Abstract
The daily production of 200 billion erythrocytes requires 20 mg of iron, accounting for nearly 80% of the iron demand in humans. Thus, erythroid precursor cells possess an efficient mechanism for iron uptake in which iron loaded transferrin (Tf) binds to the transferrin receptor (TfR) at the cell surface. The Tf:TfR complex then enters the endosome via receptor-mediated endocytosis. Upon endosomal acidification, iron is released from Tf, reduced to Fe2+ by Steap3, and transported across the endosomal membrane by divalent metal iron transporter 1. Steap3, the major ferrireductase in erythrocyte endosomes, is a member of a unique family of reductases. Steap3 is comprised of an N-terminal cytosolic oxidoreductase domain and a C-terminal heme-containing transmembrane domain. Cytosolic NADPH and a flavin are predicted cofactors, but the NADPH/flavin binding domain differs significantly from those in other eukaryotic reductases. Instead, Steap3 shows remarkable, although limited homology to FNO, an archaeal oxidoreductase. We have determined the crystal structure of the human Steap3 oxidoreductase domain in the absence and presence of NADPH. The structure reveals an FNO-like domain with an unexpected dimer interface and substrate binding sites that are well positioned to direct electron transfer from the cytosol to a heme moiety predicted to be fixed within the transmembrane domain. Here, we discuss possible gating mechanisms for electron transfer across the endosomal membrane.
Keywords: iron transport, ferric, erythrocyte
Iron plays an integral role in many biochemical processes essential to life. However, unsequestered Fe2+ is deleterious, because it catalyzes production of the hydroxyl free radical via the Fenton reaction (1). For these reasons, iron homeostasis is critical in human health, and diseases of iron transport and metabolism are among the most prevalent causes of morbidity in humans (2). Iron deficiency is thought to affect more than one billion people worldwide, particularly pregnant women and young children (3, 4) and is largely due to dietary insufficiency or excess loss. In contrast, iron overload disorders, collectively known as hereditary hemochromatosis, are among the most frequent single gene disorders in humans (5, 6). The occurrence of a single disease associated allele, HFEC282Y, is as high as 10% in individuals of Northern European descent (7). In homozygous individuals, progressive iron accumulation generates oxidative stress that results in significant cellular damage, with induction of inflammation and fibrosis that eventuates in hepatic cirrhosis, hepatocellular carcinoma, diabetes mellitus, cardiac insufficiency, and arthropathy (8). Consequently, the cellular machinery and mechanisms responsible for iron transport and homeostasis are worthy of significant investigation, because they may provide targets for pharmacological intervention to either promote or inhibit cellular iron uptake in a variety of human disorders.
The daily production of 200 billion erythrocytes requires ≈20 mg of iron, accounting for nearly 80% of the iron demand in humans (4). However, as the red blood cells senesce, they undergo phagocytosis by macrophages, and much of the erythrocyte iron is recycled, drastically reducing the need for dietary uptake of additional iron (9). To meet their iron need, erythroid precursor cells are uniquely dependent on the transferrin cycle (4, 10). In this cycle, ferric (Fe3+) iron-loaded transferrin (Tf) binds to the transferrin receptor (TfR-1) on the cell surface. The Tf:TfR-1 complex then enters the endosome via receptor-mediated endocytosis. Within the endosome, iron is released from Tf and then is reduced from Fe3+ to Fe2+ by Steap3, permitting transport across the endosomal membrane by divalent metal iron transporter 1 (DMT1), which is selective for Fe2+. The apo-Tf:TfR-1 complex is then recycled to the cell surface, where, at neutral pH, the apo-Tf is released to participate in the cycle once again (4, 10).
Ohgami et al. (11, 12) recently demonstrated that Steap3, a member of a unique family of transmembrane reductases, is the major erythroid ferrireductase. Steap (six transmembrane epithelial antigen of the prostate) family members contain a C-terminal domain composed of six transmembrane helices and are thought to coordinate a single intramembrane heme via two conserved histidine residues (12–14). With the exception of Steap1, the transmembrane domain is accompanied by an N-terminal oxidoreductase domain predicted to lie against the cytosolic face of the membrane (13). Accordingly, Steap2 and Steap4 also exhibit ferrireductase activity in vitro and, along with Steap3, have been shown to stimulate uptake of non-transferrin-bound iron and copper (13). These activities strongly suggest a greater role for Steap proteins in iron and copper metabolism.
Not surprisingly then, Steap proteins appear to play important roles in human health. Steap1 is expressed in many human cancer cell lines (15) and, with Steap2, is found at particularly high levels in prostate cancer (16), making Steap1 an appealing target for cancer immunotherapy (15–19). Mice lacking Steap3 exhibit hypochromic microcytic anemia (11, 12). Although Steap4−/− knockout mice develop spontaneous metabolic disease on a regular diet, manifesting insulin resistance, glucose intolerance, mild hyperglycemia, dyslipidemia, and fatty liver disease (20), reminiscent of “metabolic syndrome” in humans (20–22).
The Steap family oxidoreductase domain differs significantly from those of the bacterial Fre, yeast FRE, or plant FRO families of transmembrane metalloreductases. In particular, Steap3 contains an unusual oxidoreductase domain that shows limited similarity (28% identity) to F420H2:NADP+ oxidoreductase (FNO) from Archaeoglobus fulgidus (23). This archaeal enzyme utilizes F420, a unique 5′ deazaflavin derivative that is unknown in mammals. Thus, the FNO-like domain in the mammalian Steap family is likely to use a more commonly found flavin derivative such as FMN or FAD for this purpose. The reduction of iron is thought to occur by the sequential transfer of electrons from cytosolic NADPH to endosomal Fe3+ via the flavin derivative and the intramembrane heme (11–13). Importantly, the Steap3 oxidoreducatase domain is not present in yeast, nematodes or fruit flies, and in mammals is found only in other Steap family members, specifically Steap2 and Steap4. For this reason, the FNO-like domains of these proteins may represent excellent targets for pharmacological intervention (11, 12, 13).
Results
Biochemical Characterization.
The human Steap3 construct used in this study includes an N-terminal His-tag and residues 1–215 of the cytoplasmic oxidoreductase domain. Interestingly, we observe two species of purified Steap3 oxidoreductase by size exclusion chromatography (results not shown), and the relative abundance of the two species is concentration dependent. At concentrations <0.5 mg/ml, the predominant peak yields a calculated molecular mass of 20 kDa. In contrast, at concentrations >5 mg/ml the predominant peak elutes with a calculated molecular mass of 45 kDa; whereas intermediate concentrations (≈1.5 mg/ml) give a mixture of the two peaks. The calculated molecular masses are consistent with the predicted sizes for monomer and dimer forms of the oxidoreductase domain (23 and 46 kDa, respectively). Further, dimer formation is reversible, because reapplication of the dimer to the sizing column returns a mixture of the two peaks.
Structure of the Steap3 Oxidoreductase Domain.
Steap3 oxidoreductase was crystallized in both the absence (apo-Steap3) and presence of NADPH (Steap3-NADPH). The structure of apo-Steap3 was determined by multiple isomorphous replacement with anomalous scattering (MIRAS). The apo-Steap3 structure was then used as a starting point for the refinement of the Steap3-NADPH structure. Statistics on data and model quality are presented in supporting information (SI) Tables S1 and S2. The substrate-free and NADPH-bound structures each reveal a twofold symmetric dimer in the asymmetric unit (Fig. 1). A detailed comparison does not reveal any large structural differences. For example, the Cα atoms in chain A of apo-Steap3 superpose with a root mean square deviation (rmsd) of 0.26 Å on chain A of the Steap3-NADPH structure. The N-terminal His-tag and Steap3 residues 1–28 are not observed in the crystal structure. However, Western blot analyses of the crystallized material, using anti-His6 antibodies, indicate that these N-terminal residues are indeed present and are therefore disordered in the crystal. Interestingly, the disordered N-terminal residues constitute the region of greatest dissimilarity among Steap2, Steap3, and Steap4, suggesting they may be unnecessary for oxidoreductase activity. With the exception of residue 209 in chain A of the Steap3-NADPH structure, C-terminal residues 209–215 are also not observed.
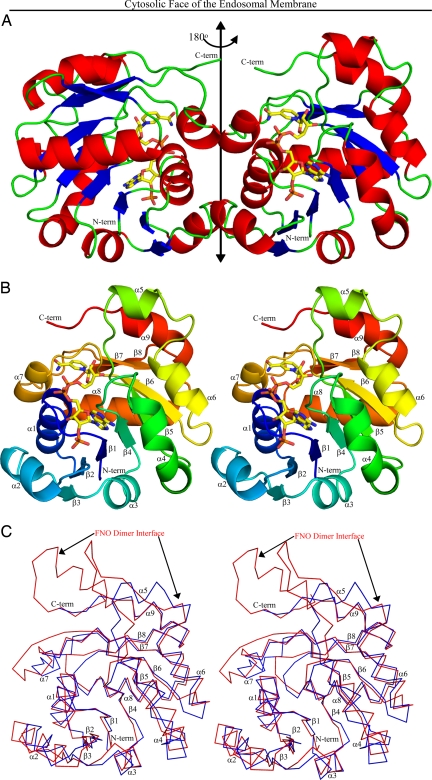
Fig. 1.
Steap3 Structure. (A) The structure of the oxidoreductase dimer is depicted with α-helices in red, β-strands in blue, and connecting loops in green. The twofold axis runs vertically within the plane of the paper (double headed arrow). The truncated C termini, which must connect to the C terminal transmembrane domain, are in green at the top of the structure. NADPH (C, yellow; N, blue; O, red; and P, orange) runs up the front side of the right subunit (back side of the left subunit) with the adenine-ribose-2′phosphate moieties near the bottom and the nicotinamide ring near the top. (B) Stereo figure of the Steap3 subunit with labeled secondary structural elements. A color gradient runs from the N terminus (blue) to the C terminus (red). Note the proximity of the NADPH binding site to the dimer interface. (C) Stereo figure depicting the superposition of FNO on the Steap3 protomer. The Steap3 Cα trace is in blue, and FNO in red. The approximate location and extent of the FNO dimer interface is indicated by black arrows along the top of FNO. In contrast, the Steap3 interface is formed by α7, α1, and the C-terminal end of α2, which is significantly shorter in Steap3. Relocation of the dimer interface, combined with the shorter β5 and α9 elements, allow the Steap3 NADPH binding site to approach the membrane.
Because full-length Steap3 is an integral membrane protein that contains a C-terminal transmembrane domain, the dimer axis might be expected to orient perpendicular to the plane of the membrane, with C-terminal residues of the truncated Steap3 dimer at the membrane proximal face. Indeed, the C termini closely approach one end of the dimer axis, with terminal Cα atoms positioned only 6 Å from the dimer axis. This suggests the orientation of the Steap3 oxidoreductase domain relative to the endosomal membrane depicted in Fig. 1.
The structure of the subunit appears as a fusion of two subdomains (Fig. 1B). At the N-terminal end, residues 29–146 form a classical nucleotide binding domain composed of a six-stranded parallel β-sheet and five flanking α-helices (α1–α4 and α6). However, strand β5 extends significantly beyond the neighboring strands, where it enters an unanticipated α-helix (α5) before returning to helix α6 and strand β6, canonical elements of the nucleotide binding domain. Residues 147–208 then form a smaller C-terminal subdomain that begins with a short α-helix (α7) that emanates from β6 of the nucleotide binding domain. The α7 helix is followed by a poorly ordered loop (Gly157-Asn162) that then connects to a β-α-β supersecondary structural element (β7-α8-β8) that joins the nucleotide binding domain to form a twisted, 8-stranded β-sheet. Although strands β7 and β8 run parallel to each other, they lie antiparallel to those in the nucleotide binding domain. A final α-helix (α9) then runs back across the top edge of the β-sheet, followed by a few C-terminal residues that approach the dimer axis. As expected, significant similarities between Steap3 and the archaeal FNO are found. The Steap3 core superposes on FNO (PDB entry 1JAX) with an rmsd of 1.44 Å [162 structurally equivalent residues, 29% identity, SSM Q score = 0.5528 (24)]. However, a number of critical differences are seen.
Structure-Function Relationships Within the Dimer.
One difference is the location of the dimer interface (Fig. 1 A and C). In FNO, also a dimer, the subunit interface is formed by a β-hairpin extending from strand β5 and elements of the C-terminal subdomain, primarily helix α8 (equivalent to Steap3 α9) and strand β9. In contrast, Steap3 lacks the β-hairpin (replacing it with helix α5), using a significantly shorter helix, and lacks the ninth β-strand (Fig. 1C). Further, it utilizes a distinctly different facet of the subunit for the dimer interface, using the α1 helix, the C-terminal end of α2, the α2–β3 loop, and the α7 helix.
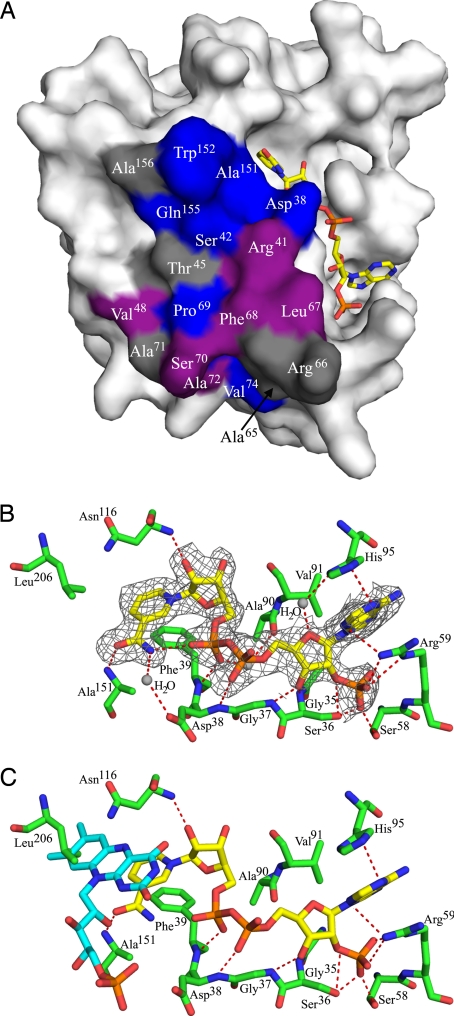
Consistent with the observed equilibrium between monomer and dimer, the interface is relatively small; dimer formation buries 717 Å2 (8.1%) of the solvent accessible surface area per subunit. The interface is composed of a mixture of hydrophobic (60%) and hydrophilic (40%) residues, with a total of 24 intersubunit hydrogen bonds. Notably, helix α2 and strand β3 are significantly shorter in Steap3 than the corresponding elements in FNO (Fig. 1C). Their role at the dimer interface provides a likely explanation for this difference. Of 18 residues present at the dimer interface, 7 are strictly conserved (Asp38, Ser42, Pro69, Val74, Ala151, Trp152, and Gln155) among human Steap2, Steap3 and Steap4 (Fig. S1). Six additional residues (Arg41, Val48, Leu67, Phe68, Ser70, and Ala72) are conserved in two of the three proteins or show conservative substitutions throughout the family. These 13 residues constitute the core of the dimer interface (Fig. 2A), and their conservation suggests that Steap2 and Steap4 may form an equivalent dimer. Further, it also suggests the possibility of Steap family heterodimers.
Fig. 2.
Dimer interface and substrate binding sites. (A) Relative to Fig. 1, the Steap3 subunit has been rotated 90° about the twofold axis to expose the dimer interface. Strictly conserved residues at the dimer interface are shown in blue, conservative substitutions in violet, and variable residues in gray. (B) The orientation is intermediate between that of Figs. 1B and 3A, with NADPH colored as in Fig. 1. For details, see NADPH Binding Site under Results. Note that Asp38 and Ala151 also serve as elements of the dimer interface. (C) FMN from biliverdin IX-beta reductase (PDB entry 1HE4) is docked to the Steap3-NADPH structure by superposition of the BVR-B nicotinamide ring onto that of the Steap3-NADPH structure. The isoalloxazine ring of the docked FMN clashes with the side chain of Leu206. FMN is colored similar to NADPH, but with carbons in cyan.
NADPH Binding Site.
As mentioned above, the structure of the binary complex with NADPH does not result in any gross structural changes. The NADPH is bound in an extended structure that slopes up the side of Steap3 (Fig. 1), with the adenine and ribose-2′ phosphate moieties bound toward the “bottom” (membrane distal) and the nicotinamide moiety near the top (membrane proximal). The interactions between protein and NADPH are shown in Fig. 2B. Phe39 lies underneath the plane of the nicotinamide ring, supporting the A- or Re-face of the nicotinamide ring. In contrast, the B or Si face is solvent exposed, with the pro-S hydrogen pointing roughly toward the membrane. This suggests that the pro-S hydrogen is used for hydride transfer and that the intermediate electron acceptor, thought to be a flavin, will bind “above” the nicotinamide ring, mediating electron flow from NADPH to the intramembrane heme. Interestingly, we identify two hydrogen bonds to the amide group of the nicotinamide ring. The first is an intramolecular interaction within NADPH as the amide NH reaches back to interact with the proximal phosphoryl group. The second is between the main chain NH of Ala151 and the amide oxygen of the nicotinamide ring (Fig. 2B). These hydrogen bonds appear to stabilize the amide oxygen in a rather unusual configuration (25), with the amide oxygen trans to C4 of the nicotinamide ring. This trans configuration is also seen in FNO (23), and, in both Steap3 and FNO, the amide edge of the nicotinamide ring faces away from the protein and toward the solvent. The mode of binding is similar in other ways to that observed for other members of the dinucleotide binding family. The diphosphate moiety caps the N-terminal end of helix α1 (Fig. 1B), interacting with the main chain NH of Ser36, Gly37, Asp38, and Phe39, which are present within the GXGXXG/A motif (Fig. S1). The 2′O of the nicotinamide ribose is caught up in a hydrogen bond to Asn116, whereas the 2′ phosphate that differentiates NADPH from NADH is recognized by Ser58 and Arg59, and the adenine ring is sandwiched in between Arg59 and His95 (Fig. 2B).
Putative Flavin Binding Site.
The A. fulgidus FNO structure was also determined as a non-Michaelis complex with NADP+ and F420 (PDB entry 1JAY). The structure of this ternary complex allows docking of F420 to Steap3 by structural superposition, and places the Si-face of the deazaflavin ring immediately “above” the Si-face (B-face) of the Steap3 nicotinamide ring, i.e., between the nicotinamide moiety and the predicted position of the lipid bilayer. However, this docking results in steric clash between the deazaflavin ring of F420 and the side chain of Leu206. This led us to consider whether F420 was the most suitable cofactor on which to model the interaction of Steap3 with FMN or FAD and led us to search the Protein Data Bank (PDB) for Steap3 homologs present as a ternary complex with a true flavin and a nicotinamide-derived cofactor. This search identified the ternary complex of Human Biliverdin IX Beta Reductase (BVR-B), FMN, and NADP+ (PDB entry 1HE4) (26). Although BVR-B, also known as flavin reductase, shows only modest structural similarity to Steap3 [rmsd = 2.954 Å for 113 Cα atoms with 17.7% identity, SSM Q score = 0.1738 (24)], the relative orientation of the isoalloxazine and nicotinamide rings are quite similar to those of the corresponding cofactors in FNO. Thus, BVR-B based modeling also predicts steric clash between Leu206 and the isoalloxazine ring of FAD or FMN (Fig. 2C). This may be an artifact of the Steap3 truncation, which disconnects the oxidoreductase and intramembrane domains. The disconnect may allow Leu206, the last well ordered C-terminal residue, to collapse in toward the active site. Alternatively, the position of Leu206 may reflect the situation in the intact protein, and conformational changes associated with electron gating might reorient Leu206, permitting the flavin to access the active site and the strictly conserved Leu206 side chain to lie over the top of the isoalloxazine ring. In this light, the Steap3 structures presented here might represent an inactive conformation. However, other than the steric clash with Leu206, the docked isoalloxazine ring would appear to be in an ideal position to mediate electron transfer between NADPH and the intramembrane heme, while, at the same time, serving to throttle-down from a two-electron process to a one-electron process (Figs. 2C and 3).
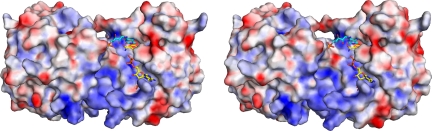
Fig. 3.
The electrostatic potential mapped to the surface of the Steap3 dimer. Positive potentials are in blue, negative potentials in red (±25 kT/e). Steap3-NADPH and the docked FMN from BVR-B are also shown. The orientation of the dimer and colors for NADPH are as in Fig. 1. FMN is similarly colored, but carbons are in cyan. Note the cleft running up the front side and across the top of the dimer interface. Because of the symmetry of the dimer, the cleft continues along the interface down the back side of the dimer.
Interestingly, the ribitol and phosphate moieties of the docked F420 and FMN cofactors project from the active site, closely approaching the neighboring Steap3 subunit (Fig. 3). Although the terminal γ-glutamyl-glutamate moiety of F420 was not observed in the FNO structure, the position of the ribitol-phosphate moiety suggests it would clash with the neighboring Steap3 subunit. Thus, the adenine-ribose moiety of a similarly docked FAD molecule also shows steric clash with the neighboring Steap3 subunit. For this reason, FMN may be the preferred flavin for Steap3. However, an alternative FAD conformation that might allow the adenine-ribose moieties to bind along the subunit interface is a distinct possibility. Alternatively, dissociation of the Steap3 dimer might result in exposure of an FAD binding site; or use of an altogether different cofactor might be indicated by the steric clash of the isoalloxazine ring with Leu206.
Proximity of Substrate Binding Sites to the Membrane.
The relative locations of the membrane proximal face of Steap3 and the location of the NADPH binding site provides additional insight into the structural differences between Steap3 and FNO. Specifically, Steap3 utilizes Pro205 to truncate the C-terminal α-helix (α9). Our model places this helix on the membrane proximal face of Steap3, where it extends at an oblique angle toward the membrane. Similarly, the β-hairpin extension in FNO is replaced by the more compact α5 helix in Steap3 (Fig. 1C). These differences allow the nicotinamide ring of Steap3 to more closely approach the membrane surface, thereby facilitating electron transfer to the intramembrane heme, presumably via the isoalloxazine ring of the flavin.
Discussion
Data from size exclusion chromatography clearly indicate two different oligomeric forms of the Steap3 oxidoreductase domain, most likely an equilibrium mixture of monomer and dimer. Approximately equal concentrations of the two species are resolved on the sizing column when Steap3 is applied at concentrations of 100–200 μM. This provides an approximate value for the dissociation constant if a negligible off rate over the time course of the chromatography experiment is assumed. However, full-length protein may exhibit an even tighter interaction. Not only may the C-terminal domain also participate in dimer formation, the orientation of the full-length protein within the membrane will result in the loss of rotational and translational degrees of freedom. Provided the C-terminal domain does not inhibit the interaction, the decreased entropy should strengthen it significantly.
Logically, the flow of electrons across the endosomal membrane should be a gated process; i.e., electron flow into the endosome is a direct response to ferric ions awaiting reduction. One possibility is that a physical interaction between Steap3 and endosomal Fe3+ might result in conformational changes within Steap3 that permit electron flow across the membrane. However, the structure of the Steap3 oxidoreductase domain suggests additional possibilities. For example, because of the proximity of the active site to the dimer interface, the oligomeric state of the enzyme might profoundly affect the structure of the catalytic site. Thus, the oligomeric state might represent a potential gating mechanism for electron transfer, serving to modulate the activity of the oxidoreductase domain. Further, Steap3 might function within a larger complex, with Steap3 activity coupled to the activity of other components of the iron transport process. The most obvious candidates are the divalent metal ion transporter 1 (DMT1) and the transferrin:transferrin receptor complex (Tf2:TfR2). We note with interest that the transferrin-transferrin receptor complex is also twofold symmetric, and a potential Steap3 homodimer, loaded with four electrons (two molecules of NADPH), nicely complements the 4 Fe+3 ions carried by the TfR2:Tf2 complex. In addition to specifying the correct stochiometry between endosomal Fe3+ and cytosolic NADPH, the complex might also ensure that Steap3 is recruited into holo-transferrin loaded endosomes. The mechanisms underlying reduction and uptake of non-transferrin bound iron and copper at the cell surface are less well understood, but it appears that Steap proteins will also play a role in this process, in which case regulation might occur with the assistance of other participating proteins.
Finally, as mentioned above, the nicotinamide ring is bound with a solvent exposed amide group, a rather uncommon orientation. In addition, the pocket harboring the nicotinamide ring opens upon a wider cleft that runs along the dimer interface (Fig. 3). The nature of the active site, combined with its proximity to the dimer interface, appears to be a unique feature of Steap3, one that may be shared with other members of the Steap family of metalloreductases. These features might be exploited for the development of inhibitors that would specifically target the various members of the Steap metalloreductase family.
Materials and Methods
Subcloning.
The human STEAP3 gene (National Center for Biotechnology Information accession no. BC042150) was obtained from the National Institutes of Health Mammalian Gene Collection. The N-terminal domain (residues 1–215) was amplified by overlapping PCR and inserted into Gateway entry vector pDONOR201. In addition to attB sites, the primers incorporated an N-terminal His6-tag and a stop codon. The resulting entry clone was sequence verified and moved into destination vector pDEST14.
Expression and Purification of Steap3.
Protein was expressed in BL21-CodonPlus (DE3)–RIL Escherichia coli, using standard protocols. The purification was done at 4°C. Cell pellets were thawed and resuspended in lysis buffer [10 mM Tris·HCl (pH 8.0), 150 mM NaCl, and 10 mM imidazole (pH 8.0)] at 5–6 ml/gm of wet pellet. PMSF (0.1 mM) was added to the cell suspension, and the cells were lysed by French press. The cleared lysate was applied to an HIS-select nickel affinity resin, washed with 10 column volumes of wash buffer (lysis buffer plus 10% glycerol), and eluted in elution buffer [10 mM Tris·HCl (pH 8.0), 150 mM NaCl, 200 mM imidazole (pH 8.0), 10% glycerol, and 0.5 mM β-mercaptoethanol]. Eluted protein was applied to a Superdex 75 gel filtration column equilibrated with 150 mM NaCl, 10% glycerol, 0.5 mM β-mercaptoethanol, and 10 mM Tris·HCl at (pH 8.0). The purified Steap3 was concentrated by centrifugal filtration to 10 mg/ml. Protein concentrations were determined by Bradford assay (27), using BSA as a standard.
Structure Determination.
Steap3 was crystallized by hanging drop vapor diffusion, using equal volumes of protein and well solution [10 mM FeCl3, 60–100 mM Na3-citrate (pH 5.6), 4% Jeffamine M600, and 15% glycerol]. Crystals also grew in the presence of 1 mM NADPH. Crystals were flash-frozen in liquid nitrogen before data collection. All data were integrated, scaled, and reduced in space group P212121 (Table S1), using HKL-2000 (28). The unit cell constants suggested one or two (66% or 33% solvent) Steap3 protomers in the asymmetric unit. Although the self-rotation function failed to provide evidence for the dimer seen by size exclusion chromatography, a 50-σ peak in the native Patterson indicated a dimer in the asymmetric unit, with the twofold axis parallel to a crystallographic screw.
The structure of apo-Steap3 was solved by MIRAS. Heavy atom soaks were for 1 h in 10 mM K2Pt(CN)4 or 10 mM mersalyl acid. SOLVE (29) was used to determine heavy atom positions and to calculate initial phases. RESOLVE (30, 31) was used for density modification and initial model-building, followed by iterative model building and refinement with COOT (32) and REFMAC (33, 34), using NCS restraints and 22 TLS groups chosen by the TLS motion determination server (35, 36, 37).
The apo-Steap3 structure was used to phase the Steap3-NADPH structure. Rigid body and positional refinement were followed by cycles of model building and refinement as described above. Statistics on model quality (Table S2) were calculated with MolProbity (38). Structural comparisons were performed with SSM (24). Figures were generated with PyMOL (39).
Supplementary Material
Acknowledgments.
Portions of this research were carried out at the Stanford Synchrotron Radiation Laboratory, a national user facility operated by Stanford University on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program, and the National Institute of General Medical Sciences. The Macromolecular Diffraction Laboratory at Montana State University was supported, in part, by a grant from the Murdock Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2VNS and 2VQ3).
This article contains supporting information online at www.pnas.org/cgi/content/full/0801318105/DCSupplemental.
References
- 1.Gutteridge JM, Rowley DA, Halliwell B. Superoxide-dependent formation of hydroxyl radicals in the presence of iron salts. Detection of “free” iron in biological systems by using bleomycin-dependent degradation of DNA. Biochem J. 1981;199:263–265. doi: 10.1042/bj1990263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrews NC. Disorders of iron metabolism. N Engl J Med. 1999;341:1986–1995. doi: 10.1056/NEJM199912233412607. [DOI] [PubMed] [Google Scholar]
- 3.Grosbois B, Decaux O, Cador B, Cazalets C, Jego P. [Human iron deficiency] Bull Acad Natl Med. 2005;189:1649–1663. discussion 1663–1664. [PubMed] [Google Scholar]
- 4.Hentze MW, Muckenthaler MU, Andrews NC. Balancing acts: Molecular control of mammalian iron metabolism. Cell. 2004;117:285–297. doi: 10.1016/s0092-8674(04)00343-5. [DOI] [PubMed] [Google Scholar]
- 5.Pietrangelo A. Hereditary hemochromatosis—a new look at an old disease. N Engl J Med. 2004;350:2383–2397. doi: 10.1056/NEJMra031573. [DOI] [PubMed] [Google Scholar]
- 6.Pietrangelo A. Non-HFE hemochromatosis. Hepatology. 2004;39:21–29. doi: 10.1002/hep.20007. [DOI] [PubMed] [Google Scholar]
- 7.Beutler E. The HFE Cys282Tyr mutation as a necessary but not sufficient cause of clinical hereditary hemochromatosis. Blood. 2003;101:3347–3350. doi: 10.1182/blood-2002-06-1747. [DOI] [PubMed] [Google Scholar]
- 8.Whitlock EP, Garlitz BA, Harris EL, Beil TL, Smith PR. Screening for hereditary hemochromatosis: A systematic review for the U.S. Preventive Services Task Force. Ann Intern Med. 2006;145:209–223. doi: 10.7326/0003-4819-145-3-200608010-00009. [DOI] [PubMed] [Google Scholar]
- 9.Knutson MD, Oukka M, Koss LM, Aydemir F, Wessling-Resnick M. Iron release from macrophages after erythrophagocytosis is up-regulated by ferroportin 1 overexpression and down-regulated by hepcidin. Proc Natl Acad Sci USA. 2005;102:1324–1328. doi: 10.1073/pnas.0409409102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Richardson DR, Ponka P. The molecular mechanisms of the metabolism and transport of iron in normal and neoplastic cells. Biochim Biophys Acta. 1997;1331:1–40. doi: 10.1016/s0304-4157(96)00014-7. [DOI] [PubMed] [Google Scholar]
- 11.Ohgami RS, et al. nm1054: A spontaneous, recessive, hypochromic, microcytic anemia mutation in the mouse. Blood. 2005;106:3625–3631. doi: 10.1182/blood-2005-01-0379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ohgami RS, et al. Identification of a ferrireductase required for efficient transferrin-dependent iron uptake in erythroid cells. Nat Genet. 2005;37:1264–1269. doi: 10.1038/ng1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohgami RS, Campagna DR, McDonald A, Fleming MD. The Steap proteins are metalloreductases. Blood. 2006;108:1388–1394. doi: 10.1182/blood-2006-02-003681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanchez-Pulido L, Rojas AM, Valencia A, Martinez AC, Andrade MA. ACRATA: A novel electron transfer domain associated to apoptosis and cancer. BMC Cancer. 2004;4:98. doi: 10.1186/1471-2407-4-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alves PM, et al. STEAP, a prostate tumor antigen, is a target of human CD8+ T cells. Cancer Immunol Immunother. 2006;55:1515–1523. doi: 10.1007/s00262-006-0165-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hubert RS, et al. STEAP: A prostate-specific cell-surface antigen highly expressed in human prostate tumors. Proc Natl Acad Sci USA. 1999;96:14523–14528. doi: 10.1073/pnas.96.25.14523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Challita-Eid PM, et al. Monoclonal antibodies to six-transmembrane epithelial antigen of the prostate-1 inhibit intercellular communication in vitro and growth of human tumor xenografts in vivo. Cancer Res. 2007;67:5798–5805. doi: 10.1158/0008-5472.CAN-06-3849. [DOI] [PubMed] [Google Scholar]
- 18.Garcia-Hernandez Mde L, Gray A, Hubby B, Kast WM. In vivo effects of vaccination with six-transmembrane epithelial antigen of the prostate: A candidate antigen for treating prostate cancer. Cancer Res. 2007;67:1344–1351. doi: 10.1158/0008-5472.CAN-06-2996. [DOI] [PubMed] [Google Scholar]
- 19.Kobayashi H, et al. Recognition of prostate and melanoma tumor cells by six-transmembrane epithelial antigen of prostate-specific helper T lymphocytes in a human leukocyte antigen class II-restricted manner. Cancer Res. 2007;67:5498–5504. doi: 10.1158/0008-5472.CAN-07-0304. [DOI] [PubMed] [Google Scholar]
- 20.Wellen KE, et al. Coordinated regulation of nutrient and inflammatory responses by STAMP2 is essential for metabolic homeostasis. Cell. 2007;129:537–548. doi: 10.1016/j.cell.2007.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Waki H, Tontonoz P. STAMPing out inflammation. Cell. 2007;129:451–452. doi: 10.1016/j.cell.2007.04.022. [DOI] [PubMed] [Google Scholar]
- 22.Nath D, Heemels M-T, Anson L. Obesity and diabetes. Nature. 2006;444:839. [Google Scholar]
- 23.Warkentin E, et al. Structures of F420H2:NADP+ oxidoreductase with and without its substrates bound. EMBO J. 2001;20:6561–6569. doi: 10.1093/emboj/20.23.6561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 25.Torres RA, Bruice TC. Theoretical investigation of the [1,2]-sigmatropic hydrogen migration in the mechanism of oxidation of 2-aminobenzoyl-CoA by 2-aminobenzoyl-CoA monooxygenase/reductase. Proc Natl Acad Sci USA. 1999;96:14748–14752. doi: 10.1073/pnas.96.26.14748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pereira PJ, et al. Structure of human biliverdin IXbeta reductase, an early fetal bilirubin IXbeta producing enzyme. Nat Struct Biol. 2001;8:215–220. doi: 10.1038/84948. [DOI] [PubMed] [Google Scholar]
- 27.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 28.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. In: Carter C, Sweet R, editors. Macromolecular Crystallography, Part A. Vol. 276. New York: Academic; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 29.Terwilliger TC, Berendzen J . Automated MAD and MIR structure solution. Acta Crystallogr D. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Terwilliger TC . Maximum likelihood density modification. Acta Crystallogr D. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Terwilliger TC . Automated main-chain model-building by template-matching and iterative fragment extension. Acta Crystallogr D. 2002;59:34–44. doi: 10.1107/S0907444902018036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 33.Bailey S . The CCP4 suite—programs for protein crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 34.Murshudov GN, Vagin AA, Dodson EJ . Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 35.Winn MD, Murshudov GN, Papiz MZ. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 2003;374:300–321. doi: 10.1016/S0076-6879(03)74014-2. [DOI] [PubMed] [Google Scholar]
- 36.Painter J, Merritt EA. TLSMD web server for the generation of multi-group TLS models. J Appl Crystallogr. 2006;39:109–111. [Google Scholar]
- 37.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr D. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 38.Davis IW, et al. MolProbity: All-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.DeLano WL. The PyMOL Molecular Graphics System. 2002 Available at www.pymol.org. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.