at solvent-exposed surface sites adopts a limited number of rotamers (29–32). For such sites, the experimental rotamer library (30–32) may be used to build R1 in a structural model of the protein and thus predict interspin distances and distributions for comparison with experimental data, as illustrated here for R (Fig. 3a). The major contribution to the width of the distance distribution from the side chain itself is likely due to rotamers of X4; for R1 residues pointing away from each other, as is the case for many pairs considered in this study (Fig. 1), this contribution is minimized (32). Second, the use of surface sites avoids structural perturbation due to R1 that likely occurs at buried sites due to overpacking of the protein core. Moreover, at buried sites, protein conformational changes could lead to rotamer changes in R1 that are not revealed in the strongly immobilized EPR spectra, giving rise to distance changes unrelated to backbone movement. Even at solvent-exposed sites, rotamer changes could contribute to distance changes, but this possibility is minimized by selecting sites where the EPR spectra do not change significantly, as is the case for the sites selected here (12, 37–40). In this case, distance changes for the nitroxide are expected to reflect positional changes of the backbone. Moreover, changes in the width of the distance distributions reflect changes in spatial disorder of the backbone.
For the relatively short-range (10–20 Å) CW method used in earlier distance mapping of rhodopsin (13, 37, 38), it was not possible to avoid the use of buried sites, and the results are subject to the concerns outlined above. Moreover, it was not possible to measure distances across the molecule to determine the global pattern of helix motion. The opportunity to overcome these problems by using the long-range capabilities of the DEER method provided the motivation for the present studies.
Structure of Inactive Rhodopsin at the Cytoplasmic Surface.
There are seven structures of R in the database, and they differ in the C-terminal sequence (residues 324–348) and the flexible cytoplasmic loop connecting TM5 and TM6 (C3); three of the sites selected for R1 reside in these sequences (sites 241, 252, and 326). In the structures derived from a tetragonal lattice (PDB entries 1F88, 1HZX, 1L9H, and 1U19), C3 is either partially resolved or folded down to lie close to the membrane surface, whereas in those derived from trigonal or rhombohedral lattices (PDB entries 1GZM, 2I35, and 2I36), C3 is formed by irregular extensions of the TM helices connected by a short loop. Previous studies on R1 mobility and accessibility (12) as well as the distance data presented above (Fig. 3) are in excellent agreement with the C3 conformation observed in the trigonal or rhombohedral but not tetragonal crystals. In particular, the nitroxide location of the 241R1 side chain modeled in the 1U19 structure (the highest-resolution structure from a tetragonal lattice) is displaced by ≈15 Å from the experimentally determined position. Thus, the DEER data suggest that R in micelles of DM has a structure most similar to that in the 1GZM, 2I35, and 2I36 structures. The only significant difference observed between the micellar and the 1GZM crystal structure is the relative position of 326R1 (Fig. 3). This is not surprising, because 326 is the last resolved residue in the structure, at the beginning of the C-terminal sequence that has been shown to be highly flexible in solution (40). Structure 1U19 resolves the entire C-terminal tail and places an R1 residue modeled at 326, in agreement with the data of Fig. 3a, and thus is a reasonable model for the micellar structure in the neighborhood of 326, even though it is highly distorted in C3.
An earlier analysis of R1 mobility in C3 showed the loop to be highly flexible, with a gradient of increasing backbone dynamics extending from the membrane intersection of either TM5 or TM6 to the center of the loop (12, 42). Interestingly, the distance distribution for 252R1, located in the regular TM6 helix at the level of the membrane, is substantially narrower than that for 241R1 in the irregular extension of the helix in the loop (Fig. 4), consistent with the flexibility gradient derived from R1 mobility in solution. The difference in conformation of C3 between the crystal forms is likely due to lattice contacts in the tetragonal lattice that distort the flexible loop (8).
Structural Changes at the Cytoplasmic Surface of Rhodopsin upon Photoactivation.
The quantitative DEER data of Figs. 3 and 4 reveal an outward radial displacement of both 241R1 and 252R1 in TM6 by 5 Å along lines passing through a central reference point of the molecule and the positions of the residues. This is interpreted as reflecting a direct outward movement of TM6 and is apparently the major event of activation at the cytoplasmic surface. Earlier CW distance measurements, although only semiquantitative for the reasons discussed above, are in general agreement with the results presented here regarding the motion of TM6 (13). A combined rotation-translation of TM6 as suggested in the earlier study could in principle account for the DEER result. However, this would require a displacement of TM6 toward TM7 or TM5, leading to steric clashes. Thus, in view of the rhodopsin crystal structure and the higher-resolution DEER data, a rectilinear displacement of TM6 is considered to be most likely.
The movements of 308R1, 63R1, and 326R1, although relatively small, clearly reveal changes in the cluster consisting of TM1, TM7, and the C-terminal domain, and earlier SDSL studies support this conclusion (24, 25, 37, 38). However, interpretation of the movements in terms of specific structural changes in the protein remains speculative because the displacement of a single residue in a helix can arise from different combinations of translations and rotations of the helix to which it is attached. The movement of 308R1 has an inward radial component of ≈2 Å (Fig. 4), but TM7 may have in addition a clockwise rotational component to explain the data in Fig. 3b. This motion would account for an earlier result showing that a residue on the opposite face of TM7 (306R1) moves farther from a residue in TM1 (65R1) (24). In that study, it was suggested that the observed motion of 306R1 away from 65R1 could simply be a movement of TM7 away from TM1, but a change in orientation of the buried 306R1 side chain was also considered as possible. The data of Fig. 3 suggest that a rotational motion of TM7 and/or a rearrangement of 306R1 are the most likely explanations of the earlier data.
The displacement of 63R1 is largely tangential and could arise from a small translation or rotation of TM1. Little can be said about the backbone change leading to the movement of 326R1 in the C-terminal domain, but based on proximity it is likely that the movements of TM1, TM7, and the C-domain are concerted. From Fig. 3b it is obvious that the widths of the distributions for 63R1, 308R1, and 326R1 increase after photoactivation, suggesting an increased disorder in these active regions; Fig. 4 further suggests that the radial width of the 241R1 distribution increases significantly.
Of equal interest to the residues that move upon photoactivation are those that do not; namely, 74R1 in TM2, 151R1 in TM4, and 225R1 in TM5 (Fig. 3b). The lack of change of distance between 74R1 and 137R1 in TM3 (Fig. 2) suggests that TM3 also does not move. Support for the lack of change at TM2, TM3, and TM4 comes from the comparative sequence analysis (33) and solid-state NMR data (19) of Smith and coworkers, who suggest that these helices are part of a well packed core of rhodopsin that is invariant under photoactivation.
One crystal structure of an “activated” rhodopsin has been published (23) in which no significant movement of helices was observed compared with R, at odds with the results reported here and other evidence mentioned in the Introduction. However, immediately after a brief illumination of an R crystal, there was a significant loss of diffraction that could be interpreted, but not measured, as a structural change. Only a continuous light exposure for 1–3 h annealed the protein into a transient conformation where a structure could be determined (see supporting information in ref. 23). Although this structure contained all-trans retinal and a deprotonated Schiff base, it does not necessarily correspond to the functionally active conformation (34). Indeed, recent work has shown that the motion of TM6 requires not only the deprotonation of the Schiff base but also the overcoming of an additional energetic barrier (35). Considering the direct and quantitative measurements presented here, it is possible that the lack of TM helix movement in the crystalline lattice reflects stabilization of a form of photoexcited rhodopsin located on the inactive side of the barrier (Meta IIa). This could be due in part to the high osmolarity of the solvent used to stabilize the crystal, a condition that can prevent conformational changes from taking place (44).
In summary, SDSL and DEER spectroscopy allowed a global distance mapping in rhodopsin not possible with the CW dipolar broadening method used previously. With proper experimental design, the results are free from potential artifacts that complicated data interpretation in the earlier studies and limited the effective resolution. The results show conclusively that the outward displacement of TM6 at the cytoplasmic surface is a hallmark of rhodopsin activation and not an artifact of the spectroscopic labeling technique. Smaller changes involving relative motions of TM1, TM7, and the C-terminal domain accompany activation, and the disorder of the molecule in this region is increased by activation.
Methods
Construction, Expression, Purification, and Spin-Labeling of the Rhodopsin Mutants.
The expression construct, pMT4-Rho, containing the base mutant Φ (C140S/C316S) is described in ref. 45. Cysteine double mutations were introduced in the Φ, and DEAE dextran-mediated transient transfections of the construct in mammalian COS1 cells were used to induce expression of the mutant opsin (46). Recombinant rhodopsin was regenerated with 11-cis-retinal (National Eye Institute), solubilized in DM (Anatrace), and purified and spin-labeled on a 1D4 monoclonal antibody (University of British Columbia) immobilized on Sepharose as described in ref. 45.
DEER Spectroscopy.
EPR measurements were conducted on a Bruker Elexsys 580 equipped with an MS-2 split-ring resonator. Rhodopsin samples of ≈15 μl were mixed with 5 μl of glycerol, placed in quartz capillary (1.8-mm O.D., 1.5-mm I.D.), and flash-frozen in liquid nitrogen. The four-pulse sequence of the Bruker-assisted DEER experiment was used with pulses of 8 ns (π/2) and 16 ns (π). The pump frequency was centered on the central resonance and the observe frequency was 70 MHz away on the lowfield side. The measuring temperature was either 50 or 80 K. To generate the photoactivated form, the sample was brought to room temperature and illuminated for 30 s with a 75-W halogen lamp fitted with a 500-nm long-pass filter; the sample was again flash-frozen for data collection. The phase-corrected dipolar evolution data were processed assuming a 3D background and Fourier-transformed, and the distance distributions were obtained with Tikhonov regularization using custom programs written in LabVIEW (National Instruments).
Modeling.
Native amino acids at investigated sites were replaced by an R1 side chain. The dihedral angles X1 and X2 were set to −60° and −60°, respectively, the preferred rotamer of R1 at helical surface sites (29–32). The second most probable rotamer (X1 = 180°, X2 = +60°) maintains a similar spatial nitroxide location (32). The dihedral X3 was set to either +90° or −90°, whichever avoided steric overlap. Dihedral X4 and X5 assume a distribution of values at solvent-exposed sites (29–32, 41) and were allowed to vary to optimize agreement with experimental data.
Geometry Optimization.
A modified Levenberg–Marquardt procedure was used to optimize coordinates of all sites, starting from a geometry taken from the crystal structure. The distance distributions obtained from Tikhonov regularization were slightly smoothed by convolution with a 1-Å-wide Gaussian. This removed any noise that could potentially interfere with the partial derivative calculations. All distances with zero probability received a value of 0.0001/(R − Rmax)2, where Rmax is the distance in Å with highest probability. This did not significantly alter the distribution function but allowed the procedure to converge toward the optimum even if the distance was outside of the probable range. Three-dimensional optimization and generation of the 2D probability maps was performed in custom programs written in LabVIEW.
The optimized geometry for the R and R* state describes a relative topology. The two datasets do not have a common reference point to each other and to the crystal structure. Each can be rotated and translated as a rigid body without changing the agreement with the DEER data. To align the data, the coordinates of the dark geometry were overlaid on the 1GZM structure to optimize the agreement with the crystal structure such that the spin locations were near the expected positions. The activated geometry was subsequently overlaid to limit significant relocations to as few residues as possible. Of course, the activated geometry could be overlaid with the structure so that TM6 remains fixed and the entire remainder of the molecule moves, but this is not a viable solution in view of a wealth of experimental data reviewed in the text.
Acknowledgments.
We thank Rob Herman for excellent technical assistance and James Trudell and Ned Van Eps for helpful discussions. This work was supported by National Institutes of Health Grant EY05216 (to W.L.H.), the Jules Stein Professorship endowment (W.L.H.), and Deutsche Forschungsgemeinschaft Sfb449 (to O.P.E.) and Sfb740 (to O.P.E. and K.P.H.).
Footnotes
The authors declare no conflict of interest.
See Commentary on page 7343.
References
- 1.Johnston C, Siderovski D. Receptor-mediated activation of heterotrimeric G-proteins: Current structural insights. Mol Pharmacol. 2007;72:219–230. doi: 10.1124/mol.107.034348. [DOI] [PubMed] [Google Scholar]
- 2.Oldham WM, Hamm HE. Heterotrimeric G protein activation by G-protein-coupled receptors. Nat Rev Mol Cell Biol. 2008;9:60–71. doi: 10.1038/nrm2299. [DOI] [PubMed] [Google Scholar]
- 3.Schwartz TW, Frimurer TM, Holst B, Rosenkilde MM, Elling CE. Molecular mechanism of 7TM receptor activation—A global toggle switch model. Annu Rev Pharmacol Toxicol. 2006;46:481–519. doi: 10.1146/annurev.pharmtox.46.120604.141218. [DOI] [PubMed] [Google Scholar]
- 4.Kobilka BK. G protein coupled receptor structure and activation. Biochim Biophys Acta. 2007;1768:794–807. doi: 10.1016/j.bbamem.2006.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palczewski K, et al. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 6.Teller DC, Okada T, Behnke CA, Palczewski K, Stenkamp RE. Advances in determination of a high-resolution three-dimensional structure of rhodopsin, a model of G protein-coupled receptors (GPCRs) Biochemistry. 2001;40:7761–7772. doi: 10.1021/bi0155091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Okada T, et al. Functional role of internal water molecules in rhodopsin revealed by x-ray crystallography. Proc Natl Acad Sci USA. 2002;99:5982–5987. doi: 10.1073/pnas.082666399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li J, Edwards PC, Burghammer M, Villa C, Schertler GF. Structure of bovine rhodopsin in a trigonal crystal form. J Mol Biol. 2004;343:1409–1438. doi: 10.1016/j.jmb.2004.08.090. [DOI] [PubMed] [Google Scholar]
- 9.Okada T, et al. The retinal conformation and its environment in rhodopsin in light of a new 2.2 Å crystal structure. J Mol Biol. 2004;342:571–583. doi: 10.1016/j.jmb.2004.07.044. [DOI] [PubMed] [Google Scholar]
- 10.Cherezov V, et al. High-resolution crystal structure of an engineered human β2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rosenbaum DM, et al. GPCR engineering yields high-resolution structural insights into β2-adrenergic receptor function. Science. 2007;318:1266–1273. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- 12.Altenbach C, et al. Structural features and light-dependent changes in the cytoplasmic interhelical E-F loop region of rhodopsin: A site-directed spin-labeling study. Biochemistry. 1996;35:12470–12478. doi: 10.1021/bi960849l. [DOI] [PubMed] [Google Scholar]
- 13.Farrens DL, Altenbach C, Yang K, Hubbell WL, Khorana HG. Requirement of rigid-body motion of transmembrane helices for light activation of rhodopsin. Science. 1996;274:768–770. doi: 10.1126/science.274.5288.768. [DOI] [PubMed] [Google Scholar]
- 14.Hubbell WL, Altenbach C, Hubbell CM, Khorana HG. Rhodopsin structure, dynamics, and activation: A perspective from crystallography, site-directed spin labeling, sulfhydryl reactivity, and disulfide cross-linking. Adv Protein Chem. 2003;63:243–290. doi: 10.1016/s0065-3233(03)63010-x. [DOI] [PubMed] [Google Scholar]
- 15.Kusnetzow AK, Altenbach C, Hubbell WL. Conformational states and dynamics of rhodopsin in micelles and bilayers. Biochemistry. 2006;45:5538–5550. doi: 10.1021/bi060101v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunham TD, Farrens DL. Conformational changes in rhodopsin. Movement of helix F detected by site-specific chemical labeling and fluorescence spectroscopy. J Biol Chem. 1999;274:1683–1690. doi: 10.1074/jbc.274.3.1683. [DOI] [PubMed] [Google Scholar]
- 17.Lin SW, Sakmar TP. Specific tryptophan UV-absorbance changes are probes of the transition of rhodopsin to its active state. Biochemistry. 1996;35:11149–11159. doi: 10.1021/bi960858u. [DOI] [PubMed] [Google Scholar]
- 18.Sheikh SP, Zvyaga TA, Lichtarge O, Sakmar TP, Bourne HR. Rhodopsin activation blocked by metal-ion binding sites linking transmembrane helices C and F. Nature. 1996;383:347–350. doi: 10.1038/383347a0. [DOI] [PubMed] [Google Scholar]
- 19.Crocker E, Eilers M, Ahuja S, Smith SO. Location of Trp265 in metarhodopsin II: Implications for the activation mechanism of the visual receptor rhodopsin. J Mol Biol. 2006;357:163–172. doi: 10.1016/j.jmb.2005.12.046. [DOI] [PubMed] [Google Scholar]
- 20.Sansom MS, Weinstein H. Hinges, swivels and switches: The role of prolines in signalling via transmembrane α-helices. Trends Pharmacol Sci. 2000;21:445–451. doi: 10.1016/s0165-6147(00)01553-4. [DOI] [PubMed] [Google Scholar]
- 21.Luo X, Zhang D, Weinstein H. Ligand-induced domain motion in the activation mechanism of a G-protein-coupled receptor. Protein Eng. 1994;7:1441–1448. doi: 10.1093/protein/7.12.1441. [DOI] [PubMed] [Google Scholar]
- 22.Shi L, et al. β2 adrenergic receptor activation. Modulation of the proline kink in transmembrane 6 by a rotamer toggle switch. J Biol Chem. 2002;277:40989–40996. doi: 10.1074/jbc.M206801200. [DOI] [PubMed] [Google Scholar]
- 23.Salom D, et al. Crystal structure of a photoactivated deprotonated intermediate of rhodopsin. Proc Natl Acad Sci USA. 2006;103:16123–16128. doi: 10.1073/pnas.0608022103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Altenbach C, Cai K, Klein-Seetharaman J, Khorana HG, Hubbell WL. Structure and function in rhodopsin: Mapping light-dependent changes in distance between residue 65 in helix TM1 and residues in the sequence 306–319 at the cytoplasmic end of helix TM7 and in helix H8. Biochemistry. 2001;40:15483–15492. doi: 10.1021/bi011546g. [DOI] [PubMed] [Google Scholar]
- 25.Altenbach C, Klein-Seetharaman J, Cai K, Khorana HG, Hubbell WL. Structure and function in rhodopsin: Mapping light-dependent changes in distance between residue 316 in helix 8 and residue in the sequence 60–75, covering the cytoplasmic end of helix TM 1 and TM 2 and their connection loop CL1. Biochemistry. 2001;40:15493–15500. doi: 10.1021/bi011545o. [DOI] [PubMed] [Google Scholar]
- 26.Altenbach C, Oh KJ, Trabanino RJ, Hideg K, Hubbell WL. Estimation of inter-residue distances in spin labeled proteins at physiological temperatures: Experimental strategies and practical limitations. Biochemistry. 2001;40:15471–15482. doi: 10.1021/bi011544w. [DOI] [PubMed] [Google Scholar]
- 27.Rabenstein MD, Shin YK. Determination of the distance between two spin labels attached to a macromolecule. Proc Natl Acad Sci USA. 1995;92:8239–8243. doi: 10.1073/pnas.92.18.8239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pannier M, Veit S, Godt A, Jeschke G, Spiess HW. Dead-time free measurement of dipole-dipole interactions between electron spins. J Magn Reson. 2000;142:331–340. doi: 10.1006/jmre.1999.1944. [DOI] [PubMed] [Google Scholar]
- 29.Langen R, Oh KJ, Cascio D, Hubbell WL. Crystal structures of spin labeled T4 lysozyme mutants: Implications for the interpretation of EPR spectra in terms of structure. Biochemistry. 2000;39:8396–8405. doi: 10.1021/bi000604f. [DOI] [PubMed] [Google Scholar]
- 30.Guo Z, Cascio D, Hideg K, Hubbell WL. Structure determinants of nitroxide motion in spin-labeled proteins: Solvent-exposed sites in helix B of T4 lysozyme. Protein Sci. 2008;17:228–239. doi: 10.1110/ps.073174008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo Z, Cascio D, Hideg K, Kalai T, Hubbell WL. Structural determinants of nitroxide motion in spin-labeled proteins: Tertiary contact and solvent-inaccessible sites in helix G of T4 lysozyme. Protein Sci. 2007;16:1069–1086. doi: 10.1110/ps.062739107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fleissner MR. Los Angeles: Univ of California; 2007. X-ray structures of nitroxide side chains in proteins: A basis for interpreting distance measurements and dynamic studies by electron paramagnetic resonance. PhD dissertation. [Google Scholar]
- 33.Liu W, Eilers M, Patel AB, Smith SO. Helix packing moments reveal diversity and conservation in membrane protein structure. J Mol Biol. 2004;337:713–729. doi: 10.1016/j.jmb.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 34.Arnis S, Hofmann KP. Two different forms of metarhodopsin II: Schiff base deprotonation precedes proton uptake and signaling. Proc Natl Acad Sci USA. 1993;90:7849–7853. doi: 10.1073/pnas.90.16.7849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Knierim B, Hofmann KP, Ernst OP, Hubbell WL. Sequence of late molecular events in the activation of rhodopsin. Proc Natl Acad Sci USA. 2007;104:20290–20295. doi: 10.1073/pnas.0710393104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ernst O, Gramse V, Kolbe M, Hofmann K, Heck M. Monomeric G protein-coupled receptor rhodopsin in solution activates its G protein transducin at the diffusion limit. Proc Natl Acad Sci USA. 2007;104:10859–10864. doi: 10.1073/pnas.0701967104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Altenbach C, Cai K, Khorana HG, Hubbell WL. Structural features and light-dependent changes in the sequence 306–322 extending from helix VII to the palmitoylation sites in rhodopsin: A site-directed spin-labeling study. Biochemistry. 1999;38:7931–7937. doi: 10.1021/bi9900121. [DOI] [PubMed] [Google Scholar]
- 38.Altenbach C, Klein-Seetharaman J, Hwa J, Khorana HG, Hubbell WL. Structural features and light-dependent changes in the sequence 59–75 connecting helices I and II in rhodopsin: A site-directed spin-labeling study. Biochemistry. 1999;38:7945–7949. doi: 10.1021/bi990014l. [DOI] [PubMed] [Google Scholar]
- 39.Farahbakhsh ZT, Ridge KD, Khorana HG, Hubbell WL. Mapping light-dependent structural changes in the cytoplasmic loop connecting helices C and D in rhodopsin: A site-directed spin labeling study. Biochemistry. 1995;34:8812–8819. doi: 10.1021/bi00027a033. [DOI] [PubMed] [Google Scholar]
- 40.Langen R, Cai K, Altenbach C, Khorana HG, Hubbell WL. Structural features of the C-terminal domain of bovine rhodopsin: A site-directed spin-labeling study. Biochemistry. 1999;38:7918–7924. doi: 10.1021/bi990010g. [DOI] [PubMed] [Google Scholar]
- 41.Columbus L, Kálai T, Jekö J, Hideg K, Hubbell WL. Molecular motion of spin labeled side chains in α-helices: Analysis by variation of side chain structure. Biochemistry. 2001;40:3828–3846. doi: 10.1021/bi002645h. [DOI] [PubMed] [Google Scholar]
- 42.Columbus L, Hubbell WL. A new spin on protein dynamics. Trends Biochem Sci. 2002;27:288–295. doi: 10.1016/s0968-0004(02)02095-9. [DOI] [PubMed] [Google Scholar]
- 43.Mchaourab HS, Lietzow MA, Hideg K, Hubbell WL. Motion of spin-labeled side chains in T4 lysozyme. Correlation with protein structure and dynamics. Biochemistry. 1996;35:7692–7704. doi: 10.1021/bi960482k. [DOI] [PubMed] [Google Scholar]
- 44.Fanucco GE, Lee JY, Cafiso DS. Spectroscopic evidence that osmolytes used in crystallization buffers inhibit a conformation change in a membrane protein. Biochemistry. 2003;42:13106–13112. doi: 10.1021/bi035439t. [DOI] [PubMed] [Google Scholar]
- 45.Resek JF, Farahbakhsh ZT, Hubbell WL, Khorana HG. Formation of the meta II photointermediate is accompanied by conformational changes in the cytoplasmic surface of rhodopsin. Biochemistry. 1993;32:12025–12032. doi: 10.1021/bi00096a012. [DOI] [PubMed] [Google Scholar]
- 46.Oprian DD, Molday RS, Kaufman RJ, Khorana HG. Expression of a synthetic bovine rhodopsin gene in monkey kidney cells. Proc Natl Acad Sci USA. 1987;84:8874–8878. doi: 10.1073/pnas.84.24.8874. [DOI] [PMC free article] [PubMed] [Google Scholar]