Abstract
Acetylcholine-binding proteins (AChBPs) from mollusks are suitable structural and functional surrogates of the nicotinic acetylcholine receptors when combined with transmembrane spans of the nicotinic receptor. These proteins assemble as a pentamer with identical ACh binding sites at the subunit interfaces and show ligand specificities resembling those of the nicotinic receptor for agonists and antagonists. A subset of ligands, termed the neonicotinoids, exhibit specificity for insect nicotinic receptors and selective toxicity as insecticides. AChBPs are of neither mammalian nor insect origin and exhibit a distinctive pattern of selectivity for the neonicotinoid ligands. We define here the binding orientation and determinants of differential molecular recognition for the neonicotinoids and classical nicotinoids by estimates of kinetic and equilibrium binding parameters and crystallographic analysis. Neonicotinoid complex formation is rapid and accompanied by quenching of the AChBP tryptophan fluorescence. Comparisons of the neonicotinoids imidacloprid and thiacloprid in the binding site from Aplysia californica AChBP at 2.48 and 1.94 Å in resolution reveal a single conformation of the bound ligands with four of the five sites occupied in the pentameric crystal structure. The neonicotinoid electronegative pharmacophore is nestled in an inverted direction compared with the nicotinoid cationic functionality at the subunit interfacial binding pocket. Characteristic of several agonists, loop C largely envelops the ligand, positioning aromatic side chains to interact optimally with conjugated and hydrophobic regions of the neonicotinoid. This template defines the association of interacting amino acids and their energetic contributions to the distinctive interactions of neonicotinoids.
Keywords: acetylcholine-binding protein, crystal structure, imidacloprid, nicotinic receptor, thiacloprid
Receptors of the Cys-loop superfamily, encompassing the nicotinic acetylcholine (ACh), γ-aminobutyric acid, glycine, and serotonin type 3 receptors in mammals, play crucial roles in central and peripheral nervous system neurotransmission (1, 2). Agonists acting to open these ligand-gated ion channels are predominantly cationic with quaternary ammonium or tertiary, secondary, or primary amines as a critical determinant. Nicotinic agonists and most competitive antagonists are simple heterocyclic compounds, with higher affinities often achieved with imines and bridged nitrogen compounds. These agonists bind in their protonated state (3), forming a hydrogen bond with the carbonyl oxygen of a conserved loop B tryptophan. In addition, a surrounding π orbital nest is formed by aromatic side chains of both contributing subunit faces, thereby stabilizing cationic ligands in the binding site. The loop C structure, characterized by a disulfide bond from vicinal cysteines at its tip, appears to envelop the bound agonist (4), and this closure may be a critical step in the agonist activation process. A sequence of conformational rearrangements in the protein structure ultimately gates ion flow by effectively widening a presumed constriction point within the transmembrane-spanning region of the receptor.
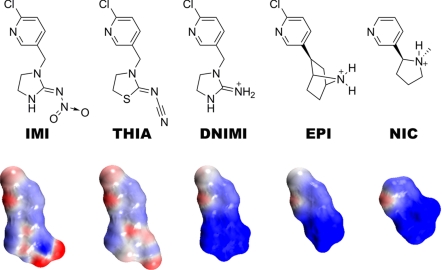
The nicotinic ACh receptor (nAChR) is a target of potential therapeutic agents for neurological dysfunction and drug abuse (1, 5). It is also the site where selective toxicity between insects and mammals is achieved with the major neonicotinoid insecticides (6). Neonicotinoid agonists imidacloprid (IMI) and thiacloprid (THIA) (Fig. 1) are exceptional in their physicochemical properties in possessing a distinctive electronegative nitroimino or cyanoimino pharmacophore, in contrast to a cationic functionality of nicotinic agonists such as desnitroimidacloprid (DNIMI), epibatidine (EPI), and nicotine (NIC) (7).
Fig. 1.
Chemical structures of neonicotinoids imidacloprid (IMI) and thiacloprid (THIA) with an electronegative nitro or cyano pharmacophore compared with cationic nicotinic agonists desnitroimidacloprid (DNIMI), epibatidine (EPI), and nicotine (NIC). Below each structure is shown its charge distribution with the electron clouds in red for negative and blue for positive. Electrostatic potentials were calculated and images rendered by using Accelrys DS Visualizer v1.7.
Crystal structures of mollusk AChBPs, as structural surrogates of the nAChR extracellular ligand-binding domain (8–10), complexed with cationic nicotinoids, EPI and NIC, provide substantial information on the recognition properties of the agonist binding site (4, 11). Similarly, structural data on the binding locations of competitive and noncompetitive antagonists reveal many determinants of ligand selectivity at the subunit interfaces. Aplysia californica AChBP binds both electronegative neonicotinoids and cationic nicotinoids with high affinity, whereas Lymnaea stagnalis AChBP is poorly sensitive to the insecticides. Therefore, these AChBPs serve as models of the insect and mammalian nAChRs defining interactions of these two chemotypes of agonists (4, 12–14). Distinctive pharmacophores of neonicotinoids, usually with a characteristic electronegative tip, undergo atypical interactions in the nAChR-binding pocket. We present here a crystallographic investigation of Aplysia AChBP liganded with the neonicotinoids IMI and THIA, thereby directly characterizing atomic interactions critical to the neonicotinoids and providing a crystallographic template for analyzing structural features that differentiate between nicotinoid and neonicotinoid selectivity.
Results
Ligand Binding Kinetics for AChBP.
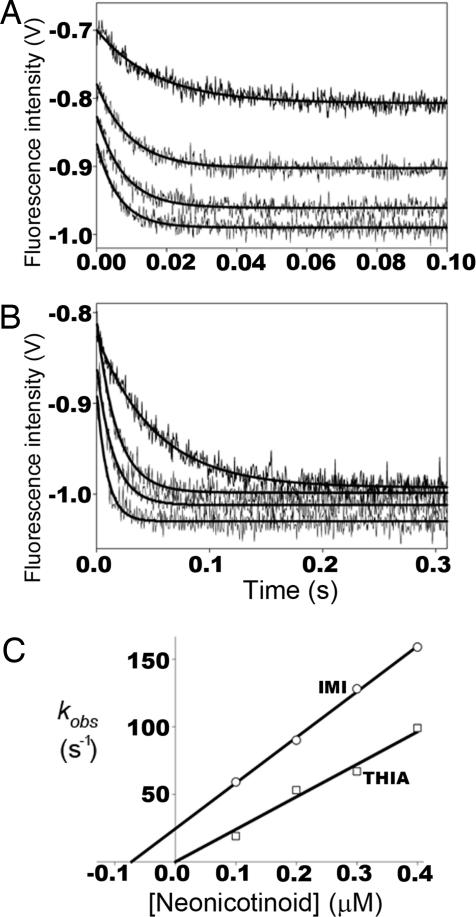
Direct measure of ligand binding through quenching of native tryptophan fluorescence of AChBP reveals several interesting features (Fig. 2 and Table 1). First, the extent of quenching is similar for the neonicotinoids, IMI and THIA, and nicotinoids, DNIMI and EPI (15), despite the different charge distribution on the two ligands. Second, the rates of association and dissociation are rapid, requiring stopped-flow instrumentation for detection (15). Association rates are similar to those for ACh (15) and approach the diffusion limit. Across a range of concentrations, the kinetics exhibit a simple bimolecular association and unimolecular dissociation mechanism for the five binding sites in the pentamer (Table 1).
Fig. 2.
Kinetic studies of ligand association with AChBP. Stopped-flow traces of tryptophan fluorescence quenching on neonicotinoid binding to Aplysia AChBP. Binding of 0.1, 0.2, 0.3, and 0.4 μM (ordered top to bottom) IMI (A) and THIA (B) to 4 nM AChBP pentamer. (C) First-order rate constants of fluorescence quenching (kobs) determined from A and B plotted against neonicotinoid concentration. Intercepts of lines on the ordinate represent first-order dissociation rate constants (koff), intercepts on the abscissa represent Kd, and line slopes represent second-order association rate constants (kon). For THIA only, kon could be determined from the association kinetics, whereas for IMI all three constants (kon, koff, and Kd) were estimated. For both IMI and THIA the first-order dissociation rate constants were measured in separate experiments by the enhancement of fluorescence after addition of excess gallamine or strychnine.
Table 1.
Kinetic constants for interaction of neonicotinoids with Aplysia AChBP
| IMI* | THIA* | DNIMI* | |
|---|---|---|---|
| Association kinetics (Fig. 2) | |||
| kon (108 M−1 sec−1) | 3.1 ± 0.1 | 2.5 ± 0.1 | 4.2 ± 0.2 |
| koff (101 sec−1) | 2.1 ± 0.7 | n.d.† | n.d.† |
| Kd (nM) | 68 ± 23 | n.d.† | n.d.† |
| Dissociation kinetics (excess competing ligand) | |||
| koff (101 sec−1) | 2.0 ± 0.3 | 0.33 ± 0.1 | 0.77 ± 0.21 |
| Kd (koff /kon), nM | 63 | 14 | 18 |
*The kon, koff, and Kd were determined from the kinetics of neonicotinoid association with AChBP (cf. Fig. 2C). koff was additionally determined in separate dissociation measurements as described in Methods. Tabulated values are the means of three to six experiments ± the standard deviations.
†n.d., not determined; immeasurable koff or kd by this method.
Crystal Structures of IMI-AChBP and THIA-AChBP Complexes.
The data collection and refinement statistics are given in Table 2. The resolved residues, alternative side-chain conformations, and atomic compositions of the final structures of the AChBP complexes with IMI (AChBP-IMI) and THIA (AChBP-THIA) at 2.48 Å and 1.94 Å, respectively, are shown in Table 3.
Table 2.
Data collection and refinement statistics
| AChBP-IMI | AChBP-THIA | |
|---|---|---|
| Data collection | ||
| Space group | P212121 | P212121 |
| Cell dimensions | ||
| a, b, c, Å | 85.4, 116.3, 132.5 | 84.9, 116.7, 132.0 |
| α, β, γ, ° | 90, 90, 90 | 90,90,90 |
| Resolution, Å | 132.45–2.48 (2.57–2.48)* | 87.37–1.94 (2.01–1.94)* |
| Rmerge, % | 0.095 (0.498)* | 0.084 (0.491)* |
| I / σ I | 18.1 (4.0)* | 21.5 (4.1)* |
| Completeness, % | 97.7 (98.1)* | 99.9 (100)* |
| Redundancy | 6.4 (6.4)* | 7.1(6.8)* |
| Refinement | ||
| Resolution, Å | 50–2.48 | 43.7–1.94 |
| No. reflections | 46,400 | 97,391 |
| Rwork/Rfree | 0.188/0.250 | 0.186/0.215 |
| No. atoms | ||
| Protein | 8645 | 8391 |
| Ligand/sugar | 80 | 92 |
| Water | 126 | 479 |
| Overall B factors, Å2 | 39.5 | 35.2 |
| R.m.s deviations | ||
| Bond lengths, Å | 0.008 | 0.006 |
| Bond angles, ° | 1.4 | 1.3 |
*Highest-resolution shell statistics are shown in parentheses.
Table 3.
Refined x-ray structures fitted into electron density
| AChBP-IMI | AChBP-THIA | |
|---|---|---|
| Resolution | 2.48 Å | 1.94 Å |
| Residues fitted | ||
| Subunit A | −2 to 208 | −2 to 207 |
| Subunit B | −3 to 208 | −3 to 208 |
| Subunit C | −1 to 17; 20 to 208 | −2 to 17; 20 to 207 |
| Subunit D | −3 to 208 | −1 to 18; 20 to 208 |
| Subunit E | −3 to 208 | −2 to 14; 20 to 207 |
| Bound ligands | 4 | 4 |
| Isopropyl alcohol molecules | 3 | 7 |
| Solvent waters | 124 | 479 |
| Side chains with alternate conformations | 3 | 10 |
| Mg2+ | — | 2 |
Atomic Interactions of Ligands with AChBP.
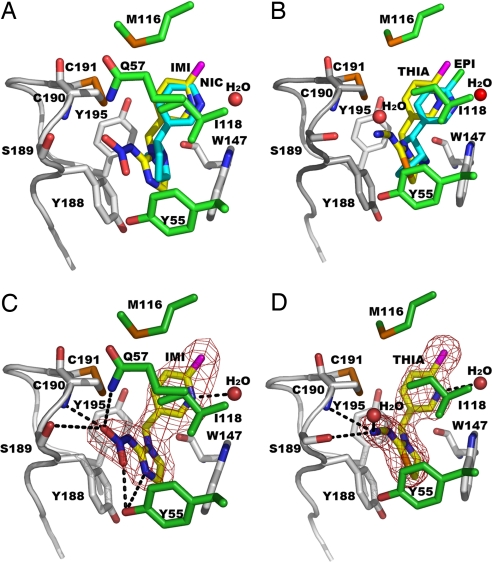
All four pyridine rings of IMI, THIA, NIC (11), and EPI (4) virtually overlap with the pyridine nitrogen in a common position (Fig. 3). The three AChBP-ligand x-ray structures with the higher resolution (IMI, THIA, and NIC) have an associated water in the same position near Ile-118. The other heterocyclic ring systems linked directly to the pyridine or through a methylene bridge also occupy similar space. All ligands interact with mostly identical AChBP residues (Table 4) making several hydrogen bonds to the residues on the two subunits forming the interface and to two incorporated water molecules. The IMI, THIA, and EPI chlorines all make van der Waals contacts with Ile106O (not displayed in Fig. 3) and Met116O (4.11 Å and 3.13 Å for IMI, 4.17 Å and 3.01 Å for THIA, and 3.61 Å and 3.71 Å for EPI, respectively).
Fig. 3.
Bound agonists shown as overlapping or individual structures. (A) AChBP-IMI (yellow) and AChBP-NIC (PDB ID code 1UW6) (11) (cyan). (B) AChBP-THIA (yellow) and AChBP-EPI (PDB ID code 2BYQ) (4) (cyan). IMI (C) and THIA (D) electron density omit maps with coefficients, Fo − Fc >3σ, showing hydrogen bonds as dashed lines. Principal face residues with distances <3.6 Å from the ligands and loop C are shown in gray. Complementary face residues are in given green, S atoms and Cys–Cys bond in orange, and Cl in magenta.
Table 4.
AChBP residues making proximal contacts shorter than 3.6 Å to ligands
| Residue | IMI | THIA | EPI | NIC* |
|---|---|---|---|---|
| Principal face | ||||
| Tyr93 | + | − | + | + |
| Trp147 | + | + | + | + |
| Thr148 | − | − | + | + |
| Tyr188 | + | + | + | − |
| Ser189 | + | + | − | − |
| Cys190 | + | + | + | − |
| Cys191 | − | − | + | + |
| Tyr195 | + | + | + | + |
| Complementary face | ||||
| Tyr55 | + | + | + | + |
| Gln57 | + | − | − | − |
| Ile106 | − | − | + | − |
| Ala107 | − | − | + | − |
| Val108 | − | − | + | + |
| Met116 | + | + | + | + |
| Ile118 | + | + | + | + |
The IMI pyridine N hydrogen bonds to one water molecule (3.13 Å) bridging to Ile118N (2.90 Å) and Ile118O (3.77 Å), and the terminal oxygens on the nitro group interact with Ser189Oγ (3.26 Å) and Cys190N (3.04 Å) of the principal subunit face, and with Tyr55OH (3.24 Å) and Gln57Nε2 (3.26 Å) of the complementary subunit face.
In an identical manner, the THIA pyridine N hydrogen bonds to a water molecule (2.91 Å) bridging to Ile118N (2.89 Å) and Ile118O (3.60 Å) and its cyano N atom makes hydrogen bonds to Ser189Oγ (3.36 Å), Cys190N (3.52 Å), and one water molecule (2.76 Å) binding to Ser189Oγ. Tyr-55 on the complementary face also appears to have a π interaction with the sulfur in the thiazolidine ring. As with the IMI guanidine plane, the THIA amidine interacts with the aromatic side chain of Tyr-188 and possibly with that of Trp-147.
In contrast to the neonicotinoids, the bridged, protonated nitrogen in EPI faces loop B in AChBP, hydrogen bonds with the carbonyl oxygen of Trp-147, and is shielded by surrounding cation–π interactions. The bridged cation, that is, functionality, of EPI is positioned closer to the carbonyl oxygen (2.8 Å) than the N1/N3 nitrogen (which is connected to the pyridinylmethyl moiety) in IMI or THIA (≈3.5 Å). In the neonicotinoids, hydrogen bond formation with the carbonyl oxygen of Trp-147 is precluded by the sp2 bond character, the perpendicular extension from the ring of the 2p orbital lobes and the poor protonation capacity of the N1/N3 nitrogen (7).
Occupied Sites in the Crystal Structure.
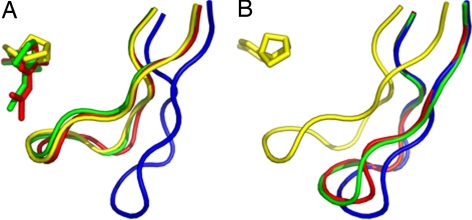
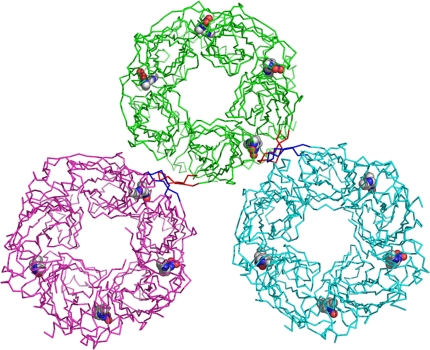
Both complex structures of AChBP-IMI and AChBP-THIA contain only four occupied sites per pentamer. Similar to other AChBP complexes, the binding surfaces are nestled between two abutting subunits. In each of the ligand-binding subunits, loop C (Gln-186 to Tyr-195) envelops the ligand by undergoing a considerable conformational change (from an “open” to “closed” loop position) upon binding of the ligands (Fig. 4). Based on the arrangement of symmetry-related molecules, when two loop C regions come within proximity, only one of the two sites at the subunit interface is occupied by ligand (Fig. 5)
Fig. 4.
Overlap of loop C and the bound ligands in AChBP, AChBP-IMI, AChBP-THIA, and AChBP-EPI (4). (A) Subunit A with closed loop C conformation for the ligand-bound AChBP-IMI (red), AChBP-THIA (green), and AChBP-EPI (yellow). Loop C has an open conformation in the native AChBP (blue) structure. (B) Subunit E in which only AChBP-EPI has a bound ligand shows a closed loop C conformation (yellow), whereas the apo-AChBP, AChBP-IMI, and AChBP-THIA sites which are unoccupied show an open loop C conformation (see also Fig. 5).
Fig. 5.
Interactions of an AChBP-IMI molecule (green) with the two symmetry-related molecules that form intermolecular contacts <3.6 Å (shown in cyan and magenta). A closed loop C (red) of subunit A (green) forms a β-sheet with an open loop C (blue) of a symmetry-related molecule (cyan) of subunit E. A similar packing interaction is seen between an open loop C (red) of subunit E to a closed loop C (blue) of another symmetry-related molecule (magenta) of subunit A. The bound IMI molecules are shown as CPK models.
Calculation of the root-mean-squared deviation in the position of the Cα carbons between two structures (Cα RMS) of subunits in AChBP, AChBP-IMI, AChBP-THIA, and AChBP-EPI (4) shows deviations ranging from 0.21 to 0.45 Å for the similar conformations of two ligand-bound subunit interfaces. Similar Cα RMS deviations are found for the two ligand-free subunit interfaces. These Cα RMS of the different trajectories of a ligand-bound subunit vs. a ligand-free subunit range between 0.79 and 1.08 Å.
Discussion
Bound Conformations and Atomic Interactions.
The high-resolution crystal structures of AChBP–neonicotinoid complexes with IMI and THIA reported here, and in earlier findings for an EPI-occupied AChBP crystal structure (4) and its nicotinic agonist relatives (11), are generally consistent with the results of photoaffinity labeling in a physiological condition with 5-azido-6-chloropyridin-3-yl IMI, THIA, and EPI probes (12–14). Collectively, the data clearly define the distinct molecular aspects of small agonist interactions with the nAChR structural surrogate, AChBP, therein accounting for the affinity and specificity of these agonists at the nAChRs. Neonicotinoids, being nonprotonated at physiological pH values and bearing the nitro or cyano electronegative functional tip (7), primarily interact with the loop C region of the binding pocket. This important aspect of the neonicotinoids with a largely neutral heterocyclic ring and a nitro- or cyanoimino moiety is an obvious deviation from that of the cationic agonists, EPI and NIC. Spectroscopic evidence from the anabaseine analogs and the pH dependence of binding of these and other nicotinic agonists reveal that the nitrogen is protonated in the bound state (3). The ammonium head of EPI or NIC principally hydrogen bonds with Trp147O (4, 11) and secondarily undergoes cation–π interaction with the loop B tryptophan aromatic indole moiety and other aromatic residues (16, 17).
Conformational Changes in Loop C Associated with Ligand Binding.
Comparing Aplysia AChBP–ligand complexes with an apo structure (Fig. 4) provides direct evidence of differential conformational rearrangements induced by antagonist and agonist occupation (4). Moreover, deuterium-hydrogen exchange rates of the amide backbone hydrogens show solvent exclusion of the majority of loop C amides after agonist binding (18). The loop C movement capping the binding pocket, characteristic of agonist occupation, is also evident from the present crystal structures, therefore suggesting stabilization of neonicotinoid interaction with the closed loop C region. This conformational change can be rationalized as an initial event for the ligand-induced channel-opening mechanism of the nAChR.
Retained Water Molecules in the Binding Pocket.
Common water molecule positions are captured in the present high-resolution crystal structures, revealing the role of water in the interactions of the ligands with the residues in the AChBP-binding pocket. A water molecule is observed near the pyridine N of NIC (11), as well as IMI and THIA, bridging to loop B and/or loop E residues. Interestingly, an additional water bridge is evident around the tip nitrogen of the THIA cyano substituent, presumably enhancing the interaction of this pharmacophore with loop C.
Crystallographic Packing and Ligand Occupation.
Both AChBP-IMI and AChBP-THIA contain four ligand-bound subunit interfaces and one apo-form subunit interface with no ligand occupation. The reason for this level of occupation of sites may be found in the crystallographic packing in the P212121 crystal form. Inspection of the symmetry-related molecules near loop C of each subunit reveals that only two of them form contacts (<3.6 Å) to the primary molecule. These are formed to subunits A and E (E being the ligand-free subunit). The loop C strand-turn-strand of subunit A forms a parallel β-sheet with loop C of subunit E of a symmetry-related molecule, that is, a closed loop makes a β-sheet surface contact with an open loop. There are 17 interactions shorter than 3.6 Å in this β-sheet formation. A similar pattern for packing is seen between the ligand-free open loop C of subunit E to the closed loop C of subunit A in the symmetry-related molecule that has a closed conformation. This β-sheet packing results in 15 contacts at <3.6 Å apart. It is plausible that such an intimate crystal packing between open and closed forms of the ligand-bound loop C results in the loss of a ligand in subunit E while retaining it in subunit A. Loop C of subunits B–D have no close contacts to symmetry-related molecules and hence retain their ligands.
Crystallographic and Solution-Based Inferences on Structure.
Kinetic profiles revealing simple bimolecular association and unimolecular dissociation of the neonicotinoids are consistent with homogeneity of the binding sites where each pentameric binding protein is not influenced by neighboring molecules in solution. Because the neonicotinoids, similar to ACh, bind at rates approaching the diffusion limit, it is likely that all of the subunit interfaces are accessible and not rigidly capped by loop C. Hence, full occupation of the five sites should be achieved in solution and the single vacant site in the crystallized molecule results from interference by the symmetry-related molecule. Similarly, AChBP is specifically photoaffinity-labeled with up to one agonist molecule for each subunit based on mass spectrometry analysis of the intact derivatized protein (12–14). Furthermore, the kinetics does not reveal the presence of two binding orientations of the bound neonicotinoid Aplysia AChBP. The present kinetics and previous labeling clearly show the presence of a single tight-binding orientation of neonicotinoids with the Aplysia AChBP, yet azido-neonicotinoid probes identify two disparate binding conformations with the Lymnaea AChBP, perhaps accounting for its inferior affinity for neonicotinoids (12, 13). However, absolute comparisons should not be overemphasized. Alternative conformational orientations might be expected when the overall binding energy of the ligand is diminished as found for Lymnaea.
Photoaffinity labeling is selective for particular reactive residues, leading to a possible bias in interpreting derivatization sites. However, crystallization may not reveal one of the binding orientations seen in solution, because nucleation and crystal growth may be forced by a single conformation. Hence, comparisons of solution and crystal-based structures provide a more comprehensive picture of the binding site with preferred and alternative binding orientations.
Concluding Remarks.
The crystal structures of soluble AChBP-neonicotinoid complexes define geometries of functional amino acids in the neonicotinoid-bound state, thereby providing an atomic scale template for uncovering the specificity differences between mammalian and insect nAChRs. Mollusk AChBP might be considered as an “intermediate” template between mammals and insects accommodating both nicotinoids and neonicotinoids. Because the position of the pyridine ring in the two classes of molecules is virtually identical, modification of surrounding residues in the vicinity of the other heterocyclic ring system and its substituents should enable one to alter specificity in either the neonicotinoid or nicotinoid direction. Such modifications not only could be monitored by changes in the binding kinetics and energetics, but also by measurement of solvent exposure to the loop C region (18) and by labeling patterns with the azido derivatives (12–14). Accordingly, selective toxicity, which forms the basis of relative efficacy and safety of insecticides, can be rationalized on the basis of precise molecular determinants on the homologous target molecules in the insect and mammal.
Methods
AChBP Preparation.
AChBP from Aplysia was expressed in HEK-293 cells, as described in ref. 3. In brief, cDNAs containing an N-terminal FLAG epitope and a preprotrypsin leader peptide for secretion were transfected by using calcium phosphate into the HEK-293 cell line deficient in the N-acetylglucosaminyltransferase I gene (19). Cells were selected for stable expression by G418 resistance. Tissue culture media containing AChBPs were harvested at 1- to 3-day intervals as necessary and supplemented with 0.02% NaN3. AChBP was purified from the medium by adsorption onto an anti-FLAG affinity column (Sigma) and eluted with FLAG peptide. Assembly as a monodisperse pentamer retaining the FLAG tag was verified by size-exclusion chromatography.
Monitoring Neonicotinoid Complex Formation.
Neonicotinoid or nicotinoid ligand complex formation with AChBP was monitored directly through tryptophan fluorescence quenching accompanying ligand association. Stopped-flow instrumentation was used to monitor association and dissociation rates from the overall rate of complex formation at various ligand concentrations (15). To monitor dissociation rates independently complexes were prepared by mixing 40 nM AChBP pentamer with 2 μM IMI or 400 nM THIA, and dissociation rates of neonicotinoids were monitored in the stopped-flow apparatus on twofold dilution of the complex into high concentration of competing ligand, gallamine, or strychnine. Because at very high concentrations of competing ligand, a ternary complex forms accelerating the apparent dissociation, the given values of koff were obtained on extrapolation of dissociation rates, measured at different competing ligand concentrations, to a plateau value on a rate versus concentration of competing ligand plot. In this concentration range, the kinetic values are limited by the intrinsic dissociation rate of the neonicotinoid.
Crystallization and Data Collection.
Ligand–AChBP complexes were formed by adding a ratio of 1.5- to 2-fold molar excess of ligand to ≈300 μM AChBP binding sites. Ligands were diluted from 80 mM stocks in dimethyl sulfoxide. The complexes were crystallized by vapor diffusion at 18°C by using a protein-to-reservoir ratio of 1:1 in 2-μl drops. The reservoir solution that yielded the best-diffracting crystals contained 0.1 M Hepes, pH 7.5, and 0.2 M MgCl2 with 22% isopropyl alcohol for IMI and 30% isopropyl alcohol for THIA.
Crystals for both complexes were flash-cooled in liquid nitrogen after a brief soak in 0.09 M Hepes, pH 7.5, 0.18 M MgCl2, 27% isopropyl alcohol, and 10% glycerol. The cryoprotecting soak for the THIA complex crystals was supplemented with 1 mM THIA. Single-wavelength diffraction data were collected at the Advanced Photon Source at Argonne National Laboratory beamline 19-ID with a wavelength of 0.979 Å. Data were processed with HKL2000 (20) giving parameters shown in Table 2.
Structure Determination and Refinement.
The structures of the AChBP complexes were solved by molecular replacement with PHASER (21), by using the apo AChBP structure (PDB ID code 2BYN) (4) as a search model. Initial electron density maps were improved by manual adjustment with COOT (22) and by refinement with CNS (23) and REFMAC5 (24) by using the maximum likelihood approach. Refinement parameters are given in Tables 2 and 3. The atomic coordinates and structure factors of the AChBP complexes have been deposited with the RCSB Protein Data Bank (PDB ID codes 3C79 for AChBP-IMI and 3C84 for AChBP-THIA). Figures of the structures were generated with PyMOL (25).
Acknowledgments.
This work was supported by National Institutes of Health Grants R37-GM18360 and UO1-NS05846 (to T.T.T. and P.T.), the William Muriece Hoskins Chair in Chemical and Molecular Entomology (J.E.C.), and National Institute of Environmental Health Sciences Grant R01 ES08424 (to M.T. and J.E.C.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors of the AChBP complexes have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3C79 for AChBP-IMI and 3C84 for AChBP-THIA).
References
- 1.Changeux JP, Edelstein SJ. Nicotinic Acetylcholine Receptors: From Molecular Biology to Cognition. New York: Odile Jacob/Johns Hopkins Univ Press; 2005. [Google Scholar]
- 2.Karlin A. Emerging structure of the nicotinic acetylcholine receptors. Nat Rev Neurosci. 2002;3:102–114. doi: 10.1038/nrn731. [DOI] [PubMed] [Google Scholar]
- 3.Talley TT, et al. Spectroscopic analysis of benzylidene anabaseine complexes with acetylcholine binding proteins as models for ligand-nicotinic receptor interactions. Biochemistry. 2006;45:8894–8902. doi: 10.1021/bi060534y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hansen SB, et al. Structures of Aplysia AChBP complexes with nicotinic agonists and antagonists reveal distinctive binding interfaces and conformations. EMBO J. 2005;24:3635–3646. doi: 10.1038/sj.emboj.7600828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gotti C, Clementi F. Neuronal nicotinic receptors: From structure to pathology. Prog Neurobiol. 2004;74:363–396. doi: 10.1016/j.pneurobio.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 6.Tomizawa M, Casida JE. Neonicotinoid insecticide toxicology: Mechanisms of selective action. Annu Rev Pharmacol Toxicol. 2005;45:247–268. doi: 10.1146/annurev.pharmtox.45.120403.095930. [DOI] [PubMed] [Google Scholar]
- 7.Tomizawa M, Zhang N, Durkin KA, Olmstead MM, Casida JE. The neonicotinoid electronegative pharmacophore plays the crucial role in the high affinity and selectivity for the Drosophila nicotinic receptor: An anomaly for the nicotinoid cation–pi interaction model. Biochemistry. 2003;42:7819–7827. doi: 10.1021/bi0300130. [DOI] [PubMed] [Google Scholar]
- 8.Brejc K, van Dijk WJ, Smit AB, Sixma TK. The 2.7 Å structure of AChBP, homologue of the ligand-binding domain of the nicotinic acetylcholine receptor. Novartis Found Symp; 2002. pp. 22–29. discussion 29–32, 165–168. [PubMed] [Google Scholar]
- 9.Smit AB, et al. A glia-derived acetylcholine-binding protein that modulates synaptic transmission. Nature. 2001;411:261–268. doi: 10.1038/35077000. [DOI] [PubMed] [Google Scholar]
- 10.Brejc K, et al. Crystal structure of an ACh-binding protein reveals the ligand-binding domain of nicotinic receptors. Nature. 2001;411:269–276. doi: 10.1038/35077011. [DOI] [PubMed] [Google Scholar]
- 11.Celie PH, et al. Nicotine and carbamylcholine binding to nicotinic acetylcholine receptors as studied in AChBP crystal structures. Neuron. 2004;41:907–914. doi: 10.1016/s0896-6273(04)00115-1. [DOI] [PubMed] [Google Scholar]
- 12.Tomizawa M, et al. Atypical nicotinic agonist bound conformations conferring subtype selectivity. Proc Natl Acad Sci USA. 2008;105:1728–1732. doi: 10.1073/pnas.0711724105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tomizawa M, et al. Mapping the elusive neonicotinoid binding site. Proc Natl Acad Sci USA. 2007;104:9075–9080. doi: 10.1073/pnas.0703309104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tomizawa M, et al. Defining nicotinic agonist binding surfaces through photoaffinity labeling. Biochemistry. 2007;46:8798–8806. doi: 10.1021/bi700667v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hansen SB, et al. Tryptophan fluorescence reveals conformational changes in the acetylcholine binding protein. J Biol Chem. 2002;277:41299–41302. doi: 10.1074/jbc.C200462200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhong W, et al. From ab initio quantum mechanics to molecular neurobiology: a cation-pi binding site in the nicotinic receptor. Proc Natl Acad Sci USA. 1998;95:12088–12093. doi: 10.1073/pnas.95.21.12088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cashin AL, Petersson EJ, Lester HA, Dougherty DA. Using physical chemistry to differentiate nicotinic from cholinergic agonists at the nicotinic acetylcholine receptor. J Am Chem Soc. 2005;127:350–356. doi: 10.1021/ja0461771. [DOI] [PubMed] [Google Scholar]
- 18.Shi J, Koeppe JR, Komives EA, Taylor P. Ligand-induced conformational changes in the acetylcholine-binding protein analyzed by hydrogen-deuterium exchange mass spectrometry. J Biol Chem. 2006;281:12170–12177. doi: 10.1074/jbc.M600154200. [DOI] [PubMed] [Google Scholar]
- 19.Reeves PJ, Callewaert N, Contreras R, Khorana HG. Structure and function in rhodopsin: High-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc Natl Acad Sci USA. 2002;99:13419–13424. doi: 10.1073/pnas.212519299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 21.Storoni LC, McCoy AJ, Read RJ. Likelihood-enhanced fast rotation functions. Acta Crystallogr D Biol Crystallogr. 2004;60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- 22.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 23.Brunger AT, et al. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 24.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 25.DeLano W. L. The PyMOL Molecular Graphics System. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]