Abstract
Homeobox (HOX) genes play a definitive role in determination of cell fate during embryogenesis and hematopoiesis. MLL-related leukemia is coincident with increased expression of a subset of HOX genes, including HOXA9. MLL functions to maintain, rather than initiate, expression of its target genes. However, the mechanism of MLL maintenance of target gene expression is not understood. Here, we demonstrate that Mll binds to specific clusters of CpG residues within the Hoxa9 locus and regulates expression of multiple transcripts. The presence of Mll at these clusters provides protection from DNA methylation. shRNA knock-down of Mll reverses the methylation protection status at the previously protected CpG clusters; methylation at these CpG residues is similar to that observed in Mll null cells. Furthermore, reconstituting MLL expression in Mll null cells can reverse DNA methylation of the same CpG residues, demonstrating a dominant effect of MLL in protecting this specific region from DNA methylation. Intriguingly, an oncogenic MLL-AF4 fusion can also reverse DNA methylation, but only for a subset of these CpGs. This method of transcriptional regulation suggests a mechanism that explains the role of Mll in transcriptional maintenance, but it may extend to other CpG DNA binding proteins. Protection from methylation may be an important mechanism of epigenetic inheritance by regulating the function of both de novo and maintenance DNA methyltransferases.
Keywords: homeodomain, leukemia, maintenance
The MLL gene (11q23) in humans causes aggressive acute leukemia when it becomes aberrantly fused via translocation in hematopoietic cells with any of >50 known partner genes (1). MLL is an ortholog of the Drosophila trithorax gene, a master regulator of Hox gene expression. Both trithorax and Mll act to maintain, rather than initiate, the appropriate Hox expression pattern (2). For example, Hoxa7 is appropriately expressed in Mll−/− mice through embryonic day (E)8.5 of development. However, starting at day E9, Hoxa7 expression is not maintained in Mll−/− mice (2). Furthermore, MLL related leukemia is often accompanied by increased expression of some HOXA cluster genes, including HOXA9 (3). Studies have shown that Hoxa9 expression is vital to the transforming ability of the MLL fusion protein MLL-ENL (4). Recent work (5, 6) has identified domains within the MLL portion of MLL fusions that are vital to leukemogenesis. These include the menin binding and CXXC domains. Deletion of the MLL CXXC domain in the context of an MLL-ENL fusion abolishes the ability to transform myeloid progenitors (5, 6). CXXC domains from multiple proteins have been found to bind specifically to CpG DNA (7–9). In vitro studies determined that the MLL CXXC domain binds preferentially to nonmethylated CpG residues in an artificial target DNA (10). Given the Mll dependence of Hoxa9 expression, the importance of HOXA9 to MLL associated leukemia, and data regarding the requirement of the CpG binding CXXC domain in MLL fusion immortalization, we investigated the role CpG islands within the Hoxa9 gene locus might play in Mll-dependent regulation of Hoxa9 transcript expression. Here, we report on a region within the Hoxa9 locus (downstream of Hoxa10), which contains a putative “ATG desert” variety promoter [supporting information (SI) Fig. S1] that demonstrates differential methylation patterns in cells either wild-type or null for Mll. The binding of Mll in this region, demonstrated both in vitro and in vivo, confers protection from DNA methylation in a subset of CpG residues. Knock-down of Mll in wild-type cells mimics the methylation pattern seen in Mll−/− cells. The binding of Mll to this region and, consequently, the methylation status of the relevant CpG residues results in differential transcript expression.
Results
The locus that encompasses the canonical Hoxa9 gene is rich in CpG islands (Fig. 1). Upstream of the two canonical Hoxa9 coding exons (exons CD and II), is a region of sequence highly conserved among a variety of species, which we refer to as the interspecies homology region (Fig. S1). Within this region, an alternative HOXA9 first exon (AB) was identified as part of a transcript from human fetal liver (11). Furthermore, a search of EST databases and data from our lab (12) reveals a variety of HOXA9 transcripts that originate from or include this upstream region (Fig. S1). Like the canonical Hoxa9, the region surrounding this upstream exon is rich in CpG islands.
Fig. 1.
MLL protects specific CpG residues in the Hoxa9 locus from methylation. (A) Schematic representation of the murine Hoxa9 region. Eight kilobases of the genomic region of Hoxa9 is represented. CpG Islands are numbered beneath. Upstream alternative first exon is labeled AB, the canonical Hoxa9 first exon is labeled CD and the canonical second exon, containing the homeodomain, is labeled II. Large asterisks indicate Mll binding as determined by ChIP. (B) MLL-dependent protection of CpG Island 1 from methylation. Direct bisulfite sequencing reveals a difference in methylation status in Mll+/+ MEF cells (circles) and Mll−/− MEF cells (X). Area under the curve analysis was performed on DNA sequence histograms and relative methylation percentage was determined for CpG residues. Each point on the graph represents a single CpG residue from the sequence below the graph. Beneath the graph are sequencing results from individual clones (10 clones each from Mll+/+ and Mll−/− MEFs) of PCR products from bisulfite treated template. Empty circles, unmethylated CpG residues; filled circles, methylated CpG residues. (C) Upon shRNA knockdown of Mll in Mll+/+ MEF cells, protection from methylation is lost. Triangles, shRNA Mll knockdown; circles, shRNA pSuper vector alone. Knockdowns were selected for 2 weeks. Four-week selection showed similar results (data not shown). (D) Upon add-back of MLL in Mll−/− MEF cells, protection from methylation returns. Filled circles, MLL add-back; x's, vector-only control. Data shown are from cells kept under selection for one week.
We examined the methylation state of CpG residues within the Hoxa9 locus, using direct sequencing of bisulfite treated genomic DNA from wild-type or Mll-null murine embryonic fibroblast (MEF) cells (13). We analyzed CpG islands (see definition used in SI Appendix) and a region within the AB exon that falls just short of the CpG island definition in mouse, but meets those criteria in human (indicated as CpG 1.5 in Fig. 1). Within CpG island 1, we determined that a small cluster of CpG residues exhibits a significantly decreased level of methylation in Mll+/+ cells compared with Mll−/− cells (P values 0.005 to 2 × 10−5) (Fig. 1B). However, CpG residues within other regions, including CpG 1.5, the canonical Hoxa9 promoter region (CpG3), and the homeodomain-containing exon II (CpG6), show no significant difference in methylation status between Mll+/+ and Mll−/− cells (Fig. S2). In contrast to the other CpG regions tested, however, CpG3 showed significantly less methylation in both Mll+/+ and Mll−/− cells. We further examined CpG islands 1 and 3 by cloning and sequencing PCR products of bisulfite treated genomic DNA (Fig. 1B and Figs. S2 and S3). The data reflect a similar Mll-dependent difference in methylation localized only to the same cluster of CpG1 residues. This suggests that Mll is required for these CpG1 residues to remain unmethylated but is irrelevant to the unmethylated status of CpG3.
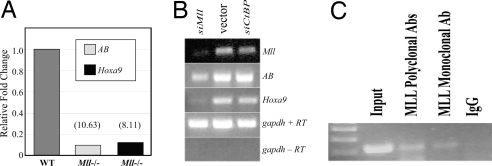
Hoxa9 is one of several Hox genes that show Mll-dependent expression. Using semiquantitative and real-time RT-PCR, we assessed expression of both canonical Hoxa9 and of transcripts containing the AB exon. A variety of transcripts have been identified that either incorporate both the AB exon and downstream canonical exons spliced into a single mRNA or that reflect a large unprocessed transcript originating in exon AB (Fig. S1). We wished to assess the Mll dependence of AB-containing transcript expression, which has not been fully explored (14). Both primers used for RT-PCR were within the AB exon; therefore, experiments included controls that ensure genomic contamination was not a factor. We show that expression of both AB-containing and canonical Hoxa9 transcripts are Mll-dependent, using both semiquantitative and real-time RT-PCR (Fig. 2A and Fig. S4).
Fig. 2.
Mll-dependent expression of Hoxa9 transcripts and Mll binding to Hoxa9 AB region. (A) RNA from Mll+/+ and Mll−/− MEF cells was analyzed by real-time RT-PCR to detect expression of upstream AB and canonical Hoxa9 transcripts. Expression was normalized to gapdh from the same samples. (B) shRNA knockdown of Mll. RNA extracted from Mll+/+ MEF cells after infection with retrovirus carrying empty vector or shRNA directed against either Mll or CtBP (an unrelated gene) were analyzed by RT-PCR to detect expression of Mll (to confirm efficient knockdown), the upstream AB transcript, the canonical Hoxa9 transcript, or gapdh control with and without reverse transcription (± RT). Cells were kept under selection for 2 weeks. (C) ChIP analysis was performed by using a mixture of polyclonal anti-Mll antibodies, a commercial anti-Mll monoclonal antibody, or IgG control antibody. Enriched chromatin was amplified by using primers specific for the AB exon.
If Mll is indeed required to protect these CpG residues from de novo methylation, the removal of Mll from Mll+/+ MEFs should permit the protected CpGs to become methylated. We used shRNAs to knock down Mll expression in Mll+/+ MEFs. RT-PCR confirmed the efficient knockdown of Mll (Fig. 2B). We also confirm that both AB and canonical Hoxa9 transcripts are Mll-dependent (Fig. 2B). For the relevant CpG residues within CpG island 1, we observed a difference in methylation between cells with knockdown of Mll and those with either a vector control (Fig. 1C) or knockdown directed against an unrelated gene, CtBP1 (data not shown). In MEFs in which Mll has been effectively knocked down, we observed increased methylation of the relevant CpG residues.
The above data demonstrate that Mll is required to maintain specific CpG residues in an unmethylated state. We further tested whether MLL could behave in a dominant manner resulting in the demethylation of previously methylated CpG DNA. We created “add-back” cell lines by introducing full-length MLL or an empty vector (an important negative control) into Mll−/− MEF cells. We analyzed DNA and RNA isolated from populations of cells after 1 week and up to 4 weeks or isolated from multiple clones from each type of add-back that had grown for >4 weeks. Reexpressing MLL in the Mll−/− MEFs completely reversed DNA methylation of the same specific CpG residues found to demonstrate Mll-dependent protection from DNA methylation (Fig. 1D). This does not occur for other CpGs, even those flanking the protected region (Fig. 1D and data not shown). This occurs rapidly (by one week), which is the earliest time point at which we could obtain sufficient cells for analysis. At this time point the Hoxa9 transcripts are not yet expressed (Fig. S4C), suggesting that in this case DNA methylation is reversed before the transcripts are reexpressed.
MLL binding to the promoter region for the canonical HOXA9 transcript has been demonstrated by chromatin immunoprecipitation (ChIP) (15, 16), which we have confirmed (data not shown). Given the Mll-dependent transcription and protection from CpG methylation in the AB region, we assessed Mll binding in this region as well. ChIP reveals the binding of endogenous Mll to the Hoxa9 AB region in Mll+/+ MEFs and an absence of binding in relevant controls (Fig. 2C). Similarly, a recent publication described MLL binding, using ChIP-on-chip analysis with tiling arrays across the human HOXA9 cluster (17). A peak of MLL binding was found in the region analogous to the region we define as having Mll-dependent protection from DNA methylation in mouse (Fig. S5).
To confirm that the regulated CpG sites we have identified are direct targets for Mll binding, we have measured in vitro binding of the MLL CXXC domain (MLL amino acids 1147–1203) to three oligonucleotides (Table 1) spanning this protected region of CpG 1 (Fig. 1). Fig. 3 shows the results of an isothermal titration calorimetry (ITC) measurement of the binding of the MLL CXXC domain to CpG1II. Table 1 lists the binding affinity, stoichiometry, and relevant thermodynamic parameters measured for each of the oligonucleotides. ΔH and ΔS refer, respectively, to the enthalpy and the entropy. Interestingly, both CpG1I and CpG1II yield a stoichiometry of 1:1 and similar binding affinities (Kd = 3.5, 7.4 μM) despite having multiple CpG elements. This is likely an indication that the contact region for the CXXC domain extends beyond the CpG element itself resulting in a steric block for binding of more than one CXXC domain and protecting these sites from methylation. CpG1III contains three CpG elements with two nucleotide spacers between them. Here, we observe a stoichiometry of 2:1 and a Kd of 24 μM, consistent with the binding of the CXXC domain to the terminal sites blocking binding of a third CXXC domain at the middle site. Similar ITC experiments measured binding of the MLL CXXC domain to CpG sequences not demonstrating Mll-dependent protection from methylation. We chose sequences from CpG3A that have a similar spacing of CpGs to those protected in CpG1 (Fig. S2D; CpGs 6–11). The binding affinities to these regions were lower (Kd = 32, 15 μM) than for the CpG1 sequences (Table 1 and Fig. S6). We have also attempted to measure binding to an oligonucleotide devoid of CG motifs but could not detect any binding; thus, although the measured binding affinity is modest, it is clearly specific.
Table 1.
Binding affinities, stoichiometries, and relevant thermodynamic parameters measured for each of the oligonucleotides
| Oligonucleotide | Sequence | Protected by MLL | Kd, μM | N | ΔH, cal/mol | ΔS, cal/mol·K |
|---|---|---|---|---|---|---|
| CpG1I | 5′-GGGTCGCGGGAG-3′ | Yes | 3.5 ± 0.4 | 1.19 ± 0.01 | −1631 ± 7 | 19.5 ± 0.2 |
| CpG1II | 5′-GAGCGCGCGCCT-3′ | Yes | 7.4 ± 1.3 | 0.91 ± 0.03 | −1187 ± 111 | 19.5 ± 0.7 |
| CpG1III | 5′-CAGCGGGCGGGCGCCT-3′ | Yes* | 23.6 ± 7.2 | 1.93 ± 0.02 | −1069 ± 183 | 17.7 ± 1.2 |
| CpG3AI | 5′-TGCCGGGCGGAC-3′ | No | 32 ± 4.2 | 1.2 ± 0.04 | −1387 ± 187 | 15.9 ± 0.3 |
| CpG3AII | 5′-TGACGCGCGTGG-3′ | No | 15 ± 3.5 | 1.1 ± 0.11 | −1776 ± 300 | 16.1 ± 1.4 |
Δ H, enthalpy (the heat of binding); Δ S entropy (the change in disorder associated with binding). The average values of thermodynamic parameters: Kd, N, ΔH, and ΔS together with the standard deviations from at least two independent ITC measurements of the CXXC domain binding to CpG1 and CpG3 oligonucleotides.
*The CpG1III cluster exhibits protection from methylation in experiments using full-length MLL or endogenous wild-type Mll but does not exhibit protection with MLL-AF4 constructs.
Fig. 3.
ITC measurement of the binding of the MLL CXXC domain to CpG1II oligonucleotide. Data shown is for addition of 10-μl aliquots of 550 μM CXXC domain to a 46 μM solution of CpG1II. The data were fit to a one-site binding model, giving Kd = 7.8 ± 1.4 μM and n = 0.93 ± 0.04.
Overexpression of HOXA9 is common in MLL related leukemia (3) and is an early requirement for the transforming potential of MLL fusions (18). MLL fusions retain the CXXC domain, and mutations that disrupt the CpG binding ability of this domain abrogate the transforming potential of an MLL fusion (5). We tested whether the MLL fusion proteins that cause leukemia would function in a manner similar to the way wild-type MLL protects specific CpG sequences from methylation. Analogous to experiments described for add-back of wild-type MLL, we created multiple cell lines with added back MLL-AF4 or MLL-AF9. Surprisingly, the effect of the leukemogenic MLL fusions is somewhat different. Only a subset of the CpG residues protected from DNA methylation by wild-type MLL are protected by the MLL fusion (Fig. 4 and data not shown). Furthermore, the MLL fusions do not reverse DNA methylation by 1 week; however, by 4 weeks, the protection of the CpG subset is observed. Introducing the MLL fusions still caused increased expression of the Hoxa9 transcripts by the later time point (Fig. S4C), even though only a core subset of the CpGs became de-methylated.
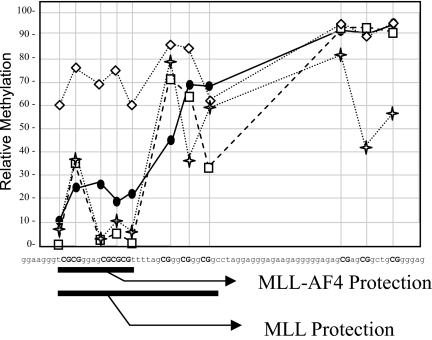
Fig. 4.
Add back of MLL-AF4 fusion reduces methylation of a portion of the protected CpG1 residues after 4 weeks and longer. Mll−/− MEF cells were transfected with either plasmid to express MLL-AF4 or empty vector. Cell populations were analyzed after 4 weeks of selection (squares, MLL-AF4; diamonds, vector control) or individual clones were selected, replated, and expanded (crosses and circles, two different representative MLL-AF4 add-back clones). DNA was subjected to bisulfite treatment and nested PCR and sequenced. Area under the curve analysis was performed on DNA sequencing histograms and relative methylation percentage was determined for CpG residues. Regions protected from DNA methylation by MLL-AF4 and by MLL (data from Fig. 1) are highlighted under the graph.
Discussion
Our results suggest a model for the Mll maintenance regulation of Hoxa9 in which the Mll CXXC domain binds to a specific cluster of CpG residues upstream of the Hoxa9 AB exon and provides a protective mechanism against de novo CpG methylation. As long as these CpG residues remain unmethylated, the locus is permissive for transcription. In the absence of Mll, these specific CpG residues become methylated, ultimately resulting in a silenced locus. Analagous models of protein binding that protect genomic regions from methylation have been suggested in other systems (19–21). CpG residues within the Lac operator of Escherichia coli can be protected from de novo Dnmt3a methylation by the DNA binding Lac repressor (LacI) protein (19). Similar results were seen by using an episomal system with the DNA binding EBNA-1 protein protecting the Epstein–Barr virus origin of replication (20). The transcription factor Sp1 has also been implicated in protecting CpG islands in the Aprt gene from methylation in a transgenic mouse model system (21). When putative Sp1 binding sites were mutated, the transgene was silenced by methylation. In those studies, in vivo footprinting demonstrated that a protein is bound to the endogenous promoter, and Sp1 was capable of binding to the target DNA sequence in vitro (21). However, to our knowledge, no direct binding of Sp1 to these sites in vivo has yet been demonstrated. Intriguingly, endogenous MLL has been found to bind to the APRT promoter (15). Additionally a motif search of the Hoxa9 region identifies a number of minimal consensus sites for Sp1 binding, including one within the area of Mll binding.
Our results demonstrate that endogenous Mll protein is capable of protecting a subset of specific CpG residues in the endogenous Hoxa9 locus from methylation by binding to that site. This protective binding of Mll to specific CpG regions within target genes could explain how Mll maintains a chromatin locus open and accessible. It also suggests how the information regarding an open locus could be passed to successive daughter cells. Mll may protect a CpG region from becoming methylated simply by blocking access to de novo DNA methyltransferases. Alternatively, a more complex mechanism may be used. This might include cooperation with additional DNA-binding proteins, such as Sp1, or chromatin-modifying proteins. HATs and HDACs have been shown to interact with MLL and could also help regulate accessibility of the target locus (22, 23). Furthermore, MLL displays a dominant effect in that it can reverse DNA methylation. Some proteins with CXXC domains, including DNMT1, preferentially bind hemi-methylated DNA (24), whereas other proteins preferentially bind methylated DNA (25). Potentially, competition of various CpG DNA binding domain proteins for the same target gene sites could contribute to remodeling that occurs during development, including the multiple cell identities within the hematopoietic cell lineage. This could also be true of the MLL fusion proteins. Our data support the ability of these oncogenic proteins to reverse DNA methylation of specific CpG sequences. The MLL fusions only protect a portion of the region from methylation, likely due to domains present in wild-type MLL that are missing in the fusions or an altered structure of the DNA binding domain caused by the fusion. This might be a critical function for keeping the HOXA9 locus open, thus contributing to continued expression of multiple transcripts from the HOXA9 locus in MLL leukemias. The differences between wild-type MLL and MLL fusion functions could potentially be exploited therapeutically in the future.
A number of alternate Hoxa9 transcripts have been identified. Some of these transcripts originate in an upstream alternate exon (AB). Many of the alternative transcripts include additional downstream sequences; therefore, it is unclear how much of the HOXA9 overexpression linked to MLL leukemogenesis is due to canonical HOXA9 and how much includes upstream elements. It is possible that both types of MLL-dependent transcripts contribute to leukemogenesis. A microRNA, mir-196b, is located within the Hoxa9 AB exon (26). Transcripts from this region might epigenetically regulate expression levels of additional genes, which would thus be controlled indirectly by MLL.
Current research suggests that there are two broad classes of promoters: the classical promoter with core promoter elements, such as TATAA and CCAAT boxes, and a nonclassical promoter devoid of any known transcriptional enhancer elements whose defining characteristic is residence within a stretch of sequence devoid of ATG triplets described as an “ATG desert” (27). The Hoxa9 canonical promoter represents the first class, whereas the upstream promoter shows all of the hallmarks of the latter class (Fig. S1). The binding and transcriptional activation activity of Mll at both locations suggests that Mll, in its role as a master regulatory gene, is able to function via different mechanisms at both promoter subtypes. Interestingly, the region upstream of the human HOXA9 gene, where transcripts have also been shown to originate (11, 28), has both an ATG desert promoter type and CpG islands. Although the homology across the entire upstream CpG island is poor between mouse and human, the core residues that show differential methylation in the mouse, are present in a similar arrangement in the human and bind MLL (17). The mechanism we elucidate here, the binding and protection of CpG clusters within the upstream promoter, is most likely directed by Mll's CXXC domain. It is interesting to note that the CXXC domain is present in all chordate and echinoderm Mll orthologs but that Drosophila and other insect MLL orthologs do not have CXXC domains. This suggests that the evolutionary appearance of the CXXC domain in Mll coincides with a new layer of epigenetic regulation important in chordates and echinoderms but apparently not in insects.
The highly conserved MLL methyltransferase homology (MT) domain (MLL amino acids 1147–1244) includes a CXXC domain. Previous measurements of binding of the MT domain to CG elements, using surface plasmon resonance (SPR), yielded a Kd of 3 × 10−8 M (10), ≈2 orders of magnitude tighter than the ITC measurements reported here. This set of experiments used a GST fusion with MT and a DNA site with six CG residues each separated by 6–8 nt. The dimerization of GST has been shown to influence fused DNA binding domains (29). The potential in these experiments for a bivalent interaction of the GST-MT with two sites on the DNA could result in the increase in affinity seen (30).
In systems where a dynamic interplay of different binding proteins is essential for proper functioning, moderate affinity, which we demonstrate in the ITC experiments, is essential for proper regulation. Dynamic binding to the CG elements mediated by the CXXC domain would assure that necessary access to these elements can be achieved when needed. A caveat of both the previous SPR and our current ITC experiments is that the binding of MLL to DNA is likely more complex than a single domain alone could constitute. The AT-hooks of MLL have been shown to bind the DNA in AT-rich regions in a structure specific manner (31). Furthermore, MLL is known to recruit other DNA binding proteins (32, 33) whose presence may stabilize or otherwise affect MLL's DNA binding capabilities.
Here, we report a mechanistic link between the CXXC domain of MLL, protection of specific clusters of CpG residues from methylation, and the maintenance of Hoxa9 expression. Recently, it was suggested from the results of genome-wide chromatin immunoprecipitation experiments that MLL might regulate expression of close to 5,000 genes (15). Several examples of MLL target genes in addition to the HOX genes are confirmed in refs. 1, 3, 14, 34, and 35. It will be important to determine whether MLL similarly maintains some or all of these genes in a state permissive for transcription via protecting specific CpG residues from methylation.
Materials and Methods
CpG Methylation Analysis by Bisulfite Treatment of DNA.
Genomic DNA was isolated by using the Puregene DNA isolation kit (Gentra Systems) per manufacturer's protocols. DNA was bisulfite treated as described in ref. 36 with some modifications. Briefly, 2 μg of genomic DNA was treated with sodium bisulfite for 10–14 h followed by desalting and concentration with the Wizard DNA resin-based clean-up system (Promega). PCR was performed in two rounds on each sample. PCR primers and conditions were optimized for each amplification and are available in Table S1. PCR products were sequenced directly with the ABI BigDye Terminator v1.1 cycle sequencing kit, or cloned into TA-Cloning vector pCR 2.1-TOPO (Invitrogen) followed by sequencing. Sequencing histograms were analyzed with Scion Image software, Version 4.0.2 Beta. Each CpG cytosine peak, representing a methylated nucleotide, was compared with its corresponding thymine peak, representing an unmethylated nucleotide. Peaks were corrected for background by measuring non-CpG cytosine conversion. Student's t test was performed on combined data to determine statistical significance of differences for individual CpGs.
Retroviral shRNA Knockdown of Mll.
Knockdowns were performed by using the pSuper retroviral system (37). Mll+/+ MEF cells at 90% confluency were infected with pSuper Mll 11487 (small hairpin targeting Mll starting at base pair 11487) or pSuper vector alone. Cells were coincubated with retrovirus on plates for 5 h with 4 μg/ml polybrene. After 5 hours, the media was changed and additional retrovirus added (no polybrene) for 12 h. Media was changed and cells given an additional 24 h to recover. Cells were then grown on selection media with 3.5 μg/ml puromycin for 2 or 4 weeks.
Add-Back Cell Lines.
Mll−/− MEF cells were transfected by using Effectene (Qiagen) with pCDNA5·FRT·TO-MLL (full-length), pCDNA5·FRT·TO-MLL-AF4 (fusion), pCDNA5·FRT·TO (vector control), or MSCVneo-MLL-AF9 (fusion). Cells were simultaneously cotransfected with pSuper (puromycin resistance) at a 1:5 molar ratio. Cells recovered for 24 h before selective pressure was initiated (4 μg/ml puromycin). Populations were selected at 1, 2, and 4 weeks. Alternatively, the same cotransfection ratios and plasmids were used for electroporation on Mll−/− MEF cells (0.4-cm gap, 500 mV, Q500, 5 × 106 cells in 500 μl of serum-free media). Cells were allowed 24 h to recover, and selective pressure (4 μg/ml puromycin) was continued until colonies could be picked (3–5 weeks). Colonies were isolated and expanded for 1–2 additional weeks while selective pressure was maintained. Both genomic DNA and total RNA were isolated from all samples.
Semiquantitative and Real-time RT-PCR.
All RNAs were isolated by using Tri-Reagent (Sigma; catalog no. T9424) according to the manufacturer's protocol. RNA was treated with DNaseI to remove genomic contamination and cDNA was prepared by using SuperScript first-strand synthesis system (Invitrogen; catalog no. 12371-019). Forward and reverse AB primers span the AB exon, whereas the canonical Hoxa9 primers span the CD and exon II junction. For quantitative RT-PCR studies, cDNA from WT and Mll −/− cells was analyzed by using an Applied Biosystems 7300 real-time PCR system. All reactions were performed in triplicate and AB and canonical Hoxa9 expression levels detected by using SYBR Green reagents. Relative expression of the AB and canonical Hoxa9 transcripts were normalized to gapdh and the fold change calculated by using the 2−ΔΔCt method. Primer sequences are provided in Table S2.
Chromatin Immnuoprecipitation (ChIP).
ChIP experiments were performed by using an EZ-ChIP Kit (Upstate Biotechnology; catalog no. 17-371) following the manufacturer's protocol. Each sample consisted of 8 × 106 cells. Antibodies used are as follows: Mix of 3 anti-MLL rabbit polyclonal antibodies (34), anti-MLL (Upstate Biotechnology; catalog no. 05-765) and IgG (Upstate Biotechnology; catalog no. 12-371B).
Expression and Purification of the MLL CXXC Domain.
DNA encoding amino acids 1147–1203 of MLL was cloned into pGEX-4T-2 (Amersham Biosciences). The plasmid was used to transform E. coli BL21 (DE3) RIPL cells (Stratagene). Protein expression was induced by addition of 1 mM IPTG. GST-fused CXXC domain was purified by glutathione affinity chromatography (Amersham), cleavage from GST with thrombin, and ion-exchange chromatography using SP-Sepharose (Amersham).
Purification of Oligonucleotides for ITC.
DNA fragments corresponding to three sequences derived from CpG1 and two sequences from CpG3A were synthesized by Integrated DNA Technologies: CpG1I, 5′-GGGTCGCGGGAG-3′; CpG1II, 5′-GAGCGCGCGCCT-3′; CpG1III, 5′-CAGCGGGCGGGCGCCT-3′; CpG3AI, 5′-TGCCGGGCGGAC-3′; and CpG3AII, 5′-TGACGCGCGTGG-3′. Complementary strands were annealed and purified by using ion-exchange chromatography on Q-Sepharose.
Isothermal Titration Calorimetry.
The CXXC domain and oligonucleotides were extensively dialyzed at 4°C against 25 mM phosphate (pH 7.2), 300 mM NaCl, and 1 mM DTT and degassed before measurement. The titrations were performed by using a VP-ITC titration calorimetric system (MicroCal) at 27°C. The calorimetric cell, containing either CpG1I, CpG1II, CpG1III, CpG3AI, or CpG3AII (concentrations in the range of 30 to 80 μM), was titrated with the CXXC domain (500–950 μM) injected in 10-μl aliquots. Data were analyzed by using Origin 7.0 (OriginLab) to obtain Kd, N (number of sites), ΔH, and ΔS values.
Supplementary Material
Acknowledgments.
Wild-type and Mll null MEF cells were kindly provided by P. Ernst (Dartmouth College, Hanover, NH) and S. Korsmeyer (Harvard University, Boston, MA); Francis Collins (National Human Genome Research Institute, National Institutes of Health, Bethesda, MD) kindly provided primary data from their work published in ref. 17. This work was supported by the Dr. Ralph and Marian Falk Medical Research Trust, National Institutes of Health/National Cancer Institute Grant CA40046 (to N.J.Z.-L.), and the Swortzel Award (to J.H.B.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0800090105/DCSupplemental.
References
- 1.Popovic R, Zeleznik-Le NJ. MLL: How complex does it get? J Cell Biochem. 2005;95:234–242. doi: 10.1002/jcb.20430. [DOI] [PubMed] [Google Scholar]
- 2.Yu BD, Hanson RD, Hess JL, Horning SE, Korsmeyer SJ. MLL, a mammalian trithorax-group gene, functions as a transcriptional maintenance factor in morphogenesis. Proc Natl Acad Sci USA. 1998;95:10632–10636. doi: 10.1073/pnas.95.18.10632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armstrong SA, et al. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat Genet. 2002;30:41–47. doi: 10.1038/ng765. [DOI] [PubMed] [Google Scholar]
- 4.Zeisig BB, et al. Hoxa9 and Meis1 are key targets for MLL-ENL-mediated cellular immortalization. Mol Cell Biol. 2004;24:617–628. doi: 10.1128/MCB.24.2.617-628.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ayton PM, Chen EH, Cleary ML. Binding to nonmethylated CpG DNA is essential for target recognition, transactivation, and myeloid transformation by an MLL oncoprotein. Mol Cell Biol. 2004;24:10470–10478. doi: 10.1128/MCB.24.23.10470-10478.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Slany RK, Lavau C, Cleary ML. The oncogenic capacity of HRX-ENL requires the transcriptional transactivation activity of ENL and the DNA binding motifs of HRX. Mol Cell Biol. 1998;18:122–129. doi: 10.1128/mcb.18.1.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee JH, Voo KS, Skalnik DG. Identification and characterization of the DNA binding domain of CpG-binding protein. J Biol Chem. 2001;276:44669–44676. doi: 10.1074/jbc.M107179200. [DOI] [PubMed] [Google Scholar]
- 8.Lee JH, Skalnik DG. CpG-binding protein (CXXC finger protein 1) is a component of the mammalian Set1 histone H3-Lys4 methyltransferase complex, the analogue of the yeast Set1/COMPASS complex. J Biol Chem. 2005;280:41725–41731. doi: 10.1074/jbc.M508312200. [DOI] [PubMed] [Google Scholar]
- 9.Jorgensen HF, Ben-Porath I, Bird AP. Mbd1 is recruited to both methylated and nonmethylated CpGs via distinct DNA binding domains. Mol Cell Biol. 2004;24:3387–3395. doi: 10.1128/MCB.24.8.3387-3395.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Birke M, et al. The MT domain of the proto-oncoprotein MLL binds to CpG-containing DNA and discriminates against methylation. Nucleic Acids Res. 2002;30:958–965. doi: 10.1093/nar/30.4.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borrow J, et al. The t(7;11)(p15;p15) translocation in acute myeloid leukaemia fuses the genes for nucleoporin NUP98 and class I homeoprotein HOXA9. Nat Genet. 1996;12:159–167. doi: 10.1038/ng0296-159. [DOI] [PubMed] [Google Scholar]
- 12.Popovic R, Erfurth F, Zeleznik-Le N. Transcriptional complexity of the HOXA9 locus. Blood Cells Mol Dis. 2008;40:156–159. doi: 10.1016/j.bcmd.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu BD, Hess JL, Horning SE, Brown GA, Korsmeyer SJ. Altered Hox expression and segmental identity in Mll-mutant mice. Nature. 1995;378:505–508. doi: 10.1038/378505a0. [DOI] [PubMed] [Google Scholar]
- 14.Milne TA, et al. MLL associates specifically with a subset of transcriptionally active target genes. Proc Natl Acad Sci USA. 2005;102:14765–14770. doi: 10.1073/pnas.0503630102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guenther MG, et al. Global and Hox-specific roles for the MLL1 methyltransferase. Proc Natl Acad Sci USA. 2005;102:8603–8608. doi: 10.1073/pnas.0503072102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Milne TA, et al. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell. 2002;10:1107–1117. doi: 10.1016/s1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]
- 17.Scacheri PC, et al. Genome-wide analysis of menin binding provides insights into MEN1 tumorigenesis. PLoS Genet. 2006;2:e51. doi: 10.1371/journal.pgen.0020051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ayton PM, Cleary ML. Transformation of myeloid progenitors by MLL oncoproteins is dependent on Hoxa7 and Hoxa9. Genes Dev. 2003;17:2298–2307. doi: 10.1101/gad.1111603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han L, Lin IG, Hsieh CL. Protein binding protects sites on stable episomes and in the chromosome from de novo methylation. Mol Cell Biol. 2001;21:3416–3424. doi: 10.1128/MCB.21.10.3416-3424.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frappier L, O'Donnell M. EBNA1 distorts oriP, the Epstein–Barr virus latent replication origin. J Virol. 1992;66:1786–1790. doi: 10.1128/jvi.66.3.1786-1790.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Macleod D, Charlton J, Mullins J, Bird AP. Sp1 sites in the mouse aprt gene promoter are required to prevent methylation of the CpG island. Genes Dev. 1994;8:2282–2292. doi: 10.1101/gad.8.19.2282. [DOI] [PubMed] [Google Scholar]
- 22.Xia ZB, Anderson M, Diaz MO, Zeleznik-Le NJ. MLL repression domain interacts with histone deacetylases, the polycomb group proteins HPC2 and BMI-1, and the corepressor C-terminal-binding protein. Proc Natl Acad Sci USA. 2003;100:8342–8347. doi: 10.1073/pnas.1436338100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ernst P, Wang J, Huang M, Goodman RH, Korsmeyer SJ. MLL and CREB bind cooperatively to the nuclear coactivator CREB-binding protein. Mol Cell Biol. 2001;21:2249–2258. doi: 10.1128/MCB.21.7.2249-2258.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Araujo FD, et al. The DNMT1 target recognition domain resides in the N terminus. J Biol Chem. 2001;276:6930–6936. doi: 10.1074/jbc.M009037200. [DOI] [PubMed] [Google Scholar]
- 25.Ballestar E, Wolffe AP. Methyl-CpG-binding proteins. Targeting specific gene repression. Eur J Biochem. 2001;268:1–6. doi: 10.1046/j.1432-1327.2001.01869.x. [DOI] [PubMed] [Google Scholar]
- 26.Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 27.Lee MP, et al. ATG deserts define a novel core promoter subclass. Genome Res. 2005;15:1189–1197. doi: 10.1101/gr.3873705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim MH, et al. Genomic structure and sequence analysis of human HOXA-9. DNA and Cell Biology. 1998;17:407–414. doi: 10.1089/dna.1998.17.407. [DOI] [PubMed] [Google Scholar]
- 29.Niedziela-Majka A, Rymarczyk G, Kochman M, Ozyhar A. GST-Induced dimerization of DNA-binding domains alters characteristics of their interaction with DNA. Protein Expression and Purification. 1998;14:208–220. doi: 10.1006/prep.1998.0932. [DOI] [PubMed] [Google Scholar]
- 30.Mammen M, Choi SK, Whitesands GM. Polyvalent interactions in biological systems: Implications for design and use of multivalent ligands and inhibitors. Angewandte Chemie International Edition. 1998;37:2754–2794. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 31.Zeleznik-Le NJ, Harden AM, Rowley JD. 11q23 translocations split the “AT-hook” cruciform DNA-binding region and the transcriptional repression domain from the activation domain of the mixed-lineage leukemia (MLL) gene. Proc Natl Acad Sci USA. 1994;91:10610–10614. doi: 10.1073/pnas.91.22.10610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yokoyama A, et al. Leukemia proto-oncoprotein MLL forms a SET1-like histone methyltransferase complex with menin to regulate Hox gene expression. Mol Cell Biol. 2004;24:5639–5649. doi: 10.1128/MCB.24.13.5639-5649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.La P, et al. Direct binding of DNA by tumor suppressor menin. J Biol Chem. 2004;279:49045–49054. doi: 10.1074/jbc.M409358200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xia ZB, et al. The MLL fusion gene, MLL-AF4, regulates cyclin-dependent kinase inhibitor CDKN1B (p27kip1) expression. Proc Natl Acad Sci USA. 2005;102:14028–14033. doi: 10.1073/pnas.0506464102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ferrando AA, et al. Gene expression signatures in MLL-rearranged T-lineage and B-precursor acute leukemias: Dominance of HOX dysregulation. Blood. 2003;102:262–268. doi: 10.1182/blood-2002-10-3221. [DOI] [PubMed] [Google Scholar]
- 36.Clark SJ, Harrison J, Paul CL, Frommer M. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 1994;22:2990–2997. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.