Abstract
Alzheimer's disease is a neurodegenerative disorder that is characterized by the cerebral deposition of amyloid fibrils formed by Aβ peptide. Despite their prevalence in Alzheimer's and other neurodegenerative diseases, important details of the structure of amyloid fibrils remain unknown. Here, we present a three-dimensional structure of a mature amyloid fibril formed by Aβ(1-40) peptide, determined by electron cryomicroscopy at ≈8-Å resolution. The fibril consists of two protofilaments, each containing ≈5-nm-long regions of β-sheet structure. A local twofold symmetry within each region suggests that pairs of β-sheets are formed from equivalent parts of two Aβ(1-40) peptides contained in each protofilament. The pairing occurs via tightly packed interfaces, reminiscent of recently reported steric zipper structures. However, unlike these previous structures, the β-sheet pairing is observed within an amyloid fibril and includes significantly longer amino acid sequences.
Keywords: Alzheimer's disease, amyloid-β, electron cryomicroscopy, neurodegeneration, protein folding
The deposition of fibrillar polypeptide aggregates is linked to several neurological disorders, including Alzheimer's, Creutzfeldt–Jakob, and Parkinson's diseases (1–3). The aggregates occurring in these diseases were often shown to be amyloid fibrils. Amyloid fibrils are defined by the presence of a characteristic structural motif that is known as cross-β. Cross-β means that the strands of a β-sheet run perpendicular to the fibril axis (4). However, obtaining structural information from amyloid fibrils at atomic resolution still represents a major challenge for current techniques in structural biology. Thus far, crystals amenable for x-ray analysis have not been obtained from fibrils of biologically relevant full-length polypeptide chains. Structures at atomic resolution were elucidated for amyloid-like microcrystals or fibrils from short peptide fragments (4–12 residues) (5–7), whereas structural models have been proposed for the fibrils from full-length Aβ(1-40) and Aβ(1-42) peptides (8–12) based on solid-state NMR data, hydrogen–deuterium exchange NMR, mutagenesis, and transmission electron microscopy (TEM) and scanning TEM (STEM) analysis. One problem associated with samples of amyloid fibrils is their structural heterogeneity (10, 11, 13). Amyloid fibrils occur in a wide variety of morphologies differing in width and helical twist. This structural diversity stands in the way of crystallization and limits the information obtained from experiments that average over different morphologies. Moreover, differences in the examined fibril morphologies may limit the comparability of results obtained with different fibril preparations.
The structure of one morphology of an Aβ(1-40) fibril has been determined by electron cryomicroscopy (cryo-EM) at 26-Å resolution (14). The examined fibril showed a polar structure of left-handed chirality and displayed all common characteristics of amyloid fibrils, including an elongated, unbranched morphology, a 4.8-Å cross-β reflection in x-ray fiber diffraction, binding of Congo red and thioflavin-T dyes, and apple-green birefringence of Congo red-bound fibrils. Here, we performed a cryo-EM analysis of the previously analyzed amyloid fibril morphology using additional data (Fig. 1 and Table 1) and a newly developed image-processing method (15). We present an improved fibril structure at ≈8-Å resolution, revealing the internal features of this fibril.
Fig. 1.
Amyloid fibrils formed from Aβ(1-40) embedded in vitreous ice and analysis of sample heterogeneity. (A) The sample was applied to a grid covered with holey-carbon support film with 1.2-μm holes and plunge-frozen in liquid ethane. (B) Quantitative cross-over distance analysis of fibrils present in the sample. A total of 46% of the fibrils (highlighted in green) were selected for image processing, thus limiting the helical twist to 142 ± 4.7 nm. (C) The averaged power spectrum of in-plane aligned fibril segments shows reflections at 4.8-Å resolution, consistent with the cross-β structure of the fibrils.
Table 1.
Image-processing statistics
| Resolution at FSC 0.5/0.143, Å | 8.8/7.1 |
| Total length of nonoverlapping segments, nm | 87,265 |
| No. of fibrils | 188 |
| No. of segments | 11,527 |
| Segment size, nm | 84.2 |
| Size of 3D reconstruction, nm | 77.7 |
| Segment step size, nm | 6.5 |
| Average cross-over distance/repeat distance, nm | 142.5/0.48 |
| Pixel size on the specimen, Å | 1.2 |
Results
Selection of Fibrils.
The careful selection of fibrils with similar morphology was an important step for the high-resolution analysis of the data. We selected fibrils based on their width and helical twist. The latter was measured as the cross-over distance. The overwhelming majority of fibrils (90%) present in the sample exhibit a width of ≈19 nm, similar to earlier studies (14). The measured crossover distances of all considered fibrils show an average of 134 nm and a standard deviation of 13.1 nm (Fig. 1B). We selected a subset of fibrils for further image processing. This subset possessed cross-over distances between 130 and 150 nm, representing ≈46% of the total fibril population. This selection significantly reduced the structural heterogeneity present in the images used for reconstruction while keeping the size of the dataset sufficiently large for high-resolution analysis. An averaged power spectrum of the processed fibril segments shows a clear 4.8-Å signal, consistent with the cross-β structure of the fibrils (Fig. 1C). The signal at 4.8-Å resolution attests to the quality of the data.
Fibril Structure.
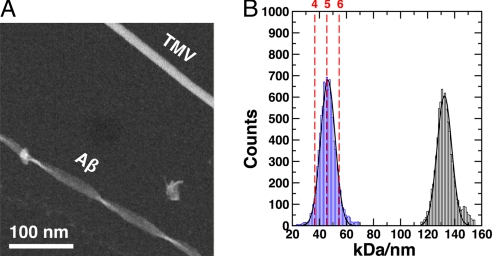
To estimate the number of Aβ(1-40) peptides in the reconstructed fibril, we used STEM to quantify the mass-per-length (MPL) value of the examined morphology. These measurements suggest that one 4.8-Å peptide layer of the fibril consists, on average, of five peptides (Fig. 2).
Fig. 2.
MPL measurements from scanning transmission electron microscopy. (A) Dry-frozen specimens of Aβ(1-40) fibrils and TMV used for mass calibration. (B) Histogram of the fibril MPL measurements shows a value of 46 kDa/nm for Aβ(1-40) with a standard deviation of 5.3 kDa/nm, indicating five Aβ(1-40) peptides per 4.8-Å repeat. The second peak at 131 kDa/nm with a standard deviation of 5.3 kDa/nm was obtained for TMV and serves as an estimate of the MPL measurement error.
The averaged 3D fibril structure possesses a width of 19 nm and a helical pitch of 285 nm (Fig. 3). Its cross-sectional structure shows quasi-2-fold symmetry that is also visible when this symmetry is not imposed (data not shown). The small asymmetry seen in the reconstructed cross-section (Fig. 4A) is due to the optimization of the image alignment procedure for one side of the fibril (see Materials and Methods). The path of the polypeptide chain is visible in most parts of the structure as a narrow density trace (Fig. 4A). The cross-section accommodates two U-shaped traces with head-to-head orientation, indicating a double-helical fibril structure consisting of two protofilaments. Hence, a protofilament represents a single filamentous substructure of the fibril. Distinct parts of the structure can be identified in cross-section within different radial regions around the central axis of the fibril.
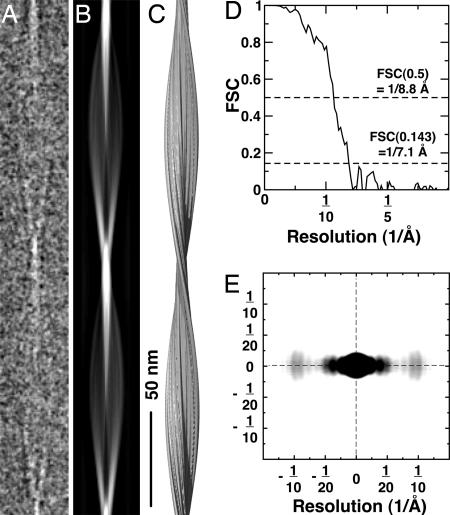
Fig. 3.
A 3D reconstruction of Aβ(1-40). (A) Ice-embedded Aβ(1-40) fibril cropped from a raw electron micrograph, displaying the distance of one helical pitch. (B) Projection of the 3D fibril reconstruction shown in the C. (C) Side-view surface rendering of the fibril reconstruction. The structure was prepared by using UCSF CHIMERA (35). (D) Fourier shell correlation curve of the reconstruction indicates a resolution of ≈8 Å. (E) Electron diffraction pattern calculated by using the fibril cryo-EM reconstruction. The equatorial reflections peak at ≈10 Å and show diffuse intensity between 8 and 12 Å, representing the β-sheet spacing. The intensity distribution at lower resolution reflects the protofilament thickness (≈20 Å nm). Possible structural disorder in the fibril and solvent effects were not included in the calculation of the pattern, leading to differences between the calculated and experimentally observed intensities.
Fig. 4.
Interpretation of the density. (A) Fibril density projected along the helical axis. The density exhibits a twofold rotational symmetry consisting of two protofilaments. (B) The β-sheet sandwich in the protofilament core is highlighted by the rectangle. It exhibits a local twofold symmetry and a corrugated core structure. (C) Comparison of Aβ models. The dashed lines represent disordered N-terminal residues of the peptide, whereas solid regions with arrows indicate β-strands. (Top) Petkova et al. (8). (Middle) Lührs et al (9). (Bottom) Model of the Aβ(1-40) peptide based on the present cryo-EM image reconstruction within one protofilament. Two oppositely directed polypeptide chains form a β-sheet core region and are bounded by the peptide N termini. The number of β-sheet segments making up the core cannot be determined from the cryo-EM structure and, therefore, individual β-strands are not indicated in this model.
The most striking feature of the structure is the presence of two elongated and juxtaposed density regions in each protofilament, at a radius between 2.5 and 7 nm (Fig. 4A). These densities are most consistent with a pair of close-packed β-sheet regions. However, the density cross-section shows a remarkable zigzag shape that appears to break up each elongated region into shorter segments (Fig. 4B). This may indicate that the fibril does not consist of β-sheets with very long and nearly straight β-strands but rather of several shorter β-segments. Alternatively, a zigzag shape similar to ours may also be produced by density corresponding to some of the larger side chains of Aβ(1-40), if these alternate between the two sides of the β-sheet. A distinction between these two possibilities will be possible at higher resolution or when an alignment of the peptide sequence with the observed density can be made.
The intersheet spacing of the individual β-segments varies between 0.9 and 1.2 nm, consistent with known sequence-dependent variations in the β-sheet of amyloid structures (16) and the diffuse equatorial reflection (spread around ≈1 nm) observed in x-ray diffraction patterns collected from Aβ(1-40) fibrils (14, 17). The x-ray fiber diffraction patterns are reproduced qualitatively by a diffraction pattern calculated by using our 3D reconstruction (Fig. 3E).
Discussion
The observation of the paired β-sheet regions is reminiscent of the recently described steric zipper structures. Steric zippers consist of two juxtaposed and often interdigitating β-sheets (7). One β-sheet side shows extensive contacts with the neighboring β-sheet, whereas the other is more solvent-exposed. Our structure also shows a sandwich of two β-sheet regions, each containing an inner and an outer side, which are equivalent to the described solvent-protected and solvent-exposed surfaces. The β-sheet regions are ≈5 nm long and can accommodate a sequence of ≈15 aa. Reported steric zippers are formed from very short peptides (4–7 aa), although the actual zipper interface is often formed by even smaller segments of these peptides. Short-sequence segments have been proposed to determine the cross-β aggregation even within much longer polypeptide chains (7, 18). Moreover, β-strands observed in native β-sheet proteins often have a relatively short size, approximately five residues (19), depending on whether they occur in an antiparallel or parallel β-sheet. By contrast, several studies provide evidence that the β-sheet regions of full-size amyloid fibrils can be much longer (8, 9, 20–23). The observed zigzag structure of the 5-nm β-sheet regions suggest that these regions may in fact consist of shorter β-segments, each forming a zipper-like structure, thus maximizing the interdigitation between the paired β-sheet regions.
At a radius of 2.5 nm (Fig. 4A), further density connects the two β-sheet regions and generates a U-shaped protofilament cross-section. This structure superficially resembles previous U-shaped Aβ models with β-arch conformation (8, 9). However, none of these previous models fits our structure in detail (Fig. 4C). At a radius of 2.5 nm, the β-sheet separation of our structure reaches 2 nm, which is significantly wider than in previously proposed β-arch conformations. Moreover, the entire protofilament cross-section seen in our structure must include considerably more than one Aβ peptide, because a single 40-residue peptide cannot explain the entire observed density. Within a 7-nm radius, a continuous trace of the strongest density features in one protofilament has a length of ≈16 nm. This value corresponds to an extended polypeptide chain of ≈50 residues, depending on the precise values of its ψ/φ dihedral angles of the backbone. The presence of more than one Aβ peptide in cross-section per protofilament is also consistent with the MPL measurements that indicate an average of 2.5 peptides per protofilament cross-section (Fig. 2).
Although the present resolution does not allow for construction of a structural model showing the position and conformation of individual residues, the strong density traces visible in cross-section (Fig. 4B) provide a clear outline for the general arrangement of peptides within a protofilament. Fig. 4C compares previously suggested models of peptides within a fibril with a model suggested by the present cryo-EM structure. An important aspect considered by our model is the local twofold symmetry of the protofilament cross-section with the symmetry axis centered between the paired β-sheet regions at a radius of ≈4.5 nm (Fig. 4B). This local symmetry relates the paired β-sheets to each other and suggests that they contain equivalent sequence regions of two peptides that run in opposite directions (Fig. 4C). The model is consistent with many other structural constraints suggested previously (7–9, 12, 24), including studies using pairwise mutations of Aβ(1-42) fibrils (9) and with solid-state NMR data from Aβ(1-40) (24). The first report assigns intermolecular contacts of F19/G38, A21/V36 and the salt bridge between D23 and K28 to intermolecular contacts between adjacently stacked peptides parallel to the fibril axis, whereas the second report interprets the interaction among L17/F19, I32/L34/L36, and I31/G37 as intermolecular. In our present model, these contacts could form between the two juxtaposed but oppositely oriented peptides in the protofilament cross-section. However, our model may not explain some intramolecular contacts reported previously (25). This difference could indicate that the presently examined fibril morphology differs from fibril morphologies examined previously. For example, Aβ(1-40) and Aβ(1-42) peptides are known to be able to form fibrils of a wide range of different morphologies (11, 26).
Despite the local symmetry of the highly ordered protofilament cross-β core, the protofilament cross-section is evidently asymmetrical, suggesting that the dihedrally paired peptides need not be strictly equivalent. Some of the asymmetry may be related to structural disorder. For example, the density between 7- and 9-nm radius does not show characteristic features of any secondary structural elements. Previous studies carried out with proteolysis experiments, hydrogen–deuterium exchange NMR, and mass spectrometry (9, 21, 27) have argued that part of the Aβ chain within the fibril adopts an ordered structure, whereas other parts of the peptide are more flexible. These studies assign the regions of structural disorder mostly to the peptide N terminus. By analogy, this would imply that the diffuse density at 7- to 9-nm radius corresponds to the N terminus of one of the two peptides forming the more ordered protofilament core (Fig. 4C). Because of the local two-fold symmetry of the protofilament cross-section, the other peptide must have its N terminus oriented toward the center of the fibril, forming the U-shaped connection <2.5-nm radius.
In addition to the protofilament core and flanking N-terminal regions interpreted by the model in Fig. 4C, our structure shows features made up of weaker density that extend parallel to the putative β-sheet core. Their interpretation is more tenuous, possibly indicating partial disorder or subtle structural polymorphism within the framework of one-fibril morphology. These features could also indicate a small fraction of peptides that associate peripherally with the highly ordered fibril core but do not occur every 4.8 Å along the fibril. This could explain the noninteger number of peptides per 4.8-Å repeat per protofilament seen in the MPL analysis (Fig. 2).
Whereas several aspects of the presented structural description correspond to earlier structural proposals on amyloid fibrils or amyloid models, the structural constraints arising from our density map show how these previously isolated properties can be combined to generate the structure of the mature fibril. One unexpected finding is the corrugated organization of the β-sheet regions. If a segmented structure is responsible for such an appearance of the β-sheet regions, this would also suggest a mechanism of fibril formation in which there is an initial association of individual β-segments that will then position adjacent segments to form the rest of the β-sheet interface. These findings could be important in the context of amyloid pathogenicity that may arise from structural precursors of these fibrils (28). The analyzed fibril structure represents only one of many different fibril morphologies. It will be interesting to learn, therefore, how different pairings of Aβ peptide are possible (7, 29), leading to the wide range of morphologies observed in a typical fibril population.
Materials and Methods
Sample Preparation.
Fibrils were formed by incubation of Aβ(1-40) peptide (Bachem) at 1 mg/ml in 50 mM sodium borate buffer (pH 7.8, 4°C) for at least 4 days.
Electron Microscopy.
Cryo-EM samples were prepared as described (14). The vitrified specimens were imaged on a Tecnai F30 microscope at 300 kV, with a magnification of 59,000 and a dose of 35 e−/A2. A total of 250 images were recorded at underfocus of 2.0, 2.5, and 3.5 μm on Kodak ISO163 film.
Image Processing.
Table 1 contains details about the processing. Sixty micrographs were selected for micrograph and fibril quality. Fibril quality was assessed according to length and straightness. Cross-over distances were limited to 142.5 ± 4.7 nm. Micrographs were scanned with a raster size of 7 μm, resulting in a pixel size of 1.19 Å. A total of 188 fibrils were segmented by using EMAN′s boxer program (30) with step size of 6.3 nm, resulting in an image stack of 11,527 squares of 84.2 × 84.2 nm. We used the SPIDER (31) image-processing procedure developed for the high-resolution study of tobacco mosaic virus (TMV) (15) and calculated a reconstruction of 77.7 × 77.7 × 77.7 nm in size. Power spectra were calculated from all segments after in-plane alignment, summed, and divided by a rotationally averaged version of the summed spectrum to amplify the weak features at high resolution (Fig. 1C). The parameters of out-of-plane tilt and helical axis angles were restrained during the alignment in addition to restraints applied to the x-shift and in-plane-rotation angle described in ref. 15. The specimen out-of-plane tilt angle was measured by CTFTILT (32). The angle around the helical axis was analyzed with respect to its segment position. We performed a linear regression based on the views, including cross-overs to predict the angles for the views between the cross-overs while obeying an ideal symmetry. Additional twofold symmetry was assumed by aligning each segment in two possible orientations differing by 180° rotation around the helical axis. However, the translational alignment along the fibril axis for a segment in each of the two possible orientations was allowed to be different. Therefore, a small asymmetry evolved during the refinement of the structure, with one protofilament (left in Fig. 4A) developing stronger density than the other. Alignment was therefore biased to match the stronger density in one protofilament, thereby optimizing its density. The protofilament with the stronger density is shown in Fig. 4B. The final reconstruction used for display and resolution assessment had a size of 30 × 30 × 30 nm. The small variations in twist, based on the measured cross-over distance, were accounted for in the symmetrization step of the reconstruction procedure. Cross-sectional views were generated by adding four x–y planes of the fibril (4 × 1.2 Å = 4.8 Å), which corresponds to a single repeat of the fibril reconstruction. We also calculated a reconstruction assuming a 9.6-Å repeat (data not shown). This reconstruction did not differ significantly from the reconstruction shown in Figs. 3 and 4. The model diffraction pattern was calculated from the superposition of a series of azimuthally rotated side-view projections of the 3D reconstruction. Furthermore, to simulate orientational disorder of experimental specimens, the power spectrum of the average was blurred rotationally by using a Gaussian distribution with 20° standard deviation. This was followed by a Gaussian low-pass filter equivalent to ≈3 × 3-pixel averaging.
STEM.
Experiments were performed at the Brookhaven National Laboratory (Upton, NY). The preparation and image acquisition conditions are described in ref. 33. Data were analyzed by using the computer program PCMASS29 (33) with the filament trace option. The MPL measurements were calibrated with TMV for each individual image as recently described (34).
Acknowledgments.
We thank Joseph Wall and Martha Simon (Brookhaven National Laboratory) for performing STEM analysis on our fibril preparations and Donald Caspar for numerous discussions and careful reading of the manuscript. N.G. gratefully acknowledges financial support by National Institutes of Health Grant 1 P01 GM-62580 and by a research fellowship from the Humboldt Foundation. M.F. was supported by the BioFuture program [Federal Ministry of Education and Research, Germany (BMBF)].
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: A density map of the Aβ(1-40) fibril reconstruction has been deposited in the Macromolecular Structure Database at the European Bioinformatics Institute (accession no. EMD-5008).
References
- 1.Cohen F, Kelly J. Therapeutic approaches to protein-misfolding diseases. Nature. 2003;426:905–909. doi: 10.1038/nature02265. [DOI] [PubMed] [Google Scholar]
- 2.Mattson MP. Pathways towards and away from Alzheimer's disease. Nature. 2004;430:631–639. doi: 10.1038/nature02621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chiti F, Dobson C. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 4.Serpell LC. Alzheimer's amyloid fibrils: Structure and assembly. Biochim Biophys Acta. 2000;1502:16–30. doi: 10.1016/s0925-4439(00)00029-6. [DOI] [PubMed] [Google Scholar]
- 5.Jaroniec C, et al. High-resolution molecular structure of a peptide in an amyloid fibril determined by magic angle spinning NMR spectroscopy. Proc Natl Acad Sci USA. 2004;101:711–716. doi: 10.1073/pnas.0304849101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Makin O, Atkins E, Sikorski P, Johansson J, Serpell L. Molecular basis for amyloid fibril formation and stability. Proc Natl Acad Sci USA. 2005;102:315–320. doi: 10.1073/pnas.0406847102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sawaya M, et al. Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Nature. 2007;447:453–457. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- 8.Petkova A, et al. A structural model for Alzheimer's β-amyloid fibrils based on experimental constraints from solid state NMR. Proc Natl Acad Sci USA. 2002;99:16742–16747. doi: 10.1073/pnas.262663499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lührs T, et al. Three-dimensional structure of Alzheimer's amyloid-β(1-42) fibrils. Proc Natl Acad Sci USA. 2005;102:17342–17347. doi: 10.1073/pnas.0506723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kodali R, Wetzel R. Polymorphism in the intermediates and products of amyloid assembly. Curr Opin Struct Biol. 2007;17:48–57. doi: 10.1016/j.sbi.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 11.Goldsbury C, et al. Studies on the in vitro assembly of a beta 1-40: Implications for the search for a beta fibril formation inhibitors. J Struct Biol. 2000;130:217–231. doi: 10.1006/jsbi.2000.4259. [DOI] [PubMed] [Google Scholar]
- 12.Petkova A, et al. Self-propagating, molecular-level polymorphism in Alzheimer's beta-amyloid fibrils. Science. 2005;307:262–265. doi: 10.1126/science.1105850. [DOI] [PubMed] [Google Scholar]
- 13.Jimenez J, et al. The protofilament structure of insulin amyloid fibrils. Proc Natl Acad Sci USA. 2002;99:9196–9201. doi: 10.1073/pnas.142459399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sachse C, et al. Quaternary structure of a mature amyloid fibril from Alzheimer's Aβ(1-40) peptide. J Mol Biol. 2006;362:347–354. doi: 10.1016/j.jmb.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 15.Sachse C, et al. High-resolution electron microscopy of helical specimens: A fresh look at tobacco mosaic virus. J Mol Biol. 2007;371:812–835. doi: 10.1016/j.jmb.2007.05.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fändrich M, Dobson C. The behaviour of polyamino acids reveals an inverse side chain effect in amyloid structure formation. EMBO J. 2002;21:5682–5690. doi: 10.1093/emboj/cdf573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malinchik SB, Inouye H, Szumowski KE, Kirschner DA. Structural analysis of Alzheimer's beta(1-40) amyloid: Protofilament assembly of tubular fibrils. Biophys J. 1998;74:537–545. doi: 10.1016/S0006-3495(98)77812-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fernandez-Escamilla A, Rousseau F, Schymkowitz J, Serrano L. Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat Biotechnol. 2004;22:1302–1306. doi: 10.1038/nbt1012. [DOI] [PubMed] [Google Scholar]
- 19.Sreerama N, Venyaminov S, Woody R. Estimation of the number of alpha-helical and beta-strand segments in proteins using circular dichroism spectroscopy. Protein Sci. 1999;8:370–380. doi: 10.1110/ps.8.2.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ritter C, et al. Correlation of structural elements and infectivity of the HET-s prion. Nature. 2005;435:844–848. doi: 10.1038/nature03793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whittemore N, et al. Hydrogen–deuterium (H/D) exchange mapping of Aβ 1-40 amyloid fibril secondary structure using nuclear magnetic resonance spectroscopy. Biochemistry. 2005;44:4434–4441. doi: 10.1021/bi048292u. [DOI] [PubMed] [Google Scholar]
- 22.Ferguson N, et al. General structural motifs of amyloid protofilaments. Proc Natl Acad Sci USA. 2006;103:16248–16253. doi: 10.1073/pnas.0607815103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iwata K, et al. Three-dimensional structure of amyloid protofilaments of β2-microglobulin fragment probed by solid-state NMR. Proc Natl Acad Sci USA. 2006;103:18119–18124. doi: 10.1073/pnas.0607180103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petkova AT, Yau WM, Tycko R. Experimental constraints on quaternary structure in Alzheimer's beta-amyloid fibrils. Biochemistry. 2006;45:498–512. doi: 10.1021/bi051952q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shivaprasad S, Wetzel R. An intersheet packing interaction in A beta fibrils mapped by disulfide cross-linking. Biochemistry. 2004;43:15310–15317. doi: 10.1021/bi048019s. [DOI] [PubMed] [Google Scholar]
- 26.Harper JD, Wong SS, Lieber CM, Lansbury PT. Observation of metastable Aβ amyloid protofibrils by atomic force microscopy. Chem Biol. 1997;4:119–125. doi: 10.1016/s1074-5521(97)90255-6. [DOI] [PubMed] [Google Scholar]
- 27.Kheterpal I, Williams A, Murphy C, Bledsoe B, Wetzel R. Structural features of the Aβ amyloid fibril elucidated by limited proteolysis. Biochemistry. 2001;40:11757–11767. doi: 10.1021/bi010805z. [DOI] [PubMed] [Google Scholar]
- 28.Caughey B, Lansbury P. Protofibrils, pores, fibrils, and neurodegeneration: separating the responsible protein aggregates from the innocent bystanders. Annu Rev Neurosci. 2003;26:267–298. doi: 10.1146/annurev.neuro.26.010302.081142. [DOI] [PubMed] [Google Scholar]
- 29.van der Wel PC, Lewandowski JR, Griffin RG. Solid-state NMR study of amyloid nanocrystals and fibrils formed by the peptide GNNQQNY from yeast prion protein Sup35p. J Am Chem Soc. 2007;129:5117–5130. doi: 10.1021/ja068633m. [DOI] [PubMed] [Google Scholar]
- 30.Ludtke SJ, Baldwin PR, Chiu W. EMAN: Semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 31.Frank J, et al. SPIDER and WEB: Processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 32.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 33.Wall J, Simon M. Scanning transmission electron microscopy of DNA–protein complexes. Methods Mol Biol. 2001;148:589–601. doi: 10.1385/1-59259-208-2:589. [DOI] [PubMed] [Google Scholar]
- 34.Diaz-Avalos R, King C, Wall J, Simon M, Caspar D. Strain-specific morphologies of yeast prion amyloid fibrils. Proc Natl Acad Sci USA. 2005;102:10165–10170. doi: 10.1073/pnas.0504599102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pettersen EF, et al. UCSF Chimera–A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]