Abstract
The family of Ap2 transcription factors comprises five members with highly conserved DNA-binding domains. Among the family members, Ap2δ is the most divergent, because it lacks highly conserved residues within the transactivation domain (TAD) and has weak affinity for known Ap2 binding sites. To identify specific Ap2δ coactivators/regulators during development, we performed a yeast two-hybrid screen, using Ap2δ's TAD. We identified the trithorax superfamily member, Ash2l, as a binding partner that interacts exclusively with Ap2δ. We showed that Ash2l positively mediates Ap2δ transactivation in a dose-dependent manner. Given the known role of Ash2l in histone modification, we determined whether Ap2δ was able to form a complex with that activity. Our results showed that Ap2δ associates with endogenous ASH2L and a member of the MLL family of histone lysine methyltransferases (HKMTs), MLL2 (ALR), forming a complex that methylates lysine 4 of histone H3 (H3K4). Additionally, we showed that Ap2δ is necessary for recruitment of Ash2l and Alr to the Hoxc8 locus and that recruitment of this complex leads to H3K4 trimethylation (H3K4me3) and subsequent gene activation. Altogether, we provide evidence of an association between a highly restricted gene-specific transcription factor and a Su(var), Enhancer of Zeste, Trithorax (SET)1/trithorax-like complex with H3K4 methyltransferase activity. Our studies also document a functional role for Ap2δ in recruiting histone methyltransferases (HMTs) to specific gene targets, such as Hoxc8. This role provides a mechanism through which these transcription factors can have diverse effects despite nearly identical DNA-binding motifs.
Keywords: development, methylation
Activating protein 2 (Ap2) transcription factors constitute a family of sequence-specific DNA-binding proteins with distinct roles in vertebrate development. Tcfap2a-e encodes five family members, Ap2α, -β, -γ, -δ, and -ε. Ap2 family members share a highly conserved basic DNA-binding domain and helix–span–helix dimerization motif at the C terminus (1). The N terminus is not as well conserved as the C terminus with the exception of a proline-rich region, the PY motif, in the TAD that is present in nearly all Ap2 proteins (2). Ap2 proteins bind a palindromic core sequence 5′-GCCN3/4GGC-3′ that has been identified in numerous cellular and viral enhancers (3). Tcfap2 genes are generally highly expressed in developing neural crest and its major derivatives (4, 5).
Ap2δ is considered the most divergent Ap2 family member because of its amino acid sequence and expression pattern. Phylogenetic analysis of vertebrate and cephalochordate Tcfap2 genes suggests that Tcfap2d diverged from the vertebrate lineage before the cooption of the Ap2 family by the neural crest (6). In support of this hypothesis, Tcfap2d is not expressed in the neural crest. Rather, its expression is restricted to the developing myocardium, central nervous system, and retina (7). Within its TAD, Ap2δ lacks the aforementioned PY motif and other highly conserved residues that are essential for transcriptional activation (8). It was suggested that Ap2δ has an alternative mechanism for transactivation or might act as a repressor (9).
Along with DNA-binding factors, histone modifiers create active or repressive states within euchromatin to regulate transcription. These modifiers include the Polycomb and trithorax superfamilies (PcG and trxG, respectively) that are important for maintaining steady states of repression and activation, respectively (10). Mixed-lineage leukemia 1 or MLL1 (ALL-1) is a trxG member and displays intrinsic histone lysine methyltransferase (HKMT) activity. Specifically, MLL1 methylates H3K4, an epigenetic mark associated with gene activation (11–13). MLL1 and its related proteins ALR (MLL2), HALR (MLL3), and MLL4 possess H3K4 histone methyltransferase (HMT) activity through a conserved SET domain and form a complex with conserved subunits ASH2L, WDR5, and RbBP5 (14–18).
Here, we report the identification of Ash2l as a binding partner of Ap2δ. We demonstrate that the interaction is specific by showing that, among Ap2 transcription factors, Ash2l binds exclusively with Ap2δ through the latter's TAD. Functionally, Ash2l synergistically activates Ap2δ-mediated gene activation in a dose-dependent manner. Ap2δ, in turn, is able to associate with endogenous ASH2L and ALR forming a complex that methylates H3K4. Finally, we show that Ap2δ plays an essential role in recruiting Ash2l and Alr to the Hoxc8 locus and that formation of this complex leads to H3K4me3 and gene activation.
Results
Identification of a trxG Member, Ash2l, as a Binding Partner of Ap2δ.
To identify uncharacterized regulators of Ap2δ, we used its TAD as bait in a yeast two-hybrid screen with a mouse embryonic cDNA library. Among 500 clones selected with high stringency, the 20 with the highest β-gal activity were isolated and sequenced. This set comprised 10 unique genes, among which three encoded proteins involved in proteasome degradation that were deemed false positives. The seven remaining candidates were Fkbp8, Sfrs2, Kifc5a, Fip1l1, Cdh11, Pom121, and Ash2l. Based on a predicted nuclear localization and involvement in transcriptional regulation, we selected Ash2l as the most probable true binding partner of Ap2δ (19, 20).
To confirm the interaction in yeast, the Ash2l clone encoding the GAL4 activation domain (AD) fused to Ash2l (amino acids 56–623) was transformed in the yeast along with constructs expressing the GAL4 DNA-binding domain (DBD) alone or GAL4 DBD fused to either the TAD or N-terminal half of Ap2δ (Fig. 1B). Under high stringency conditions, growth was observed when Ash2l was coexpressed with Ap2δ's TAD or N terminus but not with GAL4 DBD alone (Fig. 1A). Reciprocally, growth was not observed when those portions of Ap2δ were coexpressed with the GAL4 AD alone. These results demonstrated that Ash2l interacts with Ap2δ in yeast and that the TAD of Ap2δ is sufficient for the interaction.
Fig. 1.
Ap2δ interacts specifically with Ash2l in eukaryotic cells. (A) The transactivation domain (TAD) and N terminus of Ap2δ fused to a GAL4 DBD interact with clone 210 containing Ash2l fused to a GAL4 AD. Yeast were spread onto double dropout (DDO) and quadruple dropout (QDO) plates to check for the presence of plasmids and protein–protein interaction, respectively. (B) Schematic representation showing full-length and truncated Ap2δ and Ash2l used for the yeast two-hybrid and coimmunoprecipitation (co-IP) experiments. (C) Reciprocal co-IP of full-length Ap2δ and Ash2l from nuclear extracts from transiently transfected 293T cells. Immunoprecipitations were performed with the indicated antibodies, and complexes were analyzed by Western blot using HRP-conjugated anti-Myc or -V5 antibodies. (D) Ash2l coimmunoprecipitates with Ap2δ, but not -α, -β, -γ, and -ε. Methods were identical to those described in C.
We next mapped the interaction domain of Ash2l by expressing the TAD of Ap2δ fused to the GAL4 DBD with various Ash2l derivatives fused to the GAL4 AD in yeast. Ash2l contains a PHD finger and a SPRY domain at the N terminus and C terminus, respectively (Fig. 1B). The PHD finger is a known methyl-lysine binding domain found in numerous chromatin remodeling proteins, whereas the SPRY domain is a known protein–protein interaction domain (21, 22). Growth in QDO plates was observed in yeast expressing full-length Ash2l and Ash2l fragments containing amino acids 141–412 and 280–412 (Fig. 2). Yeast expressing full-length Ash2l or Ap2δ TAD with GAL4 DBD or -AD, respectively, did not survive on QDO plates similar to Fig. 1A. This indicates that the TAD of Ap2δ interacts through amino acids 280–412 of Ash2l, a peptide containing no known motif. Additionally, growth was absent in yeast expressing Ash2l fragments without the 280–412 span but containing the PHD finger or SPRY domain, excluding binding mediated through those motifs. Thus, we defined an interaction domain in Ash2l.
Fig. 2.
Ash2l interacts with Ap2δ through an interaction domain. A yeast two-hybrid assay was used to test the ability of full-length Ash2l or partial Ash2l proteins to bind the TAD of Ap2δ. Yeast with both plasmids that survived on QDO plates indicated a positive interaction (+), whereas those that failed to survive represented an inability to interact (−).
Ap2δ Coimmunoprecipitates with Ash2l in Mammalian Cells.
To establish that Ap2δ and Ash2l interact in mammalian cells, full-length tagged Ap2δ and Ash2l proteins were expressed transiently in 293T cells. After immunoprecipitating Ap2δ or Ash2l from nuclear lysates, using the appropriate anti-tag antibodies, immunoblot analyses were performed to detect the two proteins. Immunocomplexes obtained reciprocally contained Ap2δ and Ash2l (Fig. 1C), confirming the interaction observed in yeast. To exclude the possibility that Ash2l is prone to diffuse nonspecific interactions, we transiently expressed it with an unrelated nuclear protein, NOP5, but failed to coimmunoprecipitate these proteins [supporting information (SI) Fig. S1]. To confirm the biological relevance of this interaction, we determined the expression pattern of Ash2l during mid embryogenesis, which was unknown. We found that Ash2l is ubiquitously expressed at embryonic day (E)10.5 and E13.5, indicating that Ash2l and Tcfap2d have overlapping expression during embryogenesis (Fig. S2).
Ash2l Does Not Interact with Ap2α, -β, -γ, or -ε.
Although the Ap2 coactivator CBP/p300 interacts with several family members, we hypothesized that Ash2l interacts exclusively with Ap2δ because of the latter's poorly conserved TAD sequence (23). To test this, we transiently expressed Ash2l and V5-tagged, full-length Ap2α, -β, -γ, -δ, or -ε in 293T cells, immunoprecipitated complexes from nuclear lysates and then detected the proteins on immunoblots. As before, Ap2δ-containing immunocomplexes included Ash2l (Fig. 1D). In contrast, Ash2l was not detected in immunocomplexes containing the other Ap2 proteins. These results provided further confirmation that the Ap2δ–Ash2l interaction was specific and were consistent with the hypothesis that their interaction is unique among Ap2 transcription factors.
Ash2l Increases Transcriptional Activity of Ap2δ in a Dose-Dependent Manner.
Because Ash2l is associated with transcriptional activation through its interaction with MLL complexes, we hypothesized that Ash2l would function as a coactivator of Ap2δ-mediated transactivation. To test this, GAL4 DBD fused to the Ap2δ N terminus, Ash2l, and a GAL4-driven firefly luciferase reporter were expressed transiently in 293T cells. Transfection of varying amounts of the GAL4 DBD-Ap2δ expression construct without Ash2l induced a dose-dependent increase in transactivation (Fig. 3). This was attributable to the Ap2δ fragment because transfection of comparable amounts of the GAL4 DBD-only construct did not increase transactivation above the basal level. Next, we cotransfected varying amounts of the Ash2l expression construct with the GAL4 DBD-Ap2δ and luciferase reporter constructs. We observed significant increases in transactivation in a dose-responsive manner across the range of Ap2δ expression levels. Comparable coexpression of Ash2l with GAL4 DBD only did not significantly alter transactivation. These results showed that Ash2l is a coactivator of Ap2δ transactivation.
Fig. 3.
Ash2l increases Ap2δ transactivation in a dose-dependent manner. 293T cells were transfected with a luciferase reporter under the control of GAL4 UAS and a minimal promoter and varying concentrations of GAL4 DBD or Ap2δ N terminus fused to a GAL4 DBD; amounts of those constructs transfected are indicated below the graph. In addition, varying amounts of an expression construct for Ash2l were cotransfected; the shading of the bars corresponds to the dose (indicated in the key to the right of the graph). The heights of the bars represent means of ratios of the reporter luciferase activity to the control Renilla luciferase [relative light units (RLU)] for each condition. Results are expressed as means ± SEM and compared statistically, using Student's t tests. **, P < 0.005; ***, P < 0.0001.
Ap2δ Associates with Endogenous ASH2L and ALR to Form a Complex with H3K4 Methyltransferase Activity.
Ash2l is a common subunit of MLL complexes and regulates the H3K4me3 that is conferred by the SET domain of the MLL protein (24). Although this is the only functional role previously assigned to Ash2l, our results suggested that it might also recruit transcription factors such as Ap2δ to MLL complexes. To determine whether this was the case, we tested the ability of Ap2δ to bind complexes containing ASH2L and ALR/MLL2,‡‡ focusing on the latter over other SET1 proteins because of its known role in cardiac muscle. We transiently expressed V5-Ap2δ in K562 erythroleukemia cells, immunoprecipitated complexes, using anti-V5 antibodies, and then detected endogenous ASH2L and ALR, using specific antibodies. As shown, ASH2L and ALR were present in immunocomplexes that contained Ap2δ, but not those obtained with nonspecific IgG or an unrelated antibody (Fig. 4A).
Fig. 4.
Ap2δ forms a complex with endogenous ASH2L and ALR that methylates histone H3K4. (A) Endogenous ASH2L and ALR coimmunoprecipitate with transfected V5-Ap2δ in K562 cells. The V5-Ap2δ construct was transiently expressed in K562 cells. Levels of expression were documented as comparable in nuclear lysates, using anti-V5 antibodies (Upper). Immunoprecipitations were performed with those lysates using antibodies as indicated, and complexes were analyzed by Western blot, using anti-ASH2L or -ALR antibodies. (B) Complexes containing Ap2δ, ASH2L, ALR, and Su(z)12 methylate recombinant histone H3 in vitro. Nuclear lysates from K562 cells expressing V5-Ap2δ were immunoprecipitated with the indicated antibodies. Immunoprecipitates and GST fusion proteins containing G9a or ALR SET domain were assayed for HMT activity, using histone H3 (Upper). The Coomassie blue staining shows the histone H3 input (Lower). (C) Complexes containing Ap2δ, ASH2L, ALR, and Su(z)12 methylate native histone H3 in vitro. Immunocomplexes and GST fusion proteins as described in B were used for HMT assays with native histone H3. Quantities of TCA-precipitable radioactivity (cpm) are shown as the average of three experiments with the whiskers indicating standard error of the mean. (D) Complexes immunoprecipitated with Ap2δ-V5, but not an unrelated V5-tagged protein (SHP2-V5), methylate H3K4 in vitro. HMT activity is diminished upon incubation with an H3K4 (K4R) mutant but not with H3K9 (K9R) and H3K27 (K27R) mutants.
The ALR complex exhibits H3K4 methyltransferase activity through the SET domain of ALR (15, 25). To determine whether Ap2δ/ASH2L included this activity, HMT assays were performed in vitro with purified proteins and immunoprecipitated samples, using recombinant (Fig. 4B) and native (Fig. 4C) histone H3. HMT activity was determined by using the presence of radiolabeled histone H3 as an indicator of activity. We obtained complexes containing Ap2δ, ASH2L, and ALR as described above. Several positive controls were used, including immunocomplexes precipitated with anti-ALR, -ASH2L, and -Su(z)12 antibodies from K562 cells. Su(z)12 is associated with H3K27 methylation through the activity of the EZH2 lysine methyltransferase (26). Like the positive controls, the Ap2δ-containing complexes showed methylation activity (Fig. 4B). Immunocomplexes precipitated with an unrelated V5-tagged protein had no HMT activity, excluding nonspecific effects of the anti-V5 antibody (Fig. 4D). Additionally, we incubated Ap2δ-containing complexes with recombinant histone H3 proteins with a mutation at K4, K9, or K27 and found diminished HMT activity only toward the H3K4 mutant (Fig. 4D). These data show that Ap2δ associates with ASH2L and ALR/MLL2 and that, in vitro, these complexes exhibit H3K4 HMT activity. To confirm that Ap2δ-containing complexes are functional despite modest in vitro activity, we next investigated their ability to regulate downstream targets in vivo.
Ap2δ Recruits Ash2l and Alr to the Hoxc8 Locus Promoting H3K4me3 and Gene Activation.
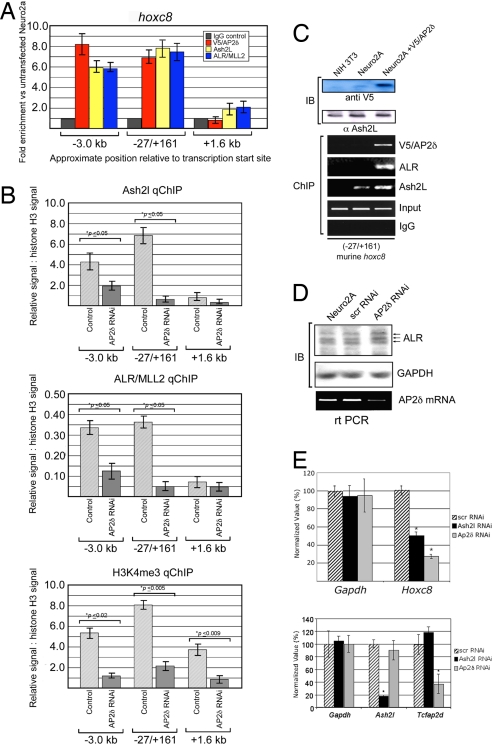
Because Ap2δ is a known gene-specific DNA-binding factor, we hypothesized that Ap2δ is necessary for the recruitment of Ash2l and Alr to specific gene loci. To identify common targets of Ap2δ and Ash2l, we treated mouse neuroblastoma (Neuro2a) cells, which express Tcfap2d and Ash2l endogenously, with siRNA against those genes and performed real-time PCR analysis on a set of candidate genes known to be regulated by MLL family members (18, 25). Hoxc8 was significantly down-regulated, with 28% and 51% of wild-type steady-state mRNA levels in cells after treatment with Tcfap2d- or Ash2l-specific siRNA, respectively (Fig. 5E).
Fig. 5.
Ap2δ recruits Ash2l and Alr to the Hoxc8 locus. (A) Ap2δ, Ash2l, and Alr cooccupy specific regions of the Hoxc8 promoter located 27 bp (−27 bp) and 3 kb (−3.0 kb) upstream of the transcriptional start site. Chromatin fragments from Neuro2a cells expressing V5-Ap2δ were immunoprecipitated with anti-V5, -Ash2l, and -Alr antibodies. DNA association was analyzed by real-time PCR, using loci-specific primers. The heights of the bars represent means of fold increases for cells expressing V5-Ap2δ compared with wild-type control. The whiskers represent standard error of the mean. (B) (Top and Middle) Ap2δ down-regulation results in a significant decrease of Ash2l and Alr at the Hoxc8 promoter. Chromatin fragments from Neuro2a cells treated with either Tcfap2d-specific siRNA or scrambled control were immunoprecipitated with anti-Ash2l and -Alr antibodies. DNA association was determined by real-time PCR, and quantities are represented as ratios of signals relative to signals obtained from a nonspecific histone H3 control. The heights of the bars represent means of ratios for each condition performed in triplicate. The whiskers represent standard error of the mean. Statistical comparisons were made between conditions, using Student's t tests. *, P ≤ 0.05. (Bottom) Ap2δ down-regulation results in a significant decrease of H3K4me3 at the Hoxc8 promoter. Chromatin fragments from Neuro2a cells treated with either Tcfap2d-specific siRNA or scrambled control were immunoprecipitated with antibodies against H3K4me3. DNA analysis was performed in a similar manner as described for Top. (C) Alr and Ash2l colocalize at the −27/+161 region of the Hoxc8 promoter when Ap2δ is present. Chromatin fragments from NIH 3T3 cells, wild-type Neuro2a cells and Neuro2a cells expressing V5-Ap2δ were immunoprecipitated with anti-V5, -Alr, and -Ash2l antibodies. The immunoprecipitated DNA was analyzed by PCR, using locus-specific primers. NIH 3T3 cells, which do not express Ap2δ, were used as negative controls for the ChIP experiments. (D) Alr protein levels are unchanged when Ap2δ is down-regulated in Neuro2a cells. Total cell extracts were obtained from wild-type Neuro2a cells or Neuro2a cells treated with Tcfap2d-specific siRNA or scrambled control and analyzed by Western blot, using anti-Alr antibodies. (E) (Upper) Ap2δ and Ash2l regulate Hoxc8 expression in Neuro2a cells. Total RNA was extracted 72 h after transfection from Neuro2a cells treated with Tcfap2d- or Ash2l-specific siRNA or scrambled control. Gapdh and Hoxc8 transcript levels were quantified by real-time PCR. Normalized values were calculated as percentages of transcript levels detected in cells treated with the scrambled control. *, P < 0.001. (Lower) Knock down of either Ap2δ or Ash2l resulted in a significant decrease of their respective transcripts. Experiments were similar to those described in Upper.
To determine whether the down-regulation of Hoxc8 was a direct effect, we performed ChIP analysis, using anti-V5, -Ash2l, or -Alr antibodies and chromatin fragments obtained from Neuro2a cells, some transfected with V5-Ap2δ. Similar regions of the Hoxc8 promoter were amplified in complexes containing V5-Ap2δ, Ash2l, and Alr by real-time PCR, indicating that these proteins are tethered to the same gene loci (Fig. 5 A and C). Ap2δ, Ash2l, and Alr were found to cooccupy specific regions of the promoter located 27 bp (−27 bp) and 3 kb (−3.0 kb) upstream of the transcriptional start site (Fig. 5A). These regions had evolutionarily conserved Ap2-binding sites that were predicted by rVista and GALA. In contrast, the region 1.6 kb (+1.6 kb) downstream of the start site did not associate with Ash2l and Alr and was not predicted to have an Ap2-binding site.
To test the hypothesis that Ash2l and Alr are tethered to the promoter through Ap2δ, we performed ChIP analysis with anti-Ash2l or -Alr antibodies and Neuro2a cells treated with Tcfap2d-specific siRNA. Down-regulation of Ap2δ significantly decreased the association of Ash2l and Alr with the −3 kb and −27 bp, but not the +1.6 kb, Hoxc8 regions (Fig. 5B). It should be noted that down-regulation of Ap2δ did not alter Alr and Ash2l protein levels (Fig. 5D and Fig. S3). These results indicate that Ap2δ plays a critical role in the recruitment of Ash2l and Alr to the Hoxc8 promoter. Because Ap2δ is a sequence-specific transcription factor, we hypothesized that Ap2δ would associate with some but not all Alr targets. To test this hypothesis, we performed additional ChIP experiments to analyze the Lamb3 locus, a known Alr target (25). Because this gene was significantly down-regulated upon treatment with Ash2l- but not Tcfap2d-specific siRNA, we predicted that Ash2l and Alr, but not Ap2δ, would be present at the promoter (data not shown). Indeed, we found Ash2l and Alr but not Ap2δ associated with the Lamb3 locus, indicating that Ap2δ associates with only a subset of Alr targets (Fig. S4).
Having demonstrated an Ap2δ-dependent recruitment of the Alr complex to the Hoxc8 locus, we determined whether this recruitment altered H3K4me3 at the Hoxc8 locus, because this epigenetic status marks transcriptional initiation (27, 28). We performed ChIP experiments, using anti-trimethylated H3K4 antibodies with chromatin fragments obtained from Neuro2a cells, some treated with Tcfap2d-specific siRNA. We found significantly reduced levels of H3K4me3 at the Hoxc8 locus when Ap2δ was down-regulated (Fig. 5B). These data indicate that Ap2δ recruits a complex that induces H3K4me3, leading to increased gene activation. Alr contributes to that HKMT activity, although we did not examine the possibility that other proteins with H3K4me3 function also contribute to this gene activation.
Discussion
In this study, we identified the trxG member Ash2l as a coactivator of Ap2δ and showed that Ash2l interacts exclusively with the TAD of Ap2δ. It had been suggested that the TAD of Ap2 proteins interact with specific coactivator complexes, resulting in divergent functions (9). We confirmed that this mechanism is operative by demonstrating that Ash2l binds Ap2δ uniquely among the Ap2 family members. Further, we showed that Ap2δ is able to function as an activator despite the lack of the PY motif and other highly conserved residues critical for transactivation for other Ap2 proteins.
Functionally, we showed that Ash2l is able to synergistically activate Ap2δ transactivation in a dose-dependent manner. We hypothesize that this is due to Ash2l's ability to associate with the basal transcriptional machinery through interactions with the MLL complex, because this complex includes active RNA polymerase II (18). In addition, we showed that Ap2δ forms a complex with ASH2L and ALR that methylates histone H3 at K4 in vitro. Studies have shown that Ash2l is a conserved subunit of MLL complexes and functions to regulate H3K4me3 (24, 29). Because it binds both Ap2δ and members of MLL complexes, our studies suggest that Ash2l also functions as an intermediary between sequence-specific DNA-binding factors and chromatin remodeling.
The role of the Ap2 family in chromatin remodeling has not been fully elucidated. Despite evidence that Ap2α, -β and -γ interact with the histone acetyltransferase (HAT) CBP/p300 to mediate Cited2-dependent coactivation, it has not been shown that complexes containing a member of the Ap2 family, CITED2, and CBP/p300 possess HAT activity (23). Additionally, CBP/p300 is a global coactivator that interacts with a host of transcription factors and components of the general transcriptional machinery (30). It was also reported that the HAT activity of CBP/p300 is dispensable, because a HAT-defective CBP is still able to coactivate through interaction with other HATs (31). Thus, Ap2δ is the first Ap2 family member to be associated with a chromatin remodeling activity per se.
Given that Ap2δ has a dual role in binding DNA in a gene-specific manner and associating with chromatin remodeling factors, we hypothesized that Ap2δ is necessary for the recruitment of Mll/Set1-like family members to specific gene loci. Indeed, we found that Ap2δ recruits both Ash2l and Alr to the Hoxc8 locus to activate gene transcription. This suggests that Ap2δ plays an essential role in the activity of these complexes. In particular, Ap2δ recruits these factors to specific targets to activate (or repress) gene expression. Because Ap2δ is a highly restricted, sequence-specific transcription factor, its ability to associate with Mll/Set1-like family members implies that it has a critical role in bringing these family members to specific gene loci in a spatiotemporal manner during development. As discussed below, this finding has implications for understanding how Mll/Set1-like proteins achieve specificity despite their global expression patterns.
Despite many recent advances in our understanding of HKMTs, the role of histone lysine methylation in dynamic gene regulation remains unclear. This role is further complicated by the capacity of such lysine residues to occupy multiple methylation states from mono- to trimethylation. The degree of H3K4 methylation increases the complexity of the histone code as it is recognized by proteins binding to methyl-lysines on histones and is regulated across actively transcribed genes (32). In fact, histone methylation was thought to be irreversible until the discovery in 2004 of the histone demethylase LSD1 (33). Histone methylation/demethylation provides a reversible dynamic state that mediates interactions with methyl-lysine-specific chromatin binding proteins, thereby effecting transcription. There are several transcription factors that associate with H3K9 HKMT activities that result in transcriptional repression within euchromatin. These include, among others, REST, CUTL1, E2F6, and PRDF1/Blimp1 (34–37). Although there are numerous examples of transcription factors that couple with arginine methyltransferases to activate transcription, there are few previous examples of that with HKMTs. Our work here links Ap2δ to gene transcriptional activation through ALR, and provides an example related to the H3K4 methylation activity of a mammalian MLL SET1-like complex.
Aside from their oncogenic properties, little is known about MLL complexes and their roles in cell differentiation and proliferation (11, 12). Mll genes are widely expressed in adult and embryonic tissues, suggesting roles as global gene regulators (16, 17, 38–40). Of note, the phenotype of Mll1-deficient mice comprises only skeletal, hematopoietic, and neural abnormalities (41). Although the absence of broader developmental defects might reflect redundant functions among the Mll proteins, the specificity is compatible with unique roles arising through interactions with highly restricted sequence-specific transcription factors. Indeed, our studies demonstrate that MLL complexes bind Ap2δ, which exhibits precisely those characteristics. This suggests that the activity of MLL complexes is context-dependent and that transcription factors with highly restricted expression patterns play an important role in their recruitment to specific targets.
Materials and Methods
Plasmid Construction.
cDNAs encoding full-length, TAD (amino acid 38–98), and N-terminal (amino acid 1–208) fragments of murine Ap2δ were PCR amplified from an EST clone containing Tcfap2d (9) and subcloned into pGBKT7 (Clontech), pcDNA6V5His (Invitrogen), and pBXG1. cDNAs encoding full-length murine Ap2α, -β, -γ, and -ε were PCR amplified from full-length Tcfap2 cDNAs and subcloned into pcDNA6V5His. Full-length human NOP5 cDNA (clone ID 5527148) and mouse Ash2l cDNA (clone ID 4211433) were obtained from the IMAGE consortium (image.llnl.gov). PCR amplification of cDNAs encoding Ash2l-FL (full-length) was performed, followed by subcloning into pCMV Myc (Clontech). Partial Ash2l cDNAs encoding the residues 1–140, 141–412, 141–280, 280–412, and 413–623 were cloned into pGADT7 (Clontech). Cloned fragments were sequenced to confirm the absence of inadvertent mutagenesis.
Yeast Two-Hybrid Screen.
The TAD of Ap2δ fused to a GAL4 DBD was used to screen a mouse embryonic cDNA library (Stratagene). Briefly, the AH109 haploid strain was transformed with the bait plasmid and mated with the Y187 haploid strain containing the library. Diploid clones were screened by nutritional selection, using Ade and His reporters, and by β-gal activities. Library plasmids were extracted with the Yeast DNA Isolation System (Stratagene). To verify the results of the screen and identify the Ash2l interaction domain, the AH109 haploid strain was transformed with constructs expressing the Ap2δ TAD or N-terminal fragment fused to the GAL4 DBD and mated with the Y187 haploid strain expressing the Ash2l library clone or various Ash2l derivatives fused to the GAL4 AD.
Cell Culture and Immunoprecipitation.
293T, K562, NIH 3T3, and Neuro2a cells [American Type Culture Collection] were maintained in DMEM supplemented with 10% FBS and antibiotics at 37°C with 5% CO2. Cells were transfected with constructs expressing Ap2 family members and/or Ash2l, using Lipofectamine 2000 (Invitrogen). Cells were harvested 48 h after transfection and incubated in hypotonic lysis buffer [20 mM Hepes (pH 7.9), 10 mM NaCl, and 0.1% Triton X-100] containing protease inhibitors (Roche Applied Science). Nuclei were pelleted and resuspended in high salt buffer [20 mM Hepes (pH 7.9), 420 mM NaCl, and 0.1% Triton X-100]. Nuclear lysates were incubated with 2.5 μg of anti-V5 or -Myc antibodies (Invitrogen) and protein G agarose beads (Roche Applied Science). Immunoprecipitates were analyzed by Western blot with anti-V5 and -Myc HRP-conjugated antibodies (Invitrogen) and anti-Ash2l antibodies (Bethyl Laboratories) and anti-ALR antibodies (a kind gift from E. Canaani, Weizmann Institute of Science, Rehovot, Israel).
Transactivation Assays.
293T cells were transfected with a firefly luciferase reporter containing several GAL4 upstream activating sequences, pBXG1 or pBXG1-AP2δ, and/or Myc-Ash2l-FL, using Lipofectamine 2000 (Invitrogen). Renilla luciferase reporter was used as an internal transfection control (Promega). The total amount of DNA in each transfection was held constant, using pGEM-7Zf (+/−). After 48 h, lysates were assayed for firefly and Renilla luciferase activities with the dual-luciferase reporter assay system (Promega), using a DYNEX Model MLX luminometer (Thermo Lab-Systems). Relative luminescence was measured as a ratio of the amount of firefly to Renilla luciferase activity. Results were expressed as means ± SEM and compared statistically, using a Student's t test with a significance threshold of P < 0.05.
In Vitro HKMT Assays.
HKMT assays were performed essentially as described in ref. 35. A construct for expressing a GST fusion protein with the C-terminal SET domain (amino acids 5112–5262) of ALR was accomplished by generating a PCR-amplified product with adapter sequences for fusion with the GST tag within pGEX-6P1 (GE Healthcare). Fragments were cloned, sequenced, and verified for expression. GST fusion proteins of GST-mG9a and GST-ALR-SET were purified by using standard affinity purification with glutathionine Sepharose (GE Healthcare) according to manufacturer's instructions. Equivalent amounts of purified GST proteins were used for the HKMT assays. Similar amounts of anti-Su(z)12 (Abcam), -ALR, -Ash2l, and -V5 antibodies were used for immunoprecipitation reactions to obtain native immunoaffinity purified proteins from nuclear extracts derived from K562 cells expressing V5-Ap2δ or -SHP2 or Neuro2a cells expressing V5-Ap2δ.
Chromatin Immunoprecipitation (ChIP).
ChIP assays were performed as described in ref. 42. Briefly, sonicated chromatin from 20 × 106 transfected, siRNA-treated, or untreated Neuro2a or NIH 3T3 cells was immunoprecipitated by using anti-V5, -Ash2l, -Alr, -trimethylated H3K4 (Millipore) or IgG antibodies. Associated DNA was analyzed by real-time PCR, using loci-specific primers (Table S1).
RNA Analysis.
Total RNA was extracted by using TRIzol reagent according to the manufacturer's protocol (Invitrogen) and reverse transcribed by using SuperScript III reverse transcriptase and oligo(dT) primers (Invitrogen). Transcript levels were determined by real-time PCR, using Gapdh as an internal control. For siRNA experiments, Neuro2a cells were transfected with Ap2δ- or Ash2l-specific siRNA or a scrambled control, using Dharmafect (Dharmacon), and total RNA was isolated 72 h after transfection.
Supplementary Material
Acknowledgments.
We thank Dr. Eli Canaani for providing antibody reagents and Drs. Michael Stallcup (University of South Carolina, Columbia, SC) and Jonathan D. Licht (Northwestern University, Evanston, IL) for plasmid DNA constructs. This work was supported by National Institutes of Health Grants HD38018 (to B.D.G.), HL067099 (to M.J.W.), and K08 HL086633-01 (to J.Z.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. D.R. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/cgi/content/full/0711896105/DCSupplemental.
A human cell line was used, so standard nomenclature with protein symbols entirely in capital letters is used here.
References
- 1.Williams T, Tjian R. Characterization of a dimerization motif in AP-2 and its function in heterologous DNA-binding proteins. Science. 1991;251:1067–1071. doi: 10.1126/science.1998122. [DOI] [PubMed] [Google Scholar]
- 2.Williams T, Tjian R. Analysis of the DNA-binding and activation properties of the human transcription factor AP-2. Genes Dev. 1991;5:670–682. doi: 10.1101/gad.5.4.670. [DOI] [PubMed] [Google Scholar]
- 3.Mohibullah N, Donner A, Ippolito JA, Williams T. SELEX and missing phosphate contact analyses reveal flexibility within the AP-2α protein: DNA binding complex. Nucleic Acids Res. 1999;27:2760–2769. doi: 10.1093/nar/27.13.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moser M, Ruschoff J, Buettner R. Comparative analysis of AP-2 alpha and AP-2 beta gene expression during murine embryogenesis. Dev Dyn. 1997;208:115–124. doi: 10.1002/(SICI)1097-0177(199701)208:1<115::AID-AJA11>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 5.Chazaud C, et al. AP-2.2, a novel gene related to AP-2, is expressed in the forebrain, limbs and face during mouse embryogenesis. Mech Dev. 1996;54:83–94. doi: 10.1016/0925-4773(95)00463-7. [DOI] [PubMed] [Google Scholar]
- 6.Meulemans D, Bronner-Fraser M. Amphioxus and lamprey AP-2 genes: Implications for neural crest evolution and migration patterns. Development. 2002;129:4953–4962. doi: 10.1242/dev.129.21.4953. [DOI] [PubMed] [Google Scholar]
- 7.Zhao F, Lufkin T, Gelb BD. Expression of Tfap2d, the gene encoding the transcription factor Ap-2 delta, during mouse embryogenesis. Gene Expr Patterns. 2003;3:213–217. doi: 10.1016/s1567-133x(02)00067-4. [DOI] [PubMed] [Google Scholar]
- 8.Wankhade S, et al. Characterization of the activation domains of AP-2 family transcription factors. J Biol Chem. 2000;275:29701–29708. doi: 10.1074/jbc.M000931200. [DOI] [PubMed] [Google Scholar]
- 9.Zhao F, et al. Cloning and characterization of a novel mouse AP-2 transcription factor, AP-2delta, with unique DNA binding and transactivation properties. J Biol Chem. 2001;276:40755–40760. doi: 10.1074/jbc.M106284200. [DOI] [PubMed] [Google Scholar]
- 10.Schuettengruber B, et al. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 11.Gu Y, et al. The t(4;11) chromosome translocation of human acute leukemias fuses the ALL-1 gene, related to Drosophila trithorax, to the AF-4 gene. Cell. 1992;71:701–708. doi: 10.1016/0092-8674(92)90603-a. [DOI] [PubMed] [Google Scholar]
- 12.Tkachuk DC, Kohler S, Cleary ML. Involvement of a homolog of Drosophila trithorax by 11q23 chromosomal translocations in acute leukemias. Cell. 1992;71:691–700. doi: 10.1016/0092-8674(92)90602-9. [DOI] [PubMed] [Google Scholar]
- 13.Nakamura T, et al. ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Mol Cell. 2002;10:1119–1128. doi: 10.1016/s1097-2765(02)00740-2. [DOI] [PubMed] [Google Scholar]
- 14.Yokoyama A, et al. Leukemia proto-oncoprotein MLL forms a SET1-like histone methyltransferase complex with menin to regulate Hox gene expression. Mol Cell Biol. 2004;24:5639–5649. doi: 10.1128/MCB.24.13.5639-5649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goo YH, et al. Activating signal cointegrator 2 belongs to a novel steady-state complex that contains a subset of trithorax group proteins. Mol Cell Biol. 2003;23:140–149. doi: 10.1128/MCB.23.1.140-149.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prasad R, et al. Structure and expression pattern of human ALR, a novel gene with strong homology to ALL-1 involved in acute leukemia and to Drosophila trithorax. Oncogene. 1997;15:549–560. doi: 10.1038/sj.onc.1201211. [DOI] [PubMed] [Google Scholar]
- 17.FitzGerald KT, Diaz MO. MLL2: A new mammalian member of the trx/MLL family of genes. Genomics. 1999;59:187–192. doi: 10.1006/geno.1999.5860. [DOI] [PubMed] [Google Scholar]
- 18.Hughes CM, et al. Menin associates with a trithorax family histone methyltransferase complex and with the hoxc8 locus. Mol Cell. 2004;13:587–597. doi: 10.1016/s1097-2765(04)00081-4. [DOI] [PubMed] [Google Scholar]
- 19.Ikegawa S, Isomura M, Koshizuka Y, Nakamura Y. Cloning and characterization of ASH2L and Ash2l, human and mouse homologs of the Drosophila ash2 gene. Cytogenet Cell Genet. 1999;84:167–172. doi: 10.1159/000015248. [DOI] [PubMed] [Google Scholar]
- 20.LaJeunesse D, Shearn A. Trans-regulation of thoracic homeotic selector genes of the Antennapedia and bithorax complexes by the trithorax group genes: Absent, small, and homeotic discs 1 and 2. Mech Dev. 1995;53:123–139. doi: 10.1016/0925-4773(95)00430-0. [DOI] [PubMed] [Google Scholar]
- 21.Mellor J. It takes a PHD to read the histone code. Cell. 2006;126:22–24. doi: 10.1016/j.cell.2006.06.028. [DOI] [PubMed] [Google Scholar]
- 22.Wang J, et al. ASH2L: Alternative splicing and down-regulation during induced megakaryocytic differentiation of multipotential leukemia cell lines. J Mol Med. 2001;79:399–405. doi: 10.1007/s001090100222. [DOI] [PubMed] [Google Scholar]
- 23.Braganca J, et al. Physical and functional interactions among AP-2 transcription factors, p300/CREB-binding protein, and CITED2. J Biol Chem. 2003;278:16021–16029. doi: 10.1074/jbc.M208144200. [DOI] [PubMed] [Google Scholar]
- 24.Steward MM, et al. Molecular regulation of H3K4 trimethylation by ASH2L, a shared subunit of MLL complexes. Nat Struct Mol Biol. 2006;13:852–854. doi: 10.1038/nsmb1131. [DOI] [PubMed] [Google Scholar]
- 25.Issaeva I, et al. Knockdown of ALR (MLL2) reveals ALR target genes and leads to alterations in cell adhesion and growth. Mol Cell Biol. 2007;27:1889–1903. doi: 10.1128/MCB.01506-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pasini D, et al. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 2004;23:4061–4071. doi: 10.1038/sj.emboj.7600402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bernstein BE, et al. Methylation of histone H3 Lys 4 in coding regions of active genes. Proc Natl Acad Sci USA. 2002;99:8695–8700. doi: 10.1073/pnas.082249499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Santos-Rosa H, et al. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 29.Dou Y, et al. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat Struct Mol Biol. 2006;13:713–719. doi: 10.1038/nsmb1128. [DOI] [PubMed] [Google Scholar]
- 30.Janknecht R, Hunter T. Transcription A growing coactivator network. Nature. 1996;383:22–23. doi: 10.1038/383022a0. [DOI] [PubMed] [Google Scholar]
- 31.Korzus E, et al. Transcription factor-specific requirements for coactivators and their acetyltransferase functions. Science. 1998;279:703–707. doi: 10.1126/science.279.5351.703. [DOI] [PubMed] [Google Scholar]
- 32.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 33.Shi Y, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 34.Gyory I, et al. PRDI-BF1 recruits the histone H3 methyltransferase G9a in transcriptional silencing. Nat Immunol. 2004;5:299–308. doi: 10.1038/ni1046. [DOI] [PubMed] [Google Scholar]
- 35.Nishio H, Walsh MJ. CCAAT displacement protein/cut homolog recruits G9a histone lysine methyltransferase to repress transcription. Proc Natl Acad Sci USA. 2004;101:11257–11262. doi: 10.1073/pnas.0401343101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roopra A, et al. Localized domains of G9a-mediated histone methylation are required for silencing of neuronal genes. Mol Cell. 2004;14:727–738. doi: 10.1016/j.molcel.2004.05.026. [DOI] [PubMed] [Google Scholar]
- 37.Ogawa H, et al. A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science. 2002;296:1132–1136. doi: 10.1126/science.1069861. [DOI] [PubMed] [Google Scholar]
- 38.Yu BD, et al. Altered Hox expression and segmental identity in Mll-mutant mice. Nature. 1995;378:505–508. doi: 10.1038/378505a0. [DOI] [PubMed] [Google Scholar]
- 39.Ruault M, et al. MLL3, a new human member of the TRX/MLL gene family, maps to 7q36, a chromosome region frequently deleted in myeloid leukaemia. Gene. 2002;284:73–81. doi: 10.1016/s0378-1119(02)00392-x. [DOI] [PubMed] [Google Scholar]
- 40.Emerling BM, et al. MLL5, a homolog of Drosophila trithorax located within a segment of chromosome band 7q22 implicated in myeloid leukemia. Oncogene. 2002;21:4849–4854. doi: 10.1038/sj.onc.1205615. [DOI] [PubMed] [Google Scholar]
- 41.Yu BD, et al. MLL, a mammalian trithorax-group gene, functions as a transcriptional maintenance factor in morphogenesis. Proc Natl Acad Sci USA. 1998;95:10632–10636. doi: 10.1073/pnas.95.18.10632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ananthanarayanan M, et al. Ligand-dependent activation of the farnesoid X-receptor directs arginine methylation of histone H3 by CARM1. J Biol Chem. 2004;279:54348–54357. doi: 10.1074/jbc.M410021200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.