Abstract
New genes can originate by the combination of sequences from unrelated genes or their duplicates to form a chimeric structure. These chimeric genes often evolve rapidly, suggesting that they undergo adaptive evolution and may therefore be involved in novel phenotypes. Their functions, however, are rarely known. Here, we describe the phenotypic effects of a chimeric gene, sphinx, that has recently evolved in Drosophila melanogaster. We show that a knockout of this gene leads to increased male–male courtship in D. melanogaster, although it leaves other aspects of mating behavior unchanged. Comparative studies of courtship behavior in other closely related Drosophila species suggest that this mutant phenotype of male–male courtship is the ancestral condition because these related species show much higher levels of male–male courtship than D. melanogaster. D. melanogaster therefore seems to have evolved in its courtship behaviors by the recruitment of a new chimeric gene.
Keywords: chimeric gene, gene duplication, new functions, phenotype evolution
Comparative genomic analyses in various eukaryotes often reveal new genes that have evolved throughout the combination of unrelated genes or their duplicates (1–3). These new genes have usually evolved rapidly with a pattern of evolution that was shaped by positive Darwinian selection (4–6), suggesting that these chimeric genes have novel functions. Previous studies of molecular functions in new gene duplicates revealed the origination of important molecular functions in animals (6–8). A new gene becomes fixed in its initial stage in natural populations and subsequently evolves under various evolutionary forces (9). Understanding of this process requires the knowledge of the phenotypic effects of new gene duplicates because such effects are usually the targets of the positive selection that would determine the fate of the new gene duplicates. However, the phenotypes of these new genes are almost never known.
In a survey of species-specific genes, we identified a courtship gene, sphinx, that originated and became fixed in a single species, Drosophila melanogaster, within the last 2–3 million years (mys) (10, 11). The sphinx gene was previously identified as a newly evolved chimeric gene (10, 11). The sphinx gene was formed by the insertion of a retroposed sequence of the ATP synthase F-chain gene (ATPS-F) from chromosome 2 into the 102F region of chromosome 4, recruiting sequences upstream to form a new exon and intron, a region we refer to as 102F-EI (10). Two alternative transcripts in adult males were detected from this locus (10). The sphinx gene appears to be functional because the gene contains only indel polymorphisms in the nonexonic sequences; it has a rate of evolution significantly above neutral expectations, suggesting rapid adaptive evolution; and it has a very specific pattern of expression (10). However, although it is derived, in part, from a protein-coding gene, it is most likely a noncoding RNA (ncRNA) because its parental-inherited coding regions are disrupted by several nonsense mutations.
A few courtship genes in Drosophila have been found in genetic and molecular analyses (12–14), significantly adding to the understanding of the genetic system that controls the courtship behaviors of Drosophila. The best studied of these genes is the fruitless gene, which shows a remarkable level of sequence conservation over at least 250 mys of divergence (15). In this article, we show that the courtship genetic system also can evolve through acquiring new genes by analyzing the origination and evolution of sphinx, the phenotypic effects of sphinx, and the evolution of the sphinx phenotype in D. melanogaster and its related species. We knocked out the D. melanogaster sphinx using a gene-replacement technique and analyzed the courtship behaviors of wild-type and knockout lines. Comparative studies of courtship behavior were performed between D. melanogaster and its close relatives. Results of these experiments and analyses cast insights into the evolutionary process of the genetic system for courtship control through the origination of a new gene.
Results
Evolution of Gene Structure and Regulatory System.
To understand the evolution of this gene and, in particular, to understand where it might have obtained its regulatory sequences, we BLASTed the 1-kb region upstream from the transcription start site of sphinx against the genomic sequences of the five melanogaster subgroup species (D. melanogaster, D. simulans, D. sechellia, D. yakuba, and D. erecta) and the genomic sequences of four additional species in the Drosophila genus (D. ananassae, D. persimilis, D. willistoni, and D. virilis) [see supporting information (SI) Fig. S1]. In each species, we identified homologous sequences on the fourth chromosome, which is where sphinx is located in D. melanogaster. We found a high degree of sequence conservation (>92%) up to ≈600 bp upstream of the transcription start site within the melanogaster subgroup (Fig. S1a) and up to ≈300 bp between the melanogaster subgroup and the more distantly related species D. virilis and D. willistoni (Fig. S1b). As an additional check, we confirmed the published genomic sequences in D. simulans and D. sechellia by PCR sequencing. Given that synonymous sites are saturated between D. melanogaster and D. virilis and D. willistoni (16), this level of conservation suggests that there may have been a regulatory element capable of promoting gene expression at this locus since these species diverged.
We also found a sequence, orthologous to the recruited 102F-EI of sphinx, in D. simulans and D. sechellia. This finding suggests that there may be a functional gene in these and other Drosophila species that might have existed soon after the Drosophila genus originated. Therefore, sphinx might have been formed by the integration of the ATP-synthase F sequence into this preexisting gene. The recruited sphinx region existed in the Drosophila lineages for at least 35–40 mys until a retroposition event that occurred <3 mys ago that combined the two previously separated gene regions, ATPS-F and 102F-EI, into the hybrid gene, sphinx.
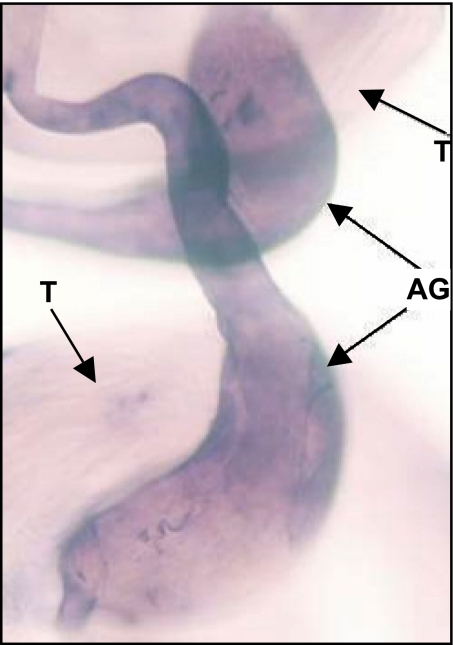
Because previous experiments detected the male-specific transcripts, we explored the male tissue-specific expression by using antisense RNA in situ hybridization (see Methods). Using an antisense riboprobe synthesized with the template of the sphinx gene region, we detected expression in the accessory glands of male adults in D. melanogaster (Fig. 1) with no expression signal detected in female reproductive organs.
Fig. 1.
Transcripts of sphinx in adult male-specific tissues detected by antisense RNA in situ hybridization. Sphinx is expressed in accessory glands (AG), but not in testis (T), as the signal of the antisense riboprobe (the deep color) shows.
Mutant Strain Generated by Gene Replacement.
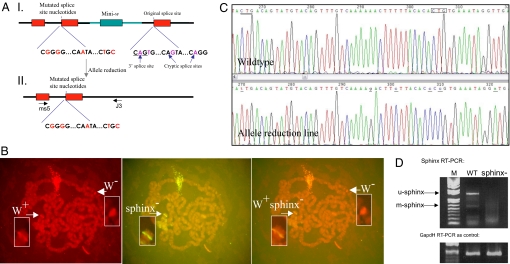
To understand the phenotypic effect of sphinx, we used a modified gene-replacement procedure derived from a standard method (17) to replace the wild-type sphinx with the sphinx sequence mutated by site-directed PCR methods (see Methods). Because sphinx is likely an ncRNA gene, the usual approach of changing the ORF by insertion/deletion cannot be applied. Therefore, we chose to change splice and cryptic splice sites in the intron–exon boundaries (Fig. 2A). We obtained two knockout lines with an identical genomic structure in the gene region of sphinx that includes a mini-w marker (Fig. 2A and Fig. S2). FISH and genomic sequencing verified the insertion of the mutated fragment on the fourth chromosome at the sphinx gene and indicated that the construct sequence had replaced the original sphinx locus (Fig. 2 B and C).
Fig. 2.
Gene replacement of sphinx. (A) Genomic structures of the sphinx knockout region. I, the structure with the marker mini-w in the initial knockout lines; II, the structure in the allele reduction line. The thick black lines represent D. melanogaster genomic DNAs; the thick cyan line represents DNA sequence from the construct. ms5 and J3 indicate the genomic positions of the two primers designed for a long-range PCR to amplify the genomic region that flanks the insert. (B) FISH with polytene chromosomes of the knockout line shows that the construct replaced the wild-type allele. This line contains mini-w (W+) and mutated sphinx− sequence that is inserted into the sphinx gene in the fourth chromosome. (Left and Center) Signals from W+ (in the background of the w− allele on the X chromosome) and sphinx−, respectively. (Right) Superimposition of the two images from Left and Center showing that the signals of mini-w and sphinx− are from the same cytological location of the sphinx gene. This line was subject to allele reduction to generate two knockout lines (allele reduction lines or reduction lines in the text) that does not contain mini-w, the downstream wild-type exon. For the genomic structures of these two lines, see Fig. S2. (C) Examples of sequencing profiles (antisense strand) show that the original splice sites and cryptic sites in sphinx were replaced by the designed different nucleotides (see the underlined letters in the reduction line profile and the boxed and double underlined letters in the wild-type line) (Oregon R). The genomic structure of the replaced region in sphinx720RW is identical to that of the wild-type line, except that the splices and cryptic splice sites are changed in sphinx720RW as shown in this figure (Fig. S2b). (D) RT-PCR experiment revealed the absence of sphinx transcripts (full-length sphinx transcript, u-sphinx and male-specific transcript, m-sphinx) in the accessory glands in the knockout male, whereas in the wild-type male the two previously detected transcripts (10) are present. WT, wild-type line; M, DNA weight marker (1-kb DNA ladder); sphinx−, knockout line. GAPDH RT-PCR was used as control of the quality of the cDNA prepared from accessory glands of males.
Because we were interested in the potential effects of sphinx on behavior (see below) and it has been observed that the activation of mini-w can impact courtship behaviors in the condition of misexpression (18), we performed an allele-reduction experiment (17) to eliminate the mini-w marker and the downstream wild-type sphinx exon 2. We obtained a final red-eye mutant strain sphinx720RW, which was identical to the wild type except for the altered splice sites and cryptic splice sites. The sequence was verified by long-range PCR-sequencing experiments (Fig. 2 A and C). We used this mini-w free sphinx729RW strain for all subsequent behavior analyses. RT-PCR results showed that the full-length and male-specific transcripts of sphinx, which are found in the wild type, are absent in the mutant (Fig. 2D), suggesting that sphinx720RW carries a loss-of-function allele, sphinx−. The absence of sphinx transcripts in the mutant strain is likely to be a consequence of the RNA surveillance system that eliminates any transcripts that are not properly spliced (19–21) because of the changed splice sites and cryptic splice sites in our engineered sphinx sequence.
Analysis of Phenotypic Effects of sphinx.
We previously detected rapid evolution of sphinx that suggested adaptive evolution with certain important new functions that arose with sphinx (10). Inspecting the mutant line sphinx720RW did not reveal obvious differences in morphology from the wild-type lines. However, we detected that sphinx is expressed in the accessory glands (Fig. 1), a male reproductive organ that also was known to regulate reproductive behavior (22). To investigate whether sphinx plays any roles in courtship, we assayed the courtship behavior of the reduction line, sphinx720RW, and compared it to that of a wild-type line that was used in the back-cross experiment in the reduction (see Methods). We compared the behavior of a wild-type line against the same line with the sphinx gene knocked out.
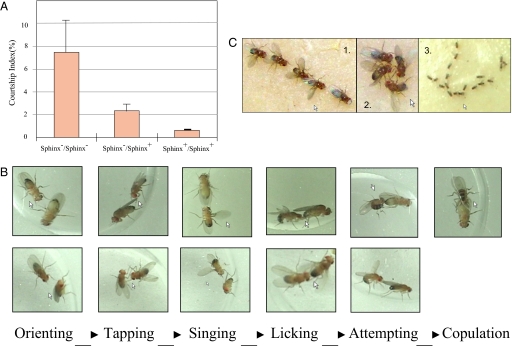
We quantified courtship using two measures: (i) the courtship time (CT), during which the two individuals showed the courtship behaviors and (ii) the courtship index (CI), which is the percentage of time a fly courts another fly within a period of 10 min (23). Initial experiments in which one male was placed with one female showed no significant difference between the mutant and wild-type lines. Both wild-type and mutant flies show the full range of normal Drosophila mating behavior, including orienting, tapping, wing extension/vibration, licking, attempting, and copulation (Fig. 3) (13, 24). Specifically, the mutant and wild-type males do not differ significantly in their courtship scores when mating with wild-type females (Table 1).
Fig. 3.
Male–male courtship behaviors. (A) Courtship indices in homozygous mutants, homozygous wild types, and the heterozygotes. The average CIs with SEMs are presented. The sample size for each genotype is 30. (B) The courtship processes of wild-type D. melanogaster (male vs. female) (Upper) and the first five steps of male–male courtship events in the homozygous mutants (male vs. male) (Lower). (C) Courtship chains (1 and 2 are high-resolution photographs of a few flies, and 3 shows a long chain). The courtship chains in 1 (4 males) and 3 (18 males) were headed by 1 female, whereas the courtship circle in 2 comprises all males.
Table 1.
Mating experiments between male and female
| Phenotype | CT, s | CI, % |
|---|---|---|
| 720rw male × WT female | 568.3 | 94.7 |
| WT male × WT female | 546.2 | 91.0 |
P = 0.3422, the Mann–Whitney test in the comparison between the two types of matings. WT, the wild-type strain (Oregon R).
However, when two males were placed together, we saw striking differences among the behavior of the wild-type, heterozygous, and mutant lines. The homozygous mutant males pursue each other for a significantly longer time compared with the homozygous wild-type males (P = 0.0012, Mann–Whitney test) (Fig. 3A). The heterozygotes show some level of male–male courtship behaviors, but much weaker than homozygote mutants, suggesting partial heterozygote insufficiency in accordance with our RT-PCR experiments, which reveal the knockout line to be a loss-of-function mutation. Mutant males go through all of the stages of normal male–female courtship except the last stage, that of copulation, but with other males (Fig. 3B). We also observed similar male–male courtship when placing two homozygous mutant males and one wild-type female together.
Furthermore, when many males are present, mutant males form courtship chains and circles (Fig. 3C), a typical male–male courtship behavior that is not seen in wild-type flies (25–27). These chains and circle of males probably arise as one male tries to court another male by approaching from behind and that male is in turn approached by another male from behind. In this way, chains of courting males appear, with the chain occasionally closing to form a circle. We did not observe any phenotypic changes in the mating behavior of homozygous and heterozygous sphinx mutant females.
Phenotype Comparison Between D. melanogaster and Its Related Species.
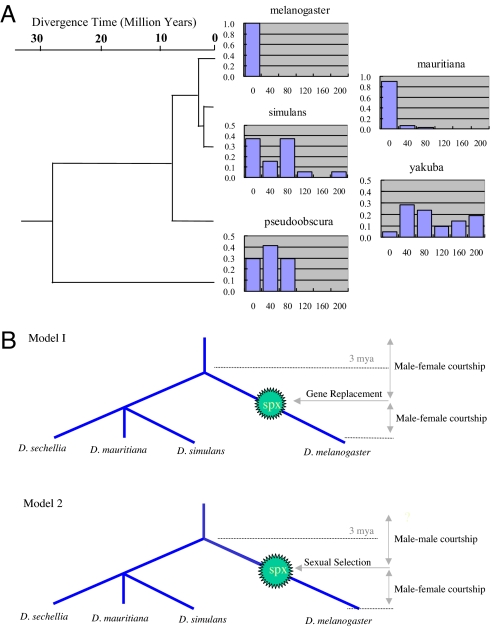
These mating behavior experiments clearly revealed male–male courtship behaviors in the sphinx knockout mutants, suggesting a functional role for sphinx in regulating courtship behaviors. Intriguingly, previous investigations of male–male mating behavior in other Drosophila species have suggested that male–male courtship is quite common (28, 29). Therefore, sphinx might have evolved to reduce such behavior and enhance male–female mating success in D. melanogaster. To investigate this idea further, we performed exactly the same male–male mating experiments as we did for D. melanogaster on three closely related species, D. simulans, D. mauritiana, and D. yakuba, and one more distantly related species, D. pseudoobscura. The males in all these species spent significantly more time courting each other than do wild-type D. melanogaster flies (Mann–Whitney tests: mel-pse, P < 0.0001; mel-yak, P < 0.0001; mel-sim, P < 0.0001; mel-mau, P = 0.0091) (Fig. 4A), although D. mauritiana's male–male courtship time (average CT = 12 s) is shorter than other relatives of D. melanogaster, it is significantly longer than the courtship time of D. melanogaster (average CT = 2 s). These data suggest that the courtship behaviors have evolved extensively since the common ancestor of these species, unexpected from the extreme conservative property of previously identified courtship genes (e.g., the fruitless gene) (14, 15). Although the male–male courtship phenotypes in the related species of D. melanogaster can be maintained by various evolutionary forces (30), it seems likely that the sphinx gene was recruited in the D. melanogaster lineage to regulate courtship behaviors.
Fig. 4.
Evolution of courtship behaviors. (A) The distribution of the male–male courtship time in Drosophila species (the vertical axis gives the frequency, the horizontal axis the CT in seconds). The phylogenetic tree of these species and the divergence time are based on refs. 16 and 35. (B) Two evolutionary models for origin of sphinx (spx) and courtship behaviors. Model 1, Gene replacement by sphinx; Model 2, Ancient behavior hypothesis: The ancestral phenotype was male–male courtship. The sphinx gene was subject to sexual selection.
Discussion
The mating behavior experiments clearly revealed a role of sphinx in the genetic control of courtship behaviors in D. melanogaster. There are two general evolutionary scenarios by which sphinx might have come to affect the level of male–male courtship (Fig. 4B): (i) sphinx might have replaced another gene with a similar function without any appreciable fitness effect, and (ii) male–male courtship might have been common in the ancestral D. melanogaster population and sphinx evolved to suppress this. In the first scenario, the sphinx gene that replaced the preexisting courtship gene (genes) would most likely have undergone neutral evolution because the function remained unchanged. Therefore, the heterosexual courtship was maintained for reproduction before and after the origination event of sphinx. It can be conceived that the male–female courtship might not be reduced by even a coexisting male–male courtship during the origination period.
However, the fact that the rate of substitution within this gene has been significantly faster than the neutral rate suggests that this process was likely driven by positive selection (10), probably sexual selection (31–34). In this model, the male–male courtship we observed in the knockout line may have been part of wild-type courtship behaviors in ancient D. melanogaster populations or even ancestral species in the melanogaster subgroup. Silencing a male-specific transcript by gene replacement would actually generate an ancestral genotype that existed before sphinx originated (Fig. 4B). This scenario is consistent with the comparative data that reveal a high level of male–male courtship common in Drosophila species other than D. melanogaster. Therefore, it seems likely that sphinx was recruited and evolved specifically to lower or inhibit male–male courtship in D. melanogaster.
The species that we used in the courtship behavior tests diverged from D. melanogaster over a range of evolutionary times (16, 35). Some diverged very recently. For example, D. simulans and D. mauritiana diverged within ≈1 mys; these two species separated from D. melanogaster within only 3 mys. Such short evolutionary distances minimize the changes in the genomic background that is associated with the compared gene region. The comparison of D. melanogaster with these species reveals that the difference in courtship behaviors of these species is correlated with the different states of the gene region, absence or presence of sphinx, supporting the ancestral behavior model. However, it is unknown yet, in addition to the role of sphinx, whether there are any other genetic and environmental factors that also contributed to the observed diversity of the courtship behavior among the species, especially among the related species of D. melanogaster. This conjecture, which was possible because of the divergence among the related species (e.g., D. mauritiana vs. the other close relatives), is to be tested for further study. Along with this line of evidence, the detected male–male courtship in D. pseudoobscura (Fig. 4) suggests that male–male courtship might have existed at least 25–30 mys ago in the most recent common ancestor of all these species. These observations suggest an evolving genetic basis for courtship behaviors, adding to the understanding of the evolution of mating behaviors among Drosophila species (36).
Methods
Analysis of Molecular Evolution and Sequence Comparison.
Homologous sequences of the sphinx gene and its upstream region were retrieved by running BLAST against the genome assemblies of the 12 Drosophila species (http://flybase.bio.indiana.edu) (37). The regions of D. simulans, D. secellia, and D. yakuba were resequenced and confirmed to be on the fourth chromosome. Multiple species alignment files of the sphinx regulatory region were generated by clustalw (www.ebi.ac.uk/Tools/clustalw/). For verification, the syntenic alignments of the sphinx region in the 12 Drosophila genomic sequences were downloaded from the University of California Santa Cruz genome browser and analyzed to detect the conserved sequence regions (http://genome.ucsc.edu).
Gene Replacement.
A P-element/FLP-mediated gene-targeting vector was constructed according to published procedures (17, 38, 39). A 5-kb DNA fragment, including the intron, exon 2, and downstream sequence of sphinx, was cloned from genomic DNA. Eight nucleotide substitutions were created by site-directed mutagenesis using the PCR method, in which the original 3′ CAG splicing and cryptic splice sites around the 3′ splice sites of Sphinx were eliminated (Fig. 2). A specific endonucleose enzyme (I-Sce1)-recognized sequence was introduced within the intron region.
Microinjection was conducted to introduce the construct into the embryos of the w1118 strain. Hatched red-eye flies were selected and crossed with w1118. Progenies were screened for insertion on X chromosome and confirmed by in situ hybridization. The transformant line was crossed with a strain carrying the FLP recombinase and I-Sce1. We heat-shocked the embryos, selected white-eye individuals from hatched embryos, and crossed them with white-eye background flies carrying constitutively expressed FLP recombinase. Then 1- to 3-day-old embryos were heat-shocked at 37°C for 1 h. The white-eye individuals from hatched embryos were then crossed with a white-eye strain that constitutively expressed FLP recombinase. We screened for uniformly red-eye individuals and test crossed with second/third chromosome balancer lines to identify those reinsertions that happened on the fourth chromosome. Finally, single fly genotyping was performed to confirm the success of targeting line named as sphinx720.
sphinx720 was further subjected to allele reduction (38, 39) to delete the marker gene mini-w and downstream sphinx sequences by crossing with the 70-I-Cre-I line carrying heat-shock-inducible I-Cre-I. Then 1- to 3-day-old heterozygous embryos were heat-shocked at 37°C for 1 h. Hatched white-eye individuals were then crossed with w1118. We obtained two independent reduction lines, sphinx720R1 and sphinx720R17, with identical sphinx regions as specified by the reduction procedure, as was shown by sequencing the sphinx regions of single flies from these lines. The reduction lines were back-crossed with an Oregon R line that, similar to Canton-S, is low or undetectable in male–male courtship behaviors measured by strict criteria, including the chaining behavior (see our tests in Figs. 3A and 4A) (25–28). Sequencing long-flanking regions of sphinx in one reduction line, sphinx720RW, by using long-range PCR amplification indicated that the mini-w with other construct-derived sequences and the wild-type exon 2 were completely deleted. The primer pairs used for the long-range PCR amplification are MS5 (forward primer), 5′-AGTGCCGGCCCTTCTCCA-3′; and J3 (reverse primer), 5′-GGCATCGGCTGTGGTTTCTA-3′ (Fig. 2).
Male–Male Courtship-Testing Experiments.
Virgin flies were collected within 8 h of enclosure at room temperature or 14 h at 18°C. Males were aged individually, females were aged in groups at room temperature in 13-h/11-h day/night cycle for 6–14 days before observation. Two male individuals of the same genotypes were placed in a glass chamber of 10-mm diameter and 4-mm height and observed under a Sony Digital Camcorder for 10–20 min or until copulation. The CT of each fly toward other flies was recorded manually. A CI was calculated for each experiments based on a 10-min video record. The statistical data were analyzed by using the Mann–Whitney test.
The standard male × female mating experiments were conducted for 15 pairs of the wild-type male × wild-type female and 15 pairs of mutant male × wild-type female, respectively.
The male–male courtship behavior was tested in the paired experiments of homozygous sphinx720RW male versus homozygous sphinx720RW male, heterozygous sphinx720RW male versus heterozygous sphinx720RW male, and wild-type male versus wild-type male, 30 pairs for each genotype. To avoid any possible bias in observation toward one of the three genotypes, blind tests were conducted to assay the male–male courtship. The courtship behaviors for each paired male–male mating experiment was scored by an observer who did not know the genotype of the paired males in a random arrangement of the movies that recorded courtship behaviors. Then the scores of this observer were compared with the courtship scores observed independently by another observer and shown to be consistent. These scores were connected to the genotype for every mating experiment for statistical analyses.
D. melanogaster (Oregon R) and its four related species (D. mauritiana, D. psedobscura, D. simulans, and D. yakuba) were tested in the male–male courtship-testing experiments. Thirty-six male–male pairs in D. melanogaster, 32 in D. mauritiana, 19 in D. simulans, 21 in D. yakuba, and 17 in D. pseudoobscura were tested and analyzed statistically by using the Mann–Whitney test.
In the tests of courtship-chaining behavior, virgin flies in D. melanogaster were collected and aged for 6–14 days as above. Then ≈50 male flies were placed in 90-mm Petri dishes with 10 females as inducers. The dishes were observed 4 h later.
Assay of the sphinx Expression in Wild Type and the Reduction Lines.
Accessory glands from males were homogenized and RNA was prepared as described by a Qiagen protocol from 60 accessory glands of 5-day-old male adult (w1118) and mutant (sphinx720RW) flies. Single-stranded cDNA was synthesized by using Superscipt III and oligo (dT) (Invitrogen). RT-PCR was carried out by using specific primers: forward, ′-CAAATAGCGTCCACCAGGAT-3′; reverse, 5′-GTGCACCTTGGGGTTGTACT-3′ for full-length transcripts and male-specific transcripts of sphinx. The extraction of RNAs and DNAs, RT-PCR, and long-range PCR that were used to amplify the targeted genomic regions and sequencing were conducted following general protocol (40).
Antisense RNA in situ hybridization was conducted by following the protocol of Invitrogen. The first exon of the sphinx gene was amplified with forward primer 5′-CCCGTGATGGCCTTTTGTTTA-3′ and reverse primer 5′-GTCAAAGGAGGGGCGTGG-3′. The amplified fragment was inserted into the construct Sphinx1stExon-pBS confirmed by sequencing. Antisense riboprobe was generated by using T7 RNA polymerase (Promega), with lacZ riboprobe as a negative control. Standard in situ hybridization was applied to the D. melanogaster line (W1118). The male reproductive organs, including testes, accessory glands, and the anterior ejaculation duct, were dissected (we also included in experiments female reproductive organs, but no expression was detected in these tissues). The tissues were treated with equal volumes of xylene and ethanol for 10 min, washed in ethanol and methanol, and fixed in 4% formaldehyde. They were then prehybridized with prehybridization solution and hybridized with 1:2,000 diluted riboprobes at 65°C. After washing with Prehyb and PBTween, the hybridized probes were detected by AP-conjugate Anti-DIG antibody (Roche), followed by an NBT/BCIP substrate reaction (Promega) visualized under the Zeiss light microscopy with Zeiss AxioCam.
Supplementary Material
Acknowledgments.
We thank Sha Sun, Mao-Lien Wu, and Jennifer Moran for assistance in fly work; Latishya Steele for technical assistance in mRNA in situ hybridization; Jean-Marc Jallon, Antonio Bernardo de Carvalho, Brian Charlesworth, Daniel Hartl, Charles Langley, Michael Ashburner, Antony Dean, Chung-I Wu, Bruce Baker, and Walter Gilbert for valuable discussions; and Bruce Walsh, Scott Roy, and Jose Ranz for critical reading of the manuscript. This work was supported by National Institutes of Health (NIH) Grants R01GM065429-01A1 and R01GM078070-01A1, National Science Foundation Career Award MCB0238168, a David and Lucile Packard Foundation Fellowship for Science and Engineering (to M.L.), a Leukemia and Lymphoma Society Scholarship, and NIH Grant R01GM074197 (to W.D.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0800693105/DCSupplemental.
References
- 1.Long M, Betrán E, Thornton K, Wang W. The origin of new genes: Glimpses from the young and old. Nat Rev Genet. 2003;4:865–874. doi: 10.1038/nrg1204. [DOI] [PubMed] [Google Scholar]
- 2.Vinckenbosch N, Dupanloup I, Kaessmann H. Evolutionary fate of retroposed gene copies in the human genome. Proc Natl Acad Sci USA. 2006;103:3220–3225. doi: 10.1073/pnas.0511307103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang W, et al. High rate of chimeric gene origination by retroposition in plant genomes. Plant Cell. 2006;18:1791–1802. doi: 10.1105/tpc.106.041905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones C, Begun D. Parallel evolution of chimeric fusion genes. Proc Natl Acad Sci USA. 2005;102:11373–11378. doi: 10.1073/pnas.0503528102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Emerson JJ, Kaessmann H, Betrán E, Long M. Extensive gene traffic on the mammalian X chromosome. Science. 2004;303:537–540. doi: 10.1126/science.1090042. [DOI] [PubMed] [Google Scholar]
- 6.Nurminsky D, Nurminskaya M, De Aguiar D, Hartl D. Selective sweep of a newly evolved sperm-specific gene in Drosophila. Nature. 1998;296:572–575. doi: 10.1038/25126. [DOI] [PubMed] [Google Scholar]
- 7.Loppin B, Lepetit D, Dorus S, Couble P, Karr TL. Origin and neofunctionalization of a Drosophila paternal effect gene essential for zygote viability. Curr Biol. 2005;15:87–93. doi: 10.1016/j.cub.2004.12.071. [DOI] [PubMed] [Google Scholar]
- 8.Long M, Langley CH. Natural selection and the origin of jingwei, a chimeric processed functional gene in Drosophila. Science. 1993;260:91–95. doi: 10.1126/science.7682012. [DOI] [PubMed] [Google Scholar]
- 9.Ohta T. Evolution by gene duplication revisited: Differentiation of regulatory elements versus proteins. Genetica. 2002;118:209–216. [PubMed] [Google Scholar]
- 10.Wang W, Brunet FG, Nevo E, Long M. Origin of sphinx, a young chimeric RNA gene in Drosophila melanogaster. Proc Natl Acad Sci USA. 2002a;99:4448–4453. doi: 10.1073/pnas.072066399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang W, Thornton K, Berry A, Long M. Nucleotide variation along the Drosophila melanogaster fourth chromosome. Science. 2002b;295:134–137. doi: 10.1126/science.1064521. [DOI] [PubMed] [Google Scholar]
- 12.Sokolowski MB. Drosophila: Genetics meets behaviour. Nat Rev Genet. 2001;2:879–890. doi: 10.1038/35098592. [DOI] [PubMed] [Google Scholar]
- 13.Bray S, Amrein H. A putative Drosophila pheromone receptor expressed in male-specific taste neurons is required for efficient courtship. Neuron. 2003;39:1019–1029. doi: 10.1016/s0896-6273(03)00542-7. [DOI] [PubMed] [Google Scholar]
- 14.Baker BS, Taylor BJ, Hall JC. Are complex behaviors specified by dedicated regulatory genes? Reasoning from Drosophila. Cell. 2001;105:13–24. doi: 10.1016/s0092-8674(01)00293-8. [DOI] [PubMed] [Google Scholar]
- 15.Gailey DA, et al. Functional conservation of the fruitless male sex-determination gene across 250 Myr of insect evolution. Mol Biol Evo l. 2006;23:633–643. doi: 10.1093/molbev/msj070. [DOI] [PubMed] [Google Scholar]
- 16.Powell JR. The Drosophila Model. New York: Oxford Univ Press; 1997. [Google Scholar]
- 17.Rong YS, Golic KG. A targeted gene knockout in Drosophila. Genetics. 2001;157:1307–1312. doi: 10.1093/genetics/157.3.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang SD, Odenwald WF. Misexpression of the white (w) gene triggers male-male courtship in Drosophila. Proc Natl Acad Sci USA. 1995;92:5525–5529. doi: 10.1073/pnas.92.12.5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vasudevan S, Peltz SW. Nuclear mRNA surveillance. Curr Opin Cell Biol. 2003;15:332–337. doi: 10.1016/s0955-0674(03)00051-6. [DOI] [PubMed] [Google Scholar]
- 20.Frischmeyer PA, et al. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science. 2002;295:2258–2261. doi: 10.1126/science.1067338. [DOI] [PubMed] [Google Scholar]
- 21.Behm-Ansmant I, Izaurralde E. NMD in Drosophila: A snapshot into the evolution of a conserved mRNA surveillance pathway. In: Maquat LE, editor. Nonsense-Mediated mRNA Decay. Austin, TX: Landes Bioscience; 2000. [Google Scholar]
- 22.Ferveur JF. Cuticular hydrocarbons: Their evolution and roles in Drosophila pheromonal communication. Behavior Genetics. 2005;35:279–295. doi: 10.1007/s10519-005-3220-5. [DOI] [PubMed] [Google Scholar]
- 23.Hall JC. Courtship among males due to a male-sterile mutation in Drosophila melanogaster. Behav Genet. 1978;8:125–141. doi: 10.1007/BF01066870. [DOI] [PubMed] [Google Scholar]
- 24.Hall JC. The mating of a fly. Science. 1994;264:1702–1714. doi: 10.1126/science.8209251. [DOI] [PubMed] [Google Scholar]
- 25.Yamamoto D, Ito H, Fujitani K. Genetic dissection of sexual orientation: Behavioral, cellular, and molecular approaches in Drosophila melanogaster. Neurosci Res. 1996;26:95–107. doi: 10.1016/s0168-0102(96)01087-5. [DOI] [PubMed] [Google Scholar]
- 26.Demir E, Dickson BJ. Fruitless splicing specifies male courtship behavior in Drosophila. Cell. 2005;121:785–794. doi: 10.1016/j.cell.2005.04.027. [DOI] [PubMed] [Google Scholar]
- 27.Stockinger P, Kvitsiani D, Rotkopf S, Tirián L, Dickson BJ. Neural circuitry that governs Drosophila male courtship behavior. Cell. 2005;121:795–807. doi: 10.1016/j.cell.2005.04.026. [DOI] [PubMed] [Google Scholar]
- 28.Cobb M, Jallon J-M. Pheromones, mate recognition and courtship stimulation in the Drosophila melanogaster species sub-group. Anim Behav. 1990;39:1058–1067. [Google Scholar]
- 29.Pailette M, Ikeda H, Jallon J-M. A new acoustic signal of the fruit-flies Drosophila simulans and D. melanogaster. Bioacoustics. 1991;3:247–254. [Google Scholar]
- 30.Gavrilets S, Rice WR. Genetic models of homosexuality: Generating testable predictions. Proc R Soc London Ser B. 2006;273:3031–3038. doi: 10.1098/rspb.2006.3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ranz JM, Castillo-Davis CI, Meiklejohn CD, Hartl DL. Sex-dependent gene expression and evolution of the Drosophila transcriptome. Science. 2003;300:1742–1745. doi: 10.1126/science.1085881. [DOI] [PubMed] [Google Scholar]
- 32.Mueller JL, et al. Cross-species comparison of Drosophila male accessory gland protein genes. Genetics. 2005;171:131–143. doi: 10.1534/genetics.105.043844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Swanson WJ, Vacquier VD. The rapid evolution of reproductive proteins. Nat Rev Genet. 2002;3:137–144. doi: 10.1038/nrg733. [DOI] [PubMed] [Google Scholar]
- 34.Clark AG, Aguadé M, Prout T, Harshman LG, Langley CH. Variation in sperm displacement and its association with accessory gland protein loci in Drosophila melanogaster. Genetics. 1995;139:189–201. doi: 10.1093/genetics/139.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lachaise D, et al. Historical biogeography of the Drosophila-melanogaster species subgroup. Evol Biol. 1988;22:159–225. [Google Scholar]
- 36.Greenspan RJ, Ferveur JF. Courtship in Drosophila. Annu Rev Genet. 2001;34:205–232. doi: 10.1146/annurev.genet.34.1.205. [DOI] [PubMed] [Google Scholar]
- 37.Clark AG, et al. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- 38.Rong YS, et al. Targeted mutagenesis by homologous recombination in D. melanogaster. Genes Dev. 2002;16:1568–1581. doi: 10.1101/gad.986602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun S, Ting CT, Wu CI. The normal function of a speciation gene, Odysseus, and its sterility effect. Science. 2004;305:81–83. doi: 10.1126/science.1093904. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2001. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.