Abstract
Responses to extracellular stress directly confer survival fitness by means of complex regulatory networks. Despite their complexity, the networks must be evolvable because of changing ecological and environmental pressures. Although the regulatory networks underlying stress responses are characterized extensively, their mechanism of evolution remains poorly understood. Here, we examine the evolution of three candidate stress response networks (chemotaxis, competence for DNA uptake, and endospore formation) by analyzing their phylogenetic distribution across several hundred diverse bacterial and archaeal lineages. We report that genes in the chemotaxis and sporulation networks group into well defined evolutionary modules with distinct functions, phenotypes, and substitution rates as compared with control sets of randomly chosen genes. The evolutionary modules vary in both number and cohesiveness among the three pathways. Chemotaxis has five coherent modules whose distribution among species shows a clear pattern of interdependence and rewiring. Sporulation, by contrast, is nearly monolithic and seems to be inherited vertically, with three weak modules constituting early and late stages of the pathway. Competence does not seem to exhibit well defined modules either at or below the pathway level. Many of the detected modules are better understood in engineering terms than in protein functional terms, as we demonstrate using a control-based ontology that classifies gene function according to roles such as “sensor,” “regulator,” and “actuator.” Moreover, we show that combinations of the modules predict phenotype, yet surprisingly do not necessarily correlate with phylogenetic inheritance. The architectures of these three pathways are therefore emblematic of different modes and constraints on evolution.
Keywords: chemotaxis, competence, module, regulatory, sporulation
Cells grow, divide, differentiate, and respond to their environment by means of an intricate regulatory program. The genetic circuitry carrying out this program is staggeringly complex; some speculate that complexity arises from the requirement for sensitive and robust response to the environment. Despite the strong coupling among components of the genetic circuitry, however, the overall system is capable of remarkable evolutionary modification for different physiological contexts and ecological niches, even resulting in altogether new phenotypes. Thus, the design of biological systems entails the seemingly incompatible objectives of complexity and evolvability. Complex circuitry might achieve the sensitive response necessary for survival, but its very intricacy could prevent modification for new functions and niches. A less complex system might be easier to modify, but the lack of intricacy could hinder its ability to respond sensitively and robustly.
An increasing number of studies suggest that modularity is one way to reconcile the seemingly incompatible objectives of complexity and evolvability. A modular system builds complexity out of simpler, repurposable units so that a minimum of rewiring among the modules can create entirely new function (1, 2). Indeed, modularity has been shown to underlie biological function at the level of transcription (3, 4), epistatic interactions (5), protein structure (6, 7), and embryonic development (8). Recent studies have examined the degree to which biological networks are modular, catalogued the types and compositions of modules, and traced how they are evolutionarily rewired and tuned by evolution for new function. Pioneering work in this field detected functional modules with shared evolutionary history by using information about gene neighborhood, gene fusions, and phylogenetic distributions of gene families (9–11). Related work confirmed that over half of all functional modules (in the form of transcriptional modules, protein complexes, and metabolic pathways) have coevolving components (12). Evidence for module rewiring is accumulating from computational studies that observe repeated domain rearrangements and module duplications (13–15), as well as from experimental studies of alternative transcriptional circuits with identical logic (16). More detailed evolutionary studies have established that modularity is hierarchical (17) and compared phylogenetic or dynamical modules among a few species: for example, the sporulation-signaling phosphorelay in five spore-formers (18) and chemotaxis regulatory dynamics in Escherichia coli vs. Bacillus subtilis (19).
Here, we investigate the level of modularity in the evolution of several representative bacterial stress responses. Responses to extracellular stress are of particular interest because they inherently represent the design tradeoff between complexity and evolvability: they directly confer survival fitness by means of complex regulatory networks, yet must encode the ability to adapt to changing ecological and environmental pressures. For our analysis, we chose three well studied stress response networks with distinct phenotypic outcomes [chemotaxis (20), spore formation (ref. 21, Ch. 33–37), and competence for DNA uptake (22)] and examined their phylogenetic variability among several hundred bacterial and archaeal lineages for which we gathered detailed phenotypic information. In particular, we chose chemotaxis because it is considered a canonical signal transduction pathway; sporulation because it is a complex developmental pathway that is closely tied to essential replication apparatus; and DNA uptake because it has wide phyletic distribution and has been provocatively linked to the evolutionary process of lateral gene transfer (23). Also, the three networks are interesting in that they do not function in isolation, but have cross-regulatory interactions (24–26) that might give rise to rewiring either within pathways (in related species inhabiting different niches) or between pathways (in species exhibiting different combinations of the three phenotypes). Because we expected to observe fine-grained differences (if any) among variants of the pathway in different species from the same habitat, and coarse-grained differences among phenotypic variants in different habitats (e.g., spores with vs. without exosporium; twitching vs. tumbling motility), bacteria proved a useful choice, because each genus has many sequenced species, strains and variants with diverse habitats and lifestyles. Finally, all three stress responses are well curated networks supported by high-quality genetic, biochemical, and phylogenetic data. This rich literature base allowed us to investigate potentially detailed inheritance patterns that might not have been discernible from large-scale, nonspecific genomic searches.
Results
Collection of Phenotypic Data.
We began by collecting detailed information on phenotype, niche, and lifestyle for 207 species of bacteria and archaea with fully sequenced genomes (supporting information (SI) Table S1 and Fig. S1). These data were gathered from literature sources that did not make use of sequence data to elucidate phenotype [Bergey's Manual of Systematic Bacteriology (27) and species-discovery papers in the International Journal of Systematic and Evolutionary Microbiology, among others], and as such provided an independent verification of the species and gene clustering discussed below. Among the 207 species surveyed, 18 were annotated as spore-formers, 85 as competent, and 101 as motile. These phenotypes correspond to the three stress response networks we chose. We also annotated each species as being Gram-positive (47 species) or not, and noted the animal pathogens (117 species), plant pathogens (17 species), strict anaerobes (97 species), and extremophiles (33 species) among the 207 species. Although the eight phenotypes listed should in theory allow for 28 = 256 possible combinations (implying that the sample size of 207 species would not even reach saturation), there were only 48 unique combinations of phenotypes, with the five most frequent combinations all being instances of disease-causing motile anaerobes, confirming the well known bias toward sequencing medically relevant intracellular pathogens.
Networks Are Composed of Distinct Evolutionary Modules.
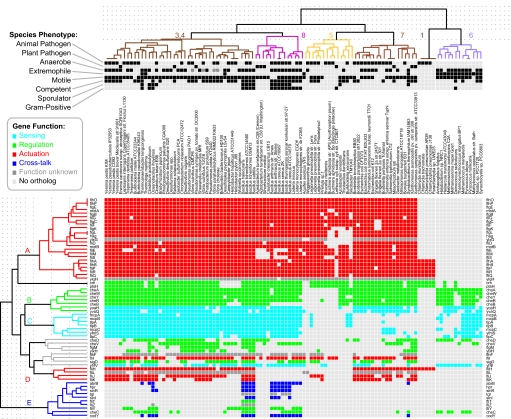
Next, we gathered lists of genes known to be expressed in each of the three pathways (61 chemotaxis genes, 153 spore-formation genes, and 62 competence genes listed in Tables S2–S4). These genes were chosen after an extensive literature search of each stress response, reflecting multiple sources of experimental evidence (genetic, biochemical, or high-throughput expression data) for each gene. The gene lists relied heavily, but not exclusively, on studies in model organisms: B. subtilis for endospore formation and chemotaxis, and B. subtilis and Haemophilus influenzae for competence (representing the two best-known cases of competence in Gram-positive and Gram-negative bacteria). A stringent version of the COG algorithm (28) was used to generate ortholog sets for each gene, with manual curation of alignments to remove paralogs and spurious BLAST hits. The resulting phylogenetic profiles were hierarchically clustered along two dimensions (genes and species). To avoid trivial clusterings resulting from identical or near-identical gene content between closely related strains or species, we benchmarked the probability of shared orthologs as a function of phylogenetic distance, and used it before the clustering step to remove 21 phylogenetically “duplicate” species sharing >90% of orthologs (Fig. S2). The optimal number of gene clusters was chosen as the partition that resulted in a maximal mean silhouette (29). Clusters with low silhouette, low statistical significance, or low cohesion were discarded (details in SI Methods). The remaining gene clusters were then denoted as evolutionary modules (Fig. 1 and Figs. S3–S6).
Fig. 1.
Evolutionary modules in chemotaxis. Orthologs of 61 B. subtilis chemotaxis genes were recovered from 207 microbial species, and the resulting gene content matrix was hierarchically clustered along both genes (rows) and species (columns). Genes were then colored according to which dynamic-control role they occupy in the network (see legend). The clustering reveals that genes group into five statistically significant evolutionary modules (A–E), and that (i) flagellar genes (flg, fli, flh) are conserved among motile bacteria but not among motile Archaea; (ii) the full complement of signal transducers (mcp, tlp) and regulators (che) is absent in many intracellular pathogens; and (iii) nonmotile bacteria that have conserved “flagellar” apparatus are in fact pathogenic Chlamydiae with orthologous type III secretion systems (omitted are 85 species with all-zero phylogenetic profiles and 21 phylogenetically “duplicate” species). For sporulation and DNA uptake phylogenetic profiles, see Figs. S3–S6.
Two of the three networks have a number of distinct and coherent evolutionary modules. The 61 genes in chemotaxis group into five evolutionary modules ranging in size from 4–24 genes found in 5–56 species, the 153 sporulation genes into three modules (30, 35, and 47 genes in 17–62 species), and the 62 DNA uptake genes group into two large clusters, one of which (cluster 2) is otherwise statistically significant (29 genes in 57–151 species) but not cohesive enough to be called an evolutionary module. This difference reveals that even within well defined functional networks, there are marked and discrete differences in the conservation patterns of individual genes. Further, as a whole, both the chemotaxis and sporulation networks have a significantly higher degree of modularity (P < 10−5), as measured by the mean distance between phylogenetic profiles of all genes in the network, than does a control set of genes randomly chosen from the genome complements of the model organisms listed above. One might expect this modularity given that the genes were chosen because they have been associated with a common phenotype; the bigger surprise was that the competence network genes as a set are no more modular than random. Please see Table S5 for module lists, member genes, and statistical measures.
Modules Are Enriched in Specific Functions and Phenotypes.
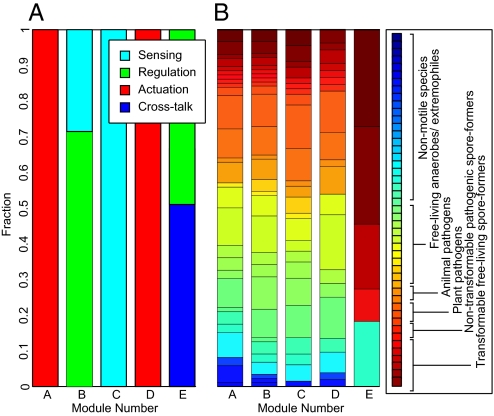
We then tested each module for over-represented functions and phenotypes. Functional annotations for genes were gathered from several manually curated functional databases (30–32). Phenotypic annotations for species were gathered as described above. Although a manual inspection of module gene annotations revealed clear functional enrichments, tests for over-represented functions did not reflect this enrichment, perhaps because standard ontologies describe functions at a “low” biochemical or molecular level. We therefore devised a previously undescribed ontology for pathways with a simpler, higher-level vocabulary based on engineered control systems. In this engineering ontology, genes are classified as sensors (signal transduction, ligand-binding, and environmental sensors), regulators (transcription factors and phosphorelay enzymes), actuators (structural proteins that actuate or realize the stress response), or cross-talk (global and master regulators that participate in two or more networks). Using a permutation test on the mutual information between classification labels and clustering (33), we confirmed that this ontology is more predictive of gene module membership than other ontologies, at least for the chemotaxis and sporulation networks. Please see Methods for z-score calculation, SI Methods for ontology classification details, and Table S6 for z-scores.
Applying these detailed descriptions of genes (engineering ontology) and species (phenotypic scoring) to modules, we found significant enrichment in functionally related genes (probability of statistical significance as compared with hypergeometric distribution, Pg) as well as in phenotypically related species (Ps). In chemotaxis, for example, module A consists entirely of flagellar genes in motile species, classified as actuators (Pg = 10−9, Ps = 10−5). Module E consists of transcriptional and cross-regulatory genes in motile and Gram-positive species, classified as regulators (Pg = 10−5, Ps = 10−7). Module C consists of the ligand-binding apparatus in free-living, nonpathogenic motile species, classified as sensors (Pg = 10−4, Ps = 10−5) (Fig. 2, Table S5). In spore-formation, module A consists of spore coat proteins, germination genes, and late stage regulatory and actuation genes (Pg = 10−5, Ps = 10−2). Sporulation module B, consisting of master regulators (abrB, sinR), early regulators (spo0B, rap phosphatases), and some germination genes, is found in sporulating bacilli, but not in all sporulators (Pg = 10−5, Ps = 10−2). Sporulation module C consists of peptide pheromones and early phosphorelay, classified as sensors (Pg = 10−5, Ps = 10−2). In competence, gene cluster 2 primarily consists of membrane-associated DNA ratchets expressed late in competence in naturally transformable bacteria, classified as actuators (Pg = 10−5, Ps = 10−2), with a surprising mix of both Gram-positive and Gram-negative genes.
Fig. 2.
Chemotaxis modules are enriched in specific functions and phenotypes. For each evolutionary module, we measured the fraction of genes in the module belonging to each of four functional categories (A), and present in each of 48 phenotypic descriptions of species (B). The relative compositions of the five modules in chemotaxis reveals significant enrichment in functionally related genes as well as in phenotypically related species. For example, the 24 genes in module A are all involved in actuation (red); the 9 genes in module E (cross-talk with other pathways) are distributed among 35 species, all of which are Firmicutes and most of which are spore-formers. Sporulation modules are also enriched in specific functions and phenotypes (Table S5).
Module Genes Have Distinct Rates of Sequence Evolution.
The degree of sequence conservation [measured as the median protein identity (mpi)] is distinct from module to module and from network to network. In chemotaxis, mpi measurements are significantly different for each module (P = 10−5, Kruskal-Wallis nonparametric analysis of variance test). Chemotaxis module A, for example (flagellar genes in motile species) has an mpi of 29.7% whereas module C (signal transduction genes in free-living motile species) has an mpi of 18.6%. This difference implies that flagellar genes are more highly conserved, whereas signal transduction genes, not required by intracellular motile pathogens, are evolving faster with more sequence variation (Table S5). In the sporulation network, although modules A and B have similar mpi values (54.6% and 58.7%), the third module C is far less conserved in sequence (mpi = 28.5%). The three modules have similar ratios of synonymous (Ks) to nonsynonymous substitutions (Ka/Ks, 0.25, 0.22, 0.31) whose variation falls well within the standard deviation for all sporulation genes (σ = 0.14). (Note that Ka/Ks ratios for sporulation genes can be calculated with reasonably minimal filtering for saturation effects because the genes are primarily conserved among a small group of closely related spore-forming bacteria. The same is not true of chemotaxis or competence genes, which are far more phylogenetically widespread and show evidence of extensive saturation; details in SI Methods.) Finally, competence cluster 2 (DNA ratchets in naturally transformable Gram-positive and Gram-negative species) is less conserved in sequence than the entire complement of DNA uptake genes (mpi = 46.5%).
Module Combinatorics Are Predictive of Phenotype.
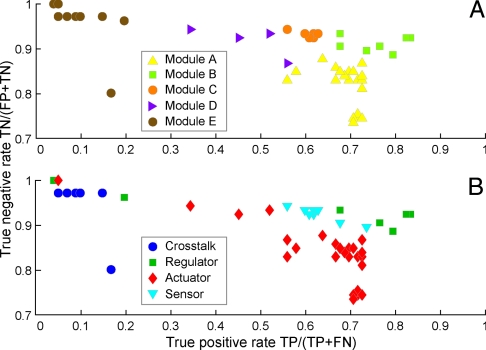
Given these findings, that is (i) the entire network is not uniformly conserved in all species with the phenotype, (ii) two of the three networks can be decomposed into distinct evolutionary modules, and (iii) certain modules have accumulated more mutations than others, we were led to examine the relationship between modules and phenotype. Are certain genes in a network “diagnostic” for the phenotype, whereas others might be present for pathway cross-talk or niche-specific sensing, and still others incidental? Alternatively, do particular combinations of genes or modules predict whether the phenotype might be found? To examine these hypotheses, we calculated the true positive (tpr) and true negative rates (tnr) of each gene's phylogenetic profile against each phenotypic profile and used linear discriminant analysis to classify tpr and tnr according to module or phenotypic membership.
For chemotaxis modules, tpr and tnr have lower classification error toward the motility phenotype (8%) than toward other phenotypes (11–17% classification error, Table S7). This trend is also the case for sporulation (0% error of sporulation genes toward sporulation phenotype vs. 1–5% error toward other phenotypes). Although the same trend exists per module, chemotaxis module E classifies better with the sporulation phenotype (7%) than with the motility phenotype (40%) or the competence phenotype (36%). This difference may be explained by the fact that chemotaxis module E consists of cross-talk regulators that participate in both sporulation and chemotaxis. In addition, tpr and tnr are remarkably distinct for each module (P = 10−4, Kruskal–Wallis nonparametric analysis of variance test), indicating that certain modules are more predictive of a phenotype, or combination of phenotypes, than other modules (Fig. 3). Modules with both high tpr and high tnr (>0.8) in the networks (that is, modules that correlate most closely with the phenotype) contain “core” genes that are actuators in sporulation (structural, coat, and cortex proteins), but in chemotaxis, are regulators (cheA-Z). Modules with low tpr (0–0.2) but high tnr (>0.8) in all three networks contain genes that sense intracellular state and act as cross-talk between two or more networks (global regulators like abrB, codY, hpr, and obg). Modules with medium tpr and tnr (0.6–0.8) in the networks contain genes that sense the extracellular environment and/or transduce signals (mcp, tlp (ligand-binding receptor) genes in chemotaxis; kin, rap, spo0 (onset) and ger (germination) genes in sporulation.
Fig. 3.
Certain modules are more predictive of phenotype than others are. Chemotaxis modules have distinct sensitivities and specificities to the motility phenotype (A); the same is true if genes are instead grouped according to the engineering ontology (B). Contrary to intuition, module genes from one phenotype do not always have decreased sensitivity and specificity to the other phenotypes (Table S7). Note that the y axis starts at 0.7.
Finally, combinations of modules are predictive of combinations of phenotypes. For example, species containing flagellar (actuation) modules, but lacking sensing, regulatory, or cross-talk modules, are all intracellular pathogens (Chlamydiales) whose highly conserved “flagellar” genes are in fact orthologs of the type III secretion system, used to inject virulent proteins into host cells (34). Similarly, species containing signal transduction and regulatory modules, but lacking flagellar modules, are all motile Archaea, which exhibit twitching rather than swimming motility, and therefore have motility organelles whose genes are orthologous to type II (pili) rather than type III (flagellar) systems (35). In sporulation, module A contains genes whose presence is both a necessary and a sufficient condition for predicting endospore formation (i.e., all species that form endospores have these genes, and no species that do not form endospores have these genes). This gene set (Table S3) should be predictive of spore formation if found in newly sequenced species, or an inability to sporulate if not found. Similarly, the presence of sporulation module B predicts spore formation, but its absence does not preclude it. These genes interact with the environment, their variability possibly due to niche adaptation, or, alternatively, merely alternative implementations of the same control strategy or the result of gene loss (Figs. S3, S4, and S7).
Discussion
We have gathered and analyzed detailed phenotypic, phylogenetic, and functional data to demonstrate that genes in some stress response pathways cluster into statistically significant evolutionary modules. These findings are in line with previous work that used phenotypic and/or phylogenetic data to demonstrate the existence of evolutionary modules (10, 11, 36, 37). Our study makes three additional observations. First, different stress response pathways are modular to different degrees, and at different levels of resolution. For pathways that are modular, each evolutionary module has a distinct, coherent conservation pattern among species related in phenotype, not phylogeny. Most of the variation (in both sequence and gene content) is in environmental sensing and signal transduction genes at the onset and completion of the response. The least variation is in structural proteins that carry out the response. Second, combinations of modules are surprisingly not maintained across phyla, but predict phenotype. Third, for a functional classification of genes to be coherent at the module-level of network organization, a particular resolution is required: one that is finer than general classifications such as “motility” or “cell cycle,” but coarser than molecular or biochemical classifications such as “ATP binding” or “methyltransferase activity.” This requirement gives rise to a new ontology that classifies gene function according to an engineering view of dynamical control, with roles such as “sensor,” “regulator,” and “actuator.”
More generally, our results show that genes functioning together to produce a certain phenotype in one species neither are all present, nor evolve at the same rate, in other species with the same phenotype. Assuming a simple relationship between phenotype and genotype, one would expect to find either species with both a phenotype and corresponding genes (true positives) or species with neither a phenotype nor the corresponding genes (true negatives). However, our analysis of evolutionary modules shows that there are also species with a phenotype but without the genes (false negatives), as well as species without a phenotype and with the genes (false positives). These exceptions to the phenotype-genotype relationship suggest module rewiring—sensors may be rewired to different signal transduction systems, in turn rewired to different actuators, and so on.
Application of the same methods yielded differing numbers of modules for each of the three networks. The numerous, extremely coherent modules found in chemotaxis, for example, together with the widespread phylogenetic distribution of the motility phenotype (Fig. S7), hints at a patchwork evolutionary history for chemotaxis. Given that the presence/absence pattern of individual chemotaxis modules correlates with neither the phenotypic nor phylogenetic clustering of species, it is interesting to speculate that the flagellar organelle may have evolved once or more, either from or preceding the type III/IV secretion systems. This speculation is supported by detailed studies of archaeal motility (38). The sensing apparatus seems to have evolved once, with extensive niche-specific gene loss, whereas the main transcriptional regulators may have been recruited from other pathways, with additional enzymatic regulators for chemotactic adaptation undergoing subsequent niche-specific tuning. This idea is supported by evidence for the rapid evolution of transcription factors (39, 40).
The three weak modules in sporulation in contrast to the strong modularity of the network as a whole, on the other hand, might imply a simpler evolutionary story. Although the sensor module (C) seems to have accumulated more substitutions than the actuator modules (A,B), the Ka/Ks measurements of the three modules do not differ significantly from each other (0.25–0.31) nor from the mean Ka/Ks for all sporulation genes (0.26), indicating that the evolution of the network has been rather uniform. Because sporulation requires building up a program of staged, sequential development, modularity may be reduced by robustness mechanisms on the core process of asymmetric cell division.
Competence presents an intriguing counterexample. First, the network has neither fine nor coarse-grained modular structure by our measures. Second, we do not consistently observe the expected clustering of Gram-positive specific competence genes with Gram-positive species, and Gram-negative specific competence genes with Gram-negative species. Although this effect could be due to poor orthology resolution for highly conserved recombinase and DNA-binding domains, it is more likely an indication that the DNA uptake machinery itself was assembled from a diverse catalog of generalized functions (41, 42). Further, the discovery of at least one instance of large-scale loss or lateral gene transfer in the nontransformable Clostridia (Table S8 and Figs. S8–S11) suggests that the evolution of this phenotype is more intricate than previously believed.
In conclusion, we found that different stress response functions have distinct evolutionary architectures. Although differences in modularity may be an artifact of how species were chosen or annotated, they more likely represent which basic functions may be reused for other cellular purposes. Some modules seem freer than others to drift and search for new functional partners. To acclimate to a new environment, sensors and actuators might drift faster or be differentially selected as compared with regulators. We show evidence for this differential diversification, at least in chemotaxis and sporulation. It is tempting to speculate that sensing new environments and accordingly changing the physics of actuation requires “sinks” for the disconnected input and output ends of signal transduction. These sinks are provided by the regulatory core, which must pass signal from input to output, and is thus constrained to maintain complex interaction structure. Finally, to close the circle between modules and pathways, we expanded pathway gene lists by probing candidate genomes for genes with similar phylogenetic profiles to module genes. This expansion yields on the order of 20–50 additional, novel genes in each pathway that remain functionally uncharacterized (Table S9); these genes are exciting potential entry points for further experimental elucidation of the stress response.
Methods
We proceeded in three phases: (i) data collection, (ii) module identification, and (iii) analysis of modules for phenotypic enrichment, functional enrichment, and evolutionary coherence, as follows (flowchart in Fig. S1).
Data Collection.
Data collection consisted of generating pathway gene lists and phylogenetic profiles, annotating genes with our engineering ontology, and annotating species with phenotypes. To build phylogenetic profiles of stress responses, we used 61 genes in chemotaxis, 153 genes in spore-formation, and 62 genes in competence (Tables S2–S4). DNA and amino acid sequences for all genes were retrieved from the MicrobesOnline website and their orthologs in 207 bacterial and archeal species identified by a 3-way bidirectional best hit algorithm as described in ref. 28, with the additional constraint that the sequence alignment coverage had to be at least 75% of the length of both genes. Orthologs were refined manually (details in SI Methods). Genes were annotated as “sensor,” “regulator,” “actuator,” or “cross-talk,” and species were annotated for phenotype and lifestyle (SI Methods, Table S1).
Identifying Evolutionary Modules.
Module identification consisted of hierarchical clustering of the phylogenetic tables, silhouette analysis to optimize clustering, and statistical significance testing of the clusters using control sets of genes randomly chosen from the same genome(s). The phylogenetic profile for each pathway was represented as a binary matrix M of size nxm, where n (rows) is the number of genes, m (columns) is the number of species, and M(i,j) = 1 if species j has an ortholog of gene i, and 0 otherwise. Before clustering the matrix, we removed 21 “duplicate” species sharing >90% of orthologs with another species in the dataset (Fig. S2). The reduced matrix M was then hierarchically clustered along both dimensions (genes and species) in Matlab (The MathWorks, Natick, MA) by using Euclidean distance and Ward's linkage. For each linkage, the clustering resulted in two dendrograms: G, which grouped genes, and S, which grouped species (Fig. 1 and Figs. S4 and S6). A first approximation of the optimal number of clusters for G was determined by calculating the mean silhouettes (29) for cuts along each gene tree producing 2–10 clusters, and choosing the partition with maximal mean silhouette over all clusters. A gene cluster was denoted an evolutionary module if both its mean cluster silhouette and coherence C were significantly higher and its mean D (Euclidean distance between phylogenetic profiles) significantly lower (P ≤ 10−3) than for a random gene cluster of the same size (1,000 iterations) drawn from either the B. subtilis genome (for sporulation and chemotaxis) or a proportional mixture of the B. subtilis and H. influenzae genomes (for competence). In the case of sporulation, this first approximation was refined by successive application of silhouette analysis (details in SI Methods).
Analyzing Modules for Evolutionary Coherence.
To measure the evolutionary coherence of each module, the mean and median percent protein alignment identity for each gene was calculated by using the Bio package of Perl (www.bioperl.org), and then averaged over all genes in each module (Tables S2–S4). The Ka/Ks ratio for each gene was calculated by an all-pairs algorithm (43) and also verified by using the codeml tool of PAML (44) with runmode = −2 and NSSites = 0 (Table S5). Saturation effects were avoided by discarding pairwise gene comparisons for which Ks > 3 (details in SI Methods).
Analyzing Modules for Functional Enrichment.
Modules were checked for over-represented functions by using various manually curated functional ontologies (Gene Ontology molecular function and biological process ontologies (30), TIGR Role Categories (32), and COG (31)), as well as the engineering ontology that we developed (SI Methods, Tables S2–S4). To determine which ontology is most predictive of gene module membership, we adapted a method that scores the mutual information between cluster membership and known gene attributes (33), as follows. For each network, let T be the n-length clustering vector such that T(i) gives the cluster number to which gene i belongs. Let L be the n-length ontology vector such that L(i) is the ontology label (e.g., “sensor,” “regulator,” “actuator,” or “cross-talk”) for gene i. Then I(T;L) is the mutual information between the clustering and the ontological classification. For 1,000 random permutations of L (resulting in the randomized ontology label vector Lr), we calculated the mutual information I(T;Lr) as well as its mean and standard deviation. We then calculated the z-score used to assess ontology meaningfulness as z = {I(T;L) − mean[I(T;Lr)]}/std[I(T;Lr)]. Z-scores for our and several other ontologies are reported in Table S6.
Analyzing Modules for Phenotypic Enrichment.
We calculated the true positive rate, tpr, of modules in a pathway to the species phenotype of that pathway (e.g., true positive rate of chemotaxis gene content matching the motility phenotype) as TP/(TP + FN), where TP = number of species with both gene and phenotype, FN = number of species with phenotype but not the gene. Similarly the true negative rate, tnr, was calculated as TN/(FP + TN), where TN = number species with neither phenotype nor gene, and FP = number of species without the phenotype but with the gene. In analogous fashion, we calculated tpr and tnr of each pathway gene to every other phenotype (e.g., chemotaxis gene content to spore-forming phenotype, or DNA uptake gene content to motility phenotype). To examine the relationship between tpr and tnr, and systematically test whether module genes are diagnostic for the corresponding phenotypes, we performed linear and quadratic discriminant analysis (ref. 45, Ch. 11) in Matlab and visualized the results on a uniform [0:1] grid with δ = 0.001 (Table S7 and Fig. S12 and Fig. S13). All programs and data are available on request from the authors.
Supplementary Material
Acknowledgments.
We thank Richard Karp, Peer Bork, Lars Jensen, Morgan Price, and the anonymous reviewers for insightful comments and discussions, all of which greatly improved the manuscript. We acknowledge support from the National Institutes of Health, the Howard Hughes Medical Institute, and the U.S. Department of Energy during this project. A.H.S. was supported by a U.S. Department of Energy Computational Science Graduate Fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0709764105/DCSupplemental.
References
- 1.Hartwell LH, Hopfield JJ, Leibler S, Murray AW. From molecular to modular cell biology. Nature. 1999;402:C47. doi: 10.1038/35011540. [DOI] [PubMed] [Google Scholar]
- 2.Schlosser G, Thieffry D. Modularity in development and evolution. BioEssays. 2000;22:1043–1045. doi: 10.1002/1521-1878(200011)22:11<1043::AID-BIES11>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 3.Ihmels J, et al. Revealing modular organization in the yeast transcriptional network. Nat Genet. 2002;31:370–377. doi: 10.1038/ng941. [DOI] [PubMed] [Google Scholar]
- 4.Ancel LW, Fontana W. Plasticity, evolvability, and modularity in RNA. J Exp Zool. 2000;288:242–283. doi: 10.1002/1097-010x(20001015)288:3<242::aid-jez5>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 5.Segre D, Deluna A, Church GM, Kishony R. Modular epistasis in yeast metabolism. Nat Genet. 2005;37:77–83. doi: 10.1038/ng1489. [DOI] [PubMed] [Google Scholar]
- 6.Hegyi H, Bork P. On the classification and evolution of protein modules. J Protein Chem. 1997;16:545–551. doi: 10.1023/a:1026382032119. [DOI] [PubMed] [Google Scholar]
- 7.Hancock JM, Simon M. Simple sequence repeats in proteins and their significance for network evolution. Gene. 2005;345:113–118. doi: 10.1016/j.gene.2004.11.023. [DOI] [PubMed] [Google Scholar]
- 8.Beldade P, Koops K, Brakefield PM. Modularity, individuality, and evo-devo in butterfly wings. Proc Natl Acad Sci USA. 2002;99:14262–14267. doi: 10.1073/pnas.222236199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Snel B, Bork P, Huynen MA. The identification of functional modules from the genomic association of genes. Proc Natl Acad Sci USA. 2002;99:5890–5895. doi: 10.1073/pnas.092632599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campillos M, von Mering C, Jensen LJ, Bork P. Identification and analysis of evolutionarily cohesive functional modules in protein networks. Genome Res. 2006;16:374–382. doi: 10.1101/gr.4336406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Korbel JO, et al. Systematic association of genes to phenotypes by genome and literature mining. PLoS Biol. 2005;3:e134. doi: 10.1371/journal.pbio.0030134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Snel B, van Noort V, Huynen MA. Gene coregulation is highly conserved in the evolution of eukaryotes and prokaryotes. Nucleic Acids Res. 2004;32:4725–4731. doi: 10.1093/nar/gkh815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amoutzias GD, Robertson DL, Oliver SG, Bornberg-Bauer E. Convergent evolution of gene networks by single-gene duplications in higher eukaryotes. EMBO Rep. 2004;5:274. doi: 10.1038/sj.embor.7400096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pereira-Leal JB, Teichmann SA. Novel specificities emerge by stepwise duplication of functional modules. Genome Res. 2005;15:552–559. doi: 10.1101/gr.3102105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alm E, Huang K, Arkin A. The evolution of two-component systems in bacteria reveals different strategies for niche adaptation. PLoS Comput Biol. 2006;2:e143. doi: 10.1371/journal.pcbi.0020143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsong AE, Tuch BB, Li H, Johnson AD. Evolution of alternative transcriptional circuits with identical logic. Nature. 2006;443:415–420. doi: 10.1038/nature05099. [DOI] [PubMed] [Google Scholar]
- 17.Ravasz E, Somera AL, Mongru DA, Oltvai ZN, et al. Hierarchical organization of modularity in metabolic networks. Science. 2002;297:1551–1555. doi: 10.1126/science.1073374. [DOI] [PubMed] [Google Scholar]
- 18.Stephenson K, Hoch JA. Evolution of signalling in the sporulation phosphorelay. Mol Microbiol. 2002;46:297–304. doi: 10.1046/j.1365-2958.2002.03186.x. [DOI] [PubMed] [Google Scholar]
- 19.Rao CV, Kirby JR, Arkin AP. Design and diversity in bacterial chemotaxis: A comparative study in Escherichia coli and Bacillus subtilis. PLoS Biol. 2004;2:E49. doi: 10.1371/journal.pbio.0020049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wadhams GH, Armitage JP. Making sense of it all: Bacterial chemotaxis. Nat Rev Mol Cell Biol. 2004;5:1024–1037. doi: 10.1038/nrm1524. [DOI] [PubMed] [Google Scholar]
- 21.Sonenshein AL, Hoch JA, Losick R. Bacillus subtilis and its closest relatives. Washington, DC: ASM Press; 2002. [Google Scholar]
- 22.Dubnau D. DNA uptake in bacteria. Annu Rev Microbiol. 1999;53:217–244. doi: 10.1146/annurev.micro.53.1.217. [DOI] [PubMed] [Google Scholar]
- 23.Redfield RJ. Do bacteria have sex? Nat Rev Genet. 2001;2:634–639. doi: 10.1038/35084593. [DOI] [PubMed] [Google Scholar]
- 24.Grossman AD. Genetic networks controlling the initiation of sporulation and the development of genetic competence in Bacillus subtilis. Annu Rev Genet. 1995;29:477–508. doi: 10.1146/annurev.ge.29.120195.002401. [DOI] [PubMed] [Google Scholar]
- 25.Liu J, Zuber P. A molecular switch controlling competence and motility: Competence regulatory factors ComS, MecA, and ComK control σD-dependent gene expression in Bacillus subtilis. J Bacteriol. 1998;180:4243–4251. doi: 10.1128/jb.180.16.4243-4251.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Msadek T. When the going gets tough: Survival strategies and environmental signaling networks in Bacillus subtilis. Trends Microbiol. 1999;7:201–207. doi: 10.1016/s0966-842x(99)01479-1. [DOI] [PubMed] [Google Scholar]
- 27.Boone DR, Castenholz RW, Garrity GM. Bergey's Manual of Systematic Bacteriology. New York: Springer; 2001. [Google Scholar]
- 28.Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997;278:631. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 29.Kaufman L, Rousseeuw PJ. Finding Groups in Data: An Introduction to Cluster Analysis. Hoboken, NJ: Wiley; 2005. [Google Scholar]
- 30.Ashburner M, et al. Gene ontology: Tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tatusov RL, et al. The COG database: An updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Selengut JD, et al. TIGRFAMs and Genome Properties: Tools for the assignment of molecular function and biological process in prokaryotic genomes. Nucleic Acids Res. 2007;35:D260–D264. doi: 10.1093/nar/gkl1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gibbons FD, Roth FP. Judging the quality of gene expression-based clustering methods using gene annotation. Genome Res. 2002;12:1574–1581. doi: 10.1101/gr.397002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Subtil A, Blocker A, Dautry-Varsat A. Type III secretion system in Chlamydia species: Identified members and candidates. Microbes Infect. 2000;2:367–369. doi: 10.1016/s1286-4579(00)00335-x. [DOI] [PubMed] [Google Scholar]
- 35.Jarrell KF, Bayley DP, Kostyukova AS. The archaeal flagellum: A unique motility structure. J Bacteriol. 1996;178:5057–5064. doi: 10.1128/jb.178.17.5057-5064.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parter M, Kashtan N, Alon U. Environmental variability and modularity of bacterial metabolic networks. BMC Evol Biol. 2007;7:169. doi: 10.1186/1471-2148-7-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Slonim N, Elemento O, Tavazoie S. Ab initio genotype-phenotype association reveals intrinsic modularity in genetic networks. Mol Syst Biol. 2006;2 doi: 10.1038/msb4100047. 2006.0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ng SY, Chaban B, Jarrell KF. Archaeal flagella, bacterial flagella and type IV pili: A comparison of genes and posttranslational modifications. J Mol Microbiol Biotechnol. 2006;11:167–191. doi: 10.1159/000094053. [DOI] [PubMed] [Google Scholar]
- 39.Price MN, Dehal PS, Arkin AP. Orthologous transcription factors in bacteria have different functions and regulate different genes. PLoS Comput Biol. 2007;3:e175. doi: 10.1371/journal.pcbi.0030175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wuichet K, Alexander RP, Zhulin IB. Comparative genomic and protein sequence analyses of a complex system controlling bacterial chemotaxis. Methods Enzymol. 2007;422:1–31. doi: 10.1016/S0076-6879(06)22001-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Claverys JP, Martin B, Havarstein LS. Competence-induced fratricide in streptococci. Mol Microbiol. 2007;65:230. doi: 10.1111/j.1365-2958.2007.05757.x. [DOI] [PubMed] [Google Scholar]
- 42.Finkel SE, Kolter R. DNA as a nutrient: Novel role for bacterial competence gene homologs. J Bacteriol. 2001;183:6288–6293. doi: 10.1128/JB.183.21.6288-6293.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Korber B. HIV sequence signatures and similarities. In: Rodrigo AG, Learn GH, editors. Computational and Evolutionary Analysis of HIV Molecular Sequences. Boston: Kluwer; 2001. pp. 55–72. [Google Scholar]
- 44.Yang Z. PAML: A program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- 45.Mardia KV, Kent JT, Bibby JM. Multivariate Analysis. London: Academic; 1979. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.