Abstract
Autoreactive T cells in patients with Goodpasture's disease are specific for epitopes in the Goodpasture antigen (the NC1 domain of the α3 chain of type IV collagen) that are rapidly destroyed during antigen processing to a degree that diminishes their presentation to T cells. We hypothesized that patients' autoreactive T cells exist because antigen processing prevents presentation of the self-epitopes they recognize, circumventing specific tolerance mechanisms. We predicted that autoreactive T cells specific for these peptides should also exist in healthy individuals, albeit at low frequency and in an unprimed state. We obtained blood from healthy unrelated donors and, using a panel of 45 α3(IV)NC1 peptides, identified α3(IV)NC1-specific T cells in all donors. Thirty-six of 45 peptides elicited a proliferative T cell response from at least one subject, and 6 of the peptides evoked a response in >50% of the individuals. This consistency was not caused by selectivity of HLA class II molecules because the donors expressed a diversity of HLA antigens, but was largely a result of the substrate-specificity of the endosomal proteases Cathepsin D and E. There was a significant correlation between high susceptibility to Cathepsin D digestion and the capacity to stimulate primary T cell responses (P = 0.00006). In summary, healthy individuals have low frequencies of unstimulated α3(IV)NC1-reactive T cells with similar specificities to the autoreactive T cells found in patients with Goodpasture disease. In both cases, existence of the α3(IV)NC1-reactive T cells can be accounted for by destructive processing.
Autoimmune diseases are uncommon because autoreactive CD4 T cells are held in check by powerful central and peripheral tolerance mechanisms. Central control is effected in the thymus where there is irrecoverable deletion of autoreactive T cells that have high avidity for any of the array of self-proteins presented by thymic epithelium.1 Peripheral controls are thought to be evoked when autoreactive T cells recognize self-epitopes presented by antigen-presenting cells (APC) in benign environments and can lead to autoreactive T cells being deleted, rendered unresponsive (anergic), or directed to develop one of probably several anti-inflammatory phenotypes (regulatory T cells).2 Importantly, central and peripheral tolerance mechanisms are tailored to self-proteins as they are constitutively presented. Consequently, T cells that recognize epitopes from self-proteins that are not usually presented (cryptic epitopes), may be less subject to control,3,4 and more likely to drive deleterious autoimmune responses. Certainly in several animal models of autoimmunity it has been shown that pathogenic T cells recognize cryptic self-epitopes.5,6
There is therefore great interest in mechanisms that diminish constitutive presentation of self-peptides, especially ones that might break under disease-initiating conditions leading to unusual presentation. One such mechanism is destructive processing,7,8 in which processing proceeds such that certain epitopes are so consistently destroyed they are not presented to T cells, even when the epitopes have high affinity for HLA molecules. We recently presented evidence that the principal epitopes in the NC1 domain of the α3 chain of type (IV) collagen (α3(IV)NC1) recognized by Goodpasture patients' T cells are destroyed early in the course of α3(IV)NC1 processing within human B cells, with diminished presentation to α3(IV)NC-specific T cell clones.9,10 It has also been reported that an epitope recognized by autoreactive T cells commonly detected in patients with multiple sclerosis is similarly rapidly destroyed during processing with consequent diminished presentation to T cells.11 In both cases it can be argued that the low constitutive processing of specific peptides allows the corresponding T cells to escape irreversible self-tolerance and persist to be available to drive autoimmune disease under initiating stimuli that perturb usual processing.
The hypothesis that proteases involved in antigen processing enable some autoreactive T cells to escape irreversible tolerance has a testable prediction: T cells specific for the destroyed peptides should be present in health as well as disease, albeit with an quiescent phenotype. In this study we tested this prediction with respect to the Goodpasture autoantigen. The results strongly support a pervasive influence of destructive processing by Cathepsin D in shaping the repertoire of α3(IV)NC1-specific autoreactive T cells in health and Goodpasture disease.
Results
T Cells Specific for α3(IV)NC1 Peptides Are Detectable in Peripheral Blood of Healthy Controls
To test the prediction that T cells specific for rapidly cleaved self-peptides should exist in a quiescent state within healthy individuals we investigated the proliferative responses of T cells from 11 healthy donors to α3(IV)NC1 peptides under conditions that measure primary responses. Previous work had screened peptides under conditions that detected responses by T cells that had been previously activated in vivo (secondary responses) and demonstrated that such responses were common in patients with Goodpasture disease but not healthy donors.9 In our study, the assay conditions were altered to detect primary responses by using relatively large culture volumes and long culture periods, extending up to 10 d.12
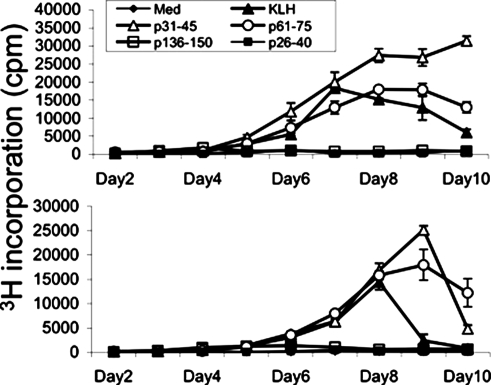
Initial experiments determined that T cells from three healthy individuals made proliferative responses to α3(IV)NC1 peptides that developed with kinetics similar to those observed for key hole limpet hemocyanin, an antigen widely used to examine primary responses (Figure 1). The responses peaked between days 7 and 10, whereas recall responses (e.g., to PPD) peaked between days 5 and 6.
Figure 1.
Representative kinetic analyses of the proliferative responses of peripheral blood T cells from healthy individuals to α3(IV)NC1 peptides (px–y, see Table 1). The slow development of the responses closely parallels that elicited by key hole limpet hemocyanin (KLH; an archetypal novel antigen), indicating that the responding T cells are previously quiescent or naive.
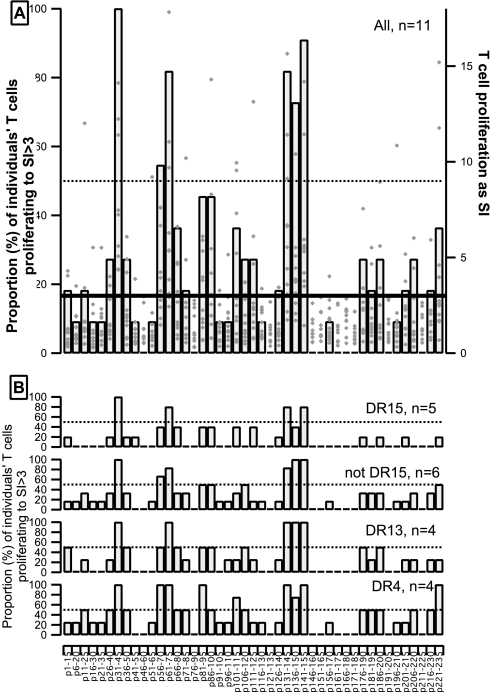
Next we examined under these conditions the responses of T cells from 11 healthy individuals to the panel of 45 α3(IV)NC1 peptides. A diversity of responses was observed as expected because the subjects were not selected for expression of HLA, with most (36 of 45) peptides eliciting proliferative responses by T cells from at least one individual (Figure 2). There was superimposed a striking conformity in that six peptides evoked significant proliferative responses of T cells from more than half the individuals tested, even though the individuals carried a range of HLA alleles (see legend to Figure 2). There was no preponderance of any one allele or even of groups of alleles known to encode class II molecules with similar peptide binding capabilities, and responses to the six peptides were observed for T cells from individuals with diverse HLA DRB1 alleles. Therefore, healthy individuals harbor quiescent unprimed α3(IV)NC1-specific T cells that exhibit consistency in peptide specificity irrespective of HLA type.
Figure 2.
Responses to α3(IV)NC1 peptides of PBMC from 11 healthy individuals. The proportion (%) of the 11 individuals whose T cells made significant proliferative responses (stimulation index (SI) >3) to the indicated peptides is shown as columns against the left hand axes. (A) All 11 healthy individuals: Diamond markers against the right axis show the SI values measured for each individual. (B) Proportion (%) of the healthy individuals with common expression of indicated HLA DRB1 alleles whose T cells made significant proliferative responses. The HLA types of the individuals were: *0311/*0801; *1101/*1101; *0401/*0701; *1501/*0101; *0401/*1302; *1502/*0401; *0101/*0401; *1501/*1301; *03xx/*1501; *15xx/*13xx; and *01xx/*13xx.
Relative Susceptibility of α3(IV)NC1 Peptides to Destruction by Cathepsin D Correlates with T cell Repertoire in Health
Patients' predominant α3(IV)NC1-reactive T cells were shown to recognize peptides destroyed by Cathepsin D/E in early processing,10 raising the possibility that processing enzymes could also account for the consistent specificities of α3(IV)NC1-reactive T cells in healthy individuals. To test this prediction we determined the relative susceptibility to Cathepsin D digestion of each of the 45 α3(IV)NC1 peptides.
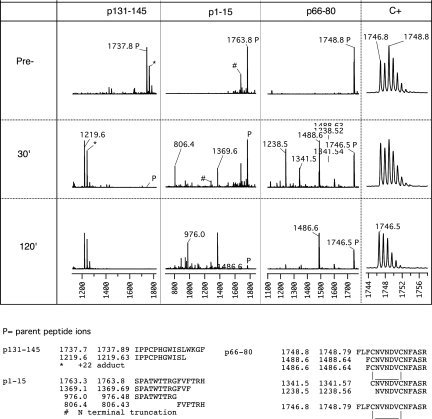
Preliminary studies found the lowest enzyme concentration at which the most scissile α3(IV)NC1 peptides were completely cut by Cathepsin D during a 30-min incubation, an interval within which T cell epitopes are known to be generated from internalized antigens by APC.13 The extent and location of cleavage within peptides were determined by matrix-assisted laser-desorption time-of-flight mass spectrometry (MALDI-TOF) analysis of the digests, as shown for three representative peptides in Figure 3.
Figure 3.
MALDI-TOF analysis of fragments generated in the course of Cathepsin D digestion of three α3(IV)NC1 peptides. The first three panels of the top section show MALDI-TOF mass spectrograms of Cathepsin D digests of three α3(IV)NC1 peptides selected to illustrate the patterns of digestion observed. For each peptide, spectrograms were taken before (top) and after 30 or 120 min incubation with protease at 37°C. Peptide p131-145 (m/z 1737.89) is highly scissile because it is completely cleaved at 30 min with disappearance of the parent ion and appearance of a single major fragment (the ions marked as +22 adducts are likely to be peptides that retain a protecting group because they were observed even after extensive desalting). The sequence of the fragment deduced from its m/z is shown in the bottom row and allows inference of SL|WK as the highly scissile peptide bond. Peptide p1-15 is just scissile because it is <50% digested at 30 min with two major fragments indicative of two scissile bonds, and some parent remains even at 120 min. Digestion of p66-80 appears only slightly more complete at 30 min than p1-15, but careful inspection of the isotope pattern (far right) of the parent ion showed the initial parent to comprise two populations with m/z 2 mass units apart with subsequent depletion of only the heavier form. Comparison of freshly oxidized and reduced peptide and acetylated p66-80 confirmed that the reduced form was highly scissile and the oxidized form (with intramolecular disulfide loop) was highly resistant (data not shown).
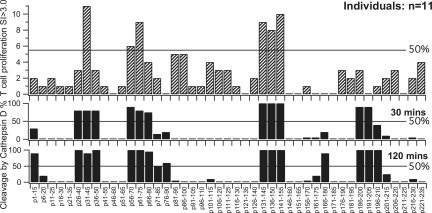
The susceptibility to Cathepsin D digestion in vitro of each of the 45 peptides is shown in Figure 4, and the locations of the scissile peptide bonds are shown against the sequences of the peptides in Table 1. All the peptides that stimulated the majority of individuals' T cells were highly susceptible to Cathepsin D. Moreover, there was overall a highly significant (P = 0.00006, Fisher exact test) correspondence between high susceptibility to Cathepsin D and capacity to stimulate primary responses by T cells from the majority of healthy individuals (Table 2).
Figure 4.
Autoreactive (to α3(IV)NC1) T cells in healthy individuals are predominantly specific for peptides rapidly cleaved by Cathepsin D. The top chart shows the proportion of healthy individuals' T cells that proliferated with SI>3 to the indicated α3(IV)NC1 peptides (sequences in Table 1). The lower charts show relative cutting by Cathepsin D at 30 and 120 min.
Table 1.
Cathepsin D cuts in α3(IV)NC1 peptidesa
| Name | Sequence | Cleavage (%)
|
Most Scissile Peptide Bonds | ||
|---|---|---|---|---|---|
| 30 min | 60 min | ||||
| p1-15 | SPATWTTRG/FVF/TRH | 30 | 90 | (G9, F12) | |
| p6-20 | TTRGFVF–TRHSQTTA | 0 | 20 | ||
| p11-25 | VFTRHSQTTAIPSCP | 0 | 0 | ||
| p16-30 | SQTTAIPSCPEGTVP | 0 | 0 | ||
| p21-35 | IPSCPEGTVPLYSGF | 0 | 0 | ||
| p26-40 | EGTVPLYSGFSF|L|FV | 80 | 100 | } | |
| p31-45 | LYSGFSF|L|FVQGNQR | 80 | 90 | } | F37, L38 |
| p36-50 | SF|L|FVQGNQRAHGQD | 80 | 100 | } | |
| p41-55 | QGNQRAHGQDLGTLG | 0 | 0 | ||
| p46-60 | AHGQDLGTLGSCLQR | 0 | 0 | ||
| p51-65 | LGTLGSCLQRFTTMP | 0 | 0 | ||
| p56-70 | SCLQRFTTMPF|L|FCN | 90 | 100 | } | F66, L67 |
| p61-75 | FTTMPF|L|FC|NVNDVC | 80 | 100 | } | C69b |
| p66-80 | FL|F|C|NVNDVCNFASR | 75 | 95 | } | |
| p71-85 | VNDVCNF/ASRNDYSY | 15 | 50 | (F77) | |
| p76-90 | NFASRNDYSY/W/LSTP | 20 | 60 | (Y85W86) | |
| p81-95 | NDYSY–W–LST–PALMPM | 0 | 5 | ||
| p86-100 | WLSTPALMPMNMAPI | 0 | 0 | ||
| p91-105 | ALMPMNMAPITGRAL | 0 | 0 | ||
| p96-110 | NMAPITGRALEPYIS | 0 | 0 | ||
| p101-115 | TGRALEPYISR|CTVC | 0 | 10 | ||
| p106-120 | EPYISRCTVCEGPAI | 0 | 0 | ||
| p111-125 | RCTVCEGPAIAIAVH | 0 | 0 | ||
| p116-130 | EGPAIAIAVHSQTTD | 0 | 0 | ||
| p121-135 | AIAVHSQTTDIPPCP | 0 | 0 | ||
| p126-140 | SQTTDIPPCPHGWIS | 0 | 0 | ||
| p131-145 | IPPCPHGW–IS–L|WKGF | 100 | 100 | } | |
| p136-150 | HGW–IS–L|WKGFSF|IMF | 100 | 100 | } | L141 |
| p141-155 | L|WKGFSF|IM–FTSAGS | 100 | 100 | } | F147 |
| p146-160 | SFIMFTSAGSEGTGQ | 0 | 0 | ||
| p151-165 | TSAGSEGTGQALASP | 0 | 0 | ||
| p156-170 | EGT–GQALASPGSCLE | 5 | 5 | ||
| p161-175 | ALASPGSCLEEF–RAS | 5 | 20 | ||
| p166-180 | GSCLEEF/RASPFLEC | 20 | 90 | (F172)b | |
| p171-185 | EFRASPFLECHGRGT | 0 | 0 | ||
| p176-190 | PFLECHGRGTCNYYS | 0 | 0 | ||
| p181-195 | HGRGTCNYYSNSYSF | 0 | 0 | ||
| p186-200 | CNYYSNSYSF|WLASL | 90 | 100 | } | |
| p191-205 | NSYSF|WL–ASLNPERM | 90 | 100 | } | F195 |
| p196-210 | WL–ASLNPERM/FRKPI | 40 | 100 | ||
| p201-215 | NPERM–FRKPIPSTVK | 10 | 25 | ||
| p206-220 | FRKPIPSTVKAGELE | 0 | 0 | ||
| p211-225 | PSTVKAGELEKIISR | 0 | 0 | ||
| p216-230 | AGELEKIISRC–QVCM | 5 | 10 | ||
| p221-235 | KIISRCQVCMKKRH | 0 | 0 | ||
The synthetic α3(IV)NC1 peptides used in the study of healthy individuals were named pfrom-to in relation to sequence of intact α3(IV)NC1, numbered relative to the sequence SPAT  at the beginning of the NC1 domain (column 1). The extent of cleavage by Cathepsin D at 30 and 120 min are shown as percentages in columns 3 and 4, respectively. Note that the peptides' sequences were chosen to overlap by 5aa so that all portions of α3(IV)NC1 were represented at least twice so as to detect substrate peptide bonds despite possible confounding effects from proximity to the ends of a peptides (e.g., F77 cut efficiently in p71-85 but less so in p66-80 and p81-95), or differences in peptide solubility (e.g., the cuts Y85W86 in p76-90 were not seen in the less water-soluble p81-95). The designations in the rightmost column are based on the greatest degree of cleavage observed. L23 peptide bonds are referenced by the name and number of the residue to its N-terminal end. “Highly scissile peptides,” peptides cleaved >50% at 30 min (11 peptides); |, highly scissile peptide cut to account for the dominant fragment ions in digests of highly scissile peptides (8 peptide bonds shown in the rightmost column in bold type); /, scissile peptide bonds cut to account for the dominant fragment ions in digests of peptides cleaved >50% at 120 min but not 30 min (six identified in the rightmost column, shown in brackets); –, other detected cleaved peptide bonds.
at the beginning of the NC1 domain (column 1). The extent of cleavage by Cathepsin D at 30 and 120 min are shown as percentages in columns 3 and 4, respectively. Note that the peptides' sequences were chosen to overlap by 5aa so that all portions of α3(IV)NC1 were represented at least twice so as to detect substrate peptide bonds despite possible confounding effects from proximity to the ends of a peptides (e.g., F77 cut efficiently in p71-85 but less so in p66-80 and p81-95), or differences in peptide solubility (e.g., the cuts Y85W86 in p76-90 were not seen in the less water-soluble p81-95). The designations in the rightmost column are based on the greatest degree of cleavage observed. L23 peptide bonds are referenced by the name and number of the residue to its N-terminal end. “Highly scissile peptides,” peptides cleaved >50% at 30 min (11 peptides); |, highly scissile peptide cut to account for the dominant fragment ions in digests of highly scissile peptides (8 peptide bonds shown in the rightmost column in bold type); /, scissile peptide bonds cut to account for the dominant fragment ions in digests of peptides cleaved >50% at 120 min but not 30 min (six identified in the rightmost column, shown in brackets); –, other detected cleaved peptide bonds.
Peptide bond resistant to Cathepsin D in oxidized (native) form but scissile in reduced form.
Table 2.
Truth table for association between rapid proteolysis and capacity to stimulate T cells from healthy individuals
| Highly scissile to Cathepsin D | Proliferation by T Cells from ≥6 of 11 Healthy Individuals
|
Total | |
|---|---|---|---|
| P-N | P-Y | ||
| C-N | 34 | 0 | 34 |
| C-Y | 5 | 6 | 11 |
| Total | 39 | 6 | 45 |
| P = 0.00006 | |||
Each peptide in the set of 45 used to interrogate healthy individuals' T cells was classified as stimulating significant proliferation (SI > 3.0) (P-Y) or not (P-N) of T cells from ≥6 of the 11 healthy subjects, and highly scissile (>50% cleaved at 30 min) (C-Y) or not (C-N) to Cathepsin D. Results are shown in a truth-table format. The proportion of T cell stimulatory peptides that also contained highly Cathepsin D–scissile peptide bonds was higher than expected from the overall frequency of highly scissile peptide bonds in the peptide set. Fisher exact test: P = 0.00006.
The results suggest that susceptibility to Cathepsin D is a factor in determining the specificity of the autoreactive T cells that escape irreversible tolerance to persist in the peripheral blood of healthy individuals. Thus, whereas numerous factors are known to interactively influence which epitopes in antigens are efficiently presented, it appears that single factors, in this case susceptibility to Cathepsin D, can be dominant determinants of low presentation and hence of the specificities of T cells that escape irreversible self-tolerance.
Relative Susceptibility of α3(IV)NC1 Peptides to Destruction by Cathepsin D and E also Correlates with Expanded T Cell Repertoire in Recent-Onset Goodpasture Disease
We reasoned that the expanded populations of T cells in patients with Goodpasture disease must be drawn from the populations that escape irreversible tolerance, so there should be similarly a correlation with high susceptibility to endosomal proteases. To test this prediction we undertook a second analysis of relative susceptibility to proteases using the set of 23 α3(IV)NC1 peptides that had been used in our study of patients' T cell responses,9 and the endosomal proteases Cathepsins D and E and asparagine endopeptidase (AEP).
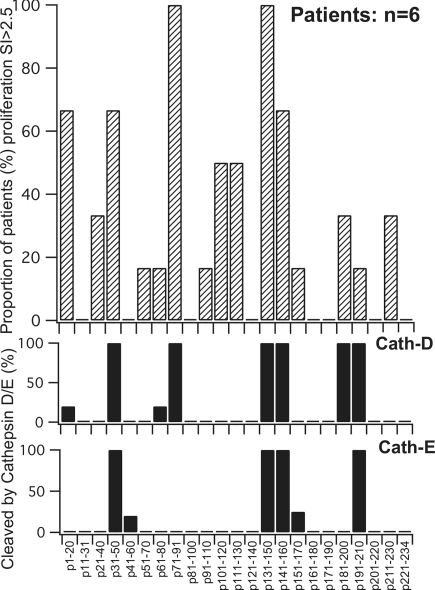
With Cathepsin D, extensive proteolysis (>50% digestion at 30 min) was observed for 6 (26%) of the 23 peptides, including 4 of the 5 dominant peptides previously found to stimulate peripheral blood T cells from the majority of Goodpasture patients (Figure 5).9 This correspondence was highly significant (P = 0.00758, Fishers exact test) (Table 3). With Cathepsin E, extensive digestion was observed for 4 peptides, all of which were peptides also extensively cleaved by Cathepsin D, in line with the recognized similarity in their substrate specificity.14 Again, the most susceptible peptides included a significant proportion of those that stimulated patients' T cells (P = 0.023) (Figure 5). The activity of AEP in relation to T cell epitopes in α3(IV)NC1 was also investigated because of its reported importance in the processing of myelin basic protein and tetanus toxoid. α3(IV)NC1 contains nine asparagine residues that are potential substrates for AEP. Fully reduced α3(IV)NC1 peptides containing any of the nine asparagines were found to be substrates for AEP.
Figure 5.
The strongest and most consistent proliferative responses by patients' peripheral blood T cells are to α3(IV)NC1 peptides rapidly cleaved by Cathepsin D/E. Peptides 20 to 22 amino acids in length spanning the sequence of α3(IV)NC1 were compared for susceptibility to Cathepsin D and E (lower 2 charts). The top panel shows the proportion of patients with recent-onset Goodpasture disease whose T cells proliferated with SI >2.5 to the indicated peptides (as reported in reference9).
Table 3.
Truth table for association between rapid proteolysis and capacity to stimulate patients' T cells
| Highly scissile to Cathepsin D | Proliferation by T Cells from ≥4 of 6 Patients with Goodpasture Disease
|
Total | |
|---|---|---|---|
| P-N | P-Y | ||
| C-N | 16 | 1 | 17 |
| C-Y | 2 | 4 | 6 |
| Total | 18 | 5 | 23 |
| P = 0.00758 | |||
Each peptide in the set of 23 used to interrogate patients' T cells was classified as in Table 2. The proportion of T cell stimulatory peptides that also contained highly Cathepsin D–scissile peptide bonds was higher than expected from the overall frequency of highly scissile peptide bonds in the peptide set. Fisher exact test: P = 0.00758.
Taken together, the results strongly suggest a pervasive influence of Cathepsin D/E and destructive processing on the repertoire of α3(IV)NC1-specific T cells in health as well as in Goodpasture disease. Indeed, for α3(IV)NC1, destructive processing appears to exert a greater influence on the specificity of T cells responses to α3(IV)NC1 peptides than does HLA type itself.
Highly Scissile Peptide Bonds Occur in Clusters in the Sequence of α3(IV)NC1
To obtain a better understanding of the relative susceptibility of peptide bonds within α3(IV)NC1 to endosomal proteases, we assembled the results of peptide digestions in this study, which interrogate chiefly the suitability of the primary sequence as a substrate for a protease, and our previously reported analysis of digestion of intact α3(IV)NC1,10 where tertiary structure exerts additional influences. There was correspondence with respect to the highly scissile peptide bonds in that all the peptide bonds assessed to be highly Cathepsin D–scissile in one or more of the three digestion studies (of 15mer peptides, 20mer peptides, or intact α3(IV)NC1) were at least scissile in all the other studies, with the exception of F66 and L67 which were cleaved in peptides but no indicative fragments were detected in digests of intact α3(IV)NC1 (Figure 6). There were also clear differences that presumably reflect differences in substrate accessibility. For example, peptides containing F10 were <50% cleaved at 30 min, but indicative fragments were prominent in α3(IV)NC1 digests, perhaps because F10 in the stalk of the NC1 domain is relatively accessible to proteases. Similarly, AEP cut intact α3(IV)NC1 only after the asparagines in cluster 3 (Figure 6), but after all the asparagines in peptides.10
Figure 6.
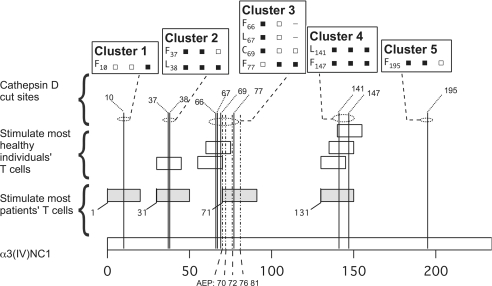
Location within α3(IV)NC1 of endosomal protease cut sites and peptides stimulating T cells of healthy individuals and patients with Goodpasture disease. The short bars depict the extent and location within intact α3(IV)NC1 (bottom bar) of the α3(IV)NC1 peptides that stimulated T cells from the majority of patients with Goodpasture disease (filled bars) or T cells from most healthy individuals (open bars). Note that four of the six peptides that stimulate T cells from most healthy individuals have sequences that are almost entirely contained within two of the peptides that stimulated T cells from most patients with Goodpasture disease, suggesting that at least some disease-associated autoreactive T cells have the same specificities as T cells that exist in health. The legend boxes show the Cathepsin D-cleaved peptide bonds (column 1) identified in the 15mer peptides, 20mer peptides, or intact α3(IV)NC1 (columns 2 through 4, respectively). Filled squares, “highly scissile” within peptide substrates or accounting for prominent early fragments in digests of intact α3(IV)NC1. Open squares, “scissile” within peptides substrates or accounting for detectable fragments in digests of intact α3(IV)NC1. The locations of the highly scissile peptide bonds within α3(IV)NC1 are depicted by vertical dotted lines. They occur in 5 clusters along the sequence of α3(IV)NC1: 1) F10T; 2) F37|L38|FVQ; 3) F66|L67|FC69|NVNDVCNF77|A; 4) L141|WKGFSF147|IM; and 5) SF195|WL (in P191–210 and P201–220). The most Cathepsin E-scissile peptide bonds were also in clusters: 1) F37|L38|FVQ; 2) not cut; 3) not cut; 4) L141|WKGFSF147|IM (in dap131–150); and 5) SF195|W196|LASL. AEP cut at CN70|VN72|DVCN76|FASRN81|D…. MN97|M. Note that all of the peptides that stimulate T cells from healthy individuals contain one or more highly scissile peptide bond.
There was a strong clustering of highly scissile peptide bonds in that all occurred within just five regions along the sequence of α3(IV)NC1 (Figure 6). All the peptides that stimulated T cells from the majority of patients or healthy individuals also localized to the scissile bond–rich regions, as expected from the strong associations demonstrated above.
Highly Scissile Peptide Bonds Are Located within the Epitopes Recognized by DR15-Restricted T Cells
The peptides used in this study were long enough (15 to 23 residues) to contain more than one register of binding to class II molecules, so it likely, but not certain, that the cleavages detected actually occurred within T cell epitopes. In the case of DR15-bearing individuals it is possible to test the hypothesis further because we have reported relative binding data for α3(IV)NC1 peptides,15 which can be refined to identify probable epitopes using published binding algorithms.
ProPred16 (http://www.imtech.res.in/raghava/propred; Institute of Microbial Technology [IMTECH], Chandigarh, India) was used to identify and rank subsequences within α3(IV)NC1 with highest predicted affinity for HLA DR15. The four peptides that stimulated T cells from the majority of five DR15-bearing healthy individuals (Figure 2), and the five peptides that stimulated T cells from the majority of patients each contained one to two subsequences with high predicted affinity for DR15 (likely epitopes). In every case, the putative epitopes contain peptide bonds that are very likely to be destroyed early in processing (Table 4).
Table 4.
Locations of Cathepsin D–scissile peptide bonds and high predicted DR15-affinity core sequences within peptides recognized by T cells from the majority of healthy and Goodpasture disease individuals
| Name and Sequence of α3(IV)NC1 | Peptides That Evoked Proliferative Responses in DR15-Bearing Patients or Healthy Controls | Rank DR15 Binding |
|---|---|---|
| dap1-20 | S P A T W T T R G / F V F / T R H S Q T T A | |
| 1 - - 4 - - - - 9 | 24 | |
| 1 - - 4 - - - - 9 | 26 | |
| dap31-50 | L Y S G F S F | L | F V Q G N Q R A H G Q D | |
| P31-45 | L Y S G F S F | L | F V Q G N Q | |
| 1 - - 4 - - - - 9 | 6 | |
| 1 - - 4 - - - - 9 | 8 | |
| P61-75 | F T T M P F | L | F C N V N D V C | |
| 1 - - 4 - - - - 9 | 13 | |
| 1 - - 4 - - - - 9 | 14 | |
| dap71-91 | V N D V C N F / A S R N D Y S Y | W - L S T P A | |
| 1 - - 4 - - - - 9 | 15 | |
| dap131-150 | I P P C P H G W - I S L | W K G F S F | I M F | |
| P131-145 | I P P C P H G W - I S L | W K G F | |
| dap141-160 | L | W K G F S F | I M F T S A G S E G T G Q | |
| P141-155 | L | W K G F S F | I M F T S A G S | |
| 1 - - 4 - - - - 9 | 3 | |
| 1 - - 4 - - - - 9 | 4 | |
| 1 - - 4 - - - - 9 | 9 |
All the peptides that stimulated proliferation in the majority of healthy individuals bearing DR15 (n = 5) or patients with Goodpasture disease (n = 5) are tabled to show their sequence overlaps (where present), the locations of the probable epitopes directed by the rank predicted DR15 binding of all possible DR15-binding subsequence determined by the ProPed algorithm,16 and the locations (using symbols defined in legend to Table 1) of peptide bonds that are highly scissile to Cathepsin D. Note that two of the bonds in these peptides, F10 and F77, are among those that were cleaved <50% at 30 min in peptide digests (marked with /) but were indicative by prominent peptides in digests of intact α3(IV)NC1.
Discussion
The data demonstrate that α3(IV)NC1-specific T cells exist in the peripheral blood of all the healthy individuals we studied. This echoes observations made for other autoantigens and adds to evidence for the existence of potentially autoreactive T cells in health. In addition, the data show an association between the α3(IV)NC1 peptides that are recognized by autoreactive T cells in man and high susceptibility to cleavage by the endosomal processing enzymes, Cathepsin D and E. The most credible explanation is that Cathepsin D/E, through efficient cleavage of α3(IV) at highly scissile peptide bonds, diminishes the constitutive presentation of certain sequences that allow corresponding T cells to escape irreversible tolerance. The proposed mechanism must operate within the complex machinery that determines the T cell repertoire, so it is not contradictory that some scissile peptides did not stimulate the subjects' T cells—corresponding T cells may never be generated or be effectively controlled by other mechanisms. The results therefore support a general importance of destructive processing to autoimmunity, as first demonstrated by the influence of AEP on presentation of one epitope from myelin basic protein and the persistence of the corresponding T cells.11
The observations strongly support a role for Cathepsin D/E in α3(IV)NC1 processing. Although both enzymes are present in the processing mechanism of APC, doubt has been cast on their importance in antigen processing,17 in particular because of the minor impact on mouse T cell responses of knocking out Cathepsin D or E.18 However, it is clear there is redundancy in the proteases arrayed against proteins in endo-lysosomes that could well prevent detection in single-protease knockout experiments of even very significant protease involvement. Our data show that Cathepsins D and E have very similar activity in processing α3(IV)NC1 as they do in substrate specificity studies.14,19 Any effects of Cathepsin D/E may not be limited to processing within APC. Collagen IV is a structural component of basement membranes and presumably requires some proteolytic processing to release soluble, or at least smaller, fragments suitable for internalization into APC. Cathepsin D is released into inflammatory sites and even reaches the urine in nephritis and so may participate in destructive processing of autoantigens such as α3(IV)NC1 before APC uptake.20
One important question is how cryptic epitopes can ever be presented to activate T cells in autoimmune disease. There are a number of possibilities. Inflammation that has a nonautoimmune cause could alter the accessibility or extracellular modification of self-antigens or their handling by APC or the responsiveness of T cells. In the case of autoimmunity against α3(IV)NC1, it is noteworthy in this regard that there are links between the onset of Goodpasture disease and inflammatory events such as renal vasculitis, membranous nephropathy, and renal trauma.21 Alternatively, antigen processing may be modulated by bound autoantibodies22 or by the actions of aberrant APC such as the malignant B cells that present cryptic epitopes from the RhD autoantigen to provoke autoimmune hemolytic anemia in chronic lymphocytic leukemia.23
Our data also allowed comparison of the specificities of α3(IV)NC1-reactive T cells that can be propagated in health and in anti-α3(IV)NC1 autoimmune disease. Although we used different peptides in the studies of T cells from patients and healthy individuals, there is substantial similarity between the sequences of stimulatory peptides (Figure 6). The results are consistent with the view that many of the autoreactive α3(IV)NC1-specific T cells that are expanded in Goodpasture patients are derived from T cells that are detectable but quiescent in health rather than arising de novo in disease. In contrast, the autoantibodies found in Goodpasture disease probably do arise de novo because the anti–glomerular basement membrane antibodies detected in healthy individuals are of different subclass.24
Last, our analysis of T cells from healthy individuals revealed a considerable concordance in their fine specificities for α3(IV)NC1, even though the donors were unselected and expressed a wide range of HLA antigens. These observations do not necessarily challenge the firmly established immunological paradigm that MHC type influences the specificity of T cell responses because the peptides investigated were long enough to contain several epitopes. However, the results do strongly indicate a tendency for helper responses to be dominated by T cells specific for peptides in particular regions of antigens. Analogous observations have been made in other human systems, including responses to the RhD protein25–27 and for exogenous antigens such as tetanus and diphtheria toxoid–based vaccines, where commonly targeted regions of antigens have been termed universal epitopes.28 Accessibility of particular regions of the antigen during processing has been suggested as a structural explanation for clustering of T cells epitopes.29,30 Our data suggest that susceptibility to processing enzymes is another factor and add credence to the view that destructive processing is a major determinant of the specificity of dominant T cell responses to self antigens.
Concise Methods
Additional details are in the online-only data supplement.
Materials
Two sets of overlapping peptides spanning the sequence of α3(IV)NC1 were used in this study. The main set comprised 45 peptides, which were 15 amino acids in length overlapping by 10 with sequences shown in Table 1. To allow comparison with previously published work, some experiments used a second panel of 23 peptides that were 20 to 22 amino acids in length overlapping by 10, as used in our study of patients' T cell proliferative responses (See Table 2 in Cairns et al.9). Recombinant monomeric Cathepsin E was supplied by Professor John Kay (University of Wales, Cardiff, UK). Cathepsin D was purchased from Calbiochem, UK. AEP was donated by Professor C. Watts (University of Dundee, Dundee, UK).
Peptide Digestions
Preliminary experiments established that a protease/peptide ratio of 10:1 resulted in complete cleavage within 30 min at 37°C of the most susceptible peptides (p136-150 and p36-50) under conditions of mild reduction and low pH (200 mM sodium acetate (pH 5), 5 mM dithiothreitol) close to those that pertain within endosomes.8 Conditions for comparative digestion experiments were varied as described above. Digestion products were identified by MALDI-TOF mass spectrometric analysis.31 Extent of digestion was assessed by comparing the abundance of parent ions in spectra of aliquots collected before and after digestion to those of any digestion products and that of a reference peptide spiked into all samples. Peptide susceptibility to protease digestion was categorized according to the degree of destruction as shown in Table 1.
PBMC Isolation and Cultures
Eleven healthy volunteers were HLA gene–typed at DRB1 as described previously.26 PBMC were obtained by density gradient centrifugation (Lymphoprep; Nycomed, Oslo, Norway) of heparinized blood from the donors. PBMC were cultures that comprised 1.25 × 106 cells in 1 ml in the Alpha Modification of Eagle's Medium (Life Technologies, Paisley, UK) supplemented with 5% autologous serum.
Proliferation and Cytokine Measurement
The α3(IV)NC1 peptides were added to cultures at 50 μg/ml and key hole limpet hemocyanin (Calbiochem, San Diego, CA) at 10 μg/ml.12 Cellular proliferation was measured in triplicate 100-μl aliquots taken from the cultures 5 to 10 d after antigen stimulation as described previously.12 The relatively large culture volumes and length of incubation favor responses by previously unprimed T cells.12 Proliferation was measured by 3H-thymidine incorporation and results are presented either as the mean counts per minute of the triplicate sample, or as the stimulation index, expressing the ratio of mean counts per minute in stimulated versus nonstimulated control cultures.
Disclosures
None.
Acknowledgments
This work was supported by grants from the Medical Research Council (UK) and Kidney Research UK.
Published online ahead of print. Publication date available online at www.jasn.org.
J.Z. and S.H. contributed equally to the work.
Supplemental information for this article is available online at http://www.jasn.org/
References
- 1.Kapler J, Roehm M, Marrack P: T cell tolerance by clonal deletion in the thymus. Cell 49: 273–280, 1987 [DOI] [PubMed] [Google Scholar]
- 2.Afzali B, Lombardi G, Lechler RI, Lord GM: The role of T helper 17 (Th17) and regulatory T cells (Treg) in human organ transplantation and autoimmune disease. Clin Exp Immunol 148: 32–46, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Elson CJ, Barker RN, Thompson SJ, Williams NA: Immunologically ignorant autoreactive T cells, epitope spreading and repertoire limitation. Immunol Today 16: 71–76, 1995 [DOI] [PubMed] [Google Scholar]
- 4.Gammon G, Sercarz E: How some T cells escape tolerance induction. Nature 342: 183–185, 1989 [DOI] [PubMed] [Google Scholar]
- 5.Sercarz EE: Processing creates the self. Nat Immunol 3: 110–112, 2002 [DOI] [PubMed] [Google Scholar]
- 6.Moudgil KD, Sercarz EE: The self-directed T cell repertoire: Its creation and activation. Rev Immunogenet 2: 26–37, 2000 [PubMed] [Google Scholar]
- 7.Watts C, Moss CX, Mazzeo D, West MA, Matthews SP, Li DN, Manoury B: Creation versus destruction of T cell epitopes in the class II MHC pathway. Ann N Y Acad Sci 987: 9–14, 2003 [DOI] [PubMed] [Google Scholar]
- 8.Trombetta ES, Mellman I: Cell biology of antigen processing in vitro and in vivo. Annu Rev Immunol 23: 975–1028, 2005 [DOI] [PubMed] [Google Scholar]
- 9.Cairns LS, Phelps RG, Bowie L, Hall AM, Saweirs WWM, Rees AJ, Barker RN: The fine specificity and cytokine profile of T helper cells responsive to a glomerular autoantigen in Goodpasture's disease. J Am Soc Nephrol 14: 2801–2812, 2003 [DOI] [PubMed] [Google Scholar]
- 10.Zou J, Thomas V, Turner AN, Phelps RG: Presentation of the Goodpasture autoantigen requires proteolytic unlocking steps that destroy prominent T cell epitopes. J Am Soc Nephrol 18: 771–779, 2007 [DOI] [PubMed] [Google Scholar]
- 11.Manoury B, Mazzeo D, Fugger L, Viner N, Ponsford M, Streeter H, Mazza G, Wraith DC, Watts C: Destructive processing by asparagine endopeptidase limits presentation of a dominant T cell epitope in MBP. Nat Immunol 3: 169–174, 2002 [DOI] [PubMed] [Google Scholar]
- 12.Barker RN, Elson CJ: Multiple self epitopes on the Rhesus polypeptides stimulate immunologically ignorant human T cells in vitro. Eur J Immunol 24: 1578–1582, 1994 [DOI] [PubMed] [Google Scholar]
- 13.Davidson HW, Watts C: Epitope-directed processing of specific antigen by B lymphocytes. J Cell Biol 109: 85–92, 1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arnold D, Keilholz W, Schild H, Dumrese T, Stevanovic S, Rammensee HG: Substrate specificity of cathepsins D and E determined by N-terminal and C-terminal sequencing of peptide pools. Eur J Biochem 249: 171–179, 1997 [DOI] [PubMed] [Google Scholar]
- 15.Phelps RG, Jones V, Coughlan M, Turner AN, Rees AJ: Presentation of the Goodpasture autoantigen to CD4 T cells is influenced more by processing constraints than by HLA class II peptide binding preferences. J Biol Chem 273: 11440–11447, 1998 [DOI] [PubMed] [Google Scholar]
- 16.Singh H, Raghava GP: ProPred: Prediction of HLA-DR binding sites. Bioinformatics 17: 1236–1237, 2001 [DOI] [PubMed] [Google Scholar]
- 17.Lennon-Dumenil AM, Bakker AH, Wolf-Bryant P, Ploegh HL, Lagaudriere-Gesbert C: A closer look at proteolysis and MHC-class-II-restricted antigen presentation. Curr Opin Immunol 14: 15–21, 2002 [DOI] [PubMed] [Google Scholar]
- 18.Deussing J, Roth W, Saftig P, Peters C, Ploegh HL, Villadangos JA: Cathepsins B and D are dispensable for major histocompatibility complex class II-mediated antigen presentation. Proc Natl Acad Sci U S A 95: 4516–4521, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dunn BM, Scarborough PE, Lowther WT, Rao-Naik C: Comparison of the active site specificity of the aspartic proteinases based on a systematic series of peptide substrates. Adv Exp Med Biol 362: 1–9, 1995 [DOI] [PubMed] [Google Scholar]
- 20.Yamamoto H, Yamada T, Itoh Y: Probable involvement of cathepsin D in the degradation of beta2-microglobulin in acidic urine. Clin Chem Lab Med 38: 495–499, 2000 [DOI] [PubMed] [Google Scholar]
- 21.Phelps RG, Turner AN: Goodpasture's syndrome: New insights into pathogenesis and clinical picture. J Nephrol 9: 111–117, 1996 [Google Scholar]
- 22.Simitsek PD, Campbell DG, Lanzavecchia A, Fairweather N, Watts C: Modulation of antigen processing by bound antibodies can boost or suppress class II major histocompatibility complex presentation of different T cell determinants. J Exp Med 181: 1957–1963, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hall AM, Vickers MA, McLeod E, Barker RN: Rh autoantigen presentation to helper T cells in chronic lymphocytic leukemia by malignant B cells. Blood 105: 2007–2015, 2005 [DOI] [PubMed] [Google Scholar]
- 24.Cui Z, Wang HY, Zhao MH: Natural autoantibodies against glomerular basement membrane exist in normal human sera. Kidney Int 69: 894–899, 2006 [DOI] [PubMed] [Google Scholar]
- 25.Barker RN, Hall AM, Standen GR, Jones J, Elson CJ: Identification of T-cell epitopes on the Rhesus polypeptides in autoimmune hemolytic anemia. Blood 90: 2701–2715, 1997 [PubMed] [Google Scholar]
- 26.Stott LM, Barker RN, Urbaniak SJ: Identification of alloreactive T-cell epitopes on the Rhesus D protein. Blood 96: 4011–4019, 2000 [PubMed] [Google Scholar]
- 27.Hall AM, Ward FJ, Vickers MA, Stott LM, Urbaniak SJ, Barker RN: Interleukin-10-mediated regulatory T-cell responses to epitopes on a human red blood cell autoantigen. Blood 100: 4529–4536, 2002 [DOI] [PubMed] [Google Scholar]
- 28.Diethelm-Okita BM, Okita DK, Banaszak L, Conti-Fine BM: Universal epitopes for human CD4+ cells on tetanus and diphtheria toxins. J Infect Dis 181: 1001–1009, 2000 [DOI] [PubMed] [Google Scholar]
- 29.Carmicle S, Dai G, Steede NK, Landry SJ: Proteolytic sensitivity and helper T-cell epitope immunodominance associated with the mobile loop in Hsp10s. J Biol Chem 277: 155–160, 2002 [DOI] [PubMed] [Google Scholar]
- 30.Dai G, Carmicle S, Steede NK, Landry SJ: Structural basis for helper T-cell and antibody epitope immunodominance in bacteriophage T4 Hsp10. Role of disordered loops. J Biol Chem 277: 161–168, 2002 [DOI] [PubMed] [Google Scholar]
- 31.Zou J, Turner AN, Phelps RG: Mass and composition (MAC) MALDI TOF analysis enables inference of the sequence of most peptides where the protein of origin is known. Analytical Chemistry 75: 2653–2662, 2003 [DOI] [PubMed] [Google Scholar]
- 32.Antoniou AN, Blackwood SL, Mazzeo D, Watts C: Control of antigen presentation by a single protease cleavage site. Immunity 12: 391–398, 2000 [DOI] [PubMed] [Google Scholar]