Abstract
Biological O2 activation often occurs after binding to a reduced metal [e.g., M(II)] in an enzyme active site. Subsequent M(II)-to-O2 electron transfer results in a reactive M(III)-superoxo species. For the extradiol aromatic ring-cleaving dioxygenases, we have proposed a different model where an electron is transferred from substrate to O2 via the M(II) center to which they are both bound, thereby obviating the need for an integral change in metal redox state. This model is tested by using homoprotocatechuate 2,3-dioxygenases from Brevibacterium fuscum (Fe-HPCD) and Arthrobacter globiformis (Mn-MndD) that share high sequence identity and very similar structures. Despite these similarities, Fe-HPCD binds Fe(II) whereas Mn-MndD incorporates Mn(II). Methods are described to incorporate the nonphysiological metal into each enzyme (Mn-HPCD and Fe-MndD). The x-ray crystal structure of Mn-HPCD at 1.7 Å is found to be indistinguishable from that of Fe-HPCD, while EPR studies show that the Mn(II) sites of Mn-MndD and Mn-HPCD, and the Fe(II) sites of the NO complexes of Fe-HPCD and Fe-MndD, are very similar. The uniform metal site structures of these enzymes suggest that extradiol dioxygenases cannot differentially compensate for the 0.7-V gap in the redox potentials of free iron and manganese. Nonetheless, all four enzymes exhibit nearly the same KM and Vmax values. These enzymes constitute an unusual pair of metallo-oxygenases that remain fully active after a metal swap, implicating a different way by which metals are used to promote oxygen activation without an integral change in metal redox state.
Keywords: bioinorganic chemistry, extradiol dioxygenase, nonheme iron, manganese
Extradiol catecholic dioxygenases catalyze the cleavage of dihydroxybenzene rings with incorporation of both atoms from O2 to yield muconic semialdehyde products (1–3). As such, these enzymes play key roles in the ability of nature to reclaim the vast quantities of organic carbon sequestered in aromatic compounds in the environment. The metal in these enzymes is coordinated by two His residues and one Glu/Asp residue that occupy one face of a (pseudo)octahedron, representing the first examples of what has become recognized as the “2-His-1-carboxylate facial triad” (2H1C triad) motif in many nonheme iron enzymes that activate dioxygen (4, 5). Indeed, a truly remarkable array of oxidative reactions is catalyzed by 2H1C triad enzymes, including C-H hydroxylation, ring expansion, and C C bond cleavage (5).
C bond cleavage (5).
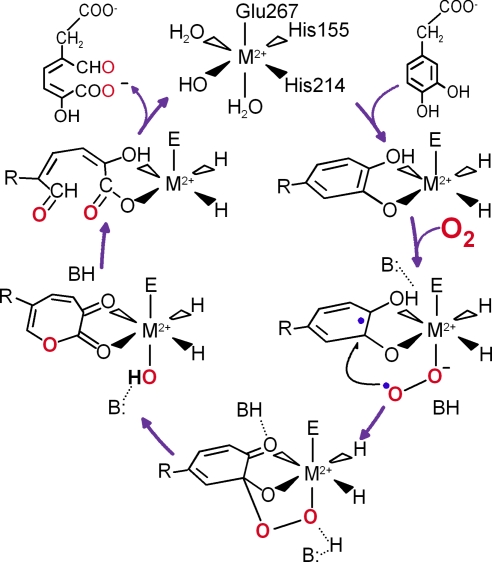
Studies from our laboratories and others have resulted in a mechanistic proposal shown in Fig. 1 for the extradiol dioxygenases that takes advantage of the 2H1C triad motif (3, 6–8). Crystal structures show that the catecholic substrate binds in a bidentate fashion to the reduced metal center trans from the histidines, displacing two or three water molecules (9–11). This primes the metal center for O2 binding in the coordination site trans to the carboxylate ligand after substrate is in place. We have proposed that electron density is transferred from the aromatic substrate to bound dioxygen via the iron, thereby giving them both radical character and activating them for reaction with each other (6, 12, 13). The attack of the bound superoxide on the oxidized catecholate ring would yield an intermediate alkylperoxy species. This intermediate would then rearrange with O O bond cleavage and ring insertion to produce an intermediate lactone that is hydrolyzed to complete the reaction. These mechanistic proposals have recently received strong support in the case of homoprotocatechuate 2,3-dioxygense from Brevibacterium fuscum (HPCD) (14), where two of the predicted intermediate species and the enzyme product complex were crystallographically characterized (15).
O bond cleavage and ring insertion to produce an intermediate lactone that is hydrolyzed to complete the reaction. These mechanistic proposals have recently received strong support in the case of homoprotocatechuate 2,3-dioxygense from Brevibacterium fuscum (HPCD) (14), where two of the predicted intermediate species and the enzyme product complex were crystallographically characterized (15).
Fig. 1.
Proposal for the catalytic cycle of extradiol dioxygenases illustrated for homoprotocatechuate 2,3-dioxygenase [M = Fe(II) or Mn(II)].
Although the mechanism proposed for the extradiol dioxygenases is compatible with experimental (6, 7, 12, 13, 15–22) and computational (23–26) studies conducted to date, it differs in at least one way from those proposed for the other members of the 2H1C triad family. Specifically, the electron required to activate O2 is proposed to derive directly from bound substrate rather than from the metal itself. Thus, under this hypothesis, the reactive species would contain an unusual, reduced metal–superoxide complex [M(II)–O2•−]. In all other enzymes in this superfamily, as well as most other types of oxygen activating metalloenzymes, O2 is proposed to bind to the reduced metal to yield an oxidized metal–superoxide complex [e.g., M(III)–O2•−], which is then the reagent used to carry out subsequent chemistry (2, 5).
The oxidation state of the metal in the substrate-bound oxy-intermediate has been difficult to test directly. However, the case of homoprotocatechuate 2,3-dioxygenase presents a possible strategy. This enzyme has been isolated in two forms: HPCD and the corresponding enzyme from Arthrobacter globiformis MndD (27, 28). HPCD and MndD are very similar enzymes, with 83% sequence identity (11). Despite this high homology, nature employs Fe(II) as the active-site metal in HPCD (Fe-HPCD) and the neighboring transition metal, Mn(II), in MndD (Mn-MndD). Both Fe-HPCD and Mn-MndD have been crystallographically characterized (11); not surprisingly, the tertiary structures of these enzymes are very similar. Indeed, the metal centers of these enzymes are effectively identical both before and after substrate binding. Fe-HPCD and Mn-MndD produce the same products from all substrates tested to date, and they both bind the probe substrate 4-nitrocatechol as a dianion with deprotonation of one OH substituent during the binding process (12, 13, 22). These two enzymes can be used to probe the role of the metal oxidation state in the oxy-complex because the reduction potentials of iron (≈0.77 V) and manganese (≈1.5 V) differ by ≈0.7 V in the absence of redox tuning by the protein structure (29). The effective identity of the active-site structures of Fe-HPCD and Mn-MndD suggests that the proteins do not differentially tune the metal potentials. Consequently, substantially different oxy-intermediates should form, and this would be expected to affect the observed catalytic parameters. However, if no significant change in metal redox state occurs during the reaction, then the relevant redox couple is established by O2 reduction and HPCA oxidation. This should be the same for each enzyme, and so the catalytic parameters would be similar for each enzyme.
In past work, Mn-MndD and Fe-HPCD have been reported to exhibit similar catalytic properties, but these studies have been carried out under different experimental conditions with enzymes of varying metal content (13, 14, 28). In addition, the highest resolution Fe-HPCD x-ray crystal structure was of a truncated form. Here, we directly compare the catalytic properties of fully activated, full-length Fe-HPCD and Mn-MndD as well as forms of each of these enzymes in which the nonnative metal has been incorporated. The x-ray crystal structure of Mn-HPCD is solved and compared with that of Fe-HPCD to allow a direct evaluation of the effect of metal substitution in the same protein matrix. The results strongly support our mechanistic model in which the metal is not required to change redox state during catalysis. This introduces a different paradigm for the role of metals in oxygen activation in which the metal serves to organize and electronically connect the organic substrate and oxygen while the potential for electron transfer is tuned by substrate rather than the protein.
Results
Characterization of Metal-Substituted Enzymes.
The homoprotocatechuate 2,3-dioxygenases cloned from B. fuscum and A. globiformis were each isolated with either iron or manganese in their active centers from Escherichia coli expression vectors grown in minimal media but augmented with an excess of the desired metal ion as described in Materials and Methods. For clarity, the individual dioxygenases will be referred to by the metal in the active site followed by the enzyme name: Fe-HPCD, Mn-HPCD, Mn-MndD, and Fe-MndD. Metal analysis showed that each of the enzymes contains 0.6–0.8 eq of metal/monomer with <0.1 eq of the other metal (Table 1). During the growth of the recombinant expression strains, supply of Fe(II) and Mn(II) in approximately equal concentrations resulted in HPCD and MndD each selecting the metal found when the enzyme was isolated from the original host organism. However, when one metal was supplied in large excess, this metal was the one mainly incorporated into either HPCD or MndD.
Table 1.
Steady-state kinetics and metal content of HPCDs and MndDs
| Enzyme | Metal | [M]/[enzyme]* |
KMHPCA, μM | KMO2, μM | kcat, min−1 | kcat/KMHPCA, μM−1·min−1 | kcat/KMO2, μM−1·min−1 | |
|---|---|---|---|---|---|---|---|---|
| Fe | Mn | |||||||
| HPCD | Fe(II) | 0.7 | <0.1 | 31 ± 6 | 60† | 470 ± 20 | 15 ± 2 | 7.8 ± 0.3 |
| Mn(II) | <0.1 | 0.8 | 35 ± 5 | 50 ± 4 | 370 ± 10 | 11 ± 2 | 7.4 ± 0.6 | |
| MndD | Mn(II) | <0.1 | 0.7 | 14 ± 4 | 62‡ | 360 ± 20 | 25 ± 4 | 5.8 ± 0.3 |
| Fe(II)§ | 0.6 | <0.1 | 15 ± 4 | 37 ± 6 | 420 ± 20 | 28 ± 4 | 11.4 ± 1.4 | |
| Fe(III)¶ | 0.6 | <0.1 | 15 ± 4 | 37 ± 6 | 29 ± 2 | 2 ± 1 | 0.8 ± 0.4 | |
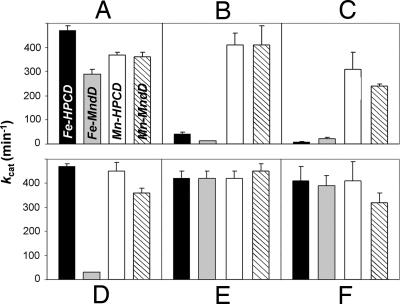
The steady-state kinetic parameters measured for purified Fe-HPCD, Mn-HPCD, Fe-MndD, and Mn-MndD are compared in Table 1 and show that all four enzymes exhibit comparably high kcat values of 350–500 min−1. Thus, iron and manganese can occupy the active sites of MndD and HPCD, respectively, and assume the role that the native metal ion plays in dioxygen activation with very little change in catalytic efficiency. To the best of our knowledge, HPCD and MndD represent the first structurally characterized pair of oxygenases that retain high catalytic activity after their distinct metal centers are swapped.
Interestingly, oxidants can distinguish the Mn-containing enzymes from those that contain iron (Fig. 2 B and C). Mn-HPCD and Mn-MndD were essentially unaffected by treatment with 1 mM H2O2 or ferricyanide, whereas both Fe-MndD and Fe-HPCD were inactivated by these reagents. This difference in oxidant susceptibility cannot be ascribed to a difference in accessibility of the reagents to the active sites, because the iron centers in both active sites are affected but not the manganese centers. Instead, the much higher susceptibility of the Fe enzymes to oxidizing reagents suggests that there remains a large difference in Mn(II) and Fe(II) redox potentials in these active sites, and that the Fe(II) potentials are much lower.
Fig. 2.
Activation and inhibition of HPCD and MndD activity in 50 mM MOPS buffer (pH 7.8). The average enzyme activity is displayed as one bar within each panel, where each enzyme is denoted as follows: Fe-HPCD, black bars; Fe-MndD, gray bars; Mn-HPCD, open bars; Mn-MndD, hatched bars. (A) Activities of as-isolated enzymes. (B and C) Data collected after the enzymes were treated with 1 mM H2O2 and potassium ferricyanide, respectively. (D) Activities of the as-isolated enzymes after a 1-h exposure to air. (E and F) Activity studies where the air-exposed enzymes were treated with 1 mM sodium ascorbate and potassium ferrocyanide, respectively.
Despite the near structural identity of HPCD and MndD in the vicinity of the active-site metal, there are minor differences in regions more remote from the metals, and these apparently cause a difference in the air stability of Fe-HPCD and Fe-MndD. As a result, Fe-HPCD is stable in air for at least several days (14), whereas Fe-MndD exhibits a high activity only initially and becomes inactivated over the course of an hour when exposed to air (Table 1 and Fig. 2D). It is possible that this indicates a slightly lower potential for Fe-MndD that accelerates metal oxidation by O2. The oxidation of Fe(II) to Fe(III) in Fe-MndD can be directly demonstrated by using EPR spectroscopy, which reveals at least three distinct sets of signals arising from high-spin Fe(III) species [supporting information (SI) Fig. S1]. This inactivation can be reversed by treatment with mild reducing agents such as ascorbate or ferrocyanide (Fig. 2 E and F). The reactivated Fe-MndD exhibits ≈30–35% higher catalytic activity than the as-isolated enzyme, indicating that partial enzyme inactivation had already occurred in the course of purification. Importantly, when treated with 1 mM ascorbate, all four enzymes show the same activity within error (Fig. 2E). Thus, any changes in the enzyme structures remote from the metal or due to the type of metal bound are not reflected in the rates of enzyme catalysis.
X-Ray Crystal Structure Comparisons.
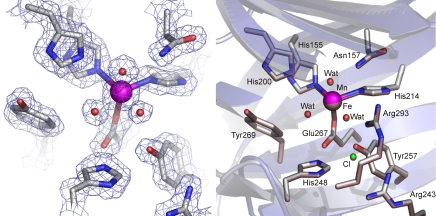
The original structures of Fe-HPCD and Mn-MndD align almost perfectly to beyond 15 Å from the metal, but they were of proteolytically truncated forms that were missing the C-terminal 43 and 42 residues, respectively (11). This truncation leaves the possibility that structural differences might be engendered by the missing regions. We recently reported a structure of the full-length Fe-HPCD, but that of Mn-MndD is still unavailable (15). The C-terminal peptide of Fe-HPCD was found to cover the entrance to the active-site region and cause minor structural shifts in this region. To determine directly whether there are any local or global changes in structure that occur as a result of the metal substitution in the full length enzyme, the x-ray crystal structure of full length Mn-HPCD was solved to 1.7 Å resolution (Table S1). As illustrated in Fig. 3, the electron density in the vicinity of the Mn(II)-substituted center is fully accounted for by the structural model. Metal substitution has no effect on the overall structure of HPCD as indicated by the RMSD of 0.20 Å for superposition of all atoms within 15 Å of the metal center and 0.4 Å for all atoms in the tetramer. More importantly, the first and second coordination spheres of the metal in Fe-HPCD and Mn-HPCD also show no changes in structure or geometry (Fig. 3). As substrate and oxygen bind, all of the solvents are displaced, so the active sites for each enzyme remain structurally identical.
Fig. 3.
Structure of the active-site region for Mn-HPCD at 1.7-Å resolution (Protein Data Bank entry 3BZA). (Left) The blue 2Fobs − Fcalc electron-density map is contoured at 1.4 σ. Atom colors: gray, carbon: blue, nitrogen; red, oxygen; magenta, manganese. (Right) Structure superposition of the metal centers in Fe-HPCD (Protein Data Bank entry 2IG9; bronze) and Mn-HPCD (Protein Data Bank entry 3BZA; colors as in Left). The blue and gray cartoon depicts the secondary structure elements for Fe-HPCD and Mn-HPCD, respectively.
EPR Spectroscopy.
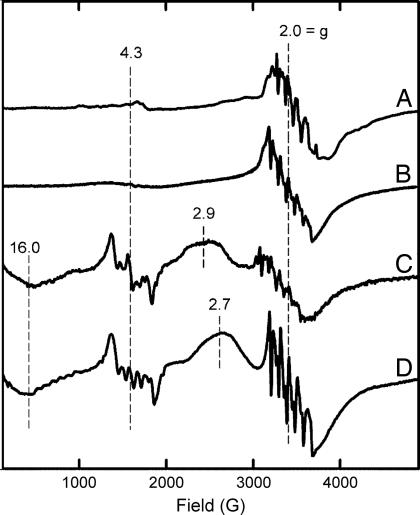
EPR spectroscopy is a more sensitive tool for detecting differences in the active centers of these homologous enzymes in solution that might not be apparent in the crystal structures. High-spin Mn(II) centers typically exhibit EPR resonances with characteristic six-line splitting patterns due to hyperfine interactions between the unpaired electrons and the I = 5/2 55Mn nucleus (30). EPR spectra of the as-isolated Mn-MndD (Fig. 4A) and Mn-HPCD (Fig. 4B) showed six-line features at g = 2.0 with nearly identical hyperfine splitting parameters and a much weaker, broad feature in the low field region near g = 4.3 (also see SI Text). Upon anaerobic addition of HPCA, their EPR spectra changed dramatically (Fig. 4 C and D), with the g = 2.0 signal decreasing in intensity and new signals appearing at g = 16, 4.3, and 2.9. The g = 4.3 signals in particular exhibited the distinctive six-line hyperfine splitting unmistakably due to manganese. Importantly, the spectra collected for Mn-MndD and Mn-HPCD were very similar, albeit not identical. Previous EPR and structural studies showed that the Mn(II) center in the Mn-MndD–HPCA complex is in an octahedral coordination environment (11, 31). The similarities found between Mn-MndD and Mn-HPCD with and without substrate bound indicate that both manganese centers are six-coordinate with the same ligand structure.
Fig. 4.
X-band EPR spectra of as-isolated Mn-MndD (A), as-isolated Mn-HPCD (B), the Mn-MndD–HPCA complex (C), and the Mn-HPCD–HPCA complex (D). Each sample contained ≈250 μM enzyme in 50 mM phosphate buffer (pH 8). 1 mM HPCA was present in the substrate complexes. EPR parameters: 4 K, 1-mW microwave power at 9.35 GHz, modulation amplitude 10 G.
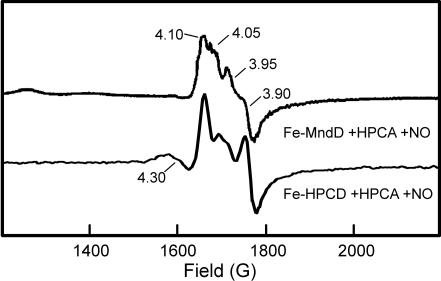
The corresponding iron enzymes were also be probed by EPR by investigating adducts of the dioxygen analogue nitric oxide (NO) and the enzyme–substrate complexes. Under reducing and anaerobic conditions (1 mM ascorbate/argon atmosphere), the Fe-MndD–HPCA complex was incubated under NO gas to produce a pale green Fe–NO complex. The EPR spectrum collected from this nitrosyl complex (Fig. 5, upper spectrum) is consistent with the [Fe NO]7 complexes previously characterized for Fe-HPCD (Fig. 5, lower spectrum) (14, 32). Both enzymes exhibit g values of 4.1, 3.90, and 2.0 (E/D = 0.01) and 4.05, 3.95, and 2.0 (E/D = 0.02), revealing the presence of at least two similar S = 3/2 species in solution. The similarity of the nitrosyl complexes of Fe-HPCD–HPCA and Fe-MndD–HPCA shows that the metal centers of these two enzymes are in nearly identical coordination environments.
NO]7 complexes previously characterized for Fe-HPCD (Fig. 5, lower spectrum) (14, 32). Both enzymes exhibit g values of 4.1, 3.90, and 2.0 (E/D = 0.01) and 4.05, 3.95, and 2.0 (E/D = 0.02), revealing the presence of at least two similar S = 3/2 species in solution. The similarity of the nitrosyl complexes of Fe-HPCD–HPCA and Fe-MndD–HPCA shows that the metal centers of these two enzymes are in nearly identical coordination environments.
Fig. 5.
EPR spectra of the NO adducts of the Fe-MndD–HPCA and Fe-HPCD–HPCA complexes. The Fe-MndD sample contained ≈250 μM enzyme in 50 mM MOPS (pH 7.8) with 1 mM ascorbate added. The sample was then exposed to NO gas for 25 min with stirring and then frozen. The data for the Fe-HPCD–HPCA–NO sample are from ref. 14. The g = 4.3 signal derives from a minor ferric impurity. EPR parameters: 4 K, 2-mW microwave power at 9.65 GHz, modulation amplitude 10 G.
Discussion
There are only a few instances in which homologous metalloenzymes are known to effectively use iron or manganese ions to catalyze a common, nonhydrolytic reaction. The best-documented of these are the mononuclear superoxide dismutases (SODs), of which some use iron and others manganese in structurally similar coordination environments to disproportionate superoxide (33). In the SOD reaction cycle, the metal is first reduced by superoxide to yield O2 and then oxidized by another molecule of superoxide to yield H2O2. Thus, the redox potential of the metal in the active site must be poised between those of O2•− acting as a reductant of the trivalent metal center and as an oxidant of the divalent metal center for this mechanism to work efficiently. Because both metal ions can readily cycle between the M(III) and M(II) oxidation states, it is not surprising that nature chose to use iron and manganese to carry out this catalysis. However, the large inherent difference in their redox potentials [+1.5 V for aqueous Mn(III)/Mn(II) versus 0.77 V for aqueous Fe(III)/Fe(II)] suggests that they cannot both function well in catalysis in the same SOD active site unless these potentials are altered. Consequently, the respective enzymes must in some way tune the potential of each metal uniquely so that the effective E° in the active site is near the midpoint of the O2•− oxidation and reduction couples. This required tuning has been directly demonstrated for a pair of SODs from E. coli (29, 34), where the ionization state of a water ligand is modulated by the different hydrogen-bonding interactions in the two enzymes (29, 34, 35). Indeed, substitution of the nonnative metal into each of these SODs results in the complete loss of catalytic activity (29, 34).
A very recently described example of Fe/Mn substitution in an enzyme is the ribonucleotide reductase from Chlamydia trachomatis in which Mn replaces one of the two irons in the usual active-site diiron cluster of the R2 subunit of class I ribonucleotide reductases (RNRs) (36–38). In RNRs, the metal sites are clearly redox-active in a reaction cycle that ultimately results in the formation of a semistable radical species later used to initiate ribonucleotide reduction (39). As noted above, the need to change oxidation state suggests that the Mn and Fe forms of C. trachomatis RNR should exhibit different activities when the metals are swapped in the same site. This appears to be true in this case because the diiron form of the C. trachomatis enzyme exhibits low activity. Thus, as with the SODs, metal substitution results in significantly altered catalytic activity.
The work presented here represents a different situation because Fe-HPCD, Mn-HPCD, Fe-MndD, and Mn-MndD all have comparable catalytic activity (Table 1). We have previously demonstrated the near structural identity of C-terminal-truncated Fe-HPCD and Mn-MndD from a comparison of their crystal structures (11). As shown more directly here, this similarity is also true of the full-length Fe-HPCD and Mn-HPCD, for which the crystal structures are also nearly identical (Fig. 3). Furthermore, the EPR spectra of the Mn-MndD– and Mn-HPCD–substrate complexes, as well as those of the nitrosyl adducts of Fe-HPCD– and Fe-MndD–substrate complexes, also indicate very similar metal sites. Most importantly, both of the Fe(II)-containing enzymes are susceptible to oxidants such as ferricyanide and H2O2, but neither of the Mn(II)-containing enzymes is affected. These results indicate that the potentials of the Mn(II) centers in these two enzyme active sites are both much higher than those of the corresponding Fe(II) centers, so that the E° values for iron and manganese are not tuned to fall within a common range by the two dioxygenases, in contrast to the SODs. Finally, there is no coordination site available for a solvent ligand to provide the redox tuning precedented by the E. coli SODs (29, 34, 35) because all three exogenous ligand sites in the ternary ESO2 complexes are occupied by the bidentate HPCA substrate and O2.
The steady-state kinetic results reported here show definitively that Fe(II) and Mn(II) are both functional in HPCD and MndD, and in fact, allow steady-state turnover with approximately the same kinetic constants for each variant. The kcat is established by the rate-limiting step in the catalytic cycle, which is a step after ring cleavage for these enzymes (12). Consequently, changes in the oxygen activation process itself might not be detected. However, the kcat/KMO2 value includes rate constants for all steps from the O2 binding up to and including the first irreversible step (40). Under the assumption that the O2 binding step is at least somewhat reversible [as is the case for 4-nitrocatechol binding to H200 mutants of HPCD (22)], the first irreversible step is O O bond-breaking and insertion (12, 22). The apparent insensitivity of kcat/KMO2 to the metal present suggests that the rates of oxygen activation and reaction with substrates are not greatly affected by the E° of the metal present. Thus, redox change at the metal center may play a much smaller role in the extradiol dioxygenase reaction mechanism than in that of the SODs. In contrast to the dioxygenation reaction, oxidative inactivation does involve an integral change in the metal redox state, and therefore it would be expected to occur much more readily in the lower potential iron sites, as observed.
O bond-breaking and insertion (12, 22). The apparent insensitivity of kcat/KMO2 to the metal present suggests that the rates of oxygen activation and reaction with substrates are not greatly affected by the E° of the metal present. Thus, redox change at the metal center may play a much smaller role in the extradiol dioxygenase reaction mechanism than in that of the SODs. In contrast to the dioxygenation reaction, oxidative inactivation does involve an integral change in the metal redox state, and therefore it would be expected to occur much more readily in the lower potential iron sites, as observed.
In the mechanism proposed for the extradiol dioxygenases, the metal center acts to facilitate the transfer of an electron from substrate to dioxygen. Although a transient redox change at the metal center may occur upon O2 binding, the lifetime of the putative M(III)–O2•− species would be at best short because substrate would rapidly transfer its electron to the M(III) center and reduce it back to its divalent state. This role differs from those ascribed to the metal centers of other oxygenases like cytochrome P450 (41) and most nonheme Fe(II) enzymes (2), where the initially formed Fe(III)–O2•− complex proceeds to a higher-valent iron oxidation state to carry out the oxidative transformation. The possible participation of high-valent species in the extradiol dioxygenase mechanism has been evaluated computationally and found to be viable in one study (23), but more recent computational studies of Siegbahn (24, 26) on the mechanisms of both iron- and manganese-dependent extradiol dioxygenases strongly support the minimal redox role we are ascribing to the dioxygenase metal center. In our view, the principal roles for the metal center are to establish the correct juxtaposition of the catecholic substrate and oxygen and to act as a conduit for facile electron transfer between them. Similar mechanisms may apply to quercetinase and acireductone dioxygenase, both of which carry out C C bond-cleavage reactions of enediolate substrates and have been shown to be effective with various divalent metal ions (42, 43).
C bond-cleavage reactions of enediolate substrates and have been shown to be effective with various divalent metal ions (42, 43).
Conclusions
The availability of two nearly identical enzymes that use different metals to catalyze the same reaction, and in which the metals can be exchanged without substantially altering the kinetic parameters, gives valuable insight into the mechanism of oxygen activation by extradiol dioxygenases. The combined use of kinetics, oxidizing and reducing agents, x-ray crystallography, and spectroscopy suggests that swapping of iron and manganese fails to cause substantial changes in activity and structure. Although the four protein–metal variants possess chemically evident differences in redox potential, they nonetheless turn over with essentially the same efficiency. Consequently, oxygen activation and substrate oxidation in the dioxygenase mechanism is likely to proceed without a requirement for an overt redox change at the metal center. This represents a different paradigm in the 2H1C triad family and more broadly for the oxygenase class. It serves to emphasize the versatility of iron in catalyzing oxygen activation reactions. Finally, these studies show that redox tuning of transition metals by enzymes is tightly linked to the reaction being catalyzed. In the current case, a ligand set is used that establishes a high potential in the absence of substrate to prevent adventitious oxygen activation. Once this is established, further tuning of the enzyme to accommodate a different metal or to promote intermediate steps in the reaction cycle is not required and has not evolved.
Materials and Methods
Reagents and General Procedures.
Reagents and buffers were the highest grade commercially available and were used as received. All solutions and media were prepared by using water purified by a Millipore Ultrapurification system.
Enzyme Preparations.
Fe-HPCD from B. fuscum was expressed in E. coli BL21(DE3) and purified as reported in ref. 27. Mn-MndD from A. globiformis CM-2 was expressed in E. coli DH5α grown in M9 minimal media and isolated as described in ref. 13.
Overexpression, isolation, and purification of Mn-HPCD were conducted as described for Fe-HPCD, except that M9 minimal media was used and 25 mg/liter MnCl2 was added to the media upon IPTG induction instead of an iron salt. Fe-MndD was expressed in E. coli DH5α cells that were grown in M9 minimal media and made anaerobic by purging sealed flasks with argon for 20 min. Upon IPTG induction, an anaerobic aliquot containing 25 mg/liter FeSO4 was added. The cells containing Fe-MndD were collected aerobically, and the isolation of Fe-MndD was carried out as described for Mn-MndD (13) except that nitrogen-purged buffers were used during protein purification.
Metal and Protein Analysis.
The metal content of all proteins was determined by inductively coupled plasma emission spectroscopy performed at the Soils Research Analytical Laboratory (University of Minnesota). Protein concentrations were determined by using the Bio-Rad protein assay, using BSA as a standard. Samples were prepared as described in ref. 31.
Enzyme Assays and Activation/Inactivation Studies.
Activity studies of all enzymes were conducted by using the rate of spectroscopic change associated with the formation of the HPCA ring cleavage product, which has a molar extinction coefficient of ε380 = 38,000 M−1·cm−1 in MOPS buffer (pH 7.8). All measurements were made at 23°C with a Beckman DU 640 spectrophotometer. All assays were run with 1 mM HPCA and 100–500 nM enzymes in air saturated 50 mM MOPS buffer (pH 7.8) unless stated otherwise. The apparent kinetic constants, Vmax, KM, and Vmax/KM, were determined from the fit of the initial velocities at varied concentrations of HPCA (S) to the expression vobs = Vmax[S]/(KM + [S]), using the nonlinear least-squares analysis with SigmaPlot 9.0 (Systat). For HPCD and MndD assays in the presence of reducing or oxidizing agents, a 5-μl aliquot of the agent (1 mM) was added to the reaction solution and allowed to incubate at room temperature for 5 min before initiating the reaction by addition of substrate.
Spectroscopic Methods.
X-band EPR spectra were measured at liquid helium temperature (4 K) on a Bruker Elexsys E-500 spectrometer equipped with an Oxford ESR-910 cryostat. The g-values and spin Hamiltonian parameters were determined as described in ref. 14.
Crystal Growth and X-Ray Crystallographic Methods.
Crystals of Mn-HPCD were grown as described for Fe-HPCD in ref. 15. X-ray data were collected at 100 K and 0.9786 Å at Beamline 19BM at the Advanced Photon Source, Argonne National Laboratory. Diffraction data were processed with HKL2000 (44). The structures were refined by cycles of restrained refinement with refmac5.2.0019 (45) and model building with Coot (46). The coordinates for Fe-HPCD [Protein Data Bank entry 2IG9 (15)] were used as a molecular replacement model.
Supplementary Material
Acknowledgments.
We thank Randy Alkire and beamline 19BM for technical support and Andrew Fielding for assistance in enzyme preparation. This work was supported by National Institutes of Health Grant GM 24689 (to J.D.L.) and GM 33162 (to L.Q.), Postdoctoral Fellowship GM 072287 (to J.P.E.), and Graduate Traineeship GM 08700 (to E.R.F.). X-ray work was performed at the Argonne National Laboratory Structural Biology Center at the Advanced Photon Source (operated by UChicago Argonne, LLC, for the U.S. Department of Energy, Office of Biological and Environmental Research under Contract DE-AC02-06CH11357).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.M.B. is a guest editor invited by the Editorial Board.
Data deposition: The x-ray crystal structure of Mn-HPCD has been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3BZA).
See Commentary on page 7341.
This article contains supporting information online at www.pnas.org/cgi/content/full/0711179105/DCSupplemental.
References
- 1.Lipscomb JD, Orville AM. Mechanistic aspects of dihydroxybenzoate dioxygenases. Metal Ions Biol Syst. 1992;28:243–298. [Google Scholar]
- 2.Costas M, Mehn MP, Jensen MP, Que L., Jr Dioxygen activation at mononuclear nonheme iron active sites: Enzymes, models, and intermediates. Chem Rev. 2004;104:939–986. doi: 10.1021/cr020628n. [DOI] [PubMed] [Google Scholar]
- 3.Vaillancourt FH, Bolin JT, Eltis LD. The ins and outs of ring-cleaving dioxygenases. Crit Rev Biochem Mol Biol. 2006;41:241–267. doi: 10.1080/10409230600817422. [DOI] [PubMed] [Google Scholar]
- 4.Hegg EL, Que L., Jr The 2-His-1-carboxylate facial triad: An emerging structural motif in mononuclear non-heme iron(II) enzymes. Eur J Biochem. 1997;250:625–629. doi: 10.1111/j.1432-1033.1997.t01-1-00625.x. [DOI] [PubMed] [Google Scholar]
- 5.Koehntop KD, Emerson JP, Que L., Jr The 2-His-1-carboxylate facial triad: A versatile platform for dioxygen activation by mononuclear non-heme iron(II) enzymes. J Biol Inorg Chem. 2005;10:87–93. doi: 10.1007/s00775-005-0624-x. [DOI] [PubMed] [Google Scholar]
- 6.Arciero DM, Lipscomb JD. Binding of 17O-labeled substrate and inhibitors to protocatechuate 4,5-dioxygenase-nitrosyl complex. J Biol Chem. 1986;261:2170–2178. [PubMed] [Google Scholar]
- 7.Shu L, et al. X-ray absorption spectroscopic studies of the Fe(II) active site of catechol 2,3-dioxygenase. Implications for the extradiol cleavage mechanism. Biochemistry. 1995;34:6649–6659. doi: 10.1021/bi00020a010. [DOI] [PubMed] [Google Scholar]
- 8.Bugg TDH. Dioxygenase enzymes: Catalytic mechanisms and chemical models. Tetrahedron. 2003;59:7075–7101. [Google Scholar]
- 9.Han S, Eltis LD, Timmis KN, Muchmore SW, Bolin JT. Crystal structure of the biphenyl-cleaving extradiol dioxygenase from a PCB-degrading pseudomonad. Science. 1995;270:976–980. doi: 10.1126/science.270.5238.976. [DOI] [PubMed] [Google Scholar]
- 10.Sato N, et al. Crystal structures of the reaction intermediate and its homologue of an extradiol-cleaving catecholic dioxygenase. J Mol Biol. 2002;321:621–636. doi: 10.1016/s0022-2836(02)00673-3. [DOI] [PubMed] [Google Scholar]
- 11.Vetting MW, Wackett LP, Que L, Jr, Lipscomb JD, Ohlendorf DH. Crystallographic comparison of manganese- and iron-dependent homoprotocatechuate 2,3-dioxygenases. J Bacteriol. 2004;186:1945–1958. doi: 10.1128/JB.186.7.1945-1958.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Groce SL, Miller-Rodeberg MA, Lipscomb JD. Single-turnover kinetics of homoprotocatechuate 2,3-dioxygenase. Biochemistry. 2004;43:15141–15153. doi: 10.1021/bi048690x. [DOI] [PubMed] [Google Scholar]
- 13.Emerson JP, et al. The role of histidine 200 in MndD, the Mn(II)-dependent 3,4-dihydroxyphenylacetate 2,3-dioxygenase from Arthrobacter globiformis CM-2, a site-directed mutagenesis study. J Biol Inorg Chem. 2005;10:751–760. doi: 10.1007/s00775-005-0017-1. [DOI] [PubMed] [Google Scholar]
- 14.Miller MA, Lipscomb JD. Homoprotocatechuate 2,3-dioxygenases from Brevibacterium fuscum. J Biol Chem. 1996;271:5524–5535. doi: 10.1074/jbc.271.10.5524. [DOI] [PubMed] [Google Scholar]
- 15.Kovaleva EG, Lipscomb JD. Crystal structures of Fe2+ dioxygenase superoxo, alkylperoxo, and bound product intermediates. Science. 2007;316:453–457. doi: 10.1126/science.1134697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arciero DM, Orville AM, Lipscomb JD. [17O]Water and nitric oxide binding by protocatechuate 4,5-dioxygenase and catechol 2,3-dioxygenase. J Biol Chem. 1985;260:14035–14044. [PubMed] [Google Scholar]
- 17.Mabrouk PA, Orville AM, Lipscomb JD, Solomon EI. Variable-temperature variable-field magnetic circular dichroism studies of the Fe(II) active site in metapyrocatechase: Implications for the molecular mechanism of extradiol dioxygenases. J Am Chem Soc. 1991;113:4053–4061. [Google Scholar]
- 18.Sanvoisin J, Langley GJ, Bugg TDH. Mechanism of extradiol catechol dioxygenases: Evidence for a lactone intermediate in the 2,3-dihydroxyphenylpropionate 1,2-dioxygenase reaction. J Am Chem Soc. 1995;117:7836–7837. [Google Scholar]
- 19.Spence EL, Langley GJ, Bugg TDH. Cis-trans isomerization of a cyclopropyl radical trap catalyzed by extradiol catechol dioxygenases: Evidence for a semiquinone intermediate. J Am Chem Soc. 1996;118:8336–8343. [Google Scholar]
- 20.Vaillancourt FH, et al. Definitive evidence for monoanionic binding of 2,3-dihydroxybiphenyl to 2,3-dihydroxybiphenyl 1,2-dioxygenase from UV resonance Raman spectroscopy, UV/Vis absorption spectroscopy, and crystallography. J Am Chem Soc. 2002;124:2485–2496. doi: 10.1021/ja0174682. [DOI] [PubMed] [Google Scholar]
- 21.Davis MI, et al. Spectroscopic and electronic structure studies of 2,3-dihydroxybiphenyl 1,2-dioxygenase: O2 reactivity of the non-heme ferrous site in extradiol dioxygenases. J Am Chem Soc. 2003;125:11214–11227. doi: 10.1021/ja029746i. [DOI] [PubMed] [Google Scholar]
- 22.Groce SL, Lipscomb JD. Aromatic ring cleavage by homoprotocatechuate 2,3-dioxygenase: Role of His200 in the kinetics of interconversion of reaction cycle intermediates. Biochemistry. 2005;44:7175–7188. doi: 10.1021/bi050180v. [DOI] [PubMed] [Google Scholar]
- 23.Deeth RJ, Bugg TDH. A density functional investigation of the extradiol cleavage mechanism in non-heme iron catechol dioxygenases. J Biol Inorg Chem. 2003;8:409–418. doi: 10.1007/s00775-002-0430-7. [DOI] [PubMed] [Google Scholar]
- 24.Siegbahn PEM, Haeffner F. Mechanism for catechol ring-cleavage by non-heme iron extradiol dioxygenases. J Am Chem Soc. 2004;126:8919–8932. doi: 10.1021/ja0493805. [DOI] [PubMed] [Google Scholar]
- 25.Bassan A, Borowski T, Siegbahn PEM. Quantum chemical studies of dioxygen activation by mononuclear non-heme iron enzymes with the 2-His-1-carboxylate facial triad. Dalton Trans. 2004:3153–3162. doi: 10.1039/B408340G. [DOI] [PubMed] [Google Scholar]
- 26.Georgiev V, Borowski T, Siegbahn PEM. Theoretical study of the catalytic reaction mechanism of MndD. J Biol Inorg Chem. 2006;11:571–585. doi: 10.1007/s00775-006-0106-9. [DOI] [PubMed] [Google Scholar]
- 27.Wang YZ, Lipscomb JD. Cloning, overexpression, and mutagenesis of the gene for homoprotocatechuate 2,3-dioxygenase from Brevibacterium fuscum. Protein Expression Purif. 1997;10:1–9. doi: 10.1006/prep.1996.0703. [DOI] [PubMed] [Google Scholar]
- 28.Boldt YR, Sadowsky MJ, Ellis LBM, Que L, Jr, Wackett LP. A manganese-dependent dioxygenase from Arthrobacter globiformis CM-2 belongs to the major extradiol dioxygenase family. J Bacteriol. 1995;177:1225–1232. doi: 10.1128/jb.177.5.1225-1232.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vance CK, Miller A-F. Novel insights into the basis for Escherichia coli superoxide dismutase's metal ion specificity from Mn-substituted FeSOD and its very high EM. Biochemistry. 2001;40:13079–13087. doi: 10.1021/bi0113317. [DOI] [PubMed] [Google Scholar]
- 30.Reed GH, Markham GD. EPR of Mn(II) complexes with enzymes and other proteins. Biol Magn Reson. 1984;6:73–142. [Google Scholar]
- 31.Whiting AK, Boldt YR, Hendrich MP, Wackett LP, Que L., Jr Manganese(II)-dependent extradiol-cleaving catechol dioxygenase from Arthrobacter globiformis CM-2. Biochemistry. 1996;35:160–170. doi: 10.1021/bi951979h. [DOI] [PubMed] [Google Scholar]
- 32.Orville AM, Lipscomb JD. Simultaneous binding of nitric oxide and isotopically labeled substrates or inhibitors by reduced protocatechuate 3,4-dioxygenase. J Biol Chem. 1993;268:8596–8607. [PubMed] [Google Scholar]
- 33.Stallings WC, Pattridge KA, Strong RK, Ludwig ML. Manganese and iron superoxide dismutases are structural homologs. J Biol Chem. 1984;259:10695–10699. [PubMed] [Google Scholar]
- 34.Vance CK, Miller A-F. A simple proposal that can explain the inactivity of metal-substituted superoxide dismutases. J Am Chem Soc. 1998;120:461–467. [Google Scholar]
- 35.Jackson TA, Brunold TC. Combined spectroscopic/computational studies on Fe- and Mn-dependent superoxide dismutases: Insights into second-sphere tuning of active site properties. Acc Chem Res. 2004;37:461–470. doi: 10.1021/ar030272h. [DOI] [PubMed] [Google Scholar]
- 36.Högbom M, et al. The radical site in Chlamydial ribonucleotide reductase defines a new R2 subclass. Science. 2004;305:245–248. doi: 10.1126/science.1098419. [DOI] [PubMed] [Google Scholar]
- 37.Jiang W, et al. A manganese(IV)/iron(III) cofactor in Chlamydia trachomatis ribonucleotide reductase. Science. 2007;316:1188–1191. doi: 10.1126/science.1141179. [DOI] [PubMed] [Google Scholar]
- 38.Voevodskaya N, Lendzian F, Ehrenberg A, Gräslund A. High catalytic activity achieved with a mixed manganese-iron site in protein R2 of Chlamydia ribonucleotide reductase. FEBS Lett. 2007;581:3351–3355. doi: 10.1016/j.febslet.2007.06.023. [DOI] [PubMed] [Google Scholar]
- 39.Nordlund P, Reichard P. Ribonucleotide reductases. Annu Rev Biochem. 2006;75:681–706. doi: 10.1146/annurev.biochem.75.103004.142443. [DOI] [PubMed] [Google Scholar]
- 40.Cleland WW. Partition analysis and the concept of net rate constants as tools in enzyme kinetics. Biochemistry. 1975;14:3220–3224. doi: 10.1021/bi00685a029. [DOI] [PubMed] [Google Scholar]
- 41.Sharrock M, et al. Cytochrome P450cam and its complexes. Mössbauer parameters of the heme iron. Biochim Biophys Acta. 1976;420:8–26. doi: 10.1016/0005-2795(76)90340-8. [DOI] [PubMed] [Google Scholar]
- 42.Schaab MR, Barney BM, Francisco WA. Kinetic and spectroscopic studies on the quercetin 2,3-dioxygenase from Bacillus subtilis. Biochemistry. 2006;45:1009–1016. doi: 10.1021/bi051571c. [DOI] [PubMed] [Google Scholar]
- 43.Dai Y, Pochapsky TC, Abeles RH. Mechanistic studies of two dioxygenases in the methionine salvage pathway of Klebsiella pneumoniae. Biochemistry. 2001;40:6379–6387. doi: 10.1021/bi010110y. [DOI] [PubMed] [Google Scholar]
- 44.Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 45.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 46.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.