SUMMARY
Bacteriophage N4 encapsidates a 3,500 amino acid-long DNA-dependent RNA polymerase (vRNAP), which is injected into the host along with the N4 genome upon infection. The three-dimensional structures of wild-type and mutant N4 viruses lacking gp17, gp50, or gp65 were determined by cryo-electron microscopy. The virion has an icosahedral capsid with T = 9 quasi-symmetry that encapsidates well-organized dsDNA and vRNAP. The tail, attached at a unique pentameric vertex of the head, consists of a neck, twelve appendages, and six ribbons that constitute a non-contractile sheath around a central tail tube. Comparison of wild-type and mutant virus structures in conjunction with bioinformatics established the identity and virion locations of the major capsid protein (gp56), a decorating protein (gp17), the vRNAP (gp50), the tail sheath (gp65), the appendages (gp66), and the portal protein (gp59). The N4 virion organization provides insights into its assembly, and suggests a mechanism for genome and vRNAP transport strategies utilized by this unique system.
Keywords: Bacteriophage N4, cryo-electron microscopy, DNA and protein transport, T = 9 quasi-symmetry, virion-encapsidated DNA-dependent RNA polymerase
Introduction
Transport of a virus’s genetic information across cell membranes into the host cytoplasm is a fundamental event in a viral life cycle. Animal viruses typically enter host cells via endocytosis, followed by disassembly of the capsid, and subsequent release of the viral genome. Tailed bacteriophages, on the other hand, leave their capsids outside the cell and have evolved elaborate DNA delivery strategies. Phages with contractile tails (Myoviridae) have a tail tube that penetrates the cell membrane followed by injection of the genome into the cytoplasm. T4, for example, attaches to the cell surface via fibers that are attached to a baseplate at the tip of the tail. Upon successful adsorption, conformational changes in the baseplate trigger contraction of a tail sheath, which then drives the tail tube through the cell envelope.1 T4 transports a 169 kb genome into the cell cytoplasm in 30 seconds. This is one of the most efficient DNA transport processes known.2 It has been proposed that the energy required for this process is provided by pressure inside the head due to the tightly-packaged, negatively-charged DNA. 3; 4 Phages with long, non-contractile tails (Siphoviridae), such as T5 with a 1,900 Å-long tail, insert a long, hollow tube into cell membranes through which DNA is delivered into the cell.5; 6 T5 initially transfers 8% of its 122 kbp genome. Then, pre-early phage-encoded proteins participate in the transport of the rest of the genome.7 For phages with short, non-contractile tails (Podoviridae), DNA injection is more problematic since the tail is too short to reach the cell’s cytoplasm.8 To overcome this problem, T7 ejects phage-encoded proteins along with the phage DNA during infection. These proteins have been proposed to form a channel that spans the entire cell envelope, providing a conduit through which DNA is transported into the cytoplasm.8; 9 However, the exact mechanism of DNA delivery into the cytoplasm utilized by tailed phages is not known.
N4, a member of Podoviridae, is a lytic dsDNA bacteriophage that infects E. coli K12 strains.10 The virus’s linear 72 kbp genome encodes three tRNAs and 72 open reading frames (ORFs) (GenBank accession number EF056008). N4 is unique among dsDNA bacteriophages in that it packages a phage-encoded RNA polymerase (vRNAP) into the capsid,11 and ejects the protein into the host cell, presumably preceding the first ~500 bp of genomic DNA. Then, vRNAP catalyzes RNA synthesis initiated at a promoter present in the first 500 bp; this process pulls the next 10 to 40 kbp of the genome out of the virus and into the host (A. A. Demidenko and L. B. Rothman-Denes, unpublished data). N4 is the only known bacteriophage that does not require the activity of the host RNA polymerase for transcription of its early genes.12 Transport of the remaining DNA requires RNA synthesis catalyzed by the N4-encoded RNA polymerase II, a product of early transcription13; 14 (A. A. Demidenko and L. B. Rothman-Denes, unpublished data).
The isometric head of N4 has a diameter of approximately 700 Å. A short, non-contractile tail is attached to the head.15 The mature virion contains at least 10 gene products (gp) (Table 1).16 Gp50, the vRNAP, consists of 3,500 amino acids and is present in the virion.16 The vRNAP N-terminal domain (~998 aa) is required for injection of the first 500 bp of the genome (A. A. Demidenko and L. B. Rothman-Denes, unpublished data), the central domain (~1100 aa, mini-vRNAP) possesses the RNA synthesizing properties of vRNAP,17 and the C-terminal domain (~1,400 aa) is required for vRNAP encapsidation.17 Gp56 is thought to be the major capsid protein based on an estimate of 520 ± 30 copies per virion.16 This number suggests a T = 9 symmetry for an icosahedral head, although no virus structure with T = 9 symmetry has been observed previously. Gp65 is the receptor-binding protein required for N4 adsorption to the host (J. McPartland and L. B. Rothman-Denes, unpublished data). Little is known about the other seven structural proteins.
Table 1.
Structural Proteins in Bacteriophage N4
| Gene Product | Amino Acids (Met) | # Copies/Virion | Proposed Role | ||
|---|---|---|---|---|---|
| 35S-Met | SDSa | CryoEMb | |||
| 17 | 279 (5 c) | 147 ± 49 | 160 ± 20 | 175 | Decorating protein |
| 50 | 3,500 (114) | 4 ± 1 | 1 or 2 | vRNA Polymerase | |
| 51 | 644 (17) | 20 ± 2 | 18 ± 1 | ||
| 52 | 150 (1 or 2d) | 49 ±7 | 33 ± 5 | ||
| 54 | 299 (7) | 17 ± 4 | 43 ± 9 | ||
| 56 | 401 (16c) | 548 ± 45 | 520 ± 30 | 535 | Major capsid protein |
| 59 | 764 (23c) | 17 ± 4 | 12 ± 2 | 12 | Portal proteind |
| 65 | 1,382 (38) | 6 ± 1 | 5 ± 1 | 6 | Non-contractile tail sheath |
| 66 | 556 (16) | 25 ± 6 | 27 ± 5 | 36 | Appendagee |
| 67 | 236 (3) | 7 ± 1 | 13 ± 5 | ||
Data are from Falco et al. 16.
Estimated from the cryoEM structures in this study.
contains N-terminal methionine.
not know if the mature protein contains the N-terminal methionine
Predicted from the sequences using bioinformatics.
Here, we report the three-dimensional, cryo-electron microscopic (cryoEM) structures of wild-type N4, as well as mutant virions lacking gp17, gp50, or gp65. Comparison of wild-type and mutant virus structures established the identity and locations of the major capsid protein (gp56), a decorating protein (gp17), the vRNAP (gp50), the tail sheath protein (gp65), the appendages (gp66), and the portal protein (gp59) in the virion. A T = 9 icosahedral head and a non-contractile tail sheath, as observed in bacteriophage N4, have not been reported previously. Based on the structure of the tail and the location of the vRNAP, we propose a mechanism for transport of the DNA and vRNAP from the virion into the host cell.
Results and Discussion
The N4 capsid has T = 9 quasi-symmetry
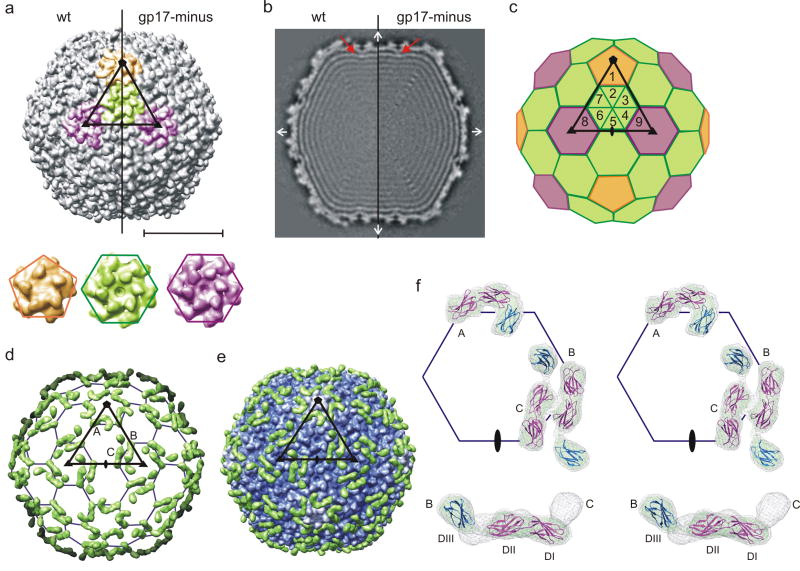
The three-dimensional, cryoEM structure of the N4 head was determined to 14 Å resolution by imposing icosahedral symmetry on mature virus particles (Table 2). The icosahedral head has a diameter of 695 Å between 2-fold vertices and 790 Å between 5-fold vertices with an average shell thickness of 27 Å. The surface of the capsid displays a complex pattern of protrusions representing the major capsid protein, gp56, and a decorating protein, gp17. The major capsid protein, gp56, forms both pentamers and hexamers with a T = 9 lattice (Fig. 1a, c). This is more apparent in the mutant that is missing the gp17 decorating protein because the latter has only three (not nine) copies per icosahedral asymmetric unit (Fig. 1a and next section). Six of the nine monomers in each icosahedral asymmetric unit reside in one hexamer located in a general position, two additional monomers are part of two neighboring hexamers coincident with the icosahedral 3-fold axes, and the ninth monomer is a part of the pentamer at each 5-fold vertex. Both the generally-positioned hexamers and the icosahedral 3-fold hexamers show good 6-fold symmetry. The centers of capsomers are approximately 140 Å apart. All the major capsid proteins have a small surface protrusion, separated by about 37 Å within pentamers and within hexamers. Based on the T = 9 triangulation, the total number of major capsid protein (gp56) copies is 535 per capsid (540 minus 5 copies since one pentamer is replaced by the portal or connector assembly). This number is in excellent agreement with the biochemically-determined values of 520 ± 3016 and 548±45 (Table 1).
Table 2.
Statistics of Image Reconstructions
| Number of Boxed Particles (Used) | Defocus, μm | Symmetry | Resolution, Å | |
|---|---|---|---|---|
| Wild-type | 2848 (2489) | 1.13 – 3.75 | icosahedral | 14 |
| C5 | 19 | |||
| C1 | 29 | |||
| gp17-minus | 2555 (2490) | 1.64 – 3.61 | icosahedral | 15 |
| vRNAP-minus | 2831 (2506) | 1.02 – 3.64 | icosahedral | 14 |
| C5 | 20 | |||
| gp65-minus | 2587 (2096) | 1.42 – 3.20 | icosahedral | 19 |
| C1 | 30 |
Figure 1.
Structures of the mature and gp17-minus N4 head. (a) Surface rendering of the N4 head viewed down an icosahedral 2-fold axis (top). The density is contoured at 3σ. The asymmetric unit is outlined by a black triangle connecting 5-, 3-, and 2-fold symmetry axes. One pentamer at the icosahedral 5-fold axis, one hexamer in a general position, and two hexamers at the icosahedral 3-fold axes are colored in orange, green, and purple, respectively. The scale bar represents 300 Å. Two independent hexamers and the pentamer in the gp17-minus virus are enlarged in order to show their similarities (bottom). (b) Central cross-section of the N4 head reconstruction. Density connecting the capsid to the most external DNA layer is indicated by a red arrow. The orthogonal icosahedral 2-fold axes are indicated with white arrows. (c) A schematic diagram of the T = 9 icosahedral lattice. The T = 9 hexagonal lattice and the icosahedral asymmetric unit are shown. The positions of nine major capsid protein subunits in an icosahedral asymmetric unit are labeled as 1-9. Subunit 1 forms one fifth of a pentamer (orange), subunit 2-7 form a general hexamer (green), each of subunit 8 and 9 form one sixth of a hexamer at the icosahedral 3-fold axis (purple). (d) Difference density between the wild-type and gp17-minus viruses. For clarity, only the front half of the map is shown. A T = 9 icosahedral lattice and asymmetric unit are shown. The three molecules of gp17 per icosahedral asymmetric unit are labeled A, B, and C. (e) The difference density between the wild-type and gp17-minus (green) superimposed with gp17-minus virus (blue to white with increasing radius). (f) Stereo view showing the fit of IgG domains into the difference density at two different contour levels. The grey caged density represents 3 σ, and a green solid density represent 5 σ above mean The three IgG domains (Protein Data Bank accession number 1FNH) were fitted as two separate rigid bodies (DI-II in magenta and DIII in blue). Three gp17 molecules are labeled as in (d). A symmetric unit (top) and a side view of the molecule B and C (bottom) are shown.
There are two major classes of icosahedral viruses both containing hexameric or pseudo-hexameric capsomers. In the first class, capsomers contain three copies of a monomer that has two independent jelly-roll folds. The centers between capsomers are separated by ~72 Å. In the second class, which includes the tailed phages T4, ϕ29, ε15, P22, and herpes simplex virus, the capsomers contain six monomers each consisting of HK97-like folds and whose centers are separated by ~135 Å.18-22 The distance between the N4 capsomer centers is approximately ~140 Å suggesting that N4 may belong to the HK97-like group of viruses. Although the N4 major capsid protein, gp56, does not have detectable sequence similarity to other tailed phage capsid proteins, the predicted secondary structure of gp56 has a similar pattern as that of the HK97 and T4 major capsid protein crystal structures.22; 23 The similarity of the fold remains uncertain given that the cryoEM reconstruction is only at 14 Å resolution, yet fitting of the HK97 capsid protein structure into the N4 capsid cryoEM density shows that the overall shape and dimensions of gp56 monomers and capsomers are consistent with the HK97 capsid protein fold. N4 gp56 is 44 kDa, larger than the HK97 capsid protein (31 kDa), but similar in size to the capsid proteins in T4 (49 kDa), ϕ29 (48 kDa), and P22 (47 kDa). The extra residues in these proteins form an insertion domain on the viral surface,20; 21; 23 making it likely that the surface protrusion on gp56 corresponds to such an insertion domain.
Structure of the gp17-minus mutant virus
The complex pattern of protruding densities on the N4 capsid surface can be attributed to two different conformations of the decorating protein, gp17. The cryoEM structure of the gp17-minus mutant virus (Fig. 2) was determined to 15 Å resolution (Table 2) showing clearly the T = 9 symmetry (Figs. 1a-c). The gp17-minus capsid, with only small protrusions in the capsomer, has a smoother surface than the wild-type virus. The difference map calculated between wild-type and gp17-minus virions shows three significant density peaks representing gp17 per asymmetric unit (or 180-5 copies per virion) (Fig. 1d), consistent with the previously-determined value of 160 ± 20 copies per virion16 or 147 ± 49 (Table 1). The gp17 density was 3σ above the maximum noise level, indicating that the capsid protein structure is not significantly affected by gp17 binding.
Figure 2.
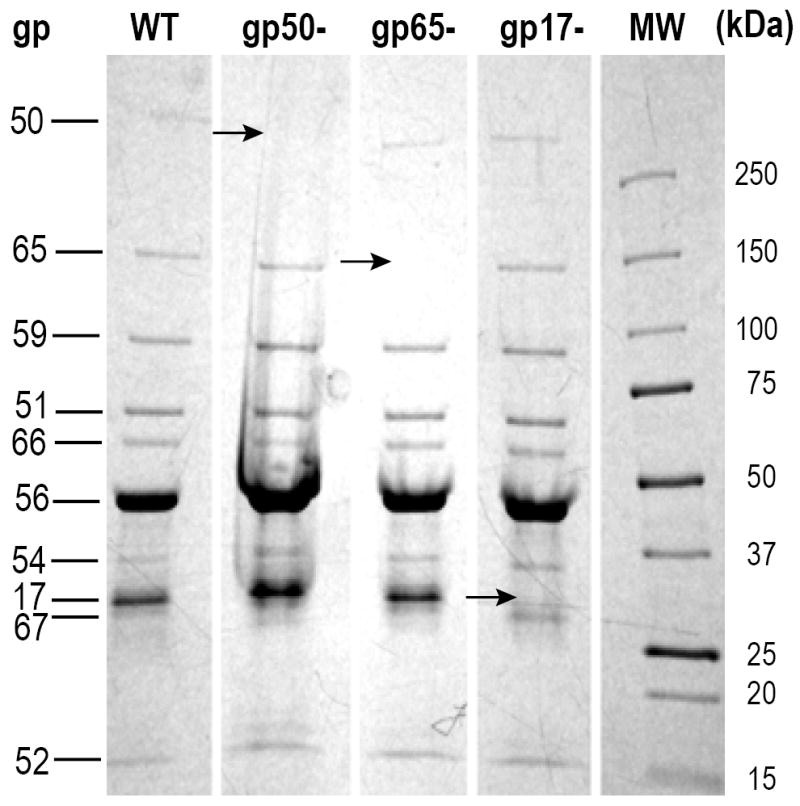
Protein composition of wild-type, gp17-minus, gp50-minus and gp65-minus virions. Arrows denote proteins absent in virions lacking gp50, gp65, or gp17. For properties of gene products, refer to Table 1.
Gp17 is an elongated molecule, located near quasi-3-fold axes relating any three capsomers, i.e, two hexamers and one pentamer, or three hexamers (Fig. 1d and e). One end of the molecule originates at the quasi-3-fold axis (thus creating a symmetry mismatch) and the other end extends towards the capsomeric boundaries. Despite the quasi T = 9 symmetry of the gp56 capsid lattice, there are only three copies of gp17 per icosahedral asymmetric unit: one at the juncture relating one pentamer and two hexamers (A in Fig. 1d), and two at the junctures relating three hexamers (B and C in Fig. 1d). As a result, the generally-positioned hexamers and the hexamers on the icosahedral 3-folds are differently decorated.
Two different conformations of the gp17 molecules were dependent on their locations. The gp17 molecule near position A has a “hook” shape, whereas the other two gp17 molecules near positions B and C have an elongated “rod” shape (Figs. 1d-f). Both densities are approximately 125 Å in length and 30 Å in width. At contour levels above 6σ, both hook- and rod-shaped densities separate into three smaller densities indicating that gp17 consists of three domains of about equal length, arbitrarily designated as DI, DII, and DIII, with DI being close to the quasi-3-fold axis. When the hook- and rod-shaped densities are compared, DI and DII superimpose well, whereas DIII from the hook-shaped gp17 is rotated about 140° with respect to domain III of the rod-shaped gp17. Thus, the difference of the two shapes is due to a change in the hinge angle between DII and DIII. Gp17 interacts with the capsid protein mainly through DI and DII, with DIII having little contact with the capsid surface in either the hook or rod conformation. DIII in the rod-shaped density has an almost radial orientation (Fig. 1f).
The fold and function of gp17
Analysis of the 279 amino acid sequence of gp17 using the program PSI-blast,24; 25 InterPro26 showed that residues 93-200 were homologous to 5 proteins that had an immunoglobulin (IgG)-like fold with greater than 30% sequence identity. The IgG-like fold of gp17 is also readily recognized in domain search programs, such as InterPro and Pfam. Domain and secondary structure predictions using the programs DomPred27 and PsiPred28 identified three domains consisting of mostly β-strands. The predicted secondary structure pattern of gp17 was further searched against known secondary structure patterns in a three-dimensional structure database using the program DomSSEA27 to identify proteins with similar folds. Two fibronectin-like proteins, each consisting of three IgG-like domains scored the highest (Protein Data Bank accession numbers 1TDQ and 1FNH), and differ from each other only in the angle between domains. Rigid-body fitting of these proteins into the cryoEM difference density produced a good fit except for the angle between DII and DIII. Thus, DI-DII and DIII were fitted as two separate rigid-bodies (Fig. 1f), resulting in an excellent fit, indicating that gp17 probably consists of three consecutive IgG domains.
Gp17 is not an essential component of the virus, since virions lacking gp17 are as infectious as wild-type virions. The location of the gp17 molecules around the periphery of capsomers might help to stabilize the capsid. A decrease in infectivity and the DNAse I sensitivity of the DNA in EDTA-treated virions (not shown) indicated that gp17-minus virions are unstable. The T4 soc, lambda gpD, and bacteriophage L Dec capsid proteins play a similar role in capsid stabilization.18; 29-31 Ig-like domain-containing proteins are found as components of many dsDNA bacteriophage virions.32 Based on the presence of Ig-like domains in bacterial surface glycohydrolases, Fraser et al. have suggested that phage Ig-like domains may aid in initial interactions with the bacterial cell surface. A similar role has been proposed for the bacteriophage ϕ29 head fibers and bacteriophage L decorating proteins;21; 33 whether gp17 plays such a role is not known. It may be of significance that the apparently unessential decorating proteins such as N4 gp17, T4 soc, the ϕ29 head fibers and the L decorating protein all bind at quasi-3-fold axes, although they form monomers in N4 and trimers in the other phages.
Interaction of capsid and DNA
The genomic DNA is organized as a series of concentric rings within the capsid shell, becoming weaker towards the center of the capsid. The central section of the icosahedrally-averaged reconstruction shows at least 8 layers of DNA with an average spacing of 24 Å (Fig. 1b). The spacing is similar to the dsDNA density observed in other bacteriophages, and is attributed to hexagonally-arranged DNA with an inter-layer spacing of 28 Å.34 There are densities connecting the outermost DNA layer with the center of the hexameric capsomers, indicating DNA-protein interaction (Fig. 1b). No such interaction is visible at the center of the pentameric capsomers. It is not clear whether the interaction between the DNA and capsid is a direct interaction with the capsid protein gp56, or whether there is an additional protein that mediates the DNA-capsomer interactions. This would require 80 copies of such a mediating protein. However, apart from gp56 and gp17, no other protein is present in a sufficient number of copies in the virus. Thus, it is likely that the interaction is mediated by the capsid protein itself.
Tail structure
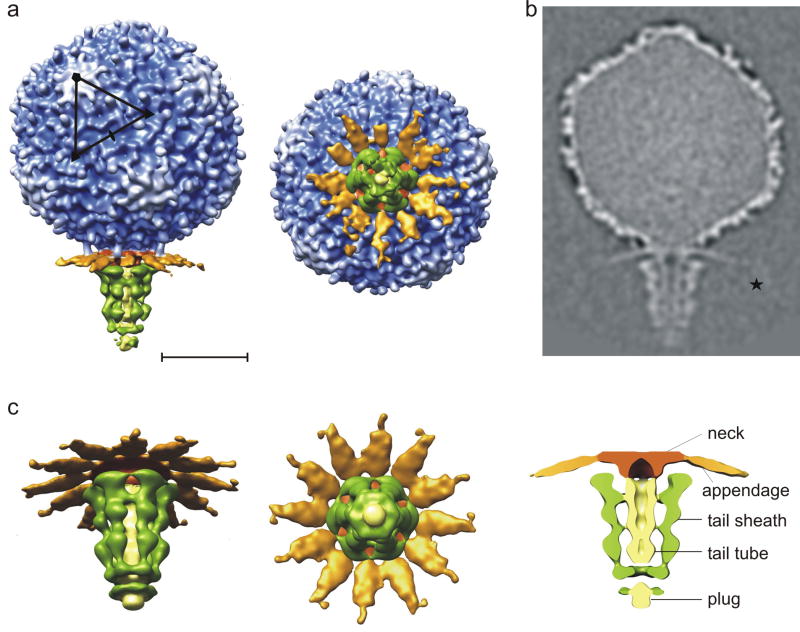
A 29 Å-resolution asymmetric cryoEM reconstruction showed that the 350 Å-long tail is assembled at the unique 5-fold vertex (Fig. 3). The tail components were named based on analogy with well-described tailed virus systems such as T4 and phi29, and the boundaries of each component were determined as a point where density disconnects and apparent symmetry breaks down. The neck protein between the head capsid and the tail also serves to attach twelve appendages, the latter being somewhat similar to the appendages of bacteriophage ϕ29.35; 36 The tail consists of a tail tube surrounded by a sheath. Each of the appendages consists of a thin, 200 Å-long stem and a distal domain. The density for the distal domain is weak and can only be seen at low contour levels, indicating flexibility.
Figure 3.
Structure of the mature N4 phage. (a) Surface representation of the asymmetric reconstruction of the mature N4 phage showing a side view (left) and an end-on view looking down the long axis of the phage (right). The head is colored from blue to white with increasing radius. The appendages (gp66), neck, non-contractile sheath (gp65), and tail tube are colored in orange, brown, green, and yellow, respectively. One icosahedral asymmetric unit is outlined in black. The scale bar represents 300 Å. (b) Central cross-section of the asymmetric reconstruction of the N4 phage. The distal domain of the appendages is marked with a star. (c) Enlarged tilted (left) view, end-on view (middle), and central section (right) of the 6-fold-averaged tail.
The 59 kDa gp66 is a good candidate for the appendage protein, although a BLAST search does not indicate homology to tail proteins in other phages. Secondary structure prediction of gp66 shows mostly α-helical structures at the N terminus, and β-structures at the C terminus. N-terminal residues between 50 and 210 contain leucine-rich regions that might form an interrupted trimeric coiled-coil. Assuming the pitch of a coiled-coil is 150 Å/100 residues, 160 residues would form a 240 Å-long coiled-coil, which would fit into the 200 Å-long and thin density of the appendage stem region. The BetaWrapPro program37 predicts the C-terminal residues 253 to 392 to form right-handed β-helices. The estimated number of copies of gp66 is 27 ± 516 or 25 ± 6 (Table 1), which is close to the 36 copies that would be present if each of the twelve appendages were trimers. Trimeric structures of coiled-coils or β-helices are common in phage tail fibers. Bacteriophage T4 fibritin is a 530 Å-long, interrupted, trimeric coiled-coil38 and the cell-puncturing device is a 3-start β-helix.39 P22, K1F, and the lactococcal phage p2 tailspikes are either three single-stranded β-helices or three-stranded β-helices.40-42 A recent cryoEM reconstruction of bacteriophage ϕ29 also suggests that the ϕ29 appendages may consist of a trimeric coiled-coil attached to P22-like β-helical structures at their distal ends.36
The central tail tube is a featureless, 310 Å-long cylinder with a 70 Å external diameter (Fig. 3c and 4). The inner diameter of the tail tube is about 25 Å, just large enough for dsDNA to pass through. In contrast to mature P22, T4, ε15, and ϕ29 phage structures,19; 36; 43; 44 no density is observed inside the tail tube. The very end of the tail tube is closed by a ‘plug’, which is presumably displaced upon infection. Surrounding the tail tube are six ribbons that form the sheath. The sheath is wider (~230 Å) near the neck and narrower (~75 Å) near the plug at the distal end of the tail. Each ribbon of the tail sheath makes only few contacts with the tail tube (Fig. 3c).
Figure 4.
Structure of gp65-minus mutant virus. (a) Surface representation of the asymmetric reconstruction of gp65-minus mutant virus showing a side view (left) and an end-on view looking down the long axis of the phage (right). The color scheme is the same as in Figure 3. One icosahedral asymmetric unit is outlined in black. The scale bar represents 300 Å. (b) Central cross-section of the asymmetric reconstruction of gp65-minus virus. (c) Tilted view of the tail for the 6-fold-averaged gp65-minus virus (left) and an end-on view (right). (d) Difference map calculated between asymmetric reconstructions of wild-type and gp65-minus viruses showing the non-contractile tail sheath (gp65).
Virions that lack gp65 do not adsorb to E. coli; moreover, purified gp65 blocks adsorption of wild-type phage to E. coli cells (J. McPartland and L. B. Rothman-Denes, unpublished data). Thus, gp65 is probably the major receptor-binding protein. An asymmetric reconstruction of a gp65-minus mutant virus (Fig. 2) lacked the tail sheath (Figs. 4a-c). A difference map calculated between the wild-type and gp65-minus mutant virions clearly showed the tail sheath consisting of six ribbons (Fig. 4d). The number of copies of gp65 in mature virions is consistent with the previously determined number.16 Each gp65 molecule is 265 Å long, and on average 35 Å wide. There are three larger lumps along the length of each ribbon that might represent separate protein domains.
It seems that the N4 tail assembles sequentially onto the DNA-filled capsids, similar to T7 or ϕ29 tail assembly,9; 45 since gp65-minus virions can successfully assemble a phage lacking only the tail sheath. When gp65 is added to the gp65-minus mutant, then the particles gain infectivity (J. McPartland and L. B. Rothman-Denes, unpublished data) suggesting that gp65 is the last protein to be added in the assembly process.
N4 is the first short-tailed phage to have a tail tube inside a non-contractile sheath. Whereas N4 uses the tail sheath to recognize the host (a function accomplished by the tail fibers of T4), the contractile sheath of T4 is part of a mechanism for inserting the tail tube into the periplasmic space of E. coli.
The portal protein and vRNAP location
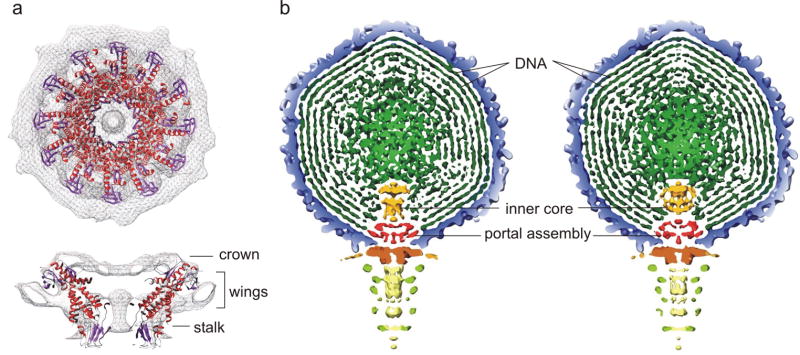
A portal protein, similar in structure to the ϕ29 portal connector assembly46 and also seen in other tailed phages,18; 19; 44; 47, is apparent at the unique 5-fold vertex associated with the tail in the cryoEM reconstruction of N4. The N4 portal is about 170 Å wide and 90 Å high and has a central channel large enough to allow the passage of dsDNA. The portal consists of three domains, similar to the crown, wing, and stalk domains observed for bacteriophages ϕ29, T7, and SPP1.46; 48-50 The internal channel diameter of the portal ranges from a minimum of 20 Å at the top, to a maximum of 80 Å in the middle, and 45 Å at the bottom (distal) end. The dimensions of the N4 portal’s length and external diameter are larger than that of the ϕ29 connector.46 Docking of the dodecameric ϕ29 connector crystal structure into the density shows a good fit in the wing and the stalk domains (Fig. 5a).
Figure 5.
The interior of bacteriophage N4. (a) The N4 portal assembly showing the crown, wing and the stalk domains. The atomic structure of the ϕ29 connector protein (Protein Data Bank accession number 1H5W) was fitted into the 5-fold-averaged cryoEM density of the N4 portal assembly. The α-helices and β-strands of the ϕ29 connector are shown in red and purple, respectively. (b) Comparison of wild-type and gp50-minus viruses. Cross section of the 5-fold-averaged wild-type (left) and gp50-minus (right) viruses viewed perpendicular to the long axis of the phage. The densities are displayed at 2.5σ above their respective means to show the molecular boundaries of portal and inner core proteins. The dsDNA, inner core proteins, and portal assembly are colored in dark green, orange, and red, respectively. All the other components of the phage are colored as in Figure 3.
Amongst phages, the connectors’ sequence identity is low and of variable size (40-100 kDa). However, secondary structure analyses of portal proteins, when compared with the ϕ29 portal structure, suggest a conserved folding motif.47 Furthermore, all known bacteriophage portal proteins are dodecameric when assembled in capsids. Based on these observations and because the predicted secondary structure for residues 295-410 matches the portal folding-motif, gp59 was identified as the N4 portal protein (Table 1). The larger molecular weight (86 kDa) of gp59, compared to 40 kDa of the ϕ29 connector, accounts for the crown and wings seen in SPP1 crystal structure.
Previously, we determined that vRNAP was present in 1-2 copies per virion16. A re-evaluation, based on the presence of 114 methionines in the vRNAP polypeptide in mature virions, indicates that vRNAP is present in 4±1 copies per virion (Table 1). Biochemical analysis of vRNAP-less particles indicates that they contain a full complement of the N4 genome (data not shown), and all virion proteins except for vRNAP (Fig. 2). The 5-fold-averaged reconstruction of a gp50-minus virus, determined to 20 Å resolution, has the same surface features as wild-type N4 (Fig. 5b), indicating that the vRNAP resides inside the capsid although this is not apparent in the asymmetric reconstruction at only 30 Å resolution. In wild-type virions, cylinder-like densities on top of the portal protein disrupt the continuity of the circular DNA density (Fig. 5b). This internal core density is ~110 Å in height and ~80 Å in width, and comprises at least two components, an upper disk-like structure that sits on top of a lower, longer cylindrical density. The internal core density in gp50-minus virus is ~80 Å in height and ~100 Å in width, smaller than that of the wild-type. One major difference is that the density corresponding to the lower end of the cylinder (closest to the portal protein) in wild-type is missing in the gp50-minus virus, suggesting that at least a part of vRNAP is located close to the internal entrance to the portal channel. The proximity of the vRNAP to the portal channel and its requirement for genome injection,17 suggest that vRNAP exits the capsid prior to the ejection of the DNA.
The process of infection
The N4 tail assembly is different from other Podoviridae viruses, which might indicate the special requirement of the N4 tail to deliver a large protein. The N4 vRNAP and DNA transport requires host proteins from several different cellular compartments, reflecting the complexity of the transport process. The N4 phage first binds to the E. coli cell surface through an interaction between the tail sheath and the outer membrane N4 receptor NfrA.51 Adsorption must trigger conformational changes in the tail, which leads to the ejection of the 3500 amino acid vRNAP and the first 500 bp of the N4 genome into the host. The narrowest part of the tail tube is 25 Å in diameter and, thus, the large size of vRNAP precludes the passage in its native form. Ejection of proteins from the bacteriophage capsid into the host cell has been reported for T7 and T4, and denatured or partially-denatured state mechanisms have been proposed.8; 9; 52 Similarly, the vRNAP would have to exit the phage head in an unfolded or semi-unfolded form, which would need to be refolded in the cytoplasm.
Materials and Methods
Bacterial and phage strains
E. coli K12 W3350 was used for growth of wild-type N4. E. coli W3350 supF was used for growth of N4 am229 (ORF 65am), N4 am29 (ORF 50am), and N4 ORF 17am. The resultant virions were used to infect E. coli W3350, yielding virions lacking gp17, gp50, or gp65 (Fig. 2).
Bacteria were grown aerobically at 37 °C in Luria-Bertani (LB) medium to 3 × 108 cells/ml. Cells were infected with phage at a multiplicity of infection of 10. The infected cells were lysed with chloroform 3 hrs. after infection. Cellular debris was pelleted and the supernatant was filtered through a 0.45 μm Millipore filter before loading onto 5%-40% glycerol step gradient. Following centrifugation at 35,000 RPM for 2 hrs. at 4°C in an SW41 Beckman rotor, the pelleted phage was resuspended in TM buffer (15 mM Tris-HCl pH 8.0, 10 mM MgCl2) and purified by CsCl buoyant density centrifugation. CsCl purified virions were resuspended in TM buffer. Samples were heated for 2 min at 100 °C in 1.6% sodium dodecyl sulphate, 60 mM Tris-HCl pH 6.8, 100 mM DTT, 5% glycerol (loading buffer), and loaded onto a precast 4-20% polyacrylamide gradient gel (Bio-Rad Laboratories, Hercules, CA). Proteins were stained with Phastgel™ Blue R (GE Healthcare Biosciences, Piscataway, NJ).
Labeling of virion proteins and determination of protein stoichiometry in virions
E. coli W3350 cells were grown arerobically at 37°C in LB medium to 3 × 108 cells/ml and infected with wild-type N4 phage at a multiplicity of infection of 10. After 6 minutes, cells were spun down, resuspended in LB medium containing 50 uCi/mL S35 L-Methionine (Perkin Elmer Life & Analytical Science, Shelton, CT), and lysed with chloroform at 3 hours post-infection. Virions were purified and processed as described above, and loaded onto a 10% SDS-polyacrylamide gel. The gels were dried, and the labeled proteins were detected by phosphorimaging and quantitated using ImageQuant Software. Several dilutions of virions were employed to assure a linear response. Virion copy number was based on the number of methionines in the mature virion protein and virion copy numbers of 6 and 535 for gp65 and coat protein, respectively.
Generation and characterization of N4 ORF 17am
An amber mutation in ORF 17 was introduced into the N4 genome. To that end, a plasmid derivative of pUC18 carrying a 2,189 bp N4 genomic fragment, spanning positions 5,430 to 7,619 (ORFs 16 and 17) and containing a TGTATA to TCTAGA change starting at position 6,922, inserted between the PstI and EcoRI sites was constructed. The sequence change, UAU (Tyr) to UAG (am) in ORF 17 (aa 56), introduced a new XbaI site overlapping with the UAG codon. E. coli W3350 pUC18 ORF 16/ORF 17am was grown in 5 ml of LB with 5 μl of 100 μg/ml ampicillin at 37 °C with aeration to 3 × 108 cell/ml and infected with N4 am15 (ORF 16am)53 at a multiplicity of 0.01. Recombination between the plasmid and the infected phage yielded phages that had incorporated the plasmid into the phage genome by homologous recombination.54 These phages grow in E. coli W3350 because they carry wild-type as well as mutated copies of ORFs 16 and 17. After 15 min. incubation, aliquots were removed, and serial dilutions were plated onto E. coli W3350 and incubated overnight at 37 °C. Large plaques (phages that had integrated the plasmid into the N4 genome and, therefore, had wild-type copies of ORF 16 and ORF 17) were spotted onto plates seeded with E. coli W3350 and E. coli W3350 supF. Plates were incubated overnight at 37 °C and individual plaques growing in E. coli W3350 supF were inoculated into tubes containing 5 ml of LB media and E. coli W3350 supF. After 3 hrs. incubation with aeration at 37 °C, cells were lysed with the addition of chloroform and cell debris was removed by centrifugation. During this second period of growth the plasmid excises to yield two types of progeny: parental N4 am15 and the newly generated N4 ORF 17am. To determine whether the phages isolated carried the amber mutation in ORF 17, ORF 17 was PCR-amplified using primers: ORF 16_F-Pst: 5′ GCA-CTGCAGGCCAATAGGCTAAGACGCGA and ORF 17_R_Eco: 5′ GCAGAATTCCATTCATAAAAACAATCACATTACTCTGT. The resulting PCR-amplified fragments were digested with XbaI to ascertain the presence of the XbaI site in ORF 17, and sequenced to confirm the presence of the TAT to TAG change in ORF 17. All phages tested plated with the same efficiency in E. coli W3350 and in E. coli W3350 supF, indicating that gp17 is not essential for phage growth. SDS-PAGE analysis of the protein composition of N4 ORF 17am virions indicated that they contain all virion proteins except gp17 (Fig. 2).
CryoEM and three-dimensional image reconstruction
All micrographs were taken at a magnification of ~39,500 using a FEI-CM200 electron microscope. Micrographs were digitized using a Zeiss scanner with a step size of 14 μm, which resulted in a 3.54 Å/pixel size. Individual particles were selected and boxed by eye using RobEM (http://cryoem.ucsd.edu/programs.shtm). The boxed particles were corrected for linear gradient and normalized means and variances. The contrast transfer function parameters were determined with the program ‘ctfit’ in the EMAN package.55
The initial model for the icosahedral head was generated from the class averages of 5-fold, 3-fold, and 2-fold views as determined by EMAN. The model for the tail was produced by putting a cylinder at one of the 5-fold vertices along the long axis of the phage. The icosahedral symmetry was released to produce an asymmetric reconstruction using a python script19 to search for 60 different orientations.35
The resolutions of each reconstruction were determined by the Fourier shell correlation method. A correlation of 0.5 was used as a cut-off between two reconstructions from independent half-data sets. The number of particles, symmetry imposed, and the final resolution for each reconstruction are summarized in Table 2.
Difference maps between the reconstructions were calculated after adjusting radial and density scale factors using RobEM (http://cryoem.ucsd.edu/programs.shtm). The program SITUS was used to fit the structure of fibronectin into the cryoEM difference density between wild-type and gp17-minus virions.
Acknowledgments
We thank Sheryl Kelly, Cheryl Towell, and Sharon Wilder for help in the preparation of the manuscript. We thank Marc Morais, Petr Leiman, Ye Xiang, Andrei Fokine, and Victor Kostyuchenko for helpful discussions; Noelia Salaberrios and Alexander Demidenko for the generation of N4 ORF 17am; and Abigail Markle for the purification of N4 phages. The work was supported by NSF grant MCB-0443899 to MGR, and NIH grant AI 12575 to LBR-D.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rossmann MG, Mesyanzhinov VV, Arisaka F, Leiman PG. The bacteriophage T4 DNA injection machine. Curr Opin Struct Biol. 2004;14:171–180. doi: 10.1016/j.sbi.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 2.Boulanger P, Letellier L. Characterization of ion channels involved in the penetration of phage T4 DNA into Escherichia coli cells. J Biol Chem. 1988;263:9767–9775. [PubMed] [Google Scholar]
- 3.Earnshaw WC, Harrison SC. DNA arrangement in isometric phage heads. Nature (London) 1977;268:598–602. doi: 10.1038/268598a0. [DOI] [PubMed] [Google Scholar]
- 4.Earnshaw WC, Casjens SR. DNA packaging by double-stranded DNA bacteriophages. Cell. 1980;21:319–331. doi: 10.1016/0092-8674(80)90468-7. [DOI] [PubMed] [Google Scholar]
- 5.Böhm J, Lambert O, Frangakis AS, Letellier L, Baumeister W, Rigaud JL. FhuA-mediated phage genome transfer into liposomes. Curr Biol. 2001;11:1168–1175. doi: 10.1016/s0960-9822(01)00349-9. [DOI] [PubMed] [Google Scholar]
- 6.Guihard G, Boulanger P, Letellier L. Involvement of phage T5 tail proteins and contact sites between the outer and inner membrane of Escherichia coli in phage T5 DNA injection. J Biol Chem. 1992;267:3173–3178. [PubMed] [Google Scholar]
- 7.Letellier L, Boulanger P, Plançon L, Jacquot P, Santamaria M. Main features on tailed phage, host recognition and DNA uptake. Front Biosci. 2004;9:1228–1239. doi: 10.2741/1333. [DOI] [PubMed] [Google Scholar]
- 8.Molineux IJ. No syringes please, ejection of phage T7 DNA from the virion is enzyme driven. Mol Microbiol. 2001;40:1–8. doi: 10.1046/j.1365-2958.2001.02357.x. [DOI] [PubMed] [Google Scholar]
- 9.Kemp P, Garcia LR, Molineux IJ. Changes in bacteriophage T7 virion structure at the initiation of infection. Virology. 2005;340:307–317. doi: 10.1016/j.virol.2005.06.039. [DOI] [PubMed] [Google Scholar]
- 10.Kazmierczak KM, Rothman-Denes LB. Bacteriophage N4. In: Calendar R, editor. The Bacteriophages. Oxford University Press; 2006. pp. 302–314. [Google Scholar]
- 11.Falco SC, VanderLaan K, Rothman-Denes LB. Virion-associated RNA polymerase required for bacteriophage N4 development. Proc Natl Acad Sci USA. 1977;74:520–523. doi: 10.1073/pnas.74.2.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rothman-Denes LB, Schito GC. Novel transcribing activities in N4-infected Escherichia coli. Virology. 1974;60:65–72. doi: 10.1016/0042-6822(74)90366-3. [DOI] [PubMed] [Google Scholar]
- 13.Zehring WA, Rothman-Denes LB. Purification and characterization of coliphage N4 RNA polymerase II activity from infected cell extracts. J Biol Chem. 1983;258:8074–8080. [PubMed] [Google Scholar]
- 14.Carter RC, Demidenko AA, Hattingh-Willis S, Rothman-Denes LB. Phage N4 RNA polymerase II recruitment to DNA by a single-stranded DNA binding protein. Genes & Dev. 2003;17:2334–2345. doi: 10.1101/gad.1121403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schito GC, Rialde G, Pesce A. Biophysical properties of N4 coliphage. Biochem Biophys Acta. 1966;129:482–490. doi: 10.1016/0005-2787(66)90063-3. [DOI] [PubMed] [Google Scholar]
- 16.Falco SC, Zehring W, Rothman-Denes LB. DNA-dependent RNA polymerase from bacteriophage N4 virions. Purification and characterization. J Biol Chem. 1980;255:4339–4347. [PubMed] [Google Scholar]
- 17.Kazmierczak KM, Davydova EK, Mustaev AA, Rothman-Denes LB. The phage N4 virion RNA polymerase catalytic domain is related to single-subunit RNA polymerases. EMBO J. 2002;21:5815–5823. doi: 10.1093/emboj/cdf584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fokine A, Chipman PR, Leiman PG, Mesyanzhinov VV, Rao VB, Rossmann MG. Molecular architecture of the prolate head of bacteriophage T4. Proc Natl Acad Sci USA. 2004;101:6003–6008. doi: 10.1073/pnas.0400444101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang W, Chang J, Jakana J, Weigele P, King J, Chiu W. Structure of epsilon15 bacteriophage reveals genome organization and DNA packaging/injection apparatus. Nature (London) 2006;439:612–616. doi: 10.1038/nature04487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang W, Li Z, Zhang Z, Baker ML, Prevelige PE, Jr, Chiu W. Coat protein fold and maturation transition of bacteriophage P22 seen at subnanometer resolutions. Nat Struct Biol. 2003;10:131–135. doi: 10.1038/nsb891. [DOI] [PubMed] [Google Scholar]
- 21.Morais MC, Choi KH, Koti JS, Chipman PR, Anderson DL, Rossmann MG. Conservation of the capsid structure in tailed dsDNA bacteriophages: the pseudoatomic structure of ϕ29. Mol Cell. 2005;18:149–159. doi: 10.1016/j.molcel.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 22.Wikoff WR, Liljas L, Duda RL, Tsuruta H, Hendrix RW, Johnson JE. Topologically linked protein rings in the bacteriophage HK97 capsid. Science. 2000;289:2129–2133. doi: 10.1126/science.289.5487.2129. [DOI] [PubMed] [Google Scholar]
- 23.Fokine A, Leiman PG, Shneider MM, Ahvazi B, Boeshans KM, Steven AC, Black LW, Mesyanzhinov VV, Rossmann MG. Structural and functional similarities between the capsid proteins of bacteriophages T4 and HK97 point to a common ancestry. Proc Natl Acad Sci USA. 2005;102:7163–7168. doi: 10.1073/pnas.0502164102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schaffer AA, Aravind L, Madden TL, Shavirin S, Spouge JL, Wolf YI, Koonin EV, Altschul SF. Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res. 2001;29:2994–3005. doi: 10.1093/nar/29.14.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, Lopez R. InterProScan: protein domains identifier. Nucleic Acids Res. 2005;33:W116–W120. doi: 10.1093/nar/gki442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marsden RL, McGuffin LJ, Jones DT. Rapid protein domain assignment from amino acid sequence using predicted secondary structure. Prot Sci. 2002;11:2814–2824. doi: 10.1110/ps.0209902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones D. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 29.Steven AC, Greenstone HL, Booy FP, Black LW, Ross PD. Conformational changes of a viral capsid protein. Thermodynamic rationale for proteolytic regulation of bacteriophage T4 capsid expansion, cooperativity, and super-stabilization by soc binding. J Mol Biol. 1992;228:870–884. doi: 10.1016/0022-2836(92)90871-g. [DOI] [PubMed] [Google Scholar]
- 30.Gilcrease EB, Winn-Stapley DA, Hewitt FC, Joss L, Casjens SR. Nucleotide sequence of the head assembly gene cluster of bacteriophage L and decoration protein characterization. J Bacteriol. 2005;187:2050–2057. doi: 10.1128/JB.187.6.2050-2057.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Imber R, Tsugita A, Wurtz M, Hohn T. Outer surface protein of bacteriophage lambda. J Mol Biol. 1980;139:277–295. doi: 10.1016/0022-2836(80)90131-x. [DOI] [PubMed] [Google Scholar]
- 32.Fraser JS, Yu Z, Maxwell KL, Davidson AR. Ig-like domains on bacteriophage: a tale of promiscuity and deceit. J Mol Biol. 2006;359:496–507. doi: 10.1016/j.jmb.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 33.Tang L, Gilcrease EB, Casjens SR, Johnson JE. Highly discriminatory binding of capsid-cementing proteins in bacteriophage L. Structure. 2006;14:837–845. doi: 10.1016/j.str.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 34.Hud NV, Downing KH. Cryoelectron microscopy of λ phage DNA condensates in vitreous ice: the fine structure of DNA toroids. Proc Natl Acad Sci USA. 2001;98:14925–14930. doi: 10.1073/pnas.261560398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morais MC, Tao Y, Olson NH, Grimes S, Jardine PJ, Anderson DL, Baker TS, Rossmann MG. Cryoelectron-microscopy image reconstruction of symmetry mismatches in bacteriophage ϕ29. J Struct Biol. 2001;135:38–46. doi: 10.1006/jsbi.2001.4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiang Y, Morais MC, Battisti AJ, Grimes S, Jardine PJ, Anderson DL, Rossmann MG. Structural changes of bacteriophage ϕ29 upon DNA packaging and release. EMBO J. 2006;25:5229–5239. doi: 10.1038/sj.emboj.7601386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McDonnell AV, Menke M, Palmer N, King J, Cowen L, Berger B. Fold recognition and accurate sequence-structure alignment of sequences directing ß-sheet proteins. Proteins. 2006;63:976–985. doi: 10.1002/prot.20942. [DOI] [PubMed] [Google Scholar]
- 38.Tao Y, Strelkov SV, Mesyanzhinov VV, Rossmann MG. Structure of bacteriophage T4 fibritin: a segmented coiled coil and the role of the C-terminal domain. Structure. 1997;5:789–798. doi: 10.1016/s0969-2126(97)00233-5. [DOI] [PubMed] [Google Scholar]
- 39.Kanamaru S, Leiman PG, Kostyuchenko VA, Chipman PR, Mesyanzhinov VV, Arisaka F, Rossmann MG. Structure of the cell-puncturing device of bacteriophage T4. Nature (London) 2002;415:553–557. doi: 10.1038/415553a. [DOI] [PubMed] [Google Scholar]
- 40.Steinbacher S, Baxa U, Miller S, Weintraub A, Seckler R, Huber R. Crystal structure of phage P22 tailspike protein complexed with Salmonella sp. O-antigen receptors. Proc Natl Acad Sci USA. 1996;93:10584–10588. doi: 10.1073/pnas.93.20.10584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stummeyer K, Dickmanns A, Mühlenhoff M, Gerardy-Schahn R, Ficner R. Crystal structure of the polysialic acid-degrading endosialidase of bacteriophage K1F. Nat Struct Biol. 2005;12:90–96. doi: 10.1038/nsmb874. [DOI] [PubMed] [Google Scholar]
- 42.Spinelli S, Desmyter A, Verrips CT, de Haard HJW, Moineau S, Cambillau C. Lactococcal bacteriophage P2 receptor-binding protein structure suggests a common ancestor gene with bacterial and mammalian viruses. Nat Struct Mol Biol. 2005;13:85–89. doi: 10.1038/nsmb1029. [DOI] [PubMed] [Google Scholar]
- 43.Leiman PG, Chipman PR, Kostyuchenko VA, Mesyanzhinov VV, Rossmann MG. Three-dimensional rearrangement of proteins in the tail of bacteriophage T4 on infection of its host. Cell. 2004;118:419–429. doi: 10.1016/j.cell.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 44.Lander GC, Tang L, Casjens SR, Gilcrease EB, Prevelige P, Poliakov A, Potter CS, Carragher B, Johnson JE. The structure of an infectious P22 virion shows the signal for headful DNA packaging. Science. 2006;312:1791–1795. doi: 10.1126/science.1127981. [DOI] [PubMed] [Google Scholar]
- 45.Studier FW. Bacteriophage T7. Science. 1972;176:367–376. doi: 10.1126/science.176.4033.367. [DOI] [PubMed] [Google Scholar]
- 46.Simpson AA, Tao Y, Leiman PG, Badasso MO, He Y, Jardine PJ, Olson NH, Morais MC, Grimes S, Anderson DL, Baker TS, Rossmann MG. Structure of the bacteriophage ϕ29 DNA packaging motor. Nature (London) 2000;408:745–750. doi: 10.1038/35047129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Agirrezabala X, Martín-Benito J, Valle M, González JM, Valencia A, Valpuesta JM, Carrascosa JL. Structure of the connector of bacteriophage T7 at 8 Å resolution: structural homologies of a basic component of a DNA translocating machinery. J Mol Biol. 2005;347:895–902. doi: 10.1016/j.jmb.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 48.Agirrezabala X, Martín-Benito J, Castón JR, Miranda R, Valpuesta JM, Carrascosa JL. Maturation of phage T7 involves structural modification of both shell and inner core components. EMBO J. 2005;24:3820–3829. doi: 10.1038/sj.emboj.7600840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lebedev AA, Krause MH, Isidro AL, Vagin AA, Orlova EV, Turner J, Dodson EJ, Tavares P, Antson AA. Structural framework for DNA translocation via the viral portal protein. EMBO J. 2007;26:1984–1994. doi: 10.1038/sj.emboj.7601643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Orlova EV, Gowen B, Dröge A, Stiege A, Weise F, Lurz R, van Heel M, Tavares P. Structure of a viral DNA gatekeeper at 10 Å resolution by cryo-electron microscopy. EMBO J. 2003;22:1255–1262. doi: 10.1093/emboj/cdg123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kiino DR, Rothman-Denes LB. Genetic analysis of bacteriophage N4 adsorption. J Bacteriol. 1989;171:4595–4602. doi: 10.1128/jb.171.9.4595-4602.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mullaney JM, Black LW. Activity of foreign proteins targeted within the bacteriophage T4 head and prohead: implications for packaged DNA structure. J Mol Biol. 1998;283:913–929. doi: 10.1006/jmbi.1998.2126. [DOI] [PubMed] [Google Scholar]
- 53.Willis SH, Kazmierczak KM, Carter RH, Rothman-Denes LB. N4 RNA polymerase II, a heterodimeric RNA polymerase with homology to the single-subunit family of RNA polymerases. J Bacteriol. 2002;184:4952–4861. doi: 10.1128/JB.184.18.4952-4961.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Willis, S. H. (1997). Analysis of bacteriophage N4 RNAPII. PhD, The University of Chicago.
- 55.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]